Abstract
Background
Reported hepatitis E virus (HEV) antibody assay performance characteristics are variable. Using a subset of surplus US blood donation samples, we compared assays for detecting anti-HEV IgM and IgG or total anti-HEV antibodies.
Methods
Samples from 5040 random blood donations, all HEV-RNA negative, collected primarily in the Midwestern US in 2015 were tested for anti-HEV IgM and IgG or total anti-HEV using assays manufactured by Diagnostic Systems, Wantai and MP Biomedicals.
Results
Overall, percent detection for anti-HEV IgG and total anti-HEV was 11.4%, and for anti-HEV IgM was 1.8%. Nine samples were reactive for anti-HEV IgM by all assays giving a recent infection rate of 0.18%. Anti-HEV IgG/total anti-HEV detection rates increased with age. Inter-assay agreement was higher among the IgG anti-HEV/total anti-HEV assays (84%) than the IgM assays (22%). Regression analyses of signal-to-cutoff ratios from IgG/total antibody assay were heteroskedastic indicating no constant variance among these assays suggesting they may detect different epitopes, or were affected by waning or less avid antibodies in the US donor population.
Conclusions
Although similar percentages of IgG anti-HEV/total anti-HEV detection were observed across the three commercial assays, each assay detected a unique sample subpopulation and were heteroskedastic when compared pairwise. Discordance was higher among anti-HEV IgM assays, but a recent HEV infection rate of at least 0.18% was estimated based on assay concordance.
Keywords: Donors, Transfusion-transmitted Disease - Hepatitis, Infectious Disease Testing
Introduction
Hepatitis E is caused by the hepatitis E virus (HEV)1. HEV is a member of the Hepeviridae family. Variants that infect humans belong to the species Orthohepevirus A2,3. This species has seven recognized genotypes of which 5, genotypes 1, 2, 3, 4 and 7 are known to infect humans3,4. Although acute hepatitis E is usually self-limiting with low mortality (about 1–3%) the mortality rate can reach levels of up to 30% in pregnant women during the second and third trimester, who are infected with genotypes 1 or 2.5,6. HEV genotypes 3 and 4 are frequently associated with chronic infection in immunocompromised individuals, most notably solid organ transplant recipients leading to chronic hepatitis, cirrhosis and liver failure7–9. Early epidemiological studies indicated the virus was restricted to developing countries where it was transmitted fecal-orally through contaminated water causing large outbreaks. Sporadic cases of hepatitis E seen in developed countries were originally thought to be associated only with travel to endemic areas. However, more recent studies have shown that autochthonous hepatitis E, genotypes 3 and 4, can be found in developed countries transmitted zoonotically from infected animals through the consumption of raw or undercooked meat and offal10–12. Most cases of acute hepatitis E in developed countries tend to be asymptomatic13–15. Because asymptomatic HEV infection in blood donors has been documented via HEV RNA detection, and HEV has been documented to be transfusion transmitted, there is increasing concern worldwide about blood safety15,16.
Antibody prevalence in blood donors varies depending on geographic location and the assays used. For example, in Europe anti-HEV IgG seroprevalence in blood donors ranges from 1.3% to 52%14, increases with age, and higher IgG positivity are usually seen in males compared to females17,18. A study in the Netherlands using a single assay showed that IgG anti-HEV prevalence had decreased from 47% in 1988 to 21% in 201118, and in the US, the National Health and Nutrition Examination Survey (NHANES) using the same anti-HEV IgG assay found decreases in IgG prevalence from 10.2% during 1988–1994 to 6% during 2009–201019 . An analysis of 1939 US blood donors at the National Institutes of Health (NIH) in 2013 found IgG prevalence of 18.8% and IgM prevalence of 0.4%. IgG prevalence increased with donor age, with prevalence decreasing from 21.8% in 2006 to 16% in 2012. The NIH study used an in-house assay for the first analysis and a commercial assay for the subsequent analysis. None of the NIH donors were HEV-RNA-positive20. A subsequent study of 4499 HEV-RNA-negative samples at the American Red Cross (ARC), which was a subset of 18,829 donation samples collected in 2013 from which HEV RNA occurred in 1 per 9500 donations (95% confidence interval [CI], 1:2850–1:56,180), the antibody detection rates using a different assay was 7.7% for IgG and 0.58% for IgM21. Similarly, antibody prevalence increased with age, and was highest in the Midwest US (12.5%; odds ratio of 2.23 versus other US regions).
In spite of documented declines in HEV seroprevance, an increase in prevalence among 18- to 21-year old Dutch blood donors was observed18. There has also been an increase in the number of reported hepatitis E cases in several countries where it is a reportable disease22–24. However, it is not known whether this increase is due to increased HEV incidence or to increased reporting. Part of the problem is that clinical assays are validated to determine the status of an analyte in a symptomatic individual and not specifically for epidemiological studies of healthy populations. This is particularly true of HEV serological assays due to the lack of concordance has led to variable findings in asymptomatic populations when commercial assays have been compared25–27. Currently, there are no FDA-approved anti-HEV or HEV-RNA assays.
This study examined samples from 5040 US blood donations using three different commercial IgG and IgM anti-HEV assays to determine the anti-HEV prevalence rates and to examine the concordance among the immunoassays used.
Methods
Sample selection and preparation
Residual samples from blood donations made to the ARC from March 22 to April 3, 2015 were obtained. Samples from donations positive for routine disease markers (e.g., hepatitis B virus, hepatitis C virus and human immunodeficiency virus) were excluded. All samples used in this study were selected from approximately 50,000 samples screened by research-use only HEV RNA assays to exclude RNA positives (the results of HEV RNA screening are not considered in the current study). A total of 5040 random samples were enrolled. Blood was collected in plasma preparation tubes; the plasma from these tubes was stored at −70°C until tested21. Samples were tested under code with donor identities not available to investigators. Where appropriate, basic demographics were obtained from an ARC research database associated with this study. The study was approved by the ARC Institutional Review Board. As part of the donation consent, all donors were provided with an information sheet describing future potential uses of their surplus samples for studies on transfusion-transmissible infections. The samples were anonymized and sent to CDC for testing.
Antibody testing
This study used six enzyme immunoassays from three commercial companies: Diagnostic Systems Incorporated (DSI S.r.l., Milan, Italy (hereafter, DSI); MP Biomedicals Asia Pacific Pte. Ltd., Singapore (MP); and Beijing Wantai Biological Pharmacy Enterprise Co., Ltd., Beijing, China (Wantai)). The detection of HEV IgM antibodies used three assays: DS-EIA-ANTI-HEV-M (batch: E-152, DSI), HEV IgM ELISA 3.0 (23162-096, MP) and HEV-IgM ELISA (WE-7192, Wantai). The detection of HEV IgG antibodies used two assays: DS-EIA-ANTI-HEV-G (E-151, DSI) and HEV-IgG ELISA (WE-7292, Wantai). The sixth assay detected total anti-HEV antibodies: HEV ELISA 4.0 (23542-096, MP). All assays were run according to the manufacturers’ instructions, except for the Wantai assays for which initially positive samples were not retested, to standardize with the DSI and MP assays that do not require retesting. The Wantai IgG assay includes a gray zone outcome in addition to positive and negative outcomes. A specific individual was designated to test assays from a specific vendor to minimize inter-operator error potentially introduced by sample or reagent handling.
The anti-HEV assays used in this study detect antibodies using different formats. For anti-HEV IgG detection, the DSI and Wantai assays use a recombinant capsid peptide to bind total antibody, then horseradish peroxidase (HRP)-conjugated anti-human IgG is used to detect captured IgG. MP uses a recombinant capsid peptide to capture total antibody and HRP-conjugated recombinant capsid peptide to bind to total bound antibody. For the detection of anti-HEV IgM, the DSI and the MP assays use a recombinant capsid peptide as the capture antigen to bind total antibody, and HRP-conjugated monoclonal mouse raised against anti-human IgM antibody to detect anti-IgM. The Wantai assay uses anti-µ antibody to capture total IgM and HRP-conjugated recombinant capsid peptide to bind to anti-HEV IgM.
Statistics
Pearson's Chi-squared test with Yates' continuity correction, 95% Confidence Intervals, odds ratios, regression analysis and Breusch-Pagan test for heteroskedasticity28 were calculated in R (ver. 2.15.3)29. Heteroskedasticity indicates that the variation in a variable is unequal across the range of values of a second variable used to predict the first.
Results
Of the 5040 samples tested, 569 (11.29%; 95% CI, 10.43% to 12.20%; DSI) and 619 (12.28%; CI, 11.39% to 13.22%; Wantai) were reactive for anti-HEV IgG, and 537 (10.65%; CI, 9.82% to 11.54%; MP) were reactive for total anti-HEV antibody, yielding an average of 11.41% (Table 1). There was no significant difference between the numbers of reactive samples detected between IgG assays. Anti-HEV IgM testing resulted in 142 reactives (2.90%; CI, 2.45% to 3.40%: DSI), 93 (1.85%; CI, 1.49% to 2.26%: MP) and 34 (0.67%; CI, 0.47% to 0.94%; Wantai), yielding an average reactivity of 1.81% (Table 1). The extent of agreement between IgG anti-HEV/total anti-HEV and IgM anti-HEV assay sample detection is shown on Supplemental Table 1.
Table 1.
Anti-HEV results by assay for 5040 blood donor samples. Results are listed as numbers per category followed by the percentage.
| Negative | Gray zone | Positive | ||||
|---|---|---|---|---|---|---|
| IgG | IgM | IgG | IgM | IgG | IgM | |
| DSI | 4471 (88.71%) | 4894 (97.10%) | 0 (0.00%) | 0 (0.00%) | 569 (11.29%) | 146 (2.90%)‡ |
| MP | 4503 (89.35%)* | 4947 (98.15%) | 0 (0.00%)* | 0 (0.00%) | 537 (10.65%)*† | 93 (1.85%)‡ |
| Wantai | 4415 (87.60%) | 5006 (99.33%) | 6 (0.1%) | 0 (0.00%) | 619 (12.28%)† | 34 (0.67%)‡ |
| mean | 88.55% | 98.19% | 0.04% | 0.00% | 11.41% | 1.81% |
Assay detects total antibody rather than IgG alone.
The MP Biomedicals (MP) and Wantai IgG assays show a significant difference at p < 0.025,
and all three IgM assays show significant differences between each other at p < 0.00075.
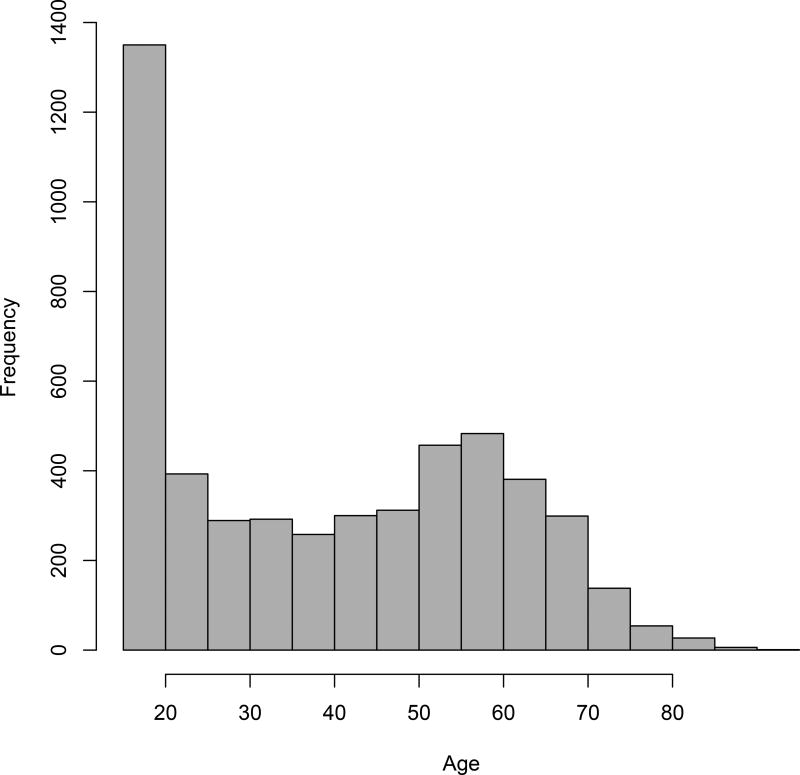
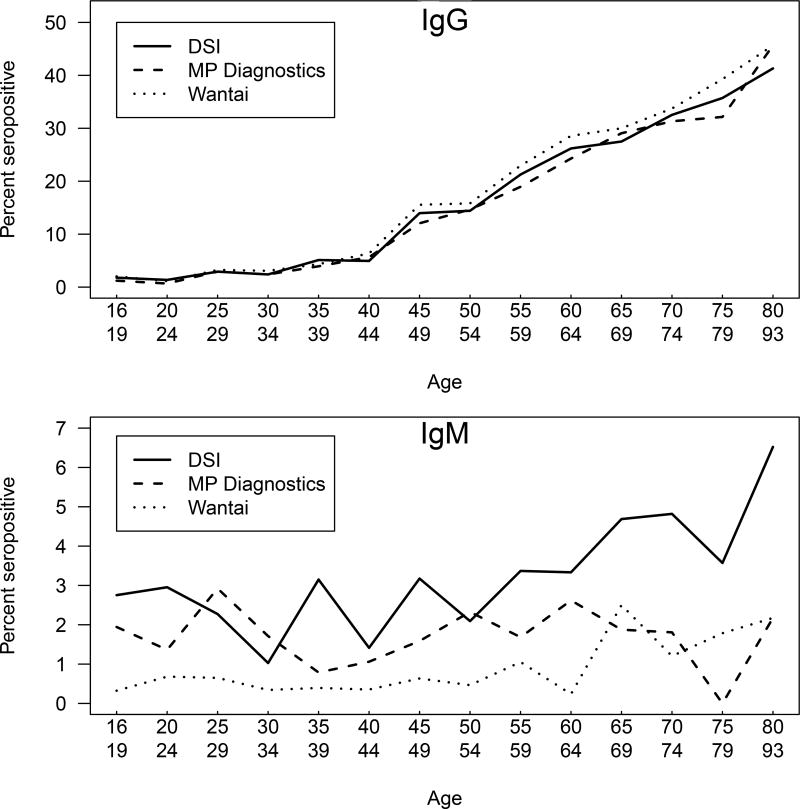
There was no statistical difference in the gender of the donors selected (male = 2682, female = 2358). The age range for donors was from 16 to 93 years of age (Figure 1). IgG anti-HEV detection rate increased with age, and all three assays exhibited similar uptrends (Figure 2, upper panel) with no significant differences seen among the trends. Age and gender adjustment of these data among the four states with the highest number of donors, Missouri, Kentucky, Illinois and Indiana, did not detect any appreciable differences in IgG anti-HEV detection rates by age group versus age and gender adjusted detection rates among the assays (Supplemental Figure 1). No trend in anti-HEV IgM seroprevalence with age was evident (Figure 2, lower panel).
Figure 1.
Histogram of donor age distribution. Frequency distribution calculated using 5 year bins, age range (16 to 93).
Figure 2.
Seroprevalence by age. Upper panel, anti-HEV IgG/total antibodies; lower panel, anti-HEV IgM. Solid line, DSI; dashed line MP Biomedicals; dotted line, Wantai. Data were plotted by averaging the seroprevalence by age range from donors 15 to 80 years of age in increments of 5 years, and all ages above 80 years of age formed the last range. Seroprevalence within each age range was plotted versus the mean age within the age range.
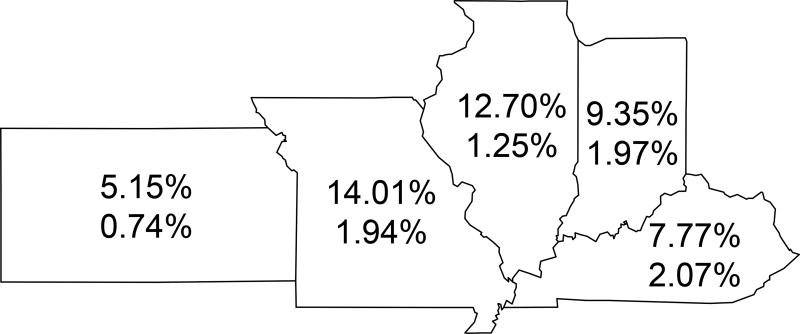
Samples came from donors residing in 21 states. Differences by state of residence were analyzed in the five states with the highest number of donors; Missouri (MO, n=2080), Kentucky (KY, n=1120), Illinois (IL, n=887), Indiana (IN, n=813) and Kansas (KS, n=97). All other states were excluded from the state by state comparison because of the low donor numbers, for example, the state with the next highest number of donations was California with 8 donors. Only anti-HEV IgG/total anti-HEV was analyzed because of the low number of anti-HEV-IgM positive samples by state. IgG anti-HEV/total anti-HEV prevalence percentages, as the mean among the three assays used to test specimens, across all assays within the five states having the highest numbers of donors ranged from Kansas having the lowest (5.15%) to Illinois and Missouri having the highest (12.70% and 14.01%, respectively) (Figure 3); although Kansas has the lowest prevalence, due to the low number of samples tested, its 95% confidence intervals overlaps those for Illinois and Missouri and thus are not significant (Table 2 and Supplemental Table 2).
Figure 3.
Anti-HEV prevalence in the 5 states with the highest number of donors. The numbers within each state boundary are IgG/total antibody prevalence (upper number) and IgM prevalence (lower number). States from left to right: Kansas, Missouri, Illinois, Indiana and Kentucky.
Table 2.
Percent IgG antibody reactive donors by state and gender. The five states with the highest number of donors were compared.
| Total* | Male | Female | |||||||
|---|---|---|---|---|---|---|---|---|---|
| State | DSI | MP | Wantai | DSI | MP | Wantai | DSI | MP | Wantai |
| Illinois | 12.06 | 11.84 | 14.21 | 13.51 | 12.89 | 16.42 | 10.34 | 10.59 | 11.58 |
| Indiana | 9.47 | 8.36 | 10.21 | 11.14 | 9.55 | 11.82 | 7.51 | 6.97 | 8.31 |
| Kansas | 5.15 | 5.15 | 5.15 | 6.67 | 6.67 | 6.67 | 3.85 | 3.85 | 3.85 |
| Kentucky | 7.77 | 7.50 | 8.04 | 8.32 | 8.16 | 9.14 | 7.10 | 6.71 | 6.71 |
| Missouri | 13.89 | 13.13 | 15.00 | 15.74‡ | 13.89 | 16.20 | 11.90‡ | 12.30 | 13.70 |
| All states† | 11.29 | 10.65 | 12.28 | 12.68‖ | 11.48 | 13.65§ | 9.71‖ | 9.71 | 10.73§ |
Missouri (MO; n=2080), Kentucky (KY; n=1120), Illinois (IL; n=887), Indiana (IN; n=813) and Kansas (KS; n=97).
Seropositivity for all 21 states in which donors resided.
p < 0.05;
p < 0.01;
p<0.005 (Chi-square for seropositivity by gender and assay).
Because this was a cross-sectional unlinked study, there is no way to estimate declines of anti-HEV over time, or when the donors were infected relative to the donation used in this study. However, looking at individuals with S/CO ratios above the cutoff for each assay allows the range of S/CO ratios for individuals within age ranges to be evaluated (Supplemental Figure 2). No significant difference or trend is observed in the range of S/CO ratios amongst these individuals by age group for any of the IgG/total anti-HEV assays used in this study.
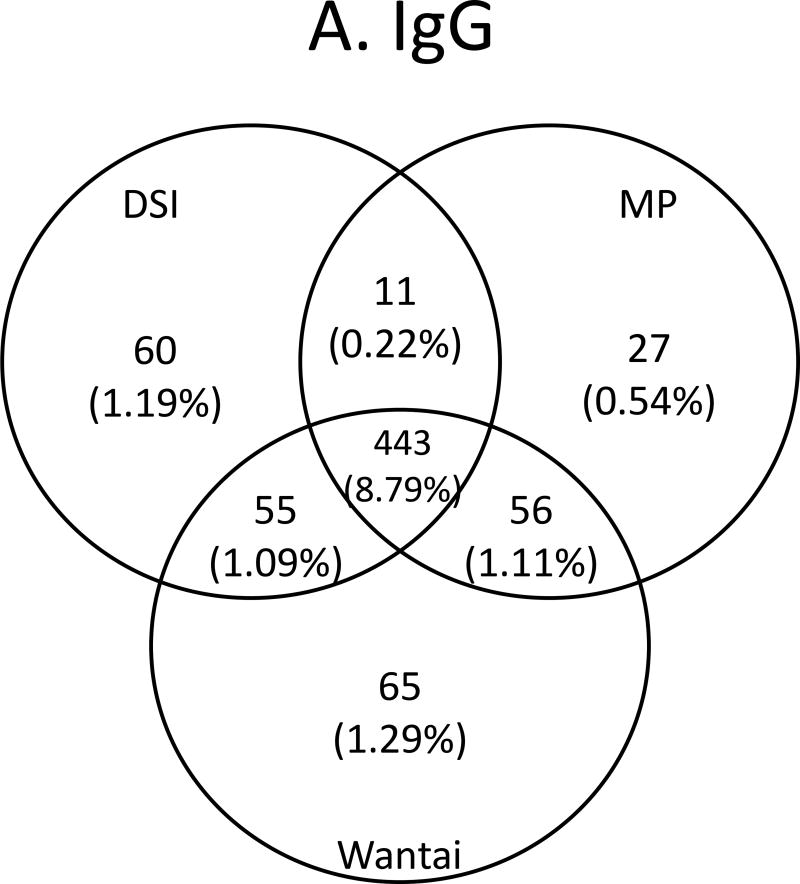
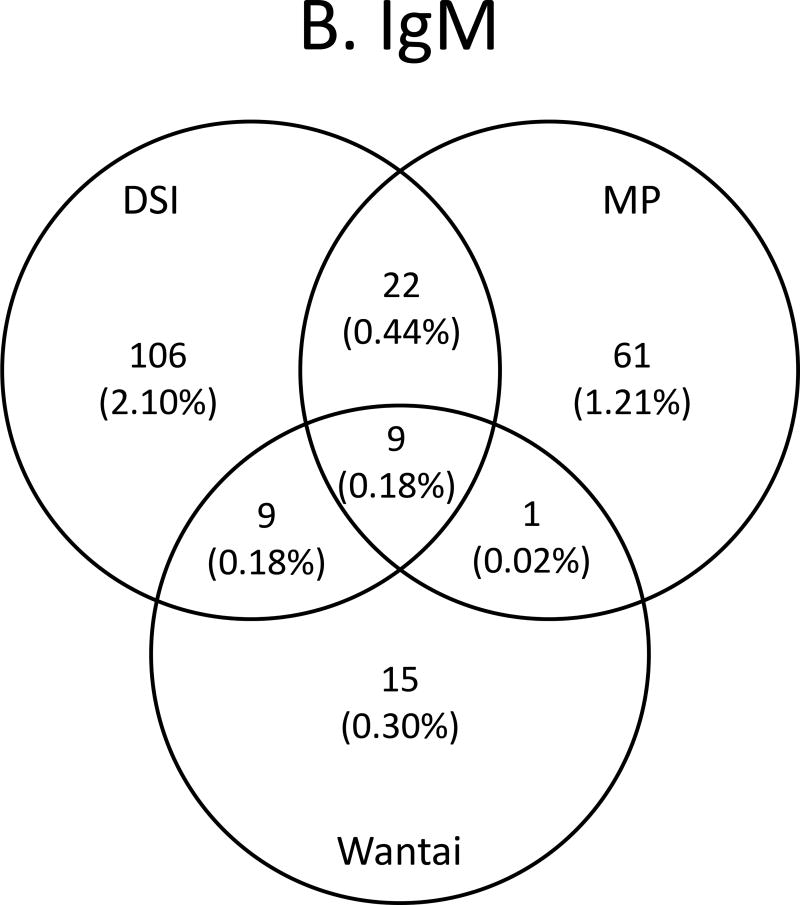
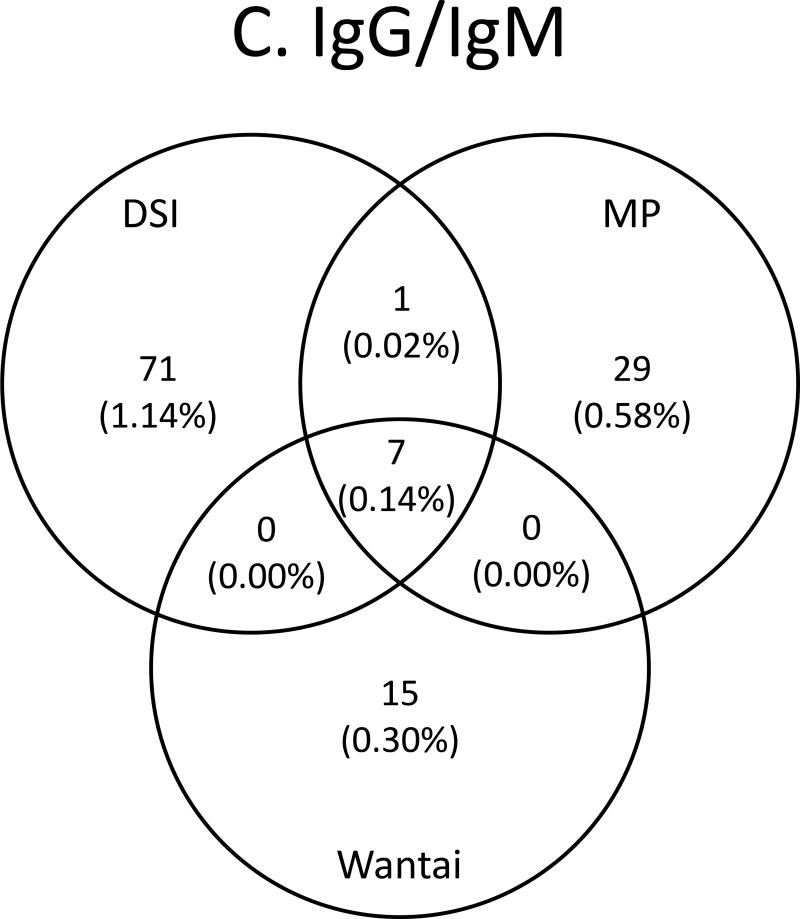
Concordance between assays is shown in Figures 4A–C and Supplemental Table 1. Concordance among IgG anti-HEV/total anti-HEV -reactive specimens ranged from 443 samples reactive by all three assays, 454 to 499 samples reactive by two assays, and from 537 to 619 samples reactive by any given assay for an overall agreement of 84%. There was a reduction in prevalence of 11.41% (reactive by any assay) to 8.79% (reactive in all three assays). The highest agreement was between MP and Wantai (Figure 4A). For IgM, concordance ranged from 34 to 146 reactive by any given assay, 10 to 31 samples reactive by two assays, and for nine samples reactive by all three assays for an overall agreement of 22%. There was a reduction in prevalence from 1.81% (reactive by any assay) to 0.18% (reactive in all three assays). The highest agreement was between DSI and MP (Figure 4B). When concordance was compared between samples having both IgG anti-HEV /total anti-HEV and IgM anti-HEV reactivity, from 15 to 71 were reactive by the assays from any given manufacturer, and for seven samples were reactive by all six assays for an overall agreement of 40%, and a prevalence of 0.14%. The highest agreement was again between DSI and MP (Figure 4C). Nine samples had concordant IgM anti-HEV reactivity (including seven with concordant IgM/IgG reactivity), giving a frequency of 1 per 560 (0.18%; Figure 4B). Since IgM is a marker of recent infection that develops within 2 to 6 weeks following infection, these nine IgM-concordantly reactive donors were considered to be likely HEV infected recently. As Table 3 shows, the nine donors came from two states: Kentucky and Missouri; all except one 18-year old donor ranged in age from 47–81 years, and six were male.
Figure 4.
Venn diagrams of anti-HEV assay concordance and discordance. A. IgG/total antibody assays, B. IgM assays and C. IgG/total antibody and IgM assays combined.
Table 3.
Characteristics of the nine IgM anti-HEV positive specimens concordant across all three vendors.
| IgG S/CO | IgM S/CO | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| sample | DSI | MP | Wantai | DSI | MP | Wantai | Age | Gender | State |
| 1 | 0.01 | 0.08 | 0.13 | 3.03 | 4.61 | 1.42 | 18 | M | KY |
| 2 | 0.83 | 8.27 | 0.34 | 6.28 | 3.23 | 5.70 | 74 | M | MO |
| 3 | 6.28 | 8.99 | 14.14 | 1.73 | 1.13 | 2.31 | 68 | M | MO |
| 4 | 5.45 | 6.40 | 10.00 | 2.88 | 1.12 | 1.13 | 59 | F | MO |
| 5 | 5.18 | 9.00 | 16.60 | 3.33 | 1.44 | 1.12 | 47 | M | KY |
| 6 | 6.40 | 8.98 | 17.27 | 4.27 | 1.99 | 1.38 | 68 | M | MO |
| 7 | 5.51 | 8.44 | 14.34 | 4.66 | 2.31 | 7.30 | 81 | F | MO |
| 8 | 5.12 | 8.91 | 16.42 | 5.41 | 2.44 | 1.27 | 57 | F | KY |
| 9 | 2.29 | 8.91 | 6.33 | 7.84 | 5.09 | 2.65 | 63 | M | MO |
S/CO, signal/cutoff; age in years; State, KY – Kentucky, MO – Missouri. Samples 3 to 9 are concordant for IgG and IgM across all three vendors.
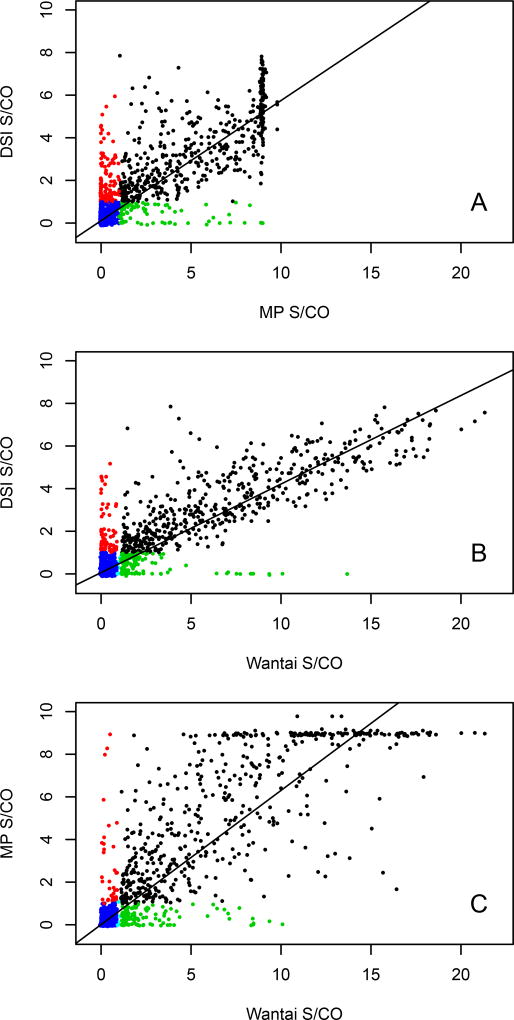
Regression analysis (Figures 5A–C) was initially used to assess correlations among the assays. Pearson’s product-moment correlation between the MP total antibody assay and the Wantai IgG assay was r(n=5038) = 0.89 (95% CI, 0.88–0.90), p < 0.00001, indicating strong correlation between these assays. However, this correlation was due to the large number of negative results (Fig 5C). If samples that are negative in both assays (“true negatives”) were removed, the correlation for the remaining samples was r(n=497) = 0.74 (95% CI, 0.70–0.80), p < 0.00001. Although this value could be considered to show reasonable agreement between these 2 assays, a plot of signal-to-cutoff (S/CO) ratios indicates that the variation between assays is heteroskedastic (Figures 5A–C; p <0.00001, using the Breusch-Pagan test for heteroskedasticity among all three IgG assays, and Supplemental Figure 3). An examination of the panels in Supplemental Figure 3 shows that no pair of assays has constant variation of data points around the regression line through the data. This result indicates that the null hypothesis of constant variance can be rejected between any two pairs of IgG assays, or, there is no relationship between S/CO ratios between pairs of assays. This outcome is similar if the DSI IgG assay was compared against either the MP or the Wantai assays (Figures 5A and 5B). The lack of constant variation is still seen when samples with saturated signal with the MP assay are removed (Supplemental Figure 3). The data suggest that each assay may be detecting an epitope not detectable by the other assays. This is further seen among the discordant samples, which are negative by one assay, but have a high S/CO by another assay (Figure 5, red and green data points). In some cases, the discrepancies are separated by more than 6 sigmas (Supplemental Figure 3).
Figure 5.
Pairwise comparison of signal to cutoff (S/CO) specimen values among the DSI, MP Biomedicals and Wantai IgG/total antibody assays. S/CO paired data are plotted as closed circles. The solid line is the regression line through the data between each pair of assays. Blue circles denote S/CO data pairs that are both non-reactive, black circles denote that both results are reactive. A. Results from DSI compared with MP Biomedicals (red circles; DSI reactive and MP non-reactive, green circles (DSI non-reactive and MP reactive)), B. DSI compared with Wantai (red circles; DSI reactive and Wantai non-reactive, green circles (DSI non-reactive and Wantai reactive)) and C. MP Biomedicals compared with Wantai (red circles; MP reactive and Wantai non-reactive, green circles (MP non-reactive and Wantai reactive)).
Discussion
It has been estimated that there are 20 million HEV infections worldwide annually, leading to about 3.3 million symptomatic cases of hepatitis E and 56,600 HEV-related deaths30,31. In the US, an analysis of clinical cases of non A-C acute hepatitis from 2005 to 2012 found that 26 (17%) of 154 cases were due to HEV infection. There was a near-even split between travel-associated and autochthonous hepatitis E cases. The autochthonous cases were all infected by HEV genotype 3 and tended to occur among older patients compared to those with travel- associated hepatitis E32.
The determination of anti-HEV serostatus remains enigmatic, and the results of various assays are often divergent. This is particularly true of healthy individuals like blood donors27. Recently pairwise concordance among three commercially available IgG anti-HEV assays and one laboratory developed assay ranged from 56 to 87% with a concordance of 52% observed in all samples tested among all four assays26. Another study of five assays for the detection of IgM and IgG found concordances of 71% and 70%. The limit of detection varied up to 19-fold for the IgM assays and 17-fold for the IgG assays33. A study of healthy US citizens using NHANES specimens found a decrease in IgG prevalence from 10.2% during 1988–1994 to 6% during 2009–2010 using the DSI assay19. IgG anti-HEV seroprevalence in NIH blood donors was 21.8% in 2006 with a decrease to 16.0% in 2012 using the Wantai assay20, and 7.7% in ARC blood donors in 2013 using the MP assay21. The decreases in anti-HEV IgG rates observed in the NHANES and NIH studies are similar to observations seen in Germany where a study of 45 subjects found that anti-HEV IgG concentrations decreased significantly after 5 years34, and a study of Dutch blood donors showed that IgG seroprevalence decreased from 19.8% in 1998 to 12.7% in 201118. These last two studies observed seroreversion and HEV reinfections in some individuals despite pre-existing HEV antibodies.
This study examined the anti-HEV IgG/total anti-HEV and IgM anti-HEV detection percentages in US blood donors and the performance of three commercial assays for IgG/total and IgM antibodies in this donor population. The assays used were from DSI, MP and Wantai. Overall the three assays yielded similar results. Within the total donor population, the IgG anti-HEV/total anti-HEV positivity was 11.41% (range by assay of 10.65% to 12.28%) and the IgM positivity was 1.81% (range by assay of 0.67% to 2.90%) (Table 1). An increase in anti-HEV IgG positivity was seen with increasing age, regardless of gender, as has been seen in other studies (Figure 2, top panel)19–21. However, no trend was seen for IgM seropositivity with increasing age (Figure 2, bottom panel). Differences were seen between genders, regardless of age, where more males than females were IgG reactive (Table 2). These results are in agreement with the earlier ARC study that only used the MP assay to test anti-HEV IgG21. However, in our study, the gender difference was significant in the total donor population with the DSI (p< 0.005) and Wantai (p< 0.01) assays, but not significant in the MP assay. When the five states with the most donors were compared individually, the differences between males and females were not significant except in Missouri, and only with the DSI assay (p< 0.05).
Agreement between the assays was better for the IgG/total anti-HEV than the IgM assays. The overall agreement among the IgG/total anti-HEV assays was 84% but was only 22% for the IgM assays (Figures 4A–C). Some of the poor agreement among the IgM assays appears to be due to the discordance between the DSI and MP assays (Fig. 4B). The difference between discordant and concordant results was 2 logs higher in the IgM assays than the IgG/total anti-HEV assays. The wide range in these values is due to the low concordance between the IgM assays (Figure 4B), which in turn is probably due to difference in the epitopes and assay formats used to detect IgM anti-HEV.
Despite the overall 84% agreement between the IgG/total antibody assays, the disparity in sample detection as well as S/CO ratios among discordant samples seen among all three assays could indicate that each assay is detecting an epitope(s) not detectable by the other assays (Figure 5, red and green data points). The presence of heteroskedasticity among the IgG/total assays indicates that the null hypothesis of constant variance should be rejected, further supporting the conclusion of differing epitope detection. Alternatively, these differences could be due in part to the varying formats used in these assays or waning or less avid antibodies in the donor population. Differences in detection rates between the IgG anti-HEV assays (DSI and Wantai) and the total antibody assay could be due to IgM-positive detection by the total antibody assay (the MP assay) not detected by the IgG-only assays. However, MP IgM anti-HEV positivity did not correlate with total antibody positivity. Detection of IgA-containing samples cannot be excluded, but there is no way to test for IgA anti-HEV. As has been previously suggested, these data indicate that a panel of well-characterized plasma samples from HEV-infected individuals needs to be created to validate anti-HEV assay performance. Additionally, some as yet undiscovered factor, as with protein Fv, may be interfering with detection of HEV epitopes35. Fv, a Fab binding factor, was found to interfere with an early in-house anti-HEV assay resulting in false-negative results.
The major limitations to this study include the fact that the assays use different antigens and detection formats with no established method to determine their absolute performance characteristics. Blood donors are a select low-risk population that does not mirror the general US population36. The assays used have been validated for clinical purposes in symptomatic persons but not for epidemiological studies in asymptomatic individuals. In addition, the performance of antibody assays is much better in patients with acute infection than in those with past infection37. Also, specimens initially positive by the Wantai assays were not retested as recommended in the manufacturer’s instructions; however, one would expect a high correlation between initially reactive and repeatedly reactive samples in a commercial assay that has been widely used for anti-HEV studies worldwide. In addition, any potential reductions in IgG or IgM reactivity due to the absence of repeat testing are not expected to reduce the heteroskedasticity seen in the assays, nor to significantly alter the lack of concordance seen among the IgM assays.
In conclusion, our data indicate that these HEV IgG/total anti-HEV assays are useful for examining seroprevalence and associated trends in seroprevalence with an inter-assay agreement of 84%; however, the disagreement between these assays indicates that there is an associated discordance rate or difference in target detection driving variability as seen by the heteroskedasticity evident among the IgG assays. The case for the IgM assays is worse since no IgM assay evaluated here demonstrated clinical utility in the blood donor population tested, although an IgM response in HEV RNA-positive blood donors has been well characterized38. These data suggest that more reliable information on prevalence may be obtained from using concordant reactive results from multiple assays. Despite these limitations, and assuming that the IgM-positive period exceeds that for HEV RNA detection, we calculated a rate of recent infection of 0.18% (1:560) based on concordance among all three anti-HEV IgM assays, noting that none of the nine IgM-reactive samples was HEV-RNA positive (data not shown). A recent study in 20,000 US blood donors found low HEV-RNA positivity (0.01%) and thus a low burden of new infection in US blood donors21.
Supplementary Material
Supplemental Figure 1. Comparison of age and gender adjusted seropositivity among the four states with the highest number of donors (Missouri, Kentucky, Illinois and Indiana)(upper panel) versus the actual data from the same four states (lower panel).
Supplemental Figure 2. Comparison of signal to cutoff (S/CO) ratios for IgG reactive specimens by age group and IgG assay. Age ranges were created as described in Figure 2. Data are shown as box plots by age group for each assay. Top panel, DSI; middle panel, MP and bottom panel, Wantai.
Supplemental Figure 3. Pairwise comparison of signal to cutoff (S/CO) specimen ratios among the DSI, MP Biomedicals and Wantai IgG assays after removal of saturated signal samples from the MP assay. Samples with an S/CO ratio greater than or equal to 8.0 with the MP assay were removed from analysis. The data from each assay were normalized with respect to the sample with the highest S/CO from each assay. Solid line, regression line through the data; dashed lined, normalized cutoff for each assay and dotted line, the six sigma lines around the regression line.
Acknowledgments
We would like to acknowledge the generous support of Roche Molecular Systems for providing the antibody reagent kits. We would like to thank Chong-Gee Teo and reviewers at CDC and the journal for reading the paper and suggesting modifications.
Funding: Antibody reagent kits were purchased by Roche Molecular Systems.
ABBREVIATIONS
- ARC
American Red Cross
- DSI
Diagnostic Systems Incorporated
- HEV
hepatitis E virus
- HRP
horseradish peroxidase
- MP
MP Biomedicals
- NHANES
National Health and Nutrition Examination Survey
- S/CO
signal-to-cutoff
Footnotes
Disclaimer
The findings and conclusions in this report have not been formally disseminated by the Centers for Disease Control and Prevention/Agency for Toxic Substances and Disease Registry. It does not represent and should not be construed to represent any agency determination or policy. Use of trade names is for identification only and does not imply endorsement by the U.S. Department of Health and Human Services, the Public Health Service, or the Centers for Disease Control and Prevention.
Conflict of interest: None
References
- 1.Khuroo MS. Discovery of hepatitis E: The epidemic non-A, non-B hepatitis 30 years down the memory lane. Virus Res. 2011;161:3–14. doi: 10.1016/j.virusres.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Purdy MA, Smith DB, Simmonds P, Emerson SU, Harrison T, Meng X-J, Okamoto H, van der Poel WHM, Jameel S. New classification scheme for Hepeviridae ICTV taxonomic proposal 2014.008a-hV.A.v6.Hepeviridae (approved) 2014 https://talk.ictvonline.org/files/ictv_official_taxonomy_updates_since_the_8th_report/m/vertebrate-official/5150.
- 3.Smith DB, Simmonds P, Jameel S, Harrison TJ, Meng X-J, Okamoto H, Van der Poel WHM, Purdy MA. Consensus Proposals for Classification of the Family Hepeviridae. J Gen Virol. 2014;95:2223–32. doi: 10.1099/vir.0.068429-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee G-H, Tan B-H, Teo EC-Y, Lim S-G, Dan Y-Y, Wee A, Aw PPK, Zhu Y, Hibberd ML, Tan C-K, Purdy MA, Teo C-G. Chronic Infection With Camelid Hepatitis E Virus in a Liver-transplant Recipient Who Regularly Consumes Camel Meat and Milk. Gastroenterology. 2016;150:355–7. doi: 10.1053/j.gastro.2015.10.048. [DOI] [PubMed] [Google Scholar]
- 5.Teo C-G. Fatal outbreaks of jaundice in pregnancy and the epidemic history of hepatitis E. Epidemiol Infect. 2012;140:767–87. doi: 10.1017/S0950268811002925. [DOI] [PubMed] [Google Scholar]
- 6.Krawczynski K, Kamili S, Aggarwal R. Global epidemiology and medical aspects of hepatitis E. Forum (Genova.) 2001;11:166–79. [PubMed] [Google Scholar]
- 7.Izopet J, Kamar N. Hepatite E: de la transmission zoonotique du virus a l'evolution chronique de l'infection chez l'immunodeprime. Med Sci (Paris) 2008;24:1023–5. doi: 10.1051/medsci/200824121023. [DOI] [PubMed] [Google Scholar]
- 8.Tavitian S, Peron JM, Huynh A, Mansuy JM, Ysebaert L, Huguet F, Vinel JP, Attal M, Izopet J, Recher C. Hepatitis E virus excretion can be prolonged in patients with hematological malignancies. J Clin Virol. 2010;49:141–4. doi: 10.1016/j.jcv.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 9.Legrand-Abravanel F, Kamar N, Sandres-Saune K, Garrouste C, Dubois M, Mansuy JM, Muscari F, Sallusto F, Rostaing L, Izopet J. Characteristics of Autochthonous Hepatitis E Virus Infection in Solid-Organ Transplant Recipients in France. J Infect Dis. 2010;202:835–44. doi: 10.1086/655899. [DOI] [PubMed] [Google Scholar]
- 10.Colson P, Borentain P, Queyriaux B, Kaba M, Moal V, Gallian P, Heyries L, Raoult D, Gerolami R. Pig Liver Sausage as a Source of Hepatitis E Virus Transmission to Humans. J Infect Dis. 2010;202:825–34. doi: 10.1086/655898. [DOI] [PubMed] [Google Scholar]
- 11.Tei S, Kitajima N, Takahashi K, Mishiro S. Zoonotic transmission of hepatitis E virus from deer to human beings. Lancet. 2003;362:371–3. doi: 10.1016/S0140-6736(03)14025-1. [DOI] [PubMed] [Google Scholar]
- 12.Mizuo H, Yazaki Y, Sugawara K, Tsuda F, Takahashi M, Nishizawa T, Okamoto H. Possible risk factors for the transmission of hepatitis E virus and for the severe form of hepatitis E acquired locally in Hokkaido, Japan. J Med Virol. 2005;76:341–9. doi: 10.1002/jmv.20364. [DOI] [PubMed] [Google Scholar]
- 13.Gotanda Y, Iwata A, Ohnuma H, Yoshikawa A, Mizoguchi H, Endo K, Takahashi M, Okamoto H. Ongoing subclinical infection of hepatitis E virus among blood donors with an elevated alanine aminotransferase level in Japan. J Med Virol. 2007;79:734–42. doi: 10.1002/jmv.20834. [DOI] [PubMed] [Google Scholar]
- 14.Lapa D, Capobianchi MR, Garbuglia AR. Epidemiology of Hepatitis E Virus in European Countries. Int J Mol Sci. 2015;16:25711–43. doi: 10.3390/ijms161025711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillois Y, Abravanel F, Miura T, Pavio N, Vaillant V, Lhomme S, Le Guyader FS, Rose N, Le Saux J-C, King LA, Izopet J, Couturier E. High proportion of asymptomatic infections in an outbreak of hepatitis E associated with a spit-roasted piglet, France, 2013. Clin Infect Dis. 2015;62:351–7. doi: 10.1093/cid/civ862. [DOI] [PubMed] [Google Scholar]
- 16.Slot E, Hogema BM, Riezebos-Brilman A, Kok TM, Molier M, Zaaijer HL. Silent hepatitis E virus infection in Dutch blood donors, 2011 to 2012. Euro Surveill. 2013;18 doi: 10.2807/1560-7917.es2013.18.31.20550. pii: 20550. [DOI] [PubMed] [Google Scholar]
- 17.Mansuy J-M, Bendall R, Legrand-Abravanel F, Saune K, Miedouge M, Ellis V, Rech H, Destruel F, Kamar N, Dalton HR, Izopet J. Hepatitis E Virus Antibodies in Blood Donors, France. Emerg Infect Dis. 2011;17:2309–12. doi: 10.3201/eid1712.110371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hogema BM, Molier M, Slot E, Zaaijer HL. Past and present of hepatitis E in the Netherlands. Transfusion. 2014;54:3092–6. doi: 10.1111/trf.12733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teshale EH, Denniston MM, Drobeniuc J, Kamili S, Teo CG, Holmberg SD. Decline in Hepatitis E Virus Antibody Prevalence in the United States from 1988–1994 to 2009–2010. J Infect Dis. 2015;211:366–73. doi: 10.1093/infdis/jiu466. [DOI] [PubMed] [Google Scholar]
- 20.Xu C, Wang RY, Schechterly CA, Ge SX, Shih JW, Xia N-S, Luban NLC, Alter HJ. An assessment of hepatitis E virus (HEV) in US blood donors and recipients: no detectable HEV RNA in 1939 donors tested and no evidence for HEV transmission to 362 prospectively followed recipients. Transfusion. 2013;53:2505–11. doi: 10.1111/trf.12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stramer SL, Moritz ED, Foster GA, Ong E, Linnen JM, Hogema BM, Mak M, Chia CP, Dodd RY. Hepatitis E virus: seroprevalence and frequency of viral RNA detection among US blood donors. Transfusion. 2015;56:481–8. doi: 10.1111/trf.13355. [DOI] [PubMed] [Google Scholar]
- 22.Chalupa P, Vasickova P, Pavlik I, Holub M. Endemic hepatitis E in the Czech Republic. Clin Infect Dis. 2014;58:509–16. doi: 10.1093/cid/cit782. [DOI] [PubMed] [Google Scholar]
- 23.Ijaz S, Said B, Boxall E, Smit E, Morgan D, Tedder RS. Indigenous Hepatitis E in England and Wales From 2003 to 2012: Evidence of an Emerging Novel Phylotype of Viruses. J Infect Dis. 2014;209:1212–8. doi: 10.1093/infdis/jit652. [DOI] [PubMed] [Google Scholar]
- 24.Centre National de Reference. Plan du rapport annuel d'activite: Virus des hepatites a transmission enterique [monograph on the internet]. http://www.cnrvha-vhe.org/wp-content/uploads/2012/03/2014-Rap-Act-VHE-VHA.pdf. 2015 Available from: http://www.cnrvha-vhe.org/wp-content/uploads/2012/03/2014-Rap-Act-VHE-VHA.pdf.
- 25.Drobeniuc J, Meng J, Reuter G, Green-Montfort T, Khudyakova N, Dimitrova Z, Kamili S, Teo CG. Serologic Assays Specific to Immunoglobulin M Antibodies against Hepatitis E Virus: Pangenotypic Evaluation of Performances. Clin Infect Dis. 2010;51:e24–e7. doi: 10.1086/654801. [DOI] [PubMed] [Google Scholar]
- 26.Kodani M, Ahmed N, Tejada-Strop A, Poe A, Denniston MM, Drobeniuc J, Kamili S. Variability in the performance characteristics of IgG anti-HEV assays and its impact on reliability of seroprevalence rates of hepatitis E. J Med Virol. 2016;89:1055–61. doi: 10.1002/jmv.24741. [DOI] [PubMed] [Google Scholar]
- 27.Wenzel JJ, Preiss J, Schemmerer M, Huber B, Jilg W. Test Performance Characteristics of Anti-HEV IgG Assays Strongly Influence Hepatitis E Seroprevalence Estimates. J Infect Dis. 2012;207:497–500. doi: 10.1093/infdis/jis688. [DOI] [PubMed] [Google Scholar]
- 28.Breusch TS, Pagan AR. A Simple Test for Heteroskedasticity and Random Coefficient Variation. Econometrica. 1979;47:1287–94. [Google Scholar]
- 29.R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 30.Rein DB, Stevens GA, Theaker J, Wittenborn JS, Wiersma ST. The global burden of hepatitis E virus genotypes 1 and 2 in 2005. Hepatology. 2012;55:988–97. doi: 10.1002/hep.25505. [DOI] [PubMed] [Google Scholar]
- 31.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, AlMazroa MA, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Barker-Collo S, Bartels DH, Bell ML, Benjamin EJ, Bennett D, Bhalla K, Bikbov B, Bin Abdulhak A, Birbeck G, Blyth F, Bolliger I, Boufous S, Bucello C, Burch M, Burney P, Carapetis J, Chen H, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, Dabhadkar KC, Dahodwala N, De Leo D, Degenhardt L, Delossantos A, Denenberg J, Des Jarlais DC, Dharmaratne SD, Dorsey ER, Driscoll T, Duber H, Ebel B, Erwin PJ, Espindola P, Ezzati M, Feigin V, Flaxman AD, Forouzanfar MH, Fowkes FGR, Franklin R, Fransen M, Freeman MK, Gabriel SE, Gakidou E, Gaspari F, Gillum RF, Gonzalez-Medina D, Halasa YA, Haring D, Harrison JE, Havmoeller R, Hay RJ, Hoen B, Hotez PJ, Hoy D, Jacobsen KH, James SL, Jasrasaria R, Jayaraman S, Johns N, Karthikeyan G, Kassebaum N, Keren A, Khoo J-P, Knowlton LM, Kobusingye O, Koranteng A, Krishnamurthi R, Lipnick M, Lipshultz SE, Ohno SL, Mabweijano J, MacIntyre MF, Mallinger L, March L, Marks GB, Marks R, Matsumori A, Matzopoulos R, Mayosi BM, McAnulty JH, McDermott MM, McGrath J, Memish ZA, Mensah GA, Merriman TR, Michaud C, Miller M, Miller TR, Mock C, Mocumbi AO, Mokdad AA, Moran A, Mulholland K, Nair MN, Naldi L, Narayan KMV, Nasseri K, Norman P, O'Donnell M, Omer SB, Ortblad K, Osborne R, Ozgediz D, Pahari B, Pandian JD, Rivero AP, Padilla RP, Perez-Ruiz F, Perico N, Phillips D, Pierce K, Pope III CA, Porrini E, Pourmalek F, Raju M, Ranganathan D, Rehm JT, Rein DB, Remuzzi G, Rivara FP, Roberts T, De Leon FR, Rosenfeld LC, Rushton L, Sacco RL, Salomon JA, Sampson U, Sanman E, Schwebel DC, Segui-Gomez M, Shepard DS, Singh D, Singleton J, Sliwa K, Smith E, Steer A, Taylor JA, Thomas B, Tleyjeh IM, Towbin JA, Truelsen T, Undurraga EA, Venketasubramanian N, Vijayakumar L, Vos T, Wagner GR, Wang M, Wang W, Watt K, Weinstock MA, Weintraub R, Wilkinson JD, Woolf AD, Wulf S, Yeh P-H, Yip P, Zabetian A, Zheng Z-J, Lopez AD, Murray CJL. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2010;380:2095–128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drobeniuc J, Greene-Montfort T, Le N-T, Mixson-Hayden TR, Ganova-Raeva L, Dong C, Novak RT, Sharapov UM, Tohma RA, Teshale E, Kamili S, Teo C-G. Laboratory-based Surveillance for Hepatitis E Virus Infection, United States, 2005–2012. Emerg Infect Dis. 2013;19:218–22. doi: 10.3201/eid1902.120961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Norder H, Karlsson M, Mellgren A, Konar J, Sandberg E, Lasson A, Castedal M, Magnius L, Lagging M. Diagnostic performance of five assays for anti-HEV IgG and IgM in a large cohort study. J Clin Microbiol. 2016;54:549–55. doi: 10.1128/JCM.02343-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schemmerer M, Rauh C, Jilg W, Wenzel JJ. Time course of hepatitis E-specific antibodies in adults. J Viral Hep. 2017;24:75–9. doi: 10.1111/jvh.12621. [DOI] [PubMed] [Google Scholar]
- 35.Diallo A, Lazizi Y, Le Guenno B, Pillot J. Hepatitis-E-virus-associated antigen: improved detection in stools by protein Fv removal. Res Virol. 1991;142:449–59. doi: 10.1016/0923-2516(91)90067-d. [DOI] [PubMed] [Google Scholar]
- 36.Goldman M, Steele WR, Di Angelantonio E, van den Hurk K, Vassallo RR, Germain M, O'Brien SF. Comparison of donor and general population demographics over time: a BEST Collaborative group study. Transfusion. 2017;57:2469–76. doi: 10.1111/trf.14307. [DOI] [PubMed] [Google Scholar]
- 37.Aggarwal R, Goel A. Advances in hepatitis E - I: Virology, pathogenesis and diagnosis. Expert Rev Gastroenterol Hepatol. 2016;10:1053–63. doi: 10.1080/17474124.2016.1185362. [DOI] [PubMed] [Google Scholar]
- 38.Tedder RS, Tettmar KI, Brailsford SU, Said B, Ushiro-Lumb I, Kitchen A, Morgan D, Lattimore S, Tossell J, Ijaz S, Hewitt PE. Virology, serology, and demography of hepatitis E viremic blood donors in South East England. Transfusion. 2016;56:1529–36. doi: 10.1111/trf.13498. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Comparison of age and gender adjusted seropositivity among the four states with the highest number of donors (Missouri, Kentucky, Illinois and Indiana)(upper panel) versus the actual data from the same four states (lower panel).
Supplemental Figure 2. Comparison of signal to cutoff (S/CO) ratios for IgG reactive specimens by age group and IgG assay. Age ranges were created as described in Figure 2. Data are shown as box plots by age group for each assay. Top panel, DSI; middle panel, MP and bottom panel, Wantai.
Supplemental Figure 3. Pairwise comparison of signal to cutoff (S/CO) specimen ratios among the DSI, MP Biomedicals and Wantai IgG assays after removal of saturated signal samples from the MP assay. Samples with an S/CO ratio greater than or equal to 8.0 with the MP assay were removed from analysis. The data from each assay were normalized with respect to the sample with the highest S/CO from each assay. Solid line, regression line through the data; dashed lined, normalized cutoff for each assay and dotted line, the six sigma lines around the regression line.