Abstract
We report the main characteristics of ‘Citricoccus massiliensis’strain Marseille-P4330, a new species within the genus Citricoccus. This strain was isolated from the skin of a healthy human man living in Dielmo, Senegal, Western Africa.
Keywords: Citricoccus massiliensis, culturomics, human microbiota, human skin
Skin was thought to harbour a limited number of microbial communities, primarily composed of aerobic cocci. Genomic studies have offered additional insight into the diversity of skin microbiota and have also allowed for quantitative analyses [1]. The earliest descriptions of skin microbiota were based on culture-dependent studies. In our laboratory, a culturomics approach, consisting of rapid identification of grown colonies using matrix-assisted desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), has been developed to isolate previously uncultured human bacteria [2], [3]. We used this technique to explore the human skin bacterial repertoire from skin swabs collected from healthy patients living in rural Senegal. The project was approved by the National Ethics Committee of Senegal (no. 53/MSAS/DPRS/CNERS, 31 March 2015).
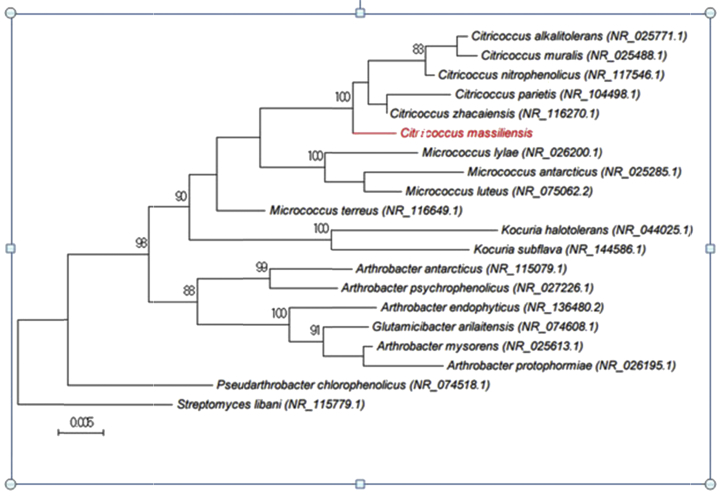
In 2017, we isolated from skin swabs a bacterial strain that could not be identified by our MALDI-TOF MS screening on a Microflex spectrometer (Bruker Daltonics, Bremen, Germany) [4]. The 16S rRNA gene was sequenced using fD1-rP2 primers as previously described using a 3130-XL sequencer (Applied Biosciences, Saint-Aubin, France) [5]. Strain Marseille-P4330 exhibited a 98.61% sequence identity with Citricoccus nitrophenolicus strain NPP1 (GenBank accession no. NR_112632) (Fig. 1). Because the strain showed a similarity of 16S rRNA gene sequence <98.65% with its phylogenetically closest species with standing in nomenclature, we here propose the creation of a new species [6].
Fig. 1.
Phylogenetic tree showing position of ‘Citricoccus massiliensis’ Marseille-P4330 relative to other phylogenetically close neighbours. 16S rRNA gene sequences were aligned using CLUSTAL W, and phylogenetic inferences were obtained using maximum-likelihood method within MEGA software. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 500 times to generate majority consensus tree. Scale bar indicates 0.5% of nucleotide sequence divergence.
Strain Marseille-P4330 was isolated in a skin swab from a 19-year-old man living in Dielmo, Senegal, Western Africa. The growth occurred in Columbia agar culture media with colistin and nalidixic acid (bioMérieux, Marcy l’Etoile, France) and 5% sheep's blood (bioMérieux). The initial agar-grown colonies were obtained after 24 to 48 hours' incubation at 37°C. Colonies were yellowish, circular, shiny, smooth and convex with regular edges and presented a mean diameter of 1.2 mm. The strain was an aerobic, Gram-positive, spore-forming and motile coccus-shaped bacterium, with positive catalase reaction and negative oxidase reaction.
Thus, we propose to classify strain Marseille-P4330 as a new species of the genus Citricoccus within the family Micrococcaceae, phylum Actinobacteria (GenBank accession no. LT960590).
Description of new species ‘Citricoccus massiliensis’
‘Citricoccus massiliensis’ had yellowish, circular, shiny, smooth and convex colonies with a mean diameter of 1.2 mm. The strain was an aerobic, Gram-positive, spore-forming and motile coccus-shaped bacterium, with positive catalase reaction and negative oxidase reaction. Strain Marseille-P4330T (= CSUR P4330) is the type strain of the new species ‘Citricoccus massiliensis’ (ma.ssi.li.en'sis, L. masc. adj. massiliensis, pertaining to Massilia, the Roman name of Marseille, where the strain was isolated).
Nucleotide sequence accession number
The 16S r-RNA gene sequence was deposited in GenBank under accession number LT960590.
Deposit in a culture collection
Strain Marseille-P4330 was deposited in the Collection de souches de l’Unité des Rickettsies (CSUR, WDCM 875) under number P4330.
Acknowledgments
Acknowledgements.
This study was funded by the Fondation Méditerranée Infection. We thank M. D. Mbogning Fonkou for technical assistance.
Conflict of interest
None declared.
References
- 1.Murillo N., Raoult D. Skin microbiota: overview and role in the skin diseases acnevulgaris and rosacea. Future Microbiol. 2013;8:209–222. doi: 10.2217/fmb.12.141. [DOI] [PubMed] [Google Scholar]
- 2.Lagier J.C., Khelifia S., Alou M.T., Ndongo S., Dione N., Hugon P. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1 doi: 10.1038/nmicrobiol.2016.203. [DOI] [PubMed] [Google Scholar]
- 3.Lagier J.C., Elkarkouri K., Rivet R., Courderc C., Raoult D., Fournier P.E. Non contiguous-finished genome sequence and description of Senegalemassilia anaerobia gen. nov., sp. nov. Stand Genomic Sci. 2013;7:343–356. doi: 10.4056/sigs.3246665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lo C.I., Fall B., Sambe-Ba B., Diawara S., Gueye M.Z., Medianikov O. MALDI–mass spectometry: a powerful tool for clinical microbiology at Hopital Principalde Dakar, Senegal (West Africa) PLoS One. 2015;10 doi: 10.1371/journal.pone.0145889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drancourt M., Bollet C., Carlioz A., Martelin R., Gyral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000;51:2182–2194. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramasamy D., Mishra A.K., Lagier J.C., Padhmanabhan R., Rossi M., Sentausa E. A polyphsic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int J Syst Evol Microbiol. 2014;64:384–391. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]