Abstract
Klebsiella spp. isolates from community-acquired infections were characterized. A total of 39 Klebsiella spp. isolates were obtained from outpatients at four rural hospitals in Mexico (2013–2014). The biochemical tests identified all as being K. pneumoniae. The molecular multiplex-PCR test identified 36 (92.4%) K. pneumoniae isolates and one (2.5%) K. variicola isolate, and phylogenetic analysis of the rpoB gene identified two isolates (5.1%) belonging to K. quasipneumoniae subsp. quasipneumoniae and K. quasivariicola. The last one was confirmed by phylogenetic analysis of six-loci concatenated genes. Mostly the isolates were multidrug resistant; however, a minority were extended-spectrum β-lactamase producing (10.2%). The extended-spectrum β-lactamase CTX-M-15 gene was identified in these isolates. Analysis of biofilm production and the hypermucoviscosity phenotype showed a total of 35 (92.3%) and seven (17.9%) of the isolates were positive for these phenotypes respectively. The K2 (4/39, 10.2%), K5 (2/39, 5.1%) and K54 (1/39, 2.5%) serotypes were identified in seven (17.9%) of the isolates, and only 28.5% (2/7) hypermucoviscous isolates were positive for the K2 and K5 serotypes. In general, the sequence type (ST) analysis and phylogenetic analysis of seven multilocus sequence typing loci were heterogeneous; however, ST29 was the most prevalent ST in the analysed isolates, accounting for 19% (4/21) of the total isolates. Two of the four ST29 isolates had the hypermucoviscosity phenotype. The virulence factors for fimbriae were the most prevalent, followed by siderophores. Community-acquired infections are caused by various species from Klebsiella genus, with different profiles of antibiotic resistance and heterogeneous virulence factors.
Keywords: Antimicrobial susceptibility, Bacterial resistance, Cephalosporin resistance, Community infection, ESBL, Hypermucoviscosity
Introduction
Klebsiella pneumoniae is a Gram-negative member of the Enterobacteriaceae family that is considered a major cause of hospital- and community-acquired infections [1]. K. pneumoniae, since it was described, has been the principal bacterial species of the Klebsiella genus. Phylogenetic analysis showed the existence of three distinct K. pneumoniae phylogroups called KpI, KpII and KpIII, with KpI being the most prevalent [2]. Subsequently, in 2004 and 2014 [2], [3], KpIII and KpII respectively were identified, which corresponded with two new bacterial species, Klebsiella variicola and K. quasipneumoniae. In the case of K. quasipneumoniae, two subspecies, K. quasipneumoniae subsp. quasipneumoniae (KpII-A) and K. quasipneumoniae subsp. similipneumoniae (KpII-B), were reported. More recently, a new bacterial species was added to the Klebsiella genus, Klebsiella quasivariicola [4], [5]. These bacterial species, like K. pneumoniae, are commonly multidrug resistant and can harbour cephalosporinase or carbapenemase genes [1], [6]. These genes confer antimicrobial resistance to β-lactam antibiotics [1]. K. pneumoniae infections have been described to have a hypermucoviscous phenotype and are susceptible to most antibiotics [7]. This phenotype has been described in K. variicola and K. quasipneumoniae clinical isolates in Mexico and Italy [8], [9], [10]. The hypervirulent K. pneumoniae clones with hypermucoviscous phenotype are considered a new variant of K. pneumoniae [11]. The virulence factors associated with hypervirulent K. pneumoniae have been described, and the siderophores, capsular serotype and regulator of mucoid have been characterized as the most important factors [12], [13]. Several K. pneumoniae–caused hospital-acquired infections have been described in Mexico; these studies have identified the extended-spectrum β-lactamase (ESBL) SHV- and CTX-M–type genes and the carbapenemase KPC- and NDM-type genes [6], [14], [15]. However, K. pneumoniae that cause community-acquired infections have not yet been characterized in Mexico.
The objectives of this work were to identify and characterize the bacterial species of community-acquired Klebsiella infections, as well as to determine the phenotypes and molecular characterization related to antimicrobial resistance and virulence.
Methods
Clinical isolates
Thirty-nine isolates were collected from Klebsiella spp. community-acquired infections between January 2013 and December 2014 from outpatients at the following four hospitals in the state of Guerrero, Mexico: Clínica ISSSTE de Chilpancingo (n = 27), Hospital General de Acapulco (n = 6), Instituto Estatal de Cancerología (n = 3) and Unidad Especializada de Gastroenterología y Endoscopia (n = 3) (Supplementary Fig. S1). The samples obtained from all outpatients were considered as an infection according to the clinical manifestations in each patient.
Biochemical and molecular determination of bacterial species
The Klebsiella spp. isolates were identified using the VITEK system (bioMérieux, Marcy l'Etoile, France). Initially a molecular test was performed to all isolates using multiplex-PCR (M-PCR-1) amplification. Briefly, M-PCR-1 identifies by PCR amplification unique genes of both K. pneumoniae (phosphohydrolase) and K. variicola (phosphoglycerate mutase) bacterial species, which were proposed by comparative genomics [16]. Considering the results of M-PCR-1, the nucleotide sequences of rpoB gene (501 bp) [17] were obtained for phylogenetic analysis from isolates 10457, 10471, 10441, 10447 and 10446. In the case of isolate 10446, the bacterial species was confirmed by phylogenetic analysis of six-loci concatenated genes (rpoB (501 bp), gapA (450 bp), mdh (477 bp), pgi (432 bp), phoE (420 bp) and infB (318 bp)) [17]. For both phylogenetic analyses, a maximum-likelihood phylogeny tree was generated by MEGA 7.0.26 [18] with a Tamura-Nei model and 1000 bootstrap replications.
Susceptibility testing and determination of ESBL production
The following susceptibility tests were conducted according to the recommendations by the Clinical and Laboratory Standards Institute (CLSI) [19] using the Mueller-Hinton disc diffusion method: clinical diameter breakpoints of ampicillin, piperacillin, ticarcillin, ticarcillin/clavulanic acid, amoxicillin/clavulanic acid, ceftazidime, cefotaxime, cefoxitin, aztreonam, gentamicin, amikacin, tetracycline, levofloxacin and nalidixic acid. The results were interpreted as resistant, intermediate or susceptible.
For the cefotaxime- and/or ceftazidime-resistant isolates, the extended-spectrum β-lactamase (ESBL) production was determined by applying ceftazidime and cefotaxime both individually and in combination with clavulanic acid. Finally, the minimal inhibitory concentration of ampicillin, amikacin, cefotaxime, nalidixic acid and ciprofloxacin was determined in the ESBL-producing positive isolates. Escherichia coli ATCC 25922 was used as a reference strain in the susceptibility testing according to the CLSI recommendations [19].
Fingerprinting by pulsed-field gel electrophoresis and multilocus sequence typing analysis
The fingerprinting of all isolates was performed using pulsed-field gel electrophoresis [20], and the results were analysed by GelCompar II software (Applied Maths, Kortrijk, Belgium). K. pneumoniae multilocus sequence typing (MLST) [17] was conducted in 21 selected isolates with different phenotypes: ESBL producers (two isolates), hypermucoviscous phenotype (seven isolates) and serotypes related to hypervirulent K. pneumoniae but with hypermucoviscous phenotype (four isolates). In addition, we selected eight K. pneumoniae isolates that were negative for the previously analysed phenotypes. The seven-loci MLST genes (rpoB (501 bp), gapA (450 bp), mdh (477 bp), pgi (432 bp), phoE (420 bp), infB (318 bp) and tonB (414 bp)) were concatenated into a single Multi-FASTA file, and a maximum-likelihood phylogeny tree was generated by MEGA 7.0.26 [18] with a Tamura-Nei model and 1000 bootstrap replications.
Identification of the β-lactamase genes by PCR and sequencing
The ESBL genes were screened via PCR in the SHV and CTX-M families using previously described PCR conditions [21]. All PCR products were sequenced using the chain termination method with the Big Dye Terminator Kit (Applied Biosystems; Thermo Fisher Scientific, Waltham, MA, USA) on an ABI PRISM 3130 (Applied Biosystems). The nucleotide sequences were analysed by the BLASTx program (https://www.ncbi.nlm.nih.gov/).
Determination of biofilm production, hypermucoviscous phenotype, virulence-determinant genes and multivariate analyses
The biofilm assay was performed according to Bandeira et al. [22]. The hypermucoviscous phenotype was analysed by the semiquantitative string test [23]. The following virulence-determinant genes were screened by PCR (Supplementary Table S1): rmpA and rmpA2, entB, iroB, irp2, iucA, fimA, fimH, ecpA, ecpRAB, mrkD, mrkA, uge, urea, allS, kfuBC, cf29a, wabG and terW.
A cluster analysis was performed by using the DendroUPGMA program [24] (http://genomes.urv.cat/UPGMA/index.php?entrada=Example2), with the average linkage based on the Jacquard similarity coefficient and the respective dendrogram (constructed using UPGMA).
Identification of serotype by PCR
The capsular serotypes K1, K2, K5, K20, K54 and K57 were identified by PCR [25], [26] (Supplementary Table S1) in all Klebsiella spp. isolates included in this study.
Results
Epidemiologic data, susceptibility and ESBL-producing isolates
The 39 isolates included in the study were obtained from patients who had community-acquired infections and who ranged in age from a few days after birth (40 days) to 91 years. However, 97.5% of the patients were older than 33 years. Most isolates (74.3%) were obtained from female patients; 25.7% were obtained from male patients. The isolates were obtained from the following sources: urine, 46.1%; vaginal secretion, 25.6%; antral biopsy samples, 15.3%; sputum, 10.2%; and cerebrospinal fluid, 2.5% (Table 1).
Table 1.
Characteristics of Klebsiella spp. community-acquired isolates
| Isolate | Biochemical and molecular identification | Hospitala | Isolation date | Sample origin | ESBL test | ESBL geneb | Biofilm index | Hypermucoviscous phenotype | Serotype | MLST ST |
|---|---|---|---|---|---|---|---|---|---|---|
| 10458 | K. pneumoniae | 1 | 01/18/2014 | Urine cultures | + | CTX-M-15 | 2.0 | − | ND | 656 |
| 10459 | K. pneumoniae | 1 | 02/23/2014 | Urine cultures | + | CTX-M-15 | 2.0 | − | ND | ND |
| 10462 | K. pneumoniae | 1 | 05/28/2014 | Vaginal secretion | + | CTX-M-15 | 2.1 | − | ND | ND |
| 10472 | K. pneumoniae | 1 | 05/09/2014 | Urine cultures | + | CTX-M-15 | 6.8 | − | ND | 392 |
| 10443 | K. pneumoniae | 2 | 08/18/2013 | Urine cultures | − | Neg | 1.7 | + | ND | 1081 |
| 10455 | K. pneumoniae | 4 | 12/12/2013 | Antral biopsy samples | − | Neg | 1.6 | + | ND | 2936 |
| 10457 | K. pneumoniae | 1 | 02/07/2013 | Urine cultures | − | Neg | 0.4 | + | ND | 2937 |
| 10481 | K. pneumoniae | 1 | 06/12/2014 | Urine cultures | − | Neg | 2.8 | + | ND | 252 |
| 10483 | K. pneumoniae | 3 | 12/08/2013 | Antral biopsy samples | − | Neg | 1.5 | + | ND | 281 |
| 10440 | K. pneumoniae | 2 | 07/31/2013 | Sputum | − | Neg | 1.5 | + | K5 | 29 |
| 10444 | K. pneumoniae | 2 | 09/13/2013 | Cerebrospinal fluid | − | Neg | 14.0 | + | K2 | 29 |
| 10445 | K. pneumoniae | 1 | 01/23/2014 | Vaginal secretion | − | Neg | 2.6 | − | ND | 2940 |
| 10476 | K. pneumoniae | 1 | 05/20/2014 | Urine cultures | − | Neg | 3.2 | − | ND | 2015 |
| 10454 | K. pneumoniae | 3 | 05/08/2013 | Antral biopsy samples | − | Neg | 0.4 | − | ND | 505 |
| 10450 | K. pneumoniae | 1 | 02/05/2014 | Vaginal secretion | − | Neg | 3.1 | − | ND | 405 |
| 10482 | K. pneumoniae | 1 | 06/23/2014 | Urine cultures | − | Neg | 3.7 | − | ND | 45 |
| 10479 | K. pneumoniae | 1 | 06/02/2014 | Urine cultures | − | Neg | 1.9 | − | ND | 45 |
| 10456 | K. pneumoniae | 4 | 12/12/2013 | Antral biopsy samples | − | Neg | 1.2 | − | ND | 37 |
| 10449 | K. pneumoniae | 1 | 02/05/2014 | Urine cultures | − | Neg | 11.2 | − | ND | 29 |
| 10478 | K. pneumoniae | 1 | 06/02/2014 | Urine cultures | − | Neg | 14.1 | − | ND | ND |
| 10477 | K. pneumoniae | 1 | 05/28/2014 | Vaginal secretion | − | Neg | 1.2 | − | ND | ND |
| 10475 | K. pneumoniae | 1 | 05/12/2014 | Urine cultures | − | Neg | 3.2 | − | ND | ND |
| 10470 | K. pneumoniae | 1 | 04/22/2014 | Vaginal secretion | − | Neg | 6.0 | − | ND | ND |
| 10469 | K. pneumoniae | 1 | 04/22/2014 | Vaginal secretion | − | Neg | 0.9 | − | ND | ND |
| 10468 | K. pneumoniae | 1 | 04/09/2014 | Vaginal secretion | − | Neg | 4.4 | − | ND | ND |
| 10465 | K. pneumoniae | 1 | 04/07/2014 | Vaginal secretion | − | Neg | 4.1 | − | ND | ND |
| 10461 | K. pneumoniae | 1 | 05/28/2014 | Vaginal secretion | − | Neg | 2.8 | − | ND | ND |
| 10452 | K. pneumoniae | 4 | 10/28/2013 | Antral biopsy samples | − | Neg | 10.3 | − | ND | ND |
| 10451 | K. pneumoniae | 1 | 02/06/2014 | Urine cultures | − | Neg | 1.6 | − | ND | ND |
| 10442 | K. pneumoniae | 2 | 07/31/2013 | Sputum | − | Neg | 1.1 | − | ND | ND |
| 10439 | K. pneumoniae | 1 | 01/07/2014 | Urine cultures | − | Neg | 3.1 | − | ND | ND |
| 10438 | K. pneumoniae | 1 | 01/07/2014 | Urine cultures | − | Neg | 2.3 | − | ND | ND |
| 10460 | K. pneumoniae | 1 | 02/23/2014 | Urine cultures | − | Neg | 3.7 | − | K54 | 2014 |
| 10471 | K. pneumoniae | 1 | 05/05/2014 | Sputum | − | Neg | 0.7 | − | K54 | 29 |
| 10453 | K. pneumoniae | 3 | 22/08/2013 | Antral biopsy samples | − | Neg | 44.2 | − | K2 | 2938 |
| 10474 | K. pneumoniae | 1 | 05/12/2014 | Sputum | − | Neg | 3.5 | − | K2 | 2939 |
| 10441 | K. quasipneumoniae subsp. quasipneumoniaec | 2 | 07/02/2013 | Urine cultures | − | Neg | 14.1 | − | K2 | ND |
| 10446 | K. quasivariicolad | 2 | 01/23/2014 | Urine cultures | − | Neg | 1.9 | − | ND | ND |
| 10447 | K. variicolae | 1 | 01/23/2014 | Vaginal secretion | − | Neg | 1.7 | − | ND | ND |
ESBL, extended-spectrum β-lactamase; MLST, multilocus sequence typing; ND, not determined; ST, sequence type.
Hospitals: 1, Clínica ISSSTE de Chilpancingo (n = 27); 2, Hospital General de Acapulco (n = 6); 3, Instituto Estatal de Cancerología (n = 3); 4, Unidad Especializada de Gastroenterología y Endoscopia (n = 3) (Supplementary Fig. S1).
Neg indicates negative for ESBL production.
K. quasipneumoniae subsp. quasipneumoniae were identified by a phylogenetic analysis of rpoB gene.
K. quasivariicola was identified by phylogenetic analysis of rpoB gene and confirmed by six-loci concatenated genes.
K. variicola was identified by multiplex-PCR test [16] and phylogenetic analysis of rpoB gene.
The susceptibility testing revealed the following resistance profiles: ampicillin and ticarcillin, 100% resistant; piperacillin, 43.5% resistant; amoxicillin/clavulanic acid, 30.7% resistant; ceftazidime, 17.9% resistant; cefotaxime, 35.8% resistant; cefoxitin, 5.1% resistant; aztreonam, 23% resistant; gentamicin, 17.9% resistant; amikacin, 2.5% resistant; tetracycline, 38.4% resistant; levofloxacin, 5.1% resistant; and nalidixic acid, 20.5% resistant (Supplementary Table S2).
The cephalosporin-resistant isolates cefotaxime and ceftazidime or both were identified in 38.4% of the samples (Supplementary Table S2). ESBL producers were identified in 10.2% of the isolates (Table 1). The susceptibility profile of these ESBL-producing isolates was 100% resistant to ampicillin (>256 mg/L), 100% resistant to ceftazidime (>64 mg/L) and 50% resistant to ciprofloxacin and nalidixic acid (>8 and >256 mg/L respectively), and all the isolates were susceptible to amikacin. The identified ESBL gene was determined to be CTX-M-15 in the four ESBL-producing isolates (10458, 10459, 10462 and 10472) (Table 1).
Identification of bacterial species in Klebsiella spp. isolates
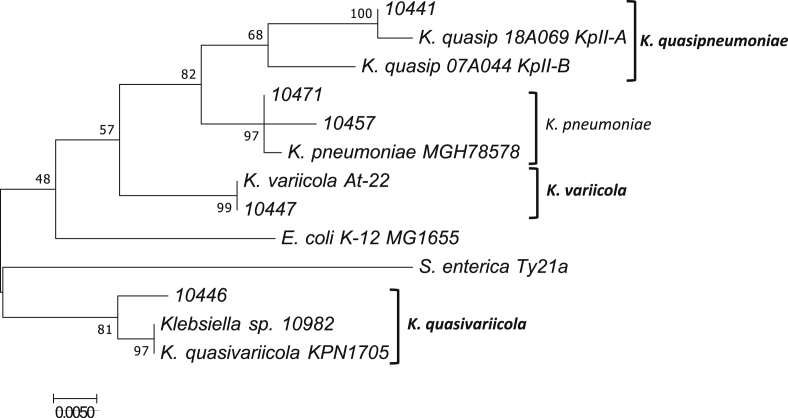
All isolates were identified biochemically as K. pneumoniae. However, using the M-PCR-1 assay, we identified 36 isolates as K. pneumoniae; isolate 10447 (2.5%) was identified as K. variicola (Table 1). In this M-PCR-1 assay, a negative result for K. pneumoniae or K. variicola bacterial species was obtained for isolates 10441 and 10446. The rpoB phylogenetic analysis confirmed as K. pneumoniae the 10457 and 10471 isolates and as K. variicola the 10447 isolate. Isolates 10441 and 10446 corresponded to K. quasipneumoniae subsp. quasipneumoniae and K. quasivariicola respectively (Fig. 1). Additional phylogenetic analysis using six-loci concatenated genes confirmed that isolate 10446 belonged to the K. quasivariicola bacterial species (Supplementary Fig. S2).
Fig. 1.
Phylogenetic analysis of rpoB genes determined of Klebsiella pneumoniae 10471 (MH003688) and 10457 (MH003687); Klebsiella quasipneumoniae 10441 (MH003686); Klebsiella variicola 10447 (MH003689); and Klebsiella quasivariicola 10446 (MH003680) isolates. Nucleotide sequence of rpoB gene was obtained from K. pneumoniae MGH78578 (CP000647.1), Klebsiella quasipneumoniae subsp. quasipneumoniae 18A069 (KpII-A) (CBZM010000001.1), Klebsiella quasipneumoniae subsp. similipneumoniae 07A044 (KpII-B) (CBZR010000001.1), Klebsiella variicola At-22 (CP001891.1), Klebsiella quasivariicola KPN1705 (CP022823.1), Klebsiella sp. 10982 (NZ_AKYX00000000), Escherichia coli K-12 MG1655 (NC_000913.3) and Salmonella enterica Ty21a (NC_021176.1) used as reference bacterial species.
Fingerprinting analysis of K. pneumoniae isolates
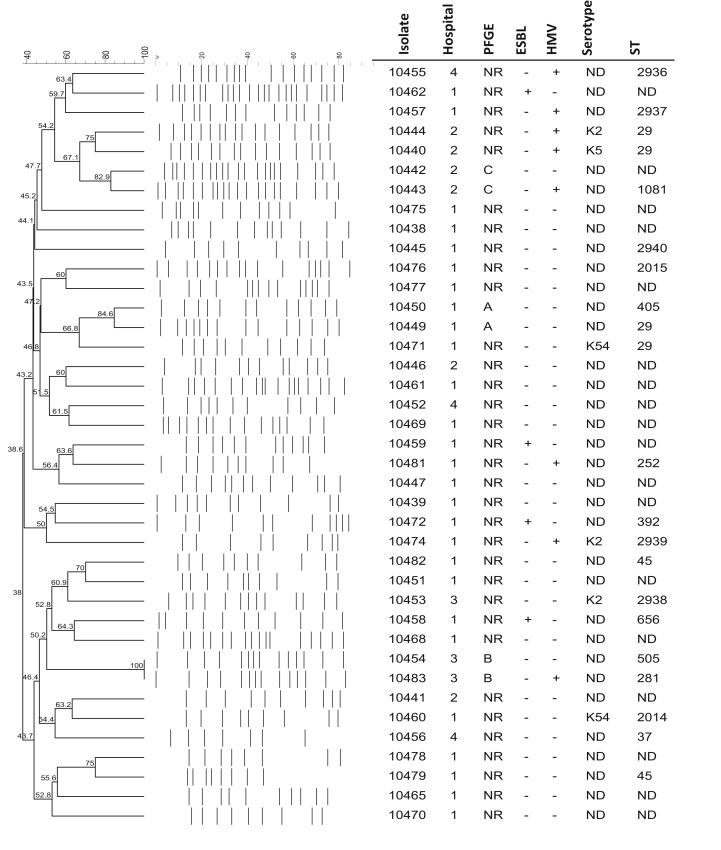
In general, most isolates obtained from the outpatients at the four hospitals were not related (<80% similarity). However, in the K. pneumoniae isolates, three clonal groups were identified (Fig. 2). Clonal group A (84.6%) corresponded to the isolates obtained from outpatients at the Clínica ISSSTE de Chilpancingo. Clonal groups B (100%) and C (82.9%) corresponded to the isolates obtained from outpatients at the Hospital General de Acapulco and the Instituto Estatal de Cancerología respectively (Fig. 2).
Fig. 2.
Identification of clonal groups by PFGE and dendrogram analysis of Klebsiella pneumoniae isolates with different phenotypic and molecular characteristics. Hospital, PFGE pattern, ESBL production ability, presence or absences of hypermucoviscous phenotype, serotype and ST are indicated. ESBL, extended-spectrum β-lactamase; HMV, hypermucoviscous phenotype; PFGE, pulsed-field gel electrophoresis; ST, sequence type.
Biofilm production, hypermucoviscous phenotype, serotype and sequence type
Biofilm production was identified in 89.7% of the Klebsiella spp. isolates, with a biofilm index of >1. Five K. pneumoniae isolates and the K. quasipneumoniae subsp. quasipneumoniae 10441 isolate showed a biofilm index of >10. Regarding the hypermucoviscous phenotype, 19.4% (7/39) isolates that corresponded to non–ESBL-producing K. pneumoniae isolates showed this phenotype, identifying a string test of >5 mm (Table 1). The main serotypes described as hypervirulent were analysed in the K. pneumoniae isolates in this study. In general, these screened serotypes were identified in seven isolates (17.9%); of these, six corresponded to K. pneumoniae, and the other corresponded to K. quasipneumoniae subsp. quasipneumoniae (10441) (Table 1). The K2 serotype was the most prevalent at 57.1% (4/7), followed by K54 at 28.5% (2/7) and K5 at 14.2% (1/7). Of these, K. pneumoniae isolates 10440 and 10444 with the hypermucoviscous phenotype corresponded to serotypes K2 and K5 respectively. However, the other five isolates (10441, 10453, 10460, 10471 and 10471) that did not have the hypermucoviscous phenotype showed the K2 and K54 serotypes.
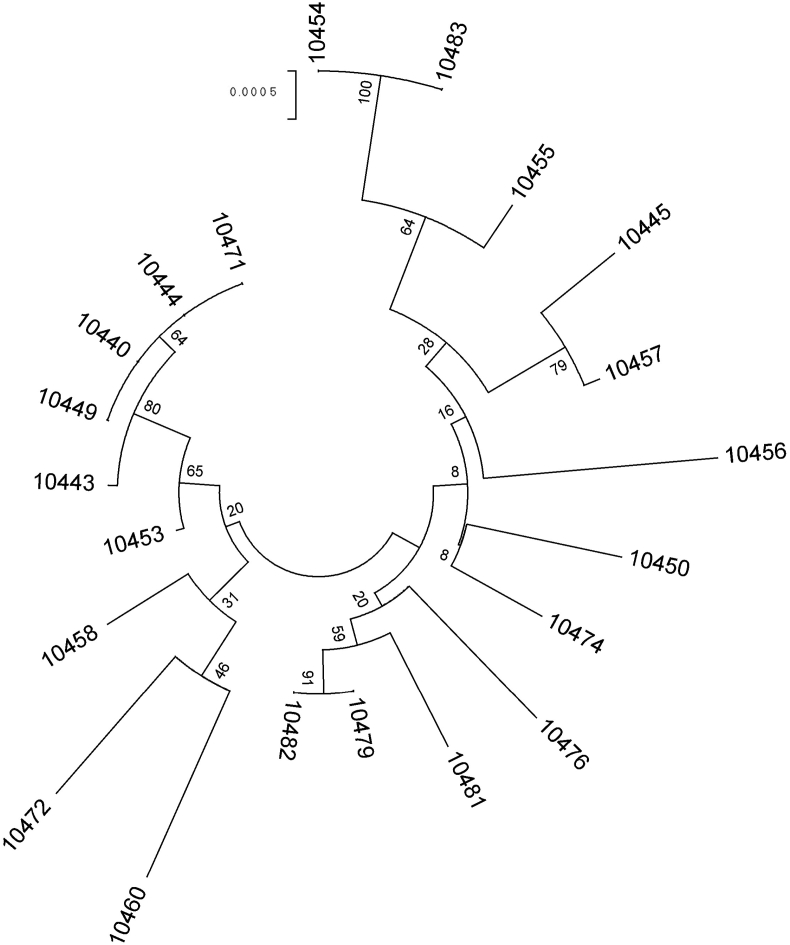
The sequence types (STs) identified in 21 K. pneumoniae isolates was heterogeneous. We identified new STs: ST2936, ST2937, ST2938, ST2939 and ST2940 (Table 1). The ESBL-producing K. pneumoniae isolates showed ST392 and ST656, and the K. pneumoniae isolates with hypermucoviscous phenotype were ST29 (two isolates), ST252, ST281, ST1081, ST2936 and ST2937. In addition, four K. pneumoniae isolates that were positive for some of the hypervirulent-related serotypes showed that ST2938, ST2939 and ST29 were associated with serotype K2, and ST2014 was associated with serotype K54 (Table 1). Finally, the K. pneumoniae isolates that did not produce ESBL and that were negative for hypermucoviscous phenotype or hypervirulent-related serotypes had the following STs: ST45 (two isolates), ST37, ST405, ST505, ST2015 and ST2940 (Table 1). Phylogenetic analysis of the concatenated sequences at the seven-loci MLST of the K. pneumoniae isolates showed clades I and II (Fig. 3). Clade I corresponded to 12 isolates, and four of these isolates belonged to ST281 and ST45. Clade II corresponded to nine isolates, and four of these isolates belonged to ST29.
Fig. 3.
Maximum-likelihood phylogeny tree from seven-loci multilocus sequence typing of Klebsiella pneumoniae clinical isolates. Bootstrapping of gene tree was implemented to evaluate support of groups. Numbers adjacent to nodes are bootstrap values. Clade I: isolates 10454, 10483, 10455, 10445, 10457, 10456, 10450, 10474, 10476, 10481, 10479 and 10482. Clade II: isolates 10471, 10444, 10440, 10449, 10443, 10453, 10458, 10472 and 10460.
Virulence factors
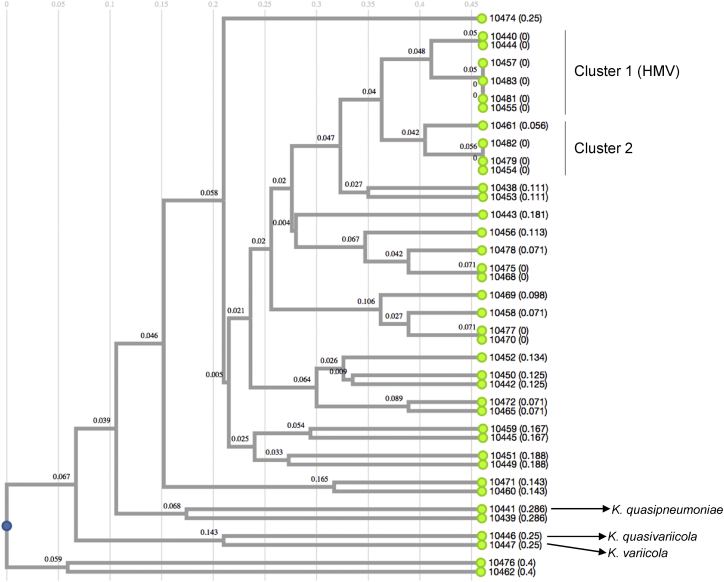
In general, eight of the 16 screened virulence factors (fimH, ecpRAB, mrkA, fimA, ureA, wabG, uge and entB) were mostly identified in the Klebsiella spp.–caused community-acquired infections. Different percentages of fimbriae fimH (94.8%), ecpRAB (84.6%), mrkA (64.1%) and fimA (58.9%) were identified. The entB siderophore was identified at a higher percentage (76.9%), followed by irp2 (7.6%), kfuBC (5.1%), iucA (5.1%) and iroB (2.5%). The ecpA (pillin) gene was identified in 2.5% of the isolates, while the cf29a (adhesin) gene was not identified. The ureA (84.6%), uge (61.5%), wabG (28.2%) and allS (5.1%) genes were identified. The mucoid regulator rmpA (2.5%) gene was identified in only one isolate (10474). A Jaccard index analysis of virulence factors showed two minor clusters. Cluster 1 was formed by six of the seven K. pneumoniae that had the hypermucoviscous phenotype. Cluster 2 differed from cluster 1 because it was negative for the hypermucoviscous phenotype (Supplementary Table S3). In particular, K. quasipneumoniae, K. variicola and K. quasivariicola isolates shared only one factor of virulence, which corresponded to the fimH (fimbriae) gene; K. quasipneumoniae and K. variicola additionally shared the ureA (urease) gene, and K. quasipneumoniae additionally contained the mrkA (fimbrial) and allS (allantoinase) genes, with this last one involved in allantoin metabolism as a nitrogen source [27].
Discussion
The present work included all Klebsiella spp. isolates obtained from community-acquired infections during this study period. They were mainly non–ESBL-producing Klebsiella spp. isolates. Nevertheless, the non–ESBL-producing isolates had resistance to other families of antibiotics. A misclassification of Klebsiella by biochemical methods has been previously described [16], [28], [29], so the biochemical identification of K. pneumoniae was not correct for all isolates because the M-PCR-1 assay identified one K. variicola isolate, and two isolates showed a negative result for this assay. These isolates could correspond to K. quasipneumoniae, as was determined previously by rpoB phylogenetic analysis [10]. The phylogenetic analysis of the rpoB gene effectively identified one K. quasipneumoniae subsp. quasipneumoniae isolate, but interestingly, isolate 10446 was found to be phylogenetically related to the new bacterial species K. quasivariicola and Klebsiella spp. 10982 (with the last one corresponding to K. quasivariicola) [5] (Fig. 1). This result was confirmed by phylogenetic analysis of six-loci concatenated genes. The K. quasivariicola represented by isolate 10446 is also a community-acquired urinary tract infection, as are the other bacterial species from the Klebsiella genus.
The phylogenetic analysis of the rpoB gene is accurate enough for proper identification and differentiation K. pneumoniae, K. quasipneumoniae and K. variicola as well as for K. quasivariicola isolates. The use of the rpoB gene and not 16S rRNA was previously proposed for Klebsiella taxonomy by Martinez et al. [30]. The growing number of new bacterial species in the Klebsiella genus in less than two decades requires the implementation of a new M-PCR-1 method to correctly differentiate among at least K. pneumoniae, K. quasipneumoniae, K. variicola and K. quasivariicola, which would immensely support the study of these bacterial species.
In general, the relationship determined by pulsed-field gel electrophoresis showed that few isolates had the same clonal origin. The clonal groups A, B and C, which were each formed by two isolates, corresponded to clones from different hospitals. However, the isolates in each cluster showed different microbiologic and molecular characteristics, such as ESBL production, hypermucoviscosity and ST.
In the present bacterial population, the prevalence of ESBL-producing isolates was low, and the β-lactamase gene, which was responsible for cephalosporin resistance, was the CTX-M-15 gene in all isolates. These isolates corresponded to ST656 and ST392, which have not been previously described in Mexico but which have been described in nosocomial infections in other regions of world. K. pneumoniae ST665 has been described in China and Philippines [31], [32] as carrying ESBL and carbapenemase genes respectively. In the case of K. pneumoniae, ST392 harbouring KPC-3 and CTX-M alleles has been documented in the United States [33], [34].
Biofilm production was identified in most isolates, and eight isolates had a high index, at >6. Biofilm production was not related to the sample origin, ESBL production, serotype or ST. This phenotypic characteristic is considered a virulence factor that helps or promotes attachment to living or abiotic surfaces, thus preventing the effects of antimicrobial agents [35]. Another important virulence factor described in K. pneumoniae is the polysaccharide matrix, which coats the bacteria and is known as the capsule [36]. Hyperproduction of the capsule has been described as a hypermucoviscous phenotype in hypervirulent K. pneumoniae [37]. The capsular serotypes that are mainly associated with hypervirulent K. pneumoniae are K1 and K2 [36] and, at a lower percentage, the K5, K20, K54 and K57 serotypes [38]. Hypervirulence has not been reported together with the hypermucoviscous phenotype in Mexico; however, the hypermucoviscous phenotype in the K. variicola and K. quasipneumoniae subsp. similipneumoniae clinical isolates has recently been described [9], [10]. When we used PCR to genotype the main serotypes described in hypervirulent K. pneumoniae, we mainly identified K2, followed by K54 and K5. Only two isolates that had the hypermucoviscous phenotype had the K2 and K5 serotypes. The other five isolates that had the hypermucoviscous phenotype showed different resistance profiles and different STs, except for isolates 10444 (K2) and 10440 (K5), which corresponded to ST29 (Table 1). The hypermucoviscous phenotype does not strictly correspond to the hypervirulent isolate; this hypothesis, recently revised [39], was previously proposed by Zhang et al. [40].
A phylogenetic analysis using the concatenate nucleotide sequences from the sequence typing showed two clades (Fig. 3). A clear phenotypic or genotypic characteristic that distinguishes the clades from one another was not identified. However, clade II members mostly correspond to STs that have the serotypes K2 (ST29, ST2938), K54 (ST29, ST2014) and K5 (ST29), are ESBL producers (ST392, ST656) and have the hypermucoviscous phenotype (ST29, ST1081).
Generally, virulence factors are poorly studied in K. pneumoniae that cause hospital-acquired infections. However, a correlation between antibiotic resistance and virulence has been proposed [41]. Holt et al. [42] explored this correlation in silico in hypervirulent and carbapenem-resistant K. pneumoniae (ST258) isolates. In the present work, the virulence factors were heterogeneous. The presence of fimbrial genes (fimH, ecpRAB, mrkA, fimA), which are involved in adhesion and subsequent gastrointestinal colonization, was common in both the non–ESBL- and ESBL-producing Klebsiella spp. [36]. The most common siderophore was enterobactin (entB), followed by yersiniabactin (irp2); however, aerobactin (iucA) was identified in two isolates (10471, 10445) compared to salmochelin (iroB), which was identified in only one isolate (10474). K. pneumoniae 10474 (ST2939) was the isolate that presented the highest number of virulence factors, including the rmpA gene (mucoid regulator), the K2 serotype and a negative hypermucoviscous phenotype (Supplementary Table S3 and Fig. 4). Interestingly, the hypermucoviscous phenotype was identified in six non–ESBL-producing K. pneumoniae isolates, whose correlation formed a cluster (cluster 1) (Fig. 4). The hypermucoviscous K. pneumoniae isolates in cluster 1 shared the fimH, fimA, ecpRAB and mrkA fimbriae genes; urease (ureA), glucuronic (wabG) and uridine (uge) metabolism genes; and the enterobactin siderophore (entB). Only two of these isolates (10440, 10444) corresponded to serotypes K2 and K5. The K. quasipneumoniae, K. quasivariicola and K. variicola isolates, together with some K. pneumoniae isolates, contain the lowest number of virulence factors included in the study. The fimbriae fimH is a characteristic common among these bacterial species, and urease is a characteristic present in K. quasipneumoniae, K. variicola and K. pneumoniae isolates and absent in K. quasivariicola. These data correlate with a recent in silico analysis of K. quasipneumoniae and K. variicola genomes which identified a mosaic distribution of different virulence-associated genes that would allow it to adapt to clinical settings [43]. In the case of K. quasivariicola, the eventual description of more isolates and genomes may later permit us to detail the potential setting for this bacterial species.
Fig. 4.
Jaccard index of phenotypic and molecular characteristics identified using microbiologic and molecular tests in Klebsiella pneumoniae, Klebsiella quasipneumoniae, Klebsiella variicola and Klebsiella quasivariicola isolates.
Conclusions
In our study, K. pneumoniae was the most represented Klebsiella species; however, K. quasipneumoniae subsp. quasipneumoniae, K. variicola and the new bacterial species K. quasivariicola also cause community-acquired infections. Major resistance was identified among ESBL-producing clinical isolates; nevertheless, we observed multidrug resistance among non–ESBL-producing isolates. The virulence factors were heterogeneous, and K. quasipneumoniae, K. variicola and K. quasivariicola in particular contain the lowest number of virulence factors, in contrast to most K. pneumoniae isolates. The hypermucoviscous phenotype and serotypes, which have been previously described in hypervirulent K. pneumoniae, were highlighted. Virulence factors associated with adhesion and subsequent tract gastrointestinal colonization are common characteristics in Klebsiella spp. isolates. In this work, we were not able to establish a correlation, molecular mechanism, or advantage to being hypermucoviscous and/or belonging to serotypes K2, K5 and K54 in these isolates. Finally, community-acquired infections are represented by various species from Klebsiella genus, with different profiles of antibiotics resistance and heterogeneous virulence factors. Nevertheless, this bacteria genus has been described to acquire plasmid-borne antibiotic resistance in combination with virulence factors and could cause serious community infections in Mexican population.
Conflict of interest
None declared.
Acknowledgements
This study is a part of the K. pneumoniae and K. variicola project, revised and approved by the ethical and scientific research commissions at the Instituto Nacional de Salud Pública (www.insp.mx) on 3 August 2016. The present study was conducted at the Instituto Nacional de Salud Pública in Cuernavaca, Morelos, México. Supported in part by grant 256988 from SEP-CONACyT (Secretaría de Educción Pública-Consejo Nacional de Ciencia y Tecnología). We thank T. Rojas-Moreno at Instituto Nacional de Salud Pública for excellent laboratory assistance.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.nmni.2018.02.002.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Paterson D.L., Bonomo R.A. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brisse S., Passet V., Grimont P.A. Description of Klebsiella quasipneumoniae sp. nov., isolated from human infections, with two subspecies, Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and demonstration that Klebsiella singaporensis is a junior heterotypic synonym of Klebsiella variicola. Int J Syst Evol Microbiol. 2014;64(Pt 9):3146–3152. doi: 10.1099/ijs.0.062737-0. [DOI] [PubMed] [Google Scholar]
- 3.Rosenblueth M., Martinez L., Silva J., Martinez-Romero E. Klebsiella variicola, a novel species with clinical and plant-associated isolates. Syst Appl Microbiol. 2004;27:27–35. doi: 10.1078/0723-2020-00261. [DOI] [PubMed] [Google Scholar]
- 4.Long S.W., Linson S.E., Ojeda S.M., Cantu C., Davis J.J., Brettin T. Whole-genome sequencing of a human clinical isolate of the novel species Klebsiella quasivariicola sp. nov. Genome Announc. 2017;5 doi: 10.1128/genomeA.01057-17. e01057–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long S.W., Linson S.E., Ojeda Saavedra M., Cantu C., Davis J.J., Brettin T. Discovery and whole genome sequencing of a human clinical isolate of the novel species Klebsiella quasivariicola sp. nov. bioRxiv. 2018 doi: 10.1128/genomeA.01057-17. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez-Zulueta P., Silva-Sanchez J., Barrios H., Reyes-Mar J., Velez-Perez F., Arroyo-Escalante S. First outbreak of KPC-3–producing Klebsiella pneumoniae (ST258) clinical isolates in a Mexican medical center. Antimicrob Agents Chemother. 2013;57:4086–4088. doi: 10.1128/AAC.02530-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shon A.S., Bajwa R.P., Russo T.A. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4:107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arena F., Henrici De A.L., Pieralli F., Di P.V., Giani T., Torricelli F. Draft genome sequence of the first hypermucoviscous Klebsiella quasipneumoniae subsp. quasipneumoniae isolate from a bloodstream infection. Genome Announc. 2015;3 doi: 10.1128/genomeA.00952-15. e00952–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garza-Ramos U., Silva-Sanchez J., Barrios H., Rodriguez-Medina N., Martinez-Barnetche J., Andrade V. Draft genome sequence of the first hypermucoviscous Klebsiella variicola clinical isolate. Genome Announc. 2015;3 doi: 10.1128/genomeA.01352-14. e01352–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garza-Ramos U., Silva-Sanchez J., Catalan-Najera J., Barrios H., Rodriguez-Medina N., Garza-Gonzalez E. Draft genome sequence of a hypermucoviscous extended-spectrum–beta-lactamase–producing Klebsiella quasipneumoniae subsp. similipneumoniae clinical isolate. Genome Announc. 2016;4 doi: 10.1128/genomeA.00475-16. e00475–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pomakova D.K., Hsiao C.B., Beanan J.M., Olson R., MacDonald U., Keynan Y. Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumoniae: an emerging and under-recognized pathogenic variant. Eur J Clin Microbiol Infect Dis. 2012;31:981–989. doi: 10.1007/s10096-011-1396-6. [DOI] [PubMed] [Google Scholar]
- 12.Lin Y.C., Lu M.C., Tang H.L., Liu H.C., Chen C.H., Liu K.S. Assessment of hypermucoviscosity as a virulence factor for experimental Klebsiella pneumoniae infections: comparative virulence analysis with hypermucoviscosity-negative strain. BMC Microbiol. 2011;11:50. doi: 10.1186/1471-2180-11-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu K.M., Li L.H., Yan J.J., Tsao N., Liao T.L., Tsai H.C. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J Bacteriol. 2009;191:4492–4501. doi: 10.1128/JB.00315-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrios H., Silva-Sanchez J., Reyna-Flores F., Sanchez-Perez A., Sanchez-Francia D., Aguirre-Torres J.A. Detection of a NDM-1–producing Klebsiella pneumoniae (ST22) clinical isolate at a pediatric hospital in Mexico. Pediatr Infect Dis J. 2014;33:335. doi: 10.1097/INF.0000000000000173. [DOI] [PubMed] [Google Scholar]
- 15.Garza-Ramos U., Barrios H., Reyna-Flores F., Sanchez-Perez A., Tamayo-Legorreta E., Ibarra-Pacheco A. Characteristics of KPC-2–producing Klebsiella pneumoniae (ST258) clinical isolates from outbreaks in 2 Mexican medical centers. Diagn Microbiol Infect Dis. 2014;79:483–485. doi: 10.1016/j.diagmicrobio.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 16.Garza-Ramos U., Silva-Sanchez J., Martinez-Romero E., Tinoco P., Pina-Gonzales M., Barrios H. Development of a multiplex-PCR probe system for the proper identification of Klebsiella variicola. BMC Microbiol. 2015;15:64. doi: 10.1186/s12866-015-0396-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diancourt L., Passet V., Verhoef J., Grimont P.A., Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute . Clinical and Laboratory Standards Institute; Wayne, PA: 2017. Performance standard for antimicrobial susceptibility testing. CLSI supplement M100. [Google Scholar]
- 20.Tenover F.C., Arbeit R.D., Goering R.V., Mickelsen P.A., Murray B.E., Persing D.H. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silva-Sanchez J., Barrios H., Reyna-Flores F., Bello-Diaz M., Sanchez-Perez A., Rojas T. Prevalence and characterization of plasmid-mediated quinolone resistance genes in extended-spectrum beta-lactamase–producing Enterobacteriaceae isolates in Mexico. Microb Drug Resist. 2011;17:497–505. doi: 10.1089/mdr.2011.0086. [DOI] [PubMed] [Google Scholar]
- 22.Bandeira M., Carvalho P.A., Duarte A., Jordao L. Exploring dangerous connections between Klebsiella pneumoniae biofilms and healthcare-associated infections. Pathogens. 2014;3:720–731. doi: 10.3390/pathogens3030720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang C.T., Chuang Y.P., Shun C.T., Chang S.C., Wang J.T. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med. 2004;199:697–705. doi: 10.1084/jem.20030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Vallve S., Palau J., Romeu A. Horizontal gene transfer in glycosyl hydrolases inferred from codon usage in Escherichia coli and Bacillus subtilis. Mol Biol Evol. 1999;16:1125–1134. doi: 10.1093/oxfordjournals.molbev.a026203. [DOI] [PubMed] [Google Scholar]
- 25.Pan Y.J., Lin T.L., Chen Y.H., Hsu C.R., Hsieh P.F., Wu M.C. Capsular types of Klebsiella pneumoniae revisited by wzc sequencing. PLoS One. 2013;8 doi: 10.1371/journal.pone.0080670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turton J.F., Perry C., Elgohari S., Hampton C.V. PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J Med Microbiol. 2010;59(Pt 5):541–547. doi: 10.1099/jmm.0.015198-0. [DOI] [PubMed] [Google Scholar]
- 27.Cusa E., Obradors N., Baldoma L., Badia J., Aguilar J. Genetic analysis of a chromosomal region containing genes required for assimilation of allantoin nitrogen and linked glyoxylate metabolism in Escherichia coli. J Bacteriol. 1999;181:7479–7484. doi: 10.1128/jb.181.24.7479-7484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen M., Li Y., Li S., Tang L., Zheng J., An Q. Genomic identification of nitrogen-fixing Klebsiella variicola, K. pneumoniae and K. quasipneumoniae. J Basic Microbiol. 2016;56:78–84. doi: 10.1002/jobm.201500415. [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Romero E., Rodriguez-Medina N., Beltrán-Rojel M., Silva-Sanchez J., Barrios-Camacho H., Perez-Rueda E. Genome misclassification of Klebsiella variicola and Klebsiella quasipneumoniae isolated from plants, animals and humans. Salud Publica Mex. 2018;60:29–40. doi: 10.21149/8149. [DOI] [PubMed] [Google Scholar]
- 30.Martinez J., Martinez L., Rosenblueth M., Silva J., Martinez-Romero E. How are gene sequence analyses modifying bacterial taxonomy? The case of Klebsiella. Int Microbiol. 2004;7:261–268. [PubMed] [Google Scholar]
- 31.Chou A., Roa M., Evangelista M.A., Sulit A.K., Lagamayo E., Torres B.C. Emergence of Klebsiella pneumoniae ST273 carrying blaNDM-7 and ST656 carrying blaNDM-1 in Manila, Philippines. Microb Drug Resist. 2016;22:585–588. doi: 10.1089/mdr.2015.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X.R., Chen J.C., Kang Y., Jiang N., An S.C., Gao Z.C. Prevalence and characterization of plasmid-mediated blaESBL with their genetic environment in Escherichia coli and Klebsiella pneumoniae in patients with pneumonia. Chin Med J (Engl) 2012;125:894–900. [PubMed] [Google Scholar]
- 33.Wang G., Huang T., Surendraiah P.K., Wang K., Komal R., Zhuge J. CTX-M beta-lactamase–producing Klebsiella pneumoniae in suburban New York City, New York, USA. Emerg Infect Dis. 2013;19:1803–1810. doi: 10.3201/eid1911.121470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Mento G., Cuscino N., Carcione C., Cardinale F., Conaldi P.G., Douradinha B. Emergence of a Klebsiella pneumoniae ST392 clone harbouring KPC-3 in an Italian transplantation hospital. J Hosp Infect. 2018 doi: 10.1016/j.jhin.2017.11.019. In press. [DOI] [PubMed] [Google Scholar]
- 35.Vuotto C., Longo F., Balice M.P., Donelli G., Varaldo P.E. Antibiotic resistance related to biofilm formation in Klebsiella pneumoniae. Pathogens. 2014;3:743–758. doi: 10.3390/pathogens3030743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paczosa M.K., Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80:629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russo T.A., Shon A.S., Beanan J.M., Olson R., MacDonald U., Pomakov A.O. Hypervirulent K. pneumoniae secretes more and more active iron-acquisition molecules than ‘classical’ K. pneumoniae thereby enhancing its virulence. PLoS One. 2011;6 doi: 10.1371/journal.pone.0026734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Z., Liu M., Cui Y., Wang L., Zhang Y., Qiu J. A novel PCR-based genotyping scheme for clinical Klebsiella pneumoniae. Future Microbiol. 2014;9:21–32. doi: 10.2217/fmb.13.137. [DOI] [PubMed] [Google Scholar]
- 39.Catalan-Najera J.C., Garza-Ramos U., Barrios-Camacho H. Hypervirulence and hypermucoviscosity: two different but complementary Klebsiella spp. phenotypes? Virulence. 2017;8:1111–1123. doi: 10.1080/21505594.2017.1317412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y., Zeng J., Liu W., Zhao F., Hu Z., Zhao C. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect. 2015;71:553–560. doi: 10.1016/j.jinf.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 41.Beceiro A., Tomas M., Bou G. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin Microbiol Rev. 2013;26:185–230. doi: 10.1128/CMR.00059-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holt K.E., Wertheim H., Zadoks R.N., Baker S., Whitehouse C.A., Dance D. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci USA. 2015;112:E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez-Romero E., Rodriguez-Medina N., Beltrán-Rojel M., Toribio-Jimenez J., Garza-Ramos U. Klebsiella variicola and Klebsiella quasipneumoniae genomic analysis provides insights into their capacity to adapt to clinical and plant settings. Salud Publica Mex. 2018;60:56–62. doi: 10.21149/8156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.