Abstract
GCN5 like-1 (GCN5L1) was identified as a novel gene with sequence homology to the histone acetyltransferase Gcn5. Subsequent protein-interaction studies identified GCN5L1 as a subunit of the multiprotein lysosome biogenesis complex, resulting in an alternative designation as biogenesis of lysosome-related organelle complex 1 subunit 1 (BLOS1 or BLOC1S1). Despite distinct nomenclatures, GCN5L1/BLOS1 has been shown to play crucial roles in mitochondria, endosomes, lysosomes and synaptic vesicle precursors. GCN5L1/BLOS1 controls mitochondrial protein acetylation, modulates metabolic pathways, and orchestrates retrograde mitochondria to nuclear signaling. It also contributes to endosome-lysosomal and vesicle trafficking and to endo-lysosomal function. Here we discuss the intracellular roles of GCN5L1/BLOS1 in hopes of linking mitochondrial-centric effects to cytosolic vesicular biology.
Keywords: Mitochondria, Retrograde signaling, Lysosome trafficking, Endosomal function, BORC complex
GCN5L1 as a regulatory protein in mitochondrial, endosome and lysosome biology
High-resolution subcellular mapping of the human proteome demonstrates that a large percentage of proteins concurrently localize to functionally unrelated organelles [1]. The consequences of this, for individual proteins, may include uniform molecular functions in spatially separate locations, or different molecular functions in distinct subcellular compartments. These location-specific functions may arise due to, for example, location-specific protein-protein interaction mediated effects or differing post-translational modifications. A novel protein, which has been labelled GCN5L1/BLOS1, epitomizes this concept. This review explores the emerging studies of this protein, with its apparent distinct functions within the mitochondria and in the modulation of endosome-lysosome trafficking and function.
General control of amino acid synthesis 5 like-1 (GCN5L1) is a 15–17 kDa protein with sequence homology to the nuclear acetyltransferase GCN5 [2, 3]. GCN5L1 has also been named RT14 and BLOS1/BLOC1S1 due, in part, to its association with a multiprotein complex linked to the biogenesis of lysosomes [4, 5]. GCN5L1/BLOS1 generates two gene products [4]. The human and mouse sequences are annotated in the Genebank by the accession numbers -NM_001487, BC130640 and BC034662. The short isoform of GCN5L1 appears to be ubiquitously expressed in mice as a ≈15 kDa protein [6] and localizes to mitochondria, while the larger (≈17 kDa) isoform is evident in the cytosol at lower levels in a limited number of tissues including liver and kidney [5–7].
Despite being cloned 25 years ago, interest in exploring the biological function of GCN5L1 has only been recently pursued, and has been driven by the finding that genetic disruption of GCN5L1/BLOS1 confers profound and diverse biological effects [7–11]. Mitochondrial localization of GCN5L1/BLOS1 first sparked interest in understanding its role in mitochondrial biology. Subsequent studies have identified its role in modulating mitochondrial protein acetylation, metabolism, mitochondrial turnover and effects on mitochondrial iniated signaling. Furthermore, GCN5L1/BLOS1 has been recently found to interact with multiple proteins in the cytoplasm and regulate endo-lysosomal trafficking and function. Here we consolidate the current findings to provide a more comprehensive overview on what is known about this protein, its distinct functions in different subcellular locations, and to discuss the future approaches required to fully elucidate its molecular function.
GCN5L1 in mitochondrial protein acetylation
Post translational modifications (PTM) play pivotal roles in the regulation of protein function. Protein phosphorylation has been the most extenstively explored PTM, although the role of acetylation has now been recognized as an abundant and functional PTM. This is most evident in mitochondria, where phosphorylation is relatively scarce [12] and acetylation is more prevalent [13, 14]. Furthermore, acetylation controls the activity of multiple metabolic and reactive oxygen species regulatory enzymes and mitochondrial quality control proteins [15–18]. Given GCN5L1’s sequence homology to the nuclear acetyltransferase Gcn5, and its mitochondrial enrichment [6] numerous groups have began exploring its role in mitochondrial protein acetylation and biology.
Initial studies found that GCN5L1 interacts with canonical substrates of the established mitochondrial deacetylase SIRT3 [6]. To date the interaction with GCN5L1 with other Sirtuin substrates does not appear to have been explored. Furthermore, the genetic disruption of GCN5L1 results in reduced acetylation of specific mitochondrial proteins, and in reduced global mitochondrial acetylation in knockdown transformed cells and GCN5L1 KO primary mouse embryonic fibroblasts (MEFs) [8, 11, 19].
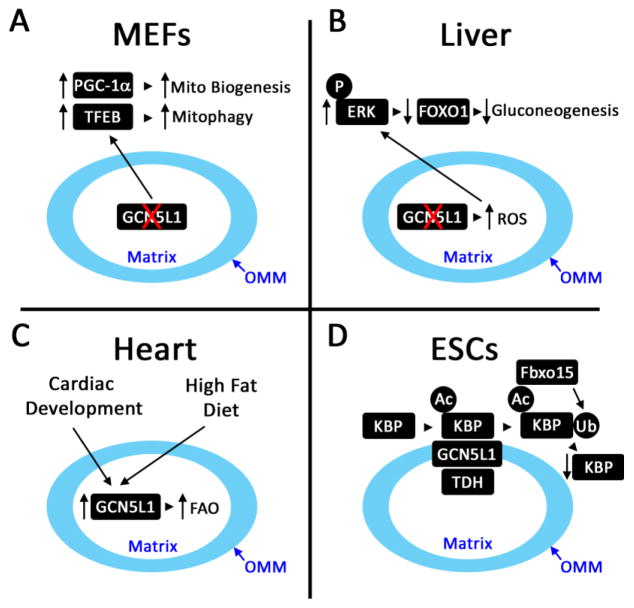
One hypothesis points to GCN5L1 functioning as a signaling rheostat, to tighly regulate mitochondrial turnover programing, in part, by controlling levels of mitochondrial protein acetylation. This hypothesis is supported by the phenotypic response to the genetic depletion or ablation of GCN5L1. First, GCN5L1 knockdown in transformed cells showed evidence of restricted mitochondrial autophagy (mitophagy) that was dependent on the canonical autophagy factors p62 and Atg5, but independent of the mitophagy mediator Parkin [11]. This mitochondrial autophagy program resulted in increased mitochondrial protein turnover, and a reduction in overall mitochondrial content and cellular oxidative metabolism [11]. The link between these observations was supported by the finding that reconstitution of GCN5L1 expression reversed both mitochondrial deacetylation and the induction of mitophagy. In contrast to the partial knockdown of GCN5L1 in cell lines, primary GCN5L1 KO MEF cells did not have a reduction in mitochondrial content [8]. Investigations into this discrepancy found a concurrent upregulation of mitochondrial turnover and biogenesis programs in GCN5L1 KO MEFs. These counteracting mechanisms appeared to be regulated by parallel upregulation of the master regulators of mitochondrial transcription, peroxisome proliferator activator receptor γ coactivator 1γ (PGC-1α) and lysosomal biogenesis, transcription factor EB (TFEB) [8]. Furthermore, genetic depletion of either of these master regulators in GCN5L1 KO MEFs resulted in the concurrent downregulation of the reciprocal master regulator, resulting in maintenance of mitochondrial content [8] (Fig. 1A). At the same time, reconstitution of GCN5L1 restored mitochondrial protein acetylation and blunted PGC-1α induction [8, 11]. Whether this mitochondrial content rheostatic function is controlled by mitochondrial acetylation as a nutrient sensing signal needs further delineation [20].
Figure 1. Schematic of Proposed Functions of Mitochondrial GCN5L1.
A. In mouse embryonic fibroblasts (MEFs) and transformed cell lines, the genetic depletion of GCN5L1 reduced mitochondrial protein acetylation in parallel with induction of mitochondrial biogenesis (PGC-1α) and lysosomal biogenesis (TFEB) regulatory programs. This effect is postulated to result from retrograde signaling from the michondria to nucleus, although the signaling pathway have not been defined. B. The role of mitochondrial signaling from the disruption of GCN5L1 levels was further expanded upon in the regulation of transcriptional regulation of gluconeogenesis. Here, in primary hepatocytes, the genetic depletion of GCN5L1 increased mitochondrial ROS-driven ERK signaling which promoted FoxO1 degradation and the subsequent downregulation in hepatic gluconeogenesis. C. Physiologic studies showed that during heart development or in response to high fat feedings cardiac GCN5L1 levels were increased and this was linked to increased fatty acid oxidation enzyme acetylation and activity. D. On the outer mitochondrial membrane (OMM) in embryonic stem cells (ESCs) GCN5L1 functions in concert with the acetyl-CoA generating enzyme L-threonine dehydrogenase (TDH) to acetylate a kinesin binding protein (KBP). This modification in turn promoted the proteasomal degradation of KBP via the ubiquitin ligase Fbxo15. In the absence of GCN5L1 the levels of KBP are maintained which drove mitochondrial biogenesis.
Alternatively, or concurrently, the disruption of mitochondrial protein acetylation may evoke metabolic changes that initiate mitochondrial to nuclear retrograde signaling [20, 21]. This could be operational as discussed above where GCN5L1 modulates the expression of both PGC-1α and TFEB in vitro, to maintain mitochondrial homeostasis. Furthermore, hepatocyte-restricted depletion of GCN5L1 reduced mitochondrial protein acetylation and diminished hepatic glucose production. Here, excess mitochondrial H2O2 production causes phosphorylation of ERK and subsequent proteosomal degradation of the canonical gluconeogenic transcription factor FoxO1, which in turn reduces the expression of gluconeogenic encoded enzymes. (Fig. 1B). This metabolic regulatory pathway was rescued following reconstitution of mitochondrial targeted GCN5L1 in GCN5L1 KO hepatocytes [22].
Given that GCN5L1 binds to mitochondrial metabolic proteins, an additional effect relates to acetylation-dependent regulation of mitochondrial metabolism. Here, the induction of fatty acid β-oxidation (FAO), either during the embryonic development of the heart or in response to a high fat diet, correlated with upregulation of cardiac GCN5L1 and increased acetylation and activity of FAO enzymes (Fig. 1C) [19, 23]. The depletion of GCN5L1 in cardiac-derived H9C2 cells reduced FAO rates and FAO enzyme acetylation [19, 23], suggesting that GCN5L1 plays an additional metabolic role by controlling FAO. The specific molecular link between GCN5L1 and FAO enzyme activity has yet to be elucidated.
The role of GCN5L1 in the regulation of mitochondrial content, and its link to mitochondrial metabolism was reinforced following the identification and characterization of the first validated lysine target of GCN5L1. Here, acetylation of the mitochondrial-associated kinesin Kif1Bα binding protein (KBP) required cooperation between GCN5L1 and the mitochondrial acetyl-CoA generating enzyme L-threonine dehydrogenase (TDH) [7]. Acetylation of KBP on lys501 was required to form a degradation motif (degron) for KBP proteolysis. Silencing of either GCN5L1 or TDH prevented proteasome-dependent KBP degradation via Fbxo15, a ubiquitin ligase that belongs to the F-box protein family [24]. The resultant KBP stability induced mitochondrial biogenesis, blunted proliferation and promoted differentiation of murine embryonic stem cells [7] (Fig. 1D). These findings supported a role of TDH-GCN5L1 in suppressing mitochondrial biogenesis to maintain proliferation of embryonic stem cells. It would be intriguing to determine whether this mechanism underpins or contributes to the accelerated mitochondrial biogenesis seen in the other GCN5L1 depletion studies [8, 11]. Interestingly, another mitochondrial response to inhibition of KBP degradation was increased mitochondrial respiration and reactive oxygen species (ROS) generation [7], and whether this contributes towards mitochondrial retrograde signaling has yet to be explored.
Together these studies support an important role of GCN5L1 in protein acetylation, and supports its role in numerous aspects of the regulation of mitochondrial function and metabolism and ROS handling/signaling.
Role in acetyl-CoA binding
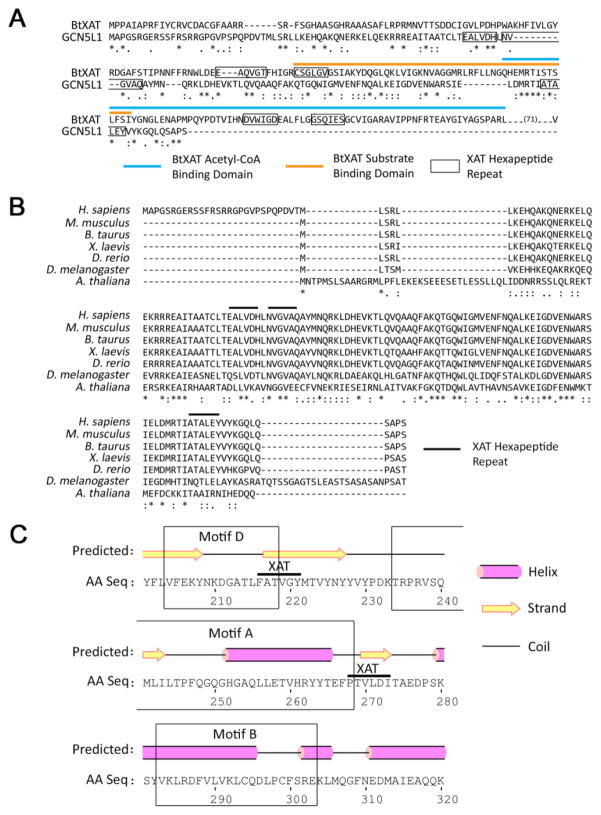
As the initial description of GCN5L1 as a component of the mitochondrial acetyltransferase machinery noted its similarity to prokaryotic proteins [6], and due to the functional mitochondrial studies described above, the sequence of GCN5L1 was compared to the Burkholderia thailandensis streptogramin xenobiotic acetyltransferase (BtXAT; accession number ZP_02389502). BtXAT is a member of the Left-handed parallel beta-Helix (LβH; cd00208) class of proteins characterized by a tandem hexapeptide repeat motif (x-[STAV]-x-[LIV]-[GAED]-x). Proteins in this class typically display acyltransferase activity, and are most commonly found in microbes [25, 26]. The GCN5L1 XAT-containing region has 53% sequence similarity to the BtXAT substrate- and acetyl-coA-binding domains. BtXAT has four XAT hexapeptide repeat motifs that largely bracket its two binding domains, suggesting that these regions are either involved directly in substrate binding, or aid the formation of the correct tertiary structure required for this process. A structural analysis of the SACOL2570 acetyltransferase from Staphylococcus aureus determined that the XAT hexapeptide repeat was integral to the folding of the LβH domains that form substrate binding pockets, suggesting that XAT hexapeptide repeat may be a necessary structure-defining feature [27]. In contrast, GCN5L1 has three XAT hexapeptide repeat motifs, with two of these regions separated by a single amino acid, and the third separated by 61 amino acids (Fig. 2A). Interestingly, this hexapeptide repeat architecture and sequence similarity is highly conserved in GCN5L1 orthologues from fruit flies to humans, with 100% motif sequence fidelity in the XAT repeat regions throughout members of the Chordata phylum (Fig. 2B).
Figure 2. The Prokaryotic Xenobiotic Acetyltransferase (XAT) Architecture of GCN5L1 is Highly Conserved in Human Lysine Acetyltransferases.
A. Alignment of the streptogramin acetyltransferase from Burkholderiathailandensis (BtXAT) and the short isoform of GCN5L1. The XAT-containing region of GCN5L1 (grey highlight), BtXAT substrate-binding (grey line), BtXAT acetyl-CoA-binding (black line) and XAT repeats (open box) are marked. B. Sequence alignment of GCN5L1 proteins from Homo sapiens, Mus musculus, Bos taurus, Xenopus laevis, Danio rerio, Drosophila melanogaster and Arabidopsis thaliana. The XAT-hexapeptide repeat regions are highlighted by horizontal bars. C. The human HAT1 acetyltransferase core domain contains two XAT repeats (grey highlight) which bracket the conserved motif A. The three motifs of the HAT1 core domain are identified by boxes.
The presence of these XAT hexapeptide repeats were assessed in other mammalian lysine acetyltransferase proteins. Using MOTIF search assessing tandem repeats of the XAT signature in the KEGG human protein database [28] showed that several members from the three major lysine acetyltransferase families – GCN5, p300 and MYST – all contain multiple XAT regions in addition to GCN5L1 (Table 1). Given the known yeast crystal structure of HAT1, a GCN5 family acetyltransferase [29], the determination of XAT domains in the human HAT1 was explored. Human HAT1 contains two XAT repeats, which bracket motif A of its conserved Gcn5 catalytic core (Fig. 2C). This motif contains the conserved [QR]-x-x-G-x-G sequence necessary for acetyltransferase activity, and is thought to be the main area of acetyl-CoA contact [29], in a similar manner to that found in the Burkholderia BtXAT. It would appear, therefore, that there is a large degree of conservation in XAT repeat placement between human and prokaryotic acetyltransferases, and suggests that GCN5L1, while not containing canonical acetyltransferase catalytic activity [6], may play a role in acetyl-CoA handling to facilitate substrate protein acetylation. This function is hypothetical to date and requires the determination of the molecular structure of GCN5L1 in association with acetyl-CoA.
Table 1.
Human Lysine Acetyltransferase Proteins Containing XAT Repeat Regions.
| Acetyltransferase Family | Gene Name | NCBI Gene ID | Protein Length (aa) | XAT Repeat Coordinates | XAT Repeat In Acetyltransferase Domain? |
|---|---|---|---|---|---|
| GCN5 | HAT1 | 8520 | 419 | 15–273 | Yes |
| ELP3 | 55140 | 547 | 240–380 | No | |
| GCN5 | 2648 | 837 | 158–248 | No | |
| PCAF | 8850 | 832 | 55–249 350–489 |
No | |
| NAT14 | 57106 | 206 | 69–173 | Yes | |
| NAT15 | 79903 | 242 | 61–93 | Yes | |
| NAT9 | 26151 | 207 | 95–196 | Yes | |
| NAT8B | 51471 | 227 | 15–177 | Yes | |
| NAT6 | 21412 | 286 | 118–269 | Yes | |
| NAT8L | 339983 | 302 | 29–255 | Yes | |
| NAT10 | 55226 | 953 | 138–384 506–611 |
Yes | |
| GCN5L1 | 2647 | 125 | 36–114 | - | |
| p300 | p300 | 2033 | 2414 | 432–473 1212–1400 2373–2413 |
Yes |
| CBP | 1387 | 2404 | 1140–1398 | Yes | |
| MYST | MOZ | 7994 | 2004 | 683–771 1349–1555 1788–1884 |
Yes |
| TIP60 | 10524 | 494 | 373–406 | Yes | |
| MOF | 84148 | 458 | 288–372 | Yes | |
| MORF | 23522 | 2073 | 894–913 1402–1462 1789–1919 |
Yes |
Role in Endosome-Lysosome Biology
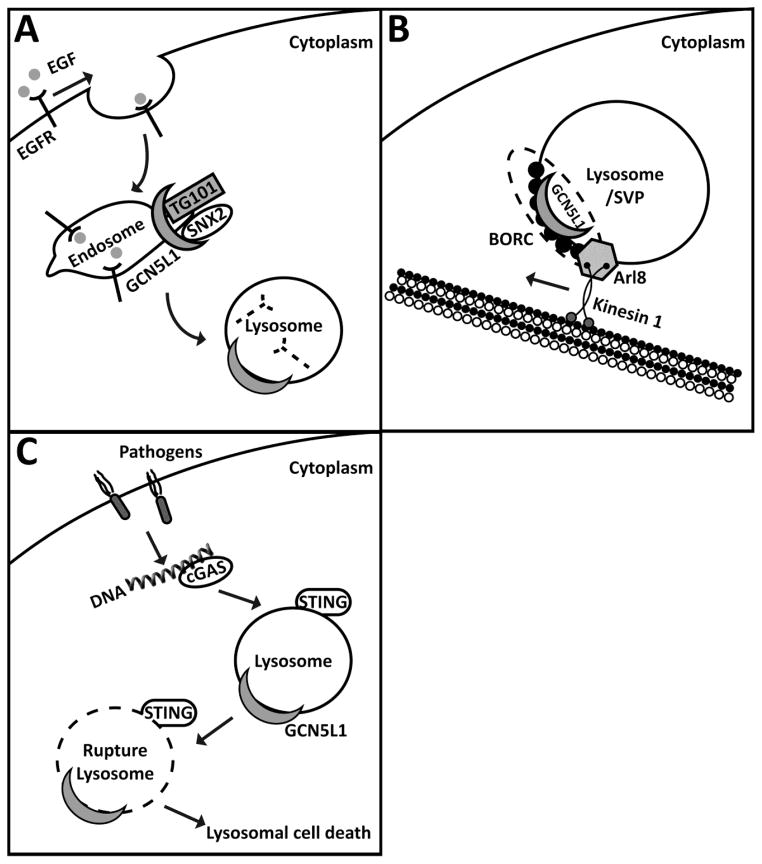
Despite the sequence homology between GCN5L1 and the transcriptional activator Gcn5, the first identified interactions with GCN5L1 identified it as a component of the multiprotein Biogenesis of Lysosome-related Organelles Complex-1 (BLOC-1). Given this finding GCN5L1 was alternatively named as a BLOC-1 subunit, i.e. BLOS1 or BLOC1S1 [5]. Mutations in numerous BLOC-1 complex proteins, but not in BLOS1 itself, manifests as the Hermansky-Pudlak syndrome [30, 31]. This syndrome results from abnormal vesicular trafficking to lysosomes and related organelles such as melanosomes and platelet dense granules, with features including albinism, prolonged bleeding, and pulmonary fibrosis. Interestingly, in drosophila BLOS1/GCN5L1 deficiency resulted in impaired intracellular protein trafficking and eye pigment defects that implicated BLOS1 in the control of intracellular trafficking [32]. Although murine germline knockout of GCN5L1/BLOS1 are embryonic lethal [8, 9], E14.5 embryos also showed defects in eye pigmentation [9]. To explore lysosomal trafficking in mammalian cells epidermal growth factor receptor (EGFR) lysosomal trafficking was interrogated in the context of the manipulation of BLOS1 levels. Moreover, BLOS1 interacts with sorting nexin 2 (SNX2) and the endosomal sorting complex required for transport 1 component, TSG101 to mediate the sorting of epidermal growth factor receptor (EGFR) into endosomal compartments in mammalian cells [9]. The silencing of BLOS1 delayed EGFR degradation, which was restored by the overexpression of BLOS1 fragments that interacted with SNX2 and TSG101 [9] (Fig. 3A). These studies suggest that GCN5L1/BLOS1 regulates vesicular trafficking to lysosomes.
Figure 3. Schematic of Poposed Functions of GCN5L1/BLOS1 in Regulating Endosome-Lysosome Trafficking and Function.
A. GCN5L1 interacted with sorting nexin 2 (SNX2) and the endosomal sorting complex required for transport 1 component TSG101 to mediate the sorting of epidermal growth factor receptor (EGFR) into endosomal compartments with its subsequent trafficking to lysomes to facilitate receptor degradation. B. As a component of the BORC complex, GCN5L1 facilitated lysosomal and synaptic vessicule precusor (SVP) trafficking in conjunction with the GTPase Arl8 and interaction with the motor protein Kinesin 1. C. Components of the BORC complex, including GCN5L1 are required to initiate STING orchestrated lysosomal rupture for the downstream activation of the NLRP3 inflammasome.
BLOS also regulates lysosome and synaptic vesicular precursor positioning. BLOS1 associates with BORC, a multiprotein complex that regulates lysosome positioning [10]. Genetic knockdown of BLOS1/GCN5L1 phenocopied the depletion of other BORC subunits and resulted in reduced dissemination of lysosomes to the periphery of cells [10]. This perturbation in the trafficking of lysosomes resulted from BORC defect-mediated impairment of the recruitment of the GTPase Arl8 to the lysosome membrane. This GTPase initiates coupling of lysosomes to kinesin plus-end-directed kinesin motors to promote trafficking of lysosomes to the cell periphery [10] (Fig. 3B). Furthermore, the axonal transport of synaptic vesicle precursors (SVPs), an additional Arl8-dependent trafficking event, was similarly disrupted following the genetic depletion of BLOS1/GCN5L1 [33]. However, biochemical studies suggest that a different BORC subunit termed SAM-4, rather than BLOS1/GCN5L1, functions as the Arl8 guanine nucleotide exchange factor [33].
BLOS1 additionally regulates lysosome death induced intracellular signaling. Here, sgRNA depletion of BLOC1S1/GCN5L1 impaired lysosome cell death signaling to impair cytosolic DNA sensing orchestrated inflammasome activation [34]. In the presence of GCN5L1 the the canonical STING-mediated lysosomal cell death program propagates the downstream activation of the NLRP3 inflammasome for the production of interleukin-1 and 18 [34] (Fig. 3C).
Together these studies clearly point to an important role of BLOS1/BLOC1S1/GCN5L1 in endosomal-lysosomal and vesicular trafficking, organelle positioning and function. These processes are operational in the cytoplasm and are linked to endosomes, lysosomes and synaptic and melanosome vessicles. Whether these functions of GCN5L1 share mechanistic overlap with its mitochondrial functions i.e. via retrograde signaling or via a direct role of acetylation in these trafficking processes require exploration.
Concluding Remarks
While only a minority of studies have explored the direct function of GCN5L1/BLOS1, it is recognized that GCN5L1 is highly regulated by developmental stage in the heart and in response to high fat feeding [11, 19, 23, 35]. Furthermore, GCN5L1/BLOS1 has been found to interact with proteins within mitochondria [6] and in the cytoplasm [9, 10, 23], and its depletion has robust effects spanning from embryonic lethality in mice [8, 9], to disruption in mitochondrial metabolism [22] and turnover [7, 8], and to effects on endosome-lysosome and vesicle trafficking [9, 10, 33] and on endo-lysosomal function [9, 34]. Despite all of these phenotypes, the sole functional partner identified to date is the mitochondrial-associated kinesin Kif1Bα binding protein KBP [7]. Here, the knockdown of TDH, the mitochondrial acetyl-CoA generating enzyme, phenocopied the effect of the loss of GCN5L1 effect on KBP, suggesting an important interaction of GCN5L1 with acetyl-CoA production.
The importance of acetyl-Coa for GCN5L1 function is further supported by the presence of a putative acetyl-CoA binding domain within the GCN5L1 sequence and the intriguing finding that the acetyl-CoA generating enzyme TDH was required for GCN5L1-dependent acetylation of KBP [7]. Together these findings warrant further biochemical investigation into whether GCN5L1 functions as an adaptor molecule in protein acetylation. At the same time, the concept of non-enzymatic acetylation of mitochondrial proteins has become firmly established [36, 37], and the genetic manipulation of GCN5L1 in these model systems, and employing advancing mass spectroscopy [38], would be intriguing approaches to explore the postulated ‘adaptor molecule’ concept of GCN5L1 in mitochondrial protein acetylation.
It will be interesting to explore additional roles of GCN5L1 level-dependent retrograde signaling from mitochondria as evident by its regulation of PGC-1α, TFEB and FoxO1. The concept that mitochondrial acetylation levels modulate transcriptional control of a metabolic pathway is compatible with the finding of increased glycolysis in SIRT3 KO mice. In that study, increased mitochondrial protein acetylation in SIRT3 KO mice stabilized HIF1α through retrograde ROS signaling [39].
Interestingly, the nuclear and cytosolic enriched Gcn5, which functions as an canonical acetyltransferase contains a functional transferase catalytic domain and operates withina multi-protein acetyltransferase complex [40]. As opposed to this, GCN5L1 may function as an adaptor molecular with an acetyl-CoA binding region as discussed above. Alternatively, additional regulatory proteins may be required to form a multiprotein complex where GCN5L1 then contributes to acetyltransferase function. These alternate concepts remain to be explored or validated.
The use of different names including GCN5L1, BLOS1 and BLOC1S1 inject a level of confusion into the field. As this protein is homologous to Gcn5, has evidence of an acetyl-Co-A interacting domain, contributes to mitochondrial protein acetylation and modulates the acetylation of regulatory proteins controlling the molecular motor protein regulatory protein KBP, we propose the coalescence around GCN5L1 as the uniform nomenclature going forward in the study of this protein.
Finally, the active identification and characterization of GCN5L1 substrates will be important to enhance our understanding of the role of this protein in mitochondrial and endosome-lysosome and vesicular homeostasis, and to reconcile the distinct molecular functions of this protein in different subcellular locations (see Outstanding Questions).
Outstanding questions.
How does GCN5L1/BLOS1 facilitate protein acetylation?
Is GCN5L1/BLOS1 a subunit of a multiprotein acetyltransferase?
What are the functional GCN5L1/BLOS1interacting partners and how does GCN5L1/BLOS1 modulate their activity?
What is the molecular structure of GCN5L1 and does it interact with acetyl-CoA
Does GCN5L1/BLOS1 have distinct mitochondrial and cytosolic mechanisms of action?
How do changes in mitochondrial acetylation regulate mitochondrial retrograde signaling?
Does GCN5L1/BLOS1 mediated mitochondrial retrograde signaling modulate endosome-lysosome trafficking or lysosomal positioning within the cell?
Is GCN5L1/BLOS1 acetylation-dependent regulation a component of its role in endosome-lysosome trafficking and function?
Trends Box.
GCN5L1, BLOS1 and BLOC1S1 are alternative names of the same gene product implicated in several distinct mitochondrial and cytosolic pathways.
This protein was initially designated GCN5L1 due to its sequence homology to the nuclear acetyltransferase GCN5.
GCN5L1 does not contain an acetyltransferase catalytic domain but may promote protein acetylation in the presence of acetyl-CoA generation pathways or contribute to acetylation as an acetyl-CoA binding protein.
The depletion of GCN5L1 has dose-dependent effects in modulating protein acetylation and retrograde signaling with profound effects on mitochondrial turnover and biogenesis.
The first functional target of GCN5L1 has been identified as the mitochondrial-associated kinesin Kif1Bα binding protein (KBP).
BLOS1/BLOC1S1 deficiency disrupts endosome-lysosome and synaptic vessicule precursor trafficking.
BLOS1/BLOC1S1 disrupts endo-lysosome and lysosome functioning.
Acknowledgments
Funding:
Supported by American Diabetes Association Research grant #1-17-IBS-197 (IS), NHLBI extramural funding grants K22-HL116728 (IS), R56-HL132917 (IS), R01-HL132917 (IS) and NHLBI intramural funding grants HL006047-06 (MNS) and HL005102-11 (MNS).
Footnotes
Conflicts of Interest:
No conflicts
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thul PJ, et al. A subcellular map of the human proteome. Science. 2017;356(6340) doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 2.Inoue M, et al. Isolation and characterization of a human cDNA clone (GCN5L1) homologous to GCN5, a yeast transcription activator. Cytogenet Cell Genet. 1996;73(1–2):134–6. doi: 10.1159/000134324. [DOI] [PubMed] [Google Scholar]
- 3.Driessen CA, et al. Cloning and structural analysis of the murine GCN5L1 gene. Gene. 1997;203(1):27–31. doi: 10.1016/s0378-1119(97)00486-1. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe TK, et al. Molecular cloning of a novel human cDNA, RT14, containing a putative ORF highly conserved between human, fruit fly, and nematode. DNA Res. 1995;2(5):235–7. doi: 10.1093/dnares/2.5.235. [DOI] [PubMed] [Google Scholar]
- 5.Starcevic M, Dell’Angelica EC. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1) J Biol Chem. 2004;279(27):28393–401. doi: 10.1074/jbc.M402513200. [DOI] [PubMed] [Google Scholar]
- 6.Scott I, et al. Identification of a molecular component of the mitochondrial acetyltransferase programme: a novel role for GCN5L1. Biochem J. 2012;443(3):655–61. doi: 10.1042/BJ20120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donato V, et al. The TDH-GCN5L1-Fbxo15-KBP axis limits mitochondrial biogenesis in mouse embryonic stem cells. Nat Cell Biol. 2017;19(4):341–351. doi: 10.1038/ncb3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott I, et al. GCN5-like protein 1 (GCN5L1) controls mitochondrial content through coordinated regulation of mitochondrial biogenesis and mitophagy. J Biol Chem. 2014;289(5):2864–72. doi: 10.1074/jbc.M113.521641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang A, et al. Biogenesis of lysosome-related organelles complex-1 subunit 1 (BLOS1) interacts with sorting nexin 2 and the endosomal sorting complex required for transport-I (ESCRT-I) component TSG101 to mediate the sorting of epidermal growth factor receptor into endosomal compartments. J Biol Chem. 2014;289(42):29180–94. doi: 10.1074/jbc.M114.576561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pu J, et al. BORC, a multisubunit complex that regulates lysosome positioning. Dev Cell. 2015;33(2):176–88. doi: 10.1016/j.devcel.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Webster BR, et al. Restricted mitochondrial protein acetylation initiates mitochondrial autophagy. J Cell Sci. 2013;126(Pt 21):4843–9. doi: 10.1242/jcs.131300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gnad F, et al. Evolutionary constraints of phosphorylation in eukaryotes, prokaryotes, and mitochondria. Mol Cell Proteomics. 2010;9(12):2642–53. doi: 10.1074/mcp.M110.001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim SC, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23(4):607–18. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 14.Wang Q, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327(5968):1004–7. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Webster BR, et al. The role of sirtuins in modulating redox stressors. Free Radic Biol Med. 2012;52(2):281–90. doi: 10.1016/j.freeradbiomed.2011.10.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sack MN, Finkel T. Mitochondrial metabolism, sirtuins, and aging. Cold Spring Harb Perspect Biol. 2012;4(12) doi: 10.1101/cshperspect.a013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samant SA, et al. SIRT3 deacetylates and activates OPA1 to regulate mitochondrial dynamics during stress. Mol Cell Biol. 2014;34(5):807–19. doi: 10.1128/MCB.01483-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y, et al. NAD+-dependent deacetylase SIRT3 regulates mitochondrial protein synthesis by deacetylation of the ribosomal protein MRPL10. J Biol Chem. 2010;285(10):7417–29. doi: 10.1074/jbc.M109.053421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukushima A, et al. Acetylation and succinylation contribute to maturational alterations in energy metabolism in the newborn heart. Am J Physiol Heart Circ Physiol. 2016;311(2):H347–63. doi: 10.1152/ajpheart.00900.2015. [DOI] [PubMed] [Google Scholar]
- 20.Webster BR, et al. Regulation of autophagy and mitophagy by nutrient availability and acetylation. Biochim Biophys Acta. 2014;1841(4):525–34. doi: 10.1016/j.bbalip.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandel NS. Evolution of Mitochondria as Signaling Organelles. Cell Metab. 2015;22(2):204–6. doi: 10.1016/j.cmet.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, et al. GCN5L1 modulates cross-talk between mitochondria and cell signaling to regulate FoxO1 stability and gluconeogenesis. Nat Commun. 2017;8(1):523. doi: 10.1038/s41467-017-00521-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thapa D, et al. Acetylation of mitochondrial proteins by GCN5L1 promotes enhanced fatty acid oxidation in the heart. Am J Physiol Heart Circ Physiol. 2017 doi: 10.1152/ajpheart.00752.2016. ajpheart 00752–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skaar JR, et al. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol. 2013;14(6):369–81. doi: 10.1038/nrm3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaara M. Eight bacterial proteins, including UDP-N-acetylglucosamine acyltransferase (LpxA) and three other transferases of Escherichia coli, consist of a six-residue periodicity theme. FEMS Microbiol Lett. 1992;76(3):249–54. doi: 10.1016/0378-1097(92)90344-n. [DOI] [PubMed] [Google Scholar]
- 26.Beaman TW, et al. Structure of the hexapeptide xenobiotic acetyltransferase from Pseudomonas aeruginosa. Biochemistry. 1998;37(19):6689–96. doi: 10.1021/bi980106v. [DOI] [PubMed] [Google Scholar]
- 27.Luo HB, et al. Biophysical analysis of the putative acetyltransferase SACOL2570 from methicillin-resistant Staphylococcus aureus. J Struct Funct Genomics. 2013;14(3):97–108. doi: 10.1007/s10969-013-9158-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M, et al. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(Database issue):D109–14. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dutnall RN, et al. Structure of the histone acetyltransferase Hat1: a paradigm for the GCN5-related N-acetyltransferase superfamily. Cell. 1998;94(4):427–38. doi: 10.1016/s0092-8674(00)81584-6. [DOI] [PubMed] [Google Scholar]
- 30.Li W, et al. Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1) Nat Genet. 2003;35(1):84–9. doi: 10.1038/ng1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiang PW, et al. The Hermansky-Pudlak syndrome 1 (HPS1) and HPS4 proteins are components of two complexes, BLOC-3 and BLOC-4, involved in the biogenesis of lysosome-related organelles. J Biol Chem. 2003;278(22):20332–7. doi: 10.1074/jbc.M300090200. [DOI] [PubMed] [Google Scholar]
- 32.Cheli VT, et al. Genetic modifiers of abnormal organelle biogenesis in a Drosophila model of BLOC-1 deficiency. Hum Mol Genet. 2010;19(5):861–78. doi: 10.1093/hmg/ddp555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niwa S, et al. BORC Regulates the Axonal Transport of Synaptic Vesicle Precursors by Activating ARL-8. Curr Biol. 2017;27(17):2569–2578e4. doi: 10.1016/j.cub.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaidt MM, et al. The DNA Inflammasome in Human Myeloid Cells Is Initiated by a STING-Cell Death Program Upstream of NLRP3. Cell. 2017;171(5):1110–1124. doi: 10.1016/j.cell.2017.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alrob OA, et al. Obesity-induced lysine acetylation increases cardiac fatty acid oxidation and impairs insulin signalling. Cardiovasc Res. 2014;103(4):485–97. doi: 10.1093/cvr/cvu156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner GR, Hirschey MD. Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol Cell. 2014;54(1):5–16. doi: 10.1016/j.molcel.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davies MN, et al. The Acetyl Group Buffering Action of Carnitine Acetyltransferase Offsets Macronutrient-Induced Lysine Acetylation of Mitochondrial Proteins. Cell Rep. 2016;14(2):243–54. doi: 10.1016/j.celrep.2015.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choudhary C, et al. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15(8):536–50. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 39.Finley LW, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell. 2011;19(3):416–28. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grant PA, et al. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999;274(9):5895–900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]