Abstract
Heterotrimeric guanine nucleotide-binding proteins (G proteins) composed of alpha (Gα), beta (Gβ), and gamma (Gγ) subunits are central signal transducers mediating the cellular response to multiple stimuli, such as cold, in eukaryotes. Plant Gγ subunits, divided into A, B, and C three structurally distinct types, provide proper cellular localization and functional specificity to the heterotrimer complex. Here, we demonstrate that a type C Gγ subunit CsGG3.2 is involved in the regulation of the CBF regulon and plant tolerance to cold stresses in cucumber (Cucumis sativus L.). We showed that CsGG3.2 transcript abundance was positively induced by cold treatments. Transgenic cucumber plants (T1) constitutively over-expressing CsGG3.2 exhibits tolerance to chilling conditions and increased expression of CBF genes and their regulon. Antioxidative enzymes, i.e., superoxide dismutase, catalase, peroxidase, and glutathione reductase activities increased in cold-stressed transgenic plants. The reactive oxygen species, oxygen free radical and H2O2, production, as well as membrane lipid peroxidation (MDA) production decreased in transgenic plants, suggesting a better antioxidant system to cope the oxidative-damages caused by cold stress. These findings provide evidence for a critical role of CsGG3.2 in mediating cold signal transduction in plant cells.
Keywords: G-protein γ subunit, CsGG3, cucumber, cold stress, CBF genes
Introduction
Cold stress, which includes chilling (<20°C) and/or freezing (<0°C) temperatures, adversely affects the growth and development of crops, and results in heavy economic losses (Chinnusamy et al., 2007). Plants exhibit an increase in chilling tolerance after being exposed to low non-freezing temperatures, which is known as cold acclimation (Medina et al., 2011). During cold acclimation, genes coding CBFs (C-repeat binding factors), HSFC1 (heat shock transcription factor C 1), ZAT12 (zinc transporter of Arabidopsis thaliana 12), and CZF1 (CCCH-type zinc finger 1) are induced, and in turn regulate downstream cold responsive gene (COR) expression making changes in metabolism and physiological processes (Jia et al., 2016; Zhao et al., 2016). These include the changes in the activity of antioxidant enzymes, such as ascorbate peroxidase (APX), superoxide dismutase (SOD), and catalase (CAT), which contributes to increased freezing tolerance (Streb et al., 2002; Janda et al., 2003; Zhang et al., 2008). CBFs, also known as dehydration-responsive element (DRE) binding factor 1 (DREB1), are key transcription factors involved in cold response (Jia et al., 2016). CBF genes are modulated by several transcriptional factors (TFs), including ICE1 (inducer of CBF expression 1), MYB15 (Myb domain protein 15), and CAMTA3 (calmodulin-binding transcription activator 3) (Jia et al., 2016; Zhao et al., 2016). Cold stress sensing leads to a cytosolic calcium (Ca2+) signal (Knight et al., 1996). The activated Ca2+ signal is postulated to activate a MAPK (mitogen-activated protein kinase) cascade, which phosphorylate TFs, such as CAMTAs and ICE1/2 (Zhu, 2016). Recently, Ma et al. (2015) reported that the interaction between heterotrimeric guanine nucleotide-binding protein (G protein) and CHILLING TOLERANCE DIVER GENCE 1 (COLD1) activates the Ca2+ signal upon cold treatment in rice. However, it is not well-established how G protein triggers Ca2+ signal.
G protein signaling pathway is an evolutionarily conserved extracellular signal transduction. G proteins comprises three distinct subunits: alpha (Gα), beta (Gβ), and gamma (Gγ) (Tuteja and Sopory, 2008). According to the canonical paradigm, ligand-bound G protein coupled receptors (GPCRs) catalyzed exchange of GDP for GTP on the Gα actives the heterotrimer, and resulting in dissociation of the two functional elements, Gα subunit and Gβγ dimer, which mediate signal transduction by interacting with multiple downstream effectors, independently (Yuri et al., 2012). In plants, G proteins and play significant roles in many stress responses. For instance, in rice and maize, Gα functions at both cell division and cellular senescence stages of plant responses to NaCl stress (Urano et al., 2014). Microarray analysis revealed that rice Gα plays an important role in the regulation of multiple abiotic stresses, such as drought, salinity, heat, and cold (Jangam et al., 2016). Arabidopsis Gβ (AGB1) positively regulates salt tolerance by affecting the expression of genes related to proline biosynthesis, oxidative stress, ion channel, and ABA-responses (Ya-Nan et al., 2015; Swain et al., 2016). It was also reported that transcripts of PsGα and PsGβ increased after heat, H2O2, and NaCl treatments in Pisum sativum, and over-expression of PsGα enhanced tolerance to salinity and heat in transgenic lines (Misra et al., 2007). However, the available set of subunits in plants are limited. In Arabidopsis genome, there is only one Gα gene (GPA1) and one Gβ gene (AGB1), while three Gγ genes (AGG1, AGG2, and AGG3) (Daisuke et al., 2013). And this is roughly the G protein inventory for plants; for example, rice genome contains only one canonical Gα gene (RGA1), one Gβ gene (RGB1), but five Gγ subunits (RGG1, RGG2, GS3, DEP1, and OsGGC2) (Yuri et al., 2012; Daisuke et al., 2013). With single Gα and Gβ subunits, the specificity of heterotrimer formation is thus solely provided by the Gγ proteins (Trusov et al., 2007, 2008). It is thus important to study the roles of Gγ subunits in triggering Ca2+ signaling for chilling tolerance in plant.
In cucumber, a typical chill-sensitive vegetable crop widely cultivated in the world, low temperatures can result in chilling injuries and lead to significant yield decreases. In the present study, we identified six Gγ proteins encoded by the cucumber genome. The transcript levels of CsGG3.2, encoding a type C Gγ, were up-regulated by cold treatment. CsGG3.2 over-expressing enhanced tolerance of cucumber to chilling stress, and positively regulated the expression of CBF genes and their regulon, as well as activity of enzymes related to reactive oxygen species (ROS) scavenging. We conclude that the type C Gγ subunit CsGG3.2 mediates cold signal transduction in cucumber.
Materials and Methods
Plant Materials and Growth Conditions
Cucumber (Cucumis sativus L.) line 9930 donated by Huang et al. (2009) was used for gene cloning and “Xintai Mici” was used for the construction of transgenic plants. Cucumber seeds germinated in darkness at 28°C were sowed in vermiculite-peat mixture [1:1, volume/volume (V/V)] in the growth chamber under a 12 h light (350 μmol m-2 s-1) at 25°C/12 h dark at 18°C cycle. Plants with two true leaves were exposed to cold stress at 8 ± 1°C for 7 days, with 350 μmol m-2 s-1 light for 12 h every day according to Wang et al. (1999).
Plant Transformation
To generate over-expression lines, the coding sequence of CsGG3.2 was amplified and cloned into the BamHI/SpeI sites of the pCAMBIA-2300 vector to obtain the Pro35S:CsGG3.2 construct. Agrobacterium LB4404 harboring the construct was used for cucumber transformation. The transformation was conducted according to the method of Mu et al. (2017). Briefly, cucumber seeds were rinsed in sterile deionized water for five times and placed on MS0 medium (MS plus 3% sucrose) for 2–3 days after being disinfected with 70% alcohol for 20 s followed by 3% sodium hypochlorite solution for 7 min. The basal cotyledons were then harvested and incubated with Agrobacterium harboring the target constructs for 15 min. The inoculated explants were then cultured on MS1 medium (MS0 medium plus 0.5 mg L-1 6-Benzylaminopurine and 1 mg L-1 ABA) for another 2 days in the dark. The explants were transferred and incubated on MS1 medium for 15–20 days until the shoots were 1–1.5 cm long. The shoots were transferred to MS2 (MS plus 200 mg L-1 cefotaxime) to develop the roots. Polymerase chain reaction (PCR) was employed to confirm the integration of the construct in regenerated plants.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was isolated using RNA prep pure Plant Kit (TANGEN) and first-strand cDNA was synthesized using Fast Quant RT Kit (TANGEN) according to the manufacturer’s instructions. PCR was then carried out using the gene-specific primers listed in Supplementary Table S1 and Super Real PreMix Plus (SYBR Green) Kit (TANGEN) with an Mx3000p Real-time PCR System (Agilent, Stratagene) according to the manufacturer’s instructions. Three biological replicates were included for each sample. The 2-ΔΔCt method was used, and the relative expression levels were normalized to Actin.
Biochemical Analysis Assays
Oxygen free radical (OFR), H2O2, and malondialdehyde (MDA) content, activity of SOD, catalase (CAT), peroxidase (POD), and glutathione reductase (GR) were determined using assay kits (COMINBIO) with a UV-1800 Spectrophotometer (SHIMADZU) according to the manufacturer’s instructions. OFR content was assayed based on the detection of the absorbance of product at 530 nm in the reaction system. H2O2 content was assayed based on the titanium superoxide synthesis method. MDA content was assayed based on the thiobarbituric acid-reactive substance assay. The SOD activity was determined based on the inhibition of formazan synthesis method. The CAT activity was determined based on the decomposition of H2O2 method. The POD activity was assayed based on the detection of the absorbance of product at 470 nm in the reaction system. The GR activity was determined based on the NADPH consumption method.
Assessment of Chilling Tolerance of Cucumber
T1 transgenic lines and control plants with/without cold-acclimated were exposed to 4 ± 1°C for 4 days, with 350 μmol m-2 s-1 light for 12 h every day. Chilling injury (CI) was indexed following Liu et al. (2010). The severity of the symptoms was assessed visually in a four-stage scale: (1) no injury; (2) slight; (3) moderate; (4) extensive. The average extent of cold damage was expressed as CI index, which was calculated using the following formula: CI index (%) = [Σ(CI level) × (number of seedlings at the CI level)/(total number of seedlings) × 4] × 100. Cold acclimation was performed at continuous 8 ± 1°C with 12 h photoperiod (350 μmol m-2 s-1) for 2 days.
Data Analyses
The results were analyzed using GraphPad Prism 6.0 (GraphPad Software) and Data Processing System (DPS) 7.05 (Tang and Zhang, 2013). Three biological replicates were included for each experiment. Data are presented as mean values ± SE (n = 3). The analyses of significant differences (p < 0.05/p < 0.01) were measured using least significant difference (LSD) test.
Results
The Cucumber Proteome Contains Six Heterotrimeric G Protein Gγ Subunits
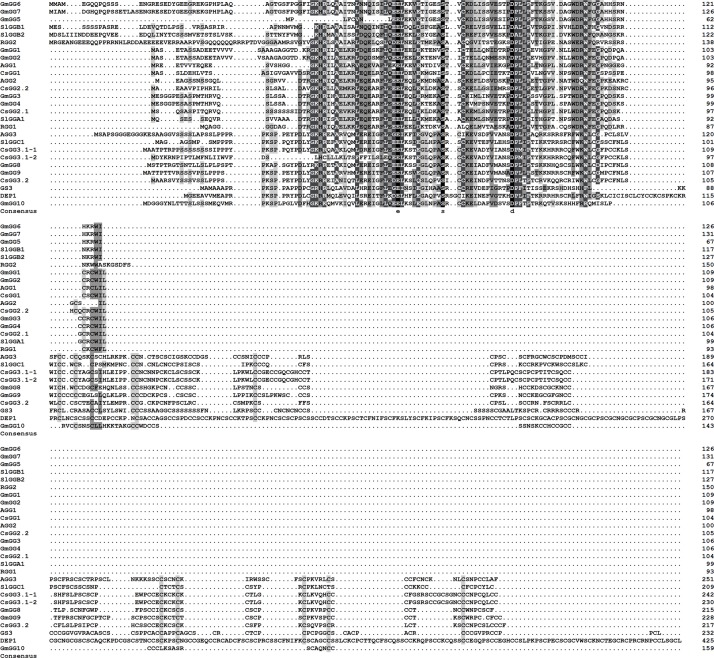
BLAST searches of the cucumber genome1 using Arabidopsis Gγ subunits as queries identified five Gγ-like genes. We named these genes CsGG1 (Csa2G228360), CsGG2.1 (Csa2G215490), CsGG2.2 (Csa3G144190), CsGG3.1 (Csa2G000110), and CsGG3.2 (Csa1G597050). CsGG3.1 alternative splicing produces two protein variants (CsGG3.1-1 and CsGG3.1-2). The six cucumber Gγ homologs were divided into two classes based on amino acid sequence alignments (Figure 1) and phylogenic methods (Figure 2). CsGG1, CsGG2.1, and CsGG2.2 belonged to the previously described type A, while CsGG3.1-1, CsGG3.1-2, and CsGG3.2 belonged to type C (Trusov et al., 2012).
FIGURE 1.
Multiple sequence alignments of CsGG with Gγ amino acids from other species. CsGG and its homologs were aligned using CLUSTAL X. Conserved regions between single classes or the whole family are shown in white text on black, with conserved substitutions shaded in gray. AGG1: Arabidopsis thaliana Q9FDX9 (Uniprot accession No.); AGG2: Arabidopsis thaliana Q93V47; AGG3: Arabidopsis thaliana Q6AWT8; RGG1: Oryza sativa subsp. Japonica Q75WU1; RGG2: Oryza sativa subsp. Japonica Q6YXX9; GS3: Oryza sativa subsp. Japonica C6L686; DEP1: Oryza sativa subsp. Japonica Q67UU9; GmGγ1: Glycine max L. Glyma10g03610 (Soybean Genome accession No.); GmGγ2: Glycine max L. Glyma02g16190; GmGγ3: Glycine max L. Glyma20g35415; GmGγ4: Glycine max L. Glyma10g32214; GmGγ5: Glycine max L. Glyma11g18050; GmGγ6: Glycine max L. Glyma14g17060; GmGγ7: Glycine max L. Glyma17g29590; GmGγ8: Glycine max L. Glyma15g19631; GmGγ9 : Glycine max L. Glyma17g05640; GmGγ10 : Glycine max L. Glyma.07G040200; SlGGA1: Solanum lycopersicum Solyc09g082940.2.1 [Tomato Genome (cv Heinz; ITAG release 2.40) accession No.]; SlGGB1: Solanum lycopersicum Solyc12g096270.1.1; SlGGB2: Solanum lycopersicum Solyc08g005950.2.1; SlGGC1: Solanum lycopersicum Solyc07g041980.2.1.
FIGURE 2.
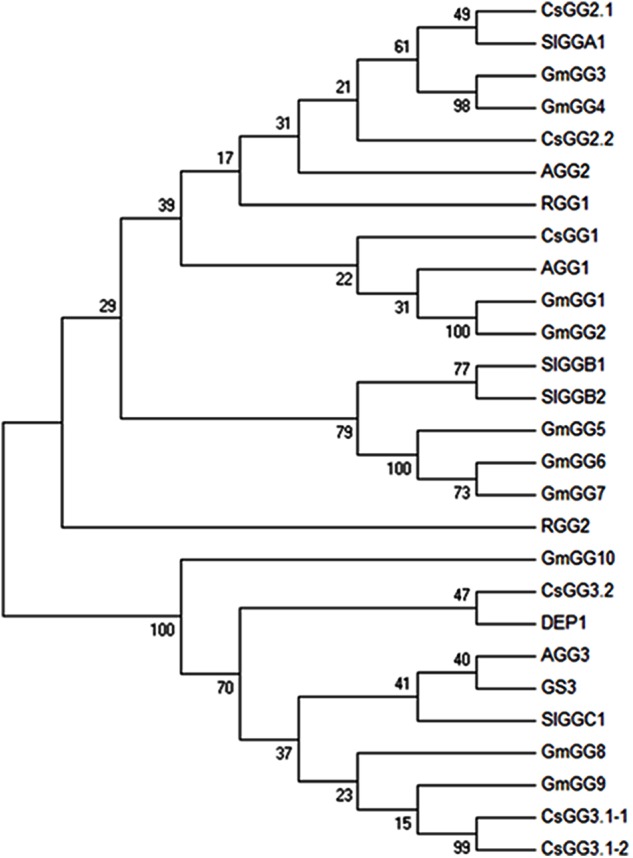
Phylogenetic analysis of CsGGs and other Gγ proteins in plants. Tree was constructed using neighbor-joining method with 1000 bootstrap replicates.
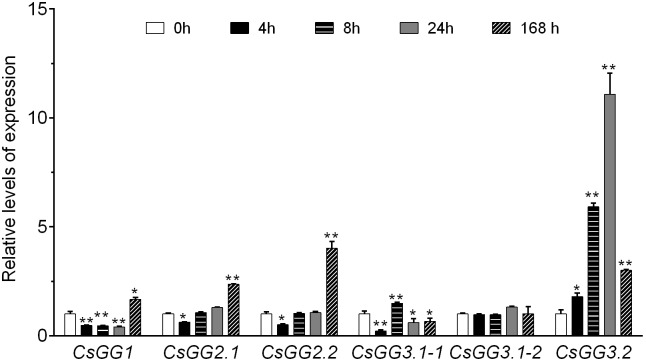
CsGG3.2 Exhibited a Cold Inductive Expression Pattern
To investigate the cold responsive characteristic of CsGG genes, the expression patterns of these genes in leaves were detected by quantitative real-time polymerase chain reaction (qRT-PCR). As shown in Figure 3, CsGG1 transcript decreased from 0 to 24 h, followed by an increase at 168 h. CsGG2.1 and CsGG2.2 showed similar expression profiles, with transcript levels decreased at 4 h, while increased apparently at 168 h. CsGG3.1-1 showed relatively low transcript levels at most of the time, while the transcript of CsGG3.1-2 showed no response to cold treatment. Among the five selected time points, the transcript levels of CsGG3.2 climbed to the peak at 24 h and descended to a lower level at 168 h. These results demonstrate that CsGG3.2 might be involved in cold tolerance in cucumber.
FIGURE 3.
Relative expression of CsGG genes in wild type (WT) cucumber under cold stress. RNA was extracted and the expression levels of the CsGG genes were analyzed by qRT-PCR. Data shown are averages ± SE (n = 3). Three biological replicates were included for each experiment and 10 seedlings were included for each line. Significant differences from plants at 0 h are indicated by asterisks (∗p < 0.05 and ∗∗p < 0.01).
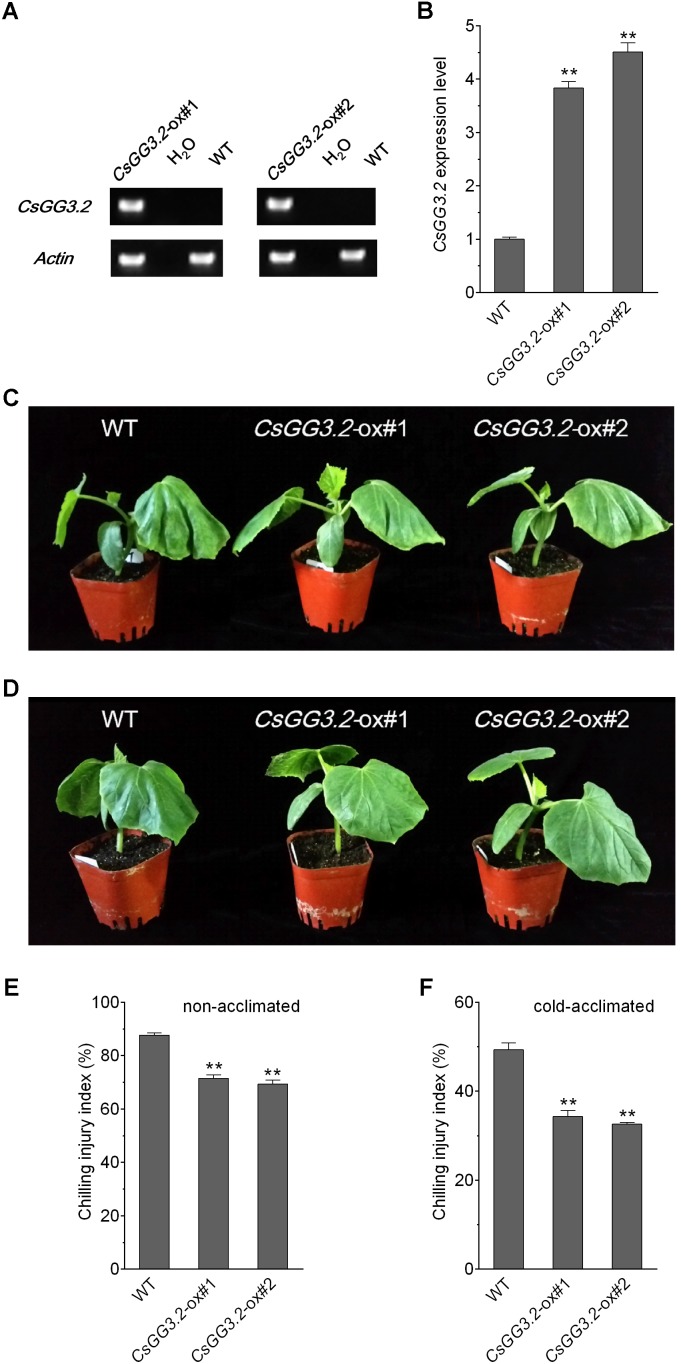
Over-Expression of CsGG3.2 Enhanced the Cold Tolerance of Transgenic Cucumbers
To investigate whether the CsGG3.2 function in chilling tolerance, transgenic cucumber plants over-expressing CsGG3.2 were constructed. Two lines (CsGG3.2-ox#1 and CsGG3.2-ox#2) with CsGG3.2 expression level about four fold higher compared with wild type (WT) cucumber (Figures 4A,B) were exposed to chilling stress to examine the chilling tolerance before and after cold acclimation. For non-acclimated plants, WT plants exhibited severe injury in their cotyledons, and even the first true leaves. Two transgenic lines CsGG3.2-ox#1 and CsGG3.2-ox#2 showed lighter injuries (Figure 4C), and their CI indices significantly lower than WT, and were decreased by 18 and 21%, respectively (Figure 4E). For cold-acclimated plants, WT plants exhibited obvious injury in their cotyledons and first true leaves. Transgenic plants showed slight or no injury, and their CI indices were decreased by 30 and 34%, respectively (Figures 4D,F) compared with WT. These results suggest that the expression of CsGG3.2 enhanced the chilling tolerance of transgenic cucumber plants.
FIGURE 4.
CsGG3.2 enhances cucumber chilling tolerance. (A) PCR analysis of the CsGG3.2 over expressing T1-transgenic lines along with WT, and negative control (H2O). (B) Expression level of the CsGG3.2 mRNA in CsGG3.2 over expressing T1-transgenic lines grown for 7 days. Chilling sensitivity of CsGG3.2 over expressing T1-transgenic and WT plants non-acclimated (C) and cold-acclimated (D). Chilling injury (CI) index of CsGG3.2 over expressing T1-transgenic and WT plants non-acclimated (E) and cold-acclimated (F). The data represent means ± SE (n = 3) of three independent replicates with 10 seedlings (20 seedlings for chilling tolerance assessment) per line per replicate. The asterisks indicate a significant difference between WT and CsGG3.2 over-expressing plants (∗∗p < 0.01).
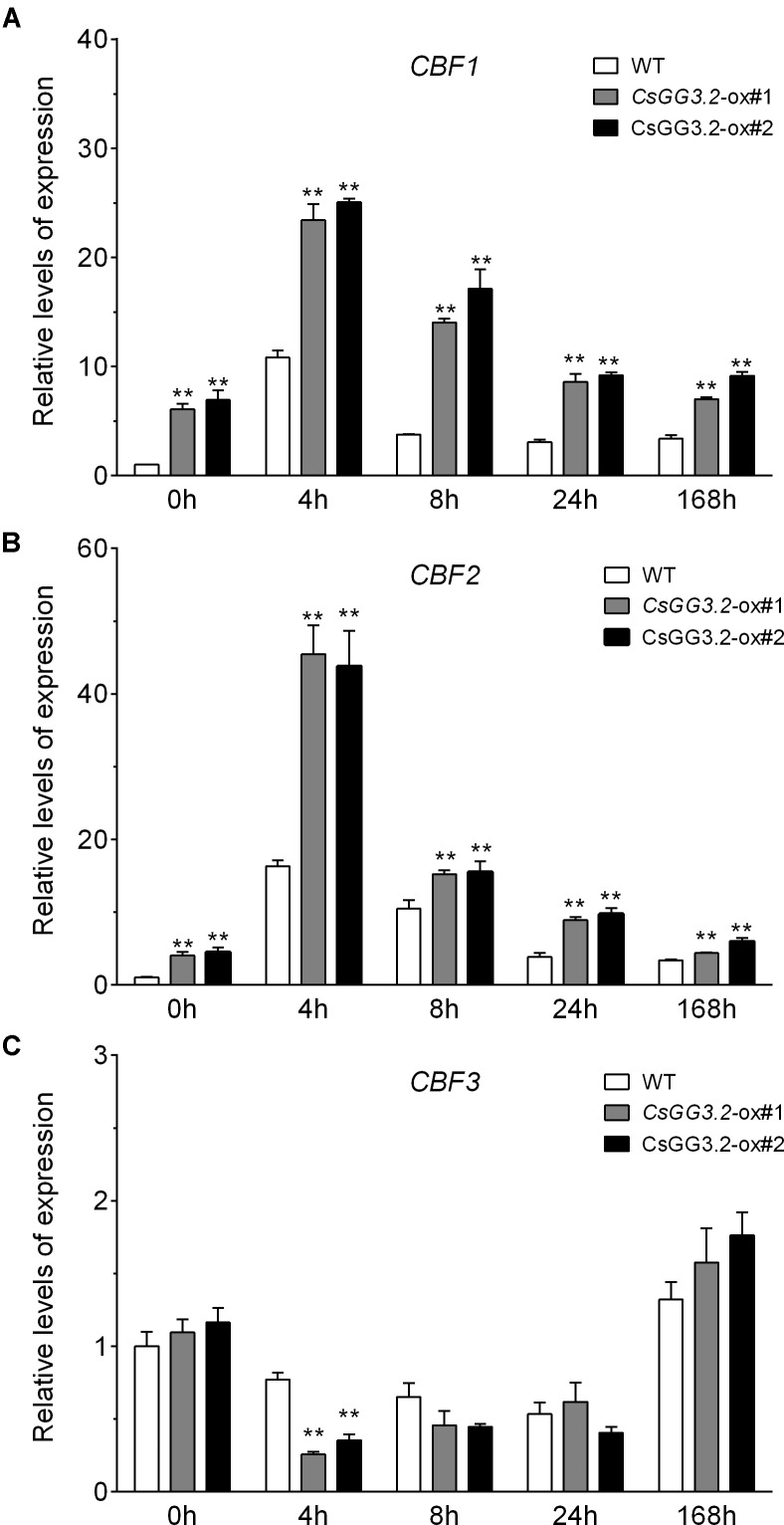
Over-Expression of CsGG3.2 Enhanced CBF and COR Genes Transcripts Upon Exposure to Cold
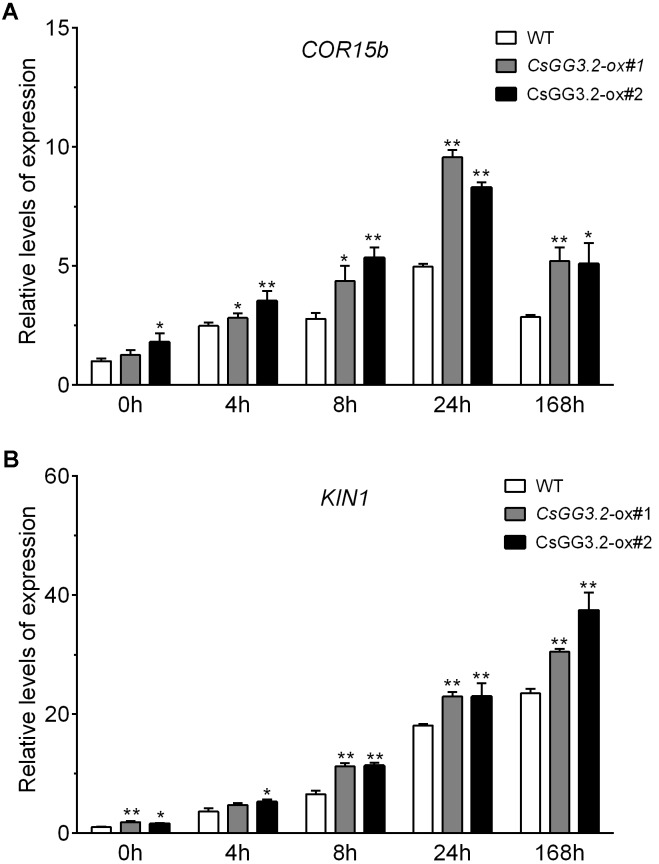
Induction of CBF genes is important for chilling stress tolerance (Zhu et al., 2007). Therefore, we investigated the expression levels of CBF genes and their downstream target genes, COR15b and KIN1 in CsGG3.2 over-expressing transgenic and WT cucumber plants during cold acclimation. Cucumber CBF and COR genes were obtained by database searching using protein sequences of Arabidopsis CBFs, COR15b, and KIN1 as queries. Csa5G174570 and Csa3G751440 corresponded to the published CsCBF1 (GenBank, DQ776899) and CsCBF3 (GenBank, JQ900769), respectively. Csa3G180260 coding a protein sharing high identities to AtCBF2, was named CsCBF2. Csa1G074950 and Csa5G165870 coding late embryogenesis abundant (LEA) proteins were designated as CsCOR15b and CsKIN1, respectively. The transcript levels for CsCBF1 and CsCBF2 increased within 4 h of plants being exposed to cold stress. The levels of CsCBF1 and CsCBF2 expression was significantly higher in CsGG3.2ox plants compared with WT plants (Figures 5A,B). Levels of CsCBF3 transcript were relatively lower compared to that of CsCBF1 and CsCBF2. CsGG3.2 over-expressing significantly reduced the transcript level of CsCBF3, but had no effect at other time points (Figure 5C). Cold increases the CsCOR15b and CsKIN1, two known CBF-inducible COR genes, expression in both WT and CsGG3.2ox plants. And their expression levels were significantly up-regulated by CsGG3.2 over-expressing (Figure 6).
FIGURE 5.
Relative gene expression of cucumber homologs CBF genes in CsGG3.2 over expressing T1-transgenic and WT cucumber plants under cold stress. Sequence data of CsCBF genes can be found in the cucumber genome database under the following accession numbers: CsCBF1, Csa5G174570 (A); CsCBF2, Csa3G180260 (B); CsCBF3, Csa3G751440 (C). Data shown are averages ± SE (n = 3). Three biological replicates were included for each experiment and 10 seedlings were included for each line. The asterisks indicate a significant difference between WT and CsGG3.2 over-expressing plants (∗∗p < 0.01).
FIGURE 6.
Relative gene expression of cucumber homologs COR15b (A), and KIN1 (B) in CsGG3.2 over-expressing T1-transgenic and WT plants under cold stress. Sequence data of CsCOR15b and CsKIN1 can be found in the cucumber genome database under the accession number Csa1G074950 and Csa5G165870 respectively. Data shown are averages ± SE (n = 3). Three biological replicates were included for each experiment and 10 seedlings were included for each line. The asterisks indicate a significant difference between WT and CsGG3.2 over-expressing plants (∗p < 0.05 and ∗∗p < 0.01).
Analysis of Antioxidant Enzymes Activity and Response of ROS and Malondialdehyde (MDA) in Transgenic Plants
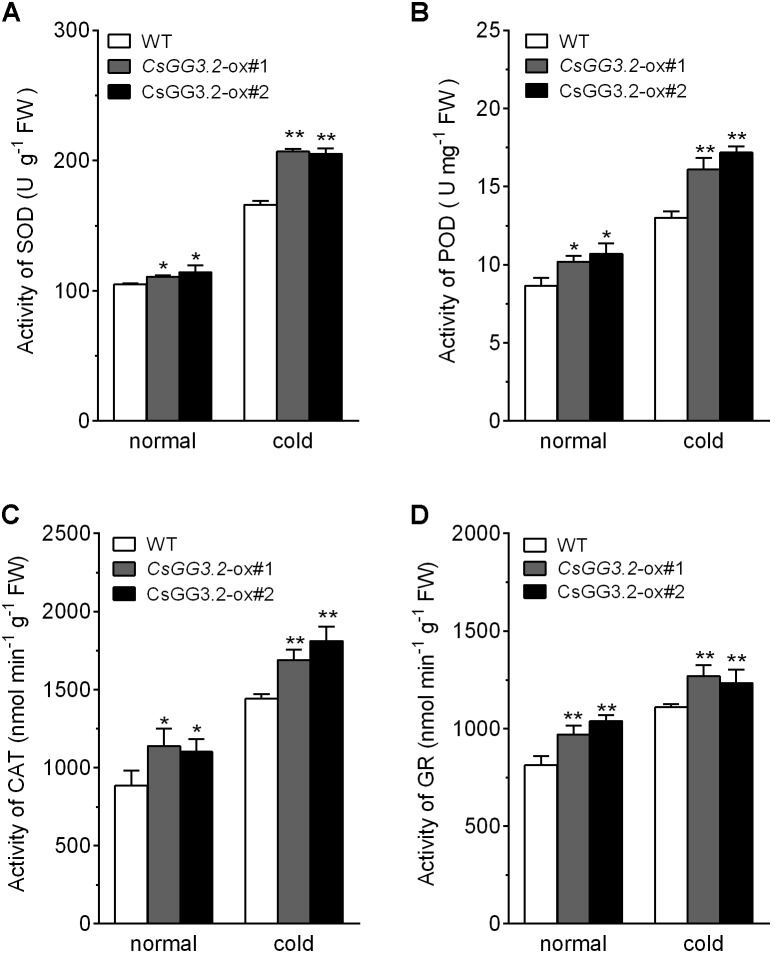
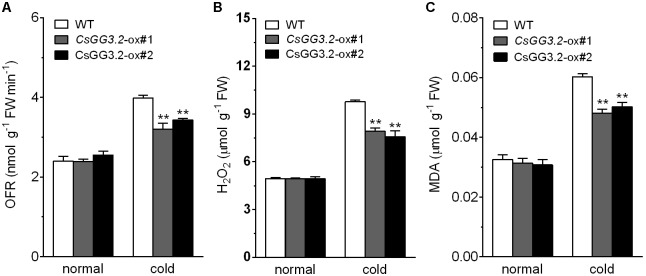
The changes induced by cold in the activities of antioxidant enzymes and production of OFR, H2O2, and malondialdehyde (MDA) in transgenic lines were compared with WT plants. Over-expression of CsGG3.2 resulted in increased enzymatic activities of SOD, POD, CAT, and GR in transgenic plants under both normal and cold conditions (Figures 7A–D). And this resulted in decreased accumulation of OFR and H2O2 in the transgenic plant in stressful environment compared with WT plants (Figures 8A,B). The increased detoxification of ROS led to reduced membrane lipid peroxidation, i.e., MDA production. MDA level was low (about 0.031 μmol g-1 FW) under normal conditions in transgenic and WT plants. MDA content in WT plants increased to 0.060 μmol g-1 FW under chilling stress, whereas with a lower increase, to only 0.048 μmol g-1 FW and 0.050 μmol g-1 FW respectively, was seen in the transgenic plants (Figure 8C).
FIGURE 7.
Antioxidant activities of CsGG3.2 over-expressing T1-transgenic and WT plants under cold stress. (A) Superoxide dismutase (SOD), one unit of enzyme activity defined as 50% formazan synthesis inhibited in the xanthine oxidation coupling reaction solution. (B) Peroxidase (POD), one unit of enzyme activity defined as the absorbency at 470 nm increased by 0.01 in 1 ml reaction solution min-1 per gram tissue. (C) Catalase (CAT) activity, one unit of enzyme activity defined as 1 nmol H2O2 oxidized min-1 per gram tissue. (D) Glutathione reductase (GR), one unit of enzyme activity defined as 1 nmol NADPH oxidized min-1 per gram tissue. Data shown are averages ± SE (n = 3). Three biological replicates were included for each experiment and 10 seedlings were included for each line. The asterisks indicate a significant difference between WT and CsGG3.2 over-expressing plants (∗p < 0.05 and ∗∗p < 0.01).
FIGURE 8.
Biochemical analysis of CsGG3.2 over-expressing T1-transgenic and WT plants under cold stress. (A) Oxygen free radical (OFR) productive rate per gram tissue. (B) Hydrogen peroxide (H2O2) content. (C) Lipid peroxidation expressed in terms of malondialdehyde (MDA) content. Data shown are averages ± SE (n = 3). Three biological replicates were included for each experiment and 10 seedlings were included for each line. The asterisks indicate a significant difference between WT and CsGG3.2 over-expressing plants (∗∗p < 0.01).
Discussion
Plant Gγ subunits are involved in a wide range of developmental and physiological processes, and have a high potential for crop improvements (Botella, 2012). In rice, DEP1 and GS3 are major quantitative trait loci for controlling seed size and panicle branching (Botella, 2012); RGG1 and RGG2 are up-regulated upon salinity, harsh temperature, and ABA treatments (Yadav et al., 2012). Arabidopsis AGG1 and AGG2 are reported to be involved in osmotic stress and root development, and AGG3 is also found to be involved in regulation of organ size and stress response (Chakravorty et al., 2011; Li et al., 2012a,b; Thung et al., 2013). Similarly, the soybean GγIII subunit plays a role in ABA-dependent lateral root development (Choudhury and Pandey, 2013). Tomato SlGGB1 also mediates auxin and ABA signaling (Subramaniam et al., 2016). Here, we demonstrated that over-expression of CsGG3.2 promotes tolerance of cucumber seedlings to chilling stress. Moreover, multiple mechanisms appear to contribute to this increase in freezing tolerance, including alterations in gene expression associated with cold acclimation and the activation of antioxidative enzymes.
Chilling tolerance of plants was enhanced by cold acclimation. DREB1/CBFs are transcription factors regulating the expression of more than 100 COLD RESPONSIVE (COR) genes, and thus important for cold acclimation and chilling tolerance (Chinnusamy et al., 2007; Park et al., 2015). Constitutive expression of any one of the CBF genes in transgenic plants gives rise to strong constitutive expression of the COR genes and hence increased freezing tolerance in plants (Jagloottosen et al., 1998; Kasuga et al., 1999; Gilmour et al., 2004; Nakashima, 2006). Expression of CsCBF1 and CsCBF2 were upregulated in CsGG3.2ox plants (Figures 5A,B), supporting the proposed role of CsGG3 in promoting chilling tolerance. However, transcription levels of CsCBF3 were lower than that of CsCBF1 and CsCBF2 in both WT and transgenic plants (Figure 5C). This results may be due to the negative interaction between CBF genes, which is important for transient and tightly controlling their expression (Knight and Knight, 2012).
The expression of CBF genes activate many downstream genes that enhance plants chilling and freezing tolerance (Park et al., 2015). Many CBF-inducible genes have been cloned and characterized from Arabidopsis and other species. COR15a encodes a chloroplast-targeted polypeptide, which functions as cryoprotective protein preventing the formation of hexagonal II-phase lipids in the chloroplast stroma (Nakayama et al., 2007). The homolog of AtCOR15a, AtCOR15b has transcript profiles similar to that of AtCOR15a under cold stress (Wilhelm and Thomashow, 1993). And the transcript level of CbCOR15b from shepherd’s purse (Capsella bursa-pastoris) was significantly upregulated under cold treatment, and over-expression of CbCOR15b enhanced cold tolerance in transgenic tobacco plants (Wu et al., 2012). KIN1 is another CBF downstream target gene from Arabidopsis encoding a 6.5 KDa polypeptide similarity to the type I fish antifreeze proteins. In Cucurbita crops, watermelon and pumpkin, transcripts of CmCOR15b, ClKIN1, and CmKIN1 significantly increased during cold stress condition, suggesting that they could contribute to the cold tolerance (Kang et al., 2009). Here, we showed that the transcript levels of cucumber CsCOR15b and CsKIN1 increased dramatically in response to low temperature (Figure 6), which are consistent with that previously found in watermelon and pumpkin (Kang et al., 2009). Meanwhile, expression of CsCOR15b and CsKIN1 was upregulated by CsGG3.2 over-expressing (Figure 6), suggesting CsGG3.2 facilitates induction of COR genes expression, and cold tolerance.
Reactive oxygen species were produced and accumulated under cold stress, which damaged cell and yielded MDA (Achard et al., 2008; Swain et al., 2016). Antioxidant enzymes are important components of the ROS scavenging system in the plant cell, thus play a significant role in plant cold tolerance (Ahmad et al., 2010; Liu et al., 2010). It has been reported that activities of antioxidant enzymes in plants are increased under low temperature stress, which might be due to the upregulation of corresponding genes (Baek and Skinner, 2003; Soltész et al., 2011). In present study, the activity of SOD, POD, CAT, and GR enzymes in WT plants increased under cold condition, but the rates of increase were higher in transgenic plants (Figure 7). The increased detoxification of ROS led to reduced membrane lipid peroxidation, and could increase the chilling tolerance of transgenic plants. These results are in agreement with the previous studies where a decreased level of ROS production under cold stress has been reported in ICE1-ox cucumber (Liu et al., 2010).
Heterotrimeric G proteins play important roles in stress responses by interacting with Ca2+ channels in animals (Wang and Chong, 2010). Recently, plant G protein signaling found to be involved in cold signal transduction: activation of G protein by COLD1, triggered Ca2+ influx upon cold treatment, leading to a cytosolic Ca2+ signal (Ma et al., 2015). Ca2+ signal is postulated to activate a MAP kinase cascade, which then phosphorylate cold responsive TFs such as CAMTAs and ICE1/2 (Zhu, 2016). The exact mechanism of G protein-mediated plant cold response is not known yet. Here, our findings showed that CsGG3.2 positively regulates the expression of CBF genes and their regulon, as well as the activities of antioxidant enzymes, which lead to cold stress tolerance. The results suggest a critical role of CsGG3.2 in cucumber cold response, which will help in understanding the G protein-mediated cold signal transduction in plants.
Author Contributions
XY, YaL, LB, and CH: conceived and designed the experiments. LB and YuL: performed the experiments. LB: analyzed the data. LB, YM, and YY: contributed reagents/materials/analysis tools. LB, YuL, and AA: wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Professor Huang for providing cucumber line 9930.
Funding. This work was financially supported by funds from the National Key Research and Development Plan of China (2016YFD0201006), the Earmarked fund for Modern Agro-industry Technology Research System in China (CARS-25-C-01), the Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-IVFCAAS), the R&D Special Fund for Public Welfare Industry (Agriculture) (201203005), and the Key Laboratory of Horticultural Crop Biology and Germplasm Innovation, Ministry of Agriculture, China.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00488/full#supplementary-material
References
- Achard P., Gong F. S., Alioua M., Hedden P., Genschik P. (2008). The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20 2117–2129. 10.1105/tpc.108.058941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad P., Jaleel C. A., Salem M. A., Nabi G., Sharma S. (2010). Roles of enzymatic and nonenzymatic antioxidants in plants during abiotic stress. Crit. Rev. Biotechnol. 30 161–175. 10.3109/07388550903524243 [DOI] [PubMed] [Google Scholar]
- Baek K. H., Skinner D. Z. (2003). Alteration of antioxidant enzyme gene expression during cold acclimation of near-isogenic wheat lines. Plant Sci. 165 1221–1227. 10.1016/S0168-9452(03)00329-7 [DOI] [Google Scholar]
- Botella J. R. (2012). Can heterotrimeric G proteins help to feed the world? Trends Plant Sci. 17 563–568. 10.1016/j.tplants.2012.06.002 [DOI] [PubMed] [Google Scholar]
- Chakravorty D., Trusov Y., Zhang W., Acharya B. R., Sheahan M. B., Mccurdy D. W., et al. (2011). An atypical heterotrimeric G-protein γ-subunit is involved in guard cell K+-channel regulation and morphological development in Arabidopsis thaliana. Plant J. 67 840–851. 10.1111/j.1365-313X.2011.04638.x [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J., Zhu J. K. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12 444–451. 10.1016/j.tplants.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Choudhury S. R., Pandey S. (2013). Specific subunits of heterotrimeric G proteins play important roles during nodulation in soybean. Plant Physiol. 162 522–533. 10.1104/pp.113.215400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daisuke U., Chen J. G., Ramón B. J., Jones A. M. (2013). Heterotrimeric G protein signalling in the plant kingdom. Open Biol. 3:120186. 10.1098/rsob.120186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour S. J., Fowler S. G., Thomashow M. F. (2004). Arabidopsis transcriptional activators CBF1. CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 54 767–781. 10.1023/B:PLAN.0000040902.06881.d4 [DOI] [PubMed] [Google Scholar]
- Huang S., Li R., Zhang Z., Li L., Gu X., Fan W., et al. (2009). The genome of the cucumber, Cucumis sativus L. Nat. Genet. 41 1275–1281. 10.1038/ng.475 [DOI] [PubMed] [Google Scholar]
- Jagloottosen K. R., Gilmour S. J., Zarka D. G., Schabenberger O., Thomashow M. F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280 104–106. 10.1126/science.280.5360.104 [DOI] [PubMed] [Google Scholar]
- Jangam A. P., Pathak R. R., Raghuram N. (2016). Microarray analysis of rice d1 (RGA1) mutant reveals the potential role of G-protein alpha subunit in regulating multiple abiotic stresses such as drought, salinity, heat, and cold. Front. Plant Sci. 7:11. 10.3389/fpls.2016.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda T., Szalai G., Rios-Gonzalez K., Veisz O., Páldi E. (2003). Comparative study of frost tolerance and antioxidant activity in cereals. Plant Sci. 164 301–306. 10.1016/S0168-9452(02)00414-4 [DOI] [Google Scholar]
- Jia Y., Ding Y., Shi Y., Zhang X., Gong Z., Yang S. (2016). The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 212 345–353. 10.1111/nph.14088 [DOI] [PubMed] [Google Scholar]
- Kang G. Z., Zhu Z. H., Guo T. C., Ren J. P. (2009). Isolation and expression pattern of COR15b and KIN1 genes in watermelon and pumpkin. Afr. J. Biotechnol. 8 5666–5672. 10.5897/AJB09.1225 [DOI] [Google Scholar]
- Kasuga M., Liu Q., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17 287–291. [DOI] [PubMed] [Google Scholar]
- Knight M. R., Knight H. (2012). Low-temperature perception leading to gene expression and cold tolerance in higher plants. New Phytol. 195 737–751. 10.1111/j.1469-8137.2012.04239.x [DOI] [PubMed] [Google Scholar]
- Knight H., Trewavas A. J., Knight M. R. (1996). Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 8 489–503. 10.1105/tpc.8.3.489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Liu W., Zhang X., Liu Y., Li N., Li Y. (2012a). Roles of the Arabidopsis G protein γ subunit AGG3 and its rice homologs GS3 and DEP1 in seed and organ size control. Plant Signal. Behav. 7 1357–1359. 10.4161/psb.21620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Liu Y., Zheng L., Chen L., Li N., Corke F., et al. (2012b). The plant-specific G protein γ subunit AGG3 influences organ size and shape in Arabidopsis thaliana. New Phytol. 194 690–703. 10.1111/j.1469-8137.2012.04083.x [DOI] [PubMed] [Google Scholar]
- Liu L., Duan L., Zhang J., Zhang Z., Mi G., Ren H. (2010). Cucumber (Cucumis sativus L.) over-expressing cold-induced transcriptome regulator ICE1 exhibits changed morphological characters and enhances chilling tolerance. Sci. Hortic. 124 29–33. 10.1016/j.scienta.2009.11.018 [DOI] [Google Scholar]
- Ma Y., Dai X., Xu Y., Luo W., Zheng X., Zeng D., et al. (2015). COLD1 confers chilling tolerance in rice. Cell 160 1209–1221. 10.1016/j.cell.2015.01.046 [DOI] [PubMed] [Google Scholar]
- Medina J., Catalá R., Salinas J. (2011). The CBFs: three arabidopsis transcription factors to cold acclimate. Plant Sci. 180 3–11. 10.1016/j.plantsci.2010.06.019 [DOI] [PubMed] [Google Scholar]
- Misra S., Wu Y., Venkataraman G., Sopory S. K., Tuteja N. (2007). Heterotrimeric G-protein complex and G-protein-coupled receptor from a legume (Pisum sativum): role in salinity and heat stress and cross-talk with phospholipase C. Plant J. 51 656–669. 10.1111/j.1365-313X.2007.03169.x [DOI] [PubMed] [Google Scholar]
- Mu Y., Liu Y., Bai L., Li S., He C., Yan Y., et al. (2017). Cucumber CsBPCs regulate the expression of CsABI3 during seed germination. Front. Plant Sci. 8:459. 10.3389/fpls.2017.00459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K. (2006). Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plants. Physiol. Plant. 126 62–71. 10.1111/j.1399-3054.2005.00592.x [DOI] [Google Scholar]
- Nakayama K., Okawa K., Kakizaki T., Honma T., Itoh H., Inaba T. (2007). Arabidopsis Cor15am is a chloroplast stromal protein that has cryoprotective activity and forms oligomers. Plant Physiol. 144 513–523. 10.1104/pp.106.094581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Lee C. M., Doherty C. J., Gilmour S. J., Kim Y. S., Thomashow M. F. (2015). Regulation of the Arabidopsis CBF regulon by a complex low - temperature regulatory network. Plant J. 82 193–207. 10.1111/tpj.12796 [DOI] [PubMed] [Google Scholar]
- Soltész A., Tímár I., Vashegyi I., Tóth B., Kellos T., Szalai G., et al. (2011). Redox changes during cold acclimation affect freezing tolerance but not the vegetative/reproductive transition of the shoot apex in wheat. Plant Biol. 13 757–766. 10.1111/j.1438-8677.2010.00429.x [DOI] [PubMed] [Google Scholar]
- Streb P., Shang W., Feierabend J. (2002). Resistance of cold-hardened winter rye leaves (Secale cereale L.) to photo-oxidative stress. Plant Cell Environ. 22 1211–1223. 10.1046/j.1365-3040.1999.00483.x [DOI] [Google Scholar]
- Subramaniam G., Trusov Y., Encina C. L., Hayashi S., Batley J., Botella J. R. (2016). Type B heterotrimeric G protein γ subunit regulates auxin and ABA signaling in tomato. Plant Physiol. 170 1117–1134. 10.1104/pp.15.01675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain D. M., Sahoo R. K., Srivastava V. K., Tripathy B. C., Tuteja R., Tuteja N. (2016). Function of heterotrimeric G-protein γ subunit RGG1 in providing salinity stress tolerance in rice by elevating detoxification of ROS. Planta 245 1–17. 10.1007/s00425-016-2614-3 [DOI] [PubMed] [Google Scholar]
- Tang Q. Y., Zhang C. X. (2013). Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci. 20 254–260. 10.1111/j.1744-7917.2012.01519.x [DOI] [PubMed] [Google Scholar]
- Thung L., Chakravorty D., Trusov Y., Jones A. M., Botella J. R. (2013). Signaling specificity provided by the Arabidopsis thaliana heterotrimeric G-protein γ subunits AGG1 and AGG2 is partially but not exclusively provided through transcriptional regulation. PLoS One 8:3. 10.1371/journal.pone.0058503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusov Y., Chakravorty D., Botella J. R. (2012). Diversity of heterotrimeric G-protein γ subunits in plants. BMC Res. Notes 5:608. 10.1186/1756-0500-5-608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusov Y., Rookes J. E., Tilbrook K., Chakravorty D., Mason M. G., Anderson D., et al. (2007). Heterotrimeric G protein gamma subunits provide functional selectivity in Gbetagamma dimer signaling in Arabidopsis. Plant Cell 19 1235–1250. 10.1105/tpc.107.050096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusov Y., Zhang W., Assmann S. M., Botella J. R. (2008). Gγ1 + Gγ2 ≠ Gβ: heterotrimeric G protein Gγ-deficient mutants do not recapitulate all phenotypes of Gβ-deficient mutants. Plant Physiol. 147 636–649. 10.1104/pp.108.117655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja N., Sopory S. K. (2008). Plant signaling in stress: G-protein coupled receptors, heterotrimeric G-proteins and signal coupling via phospholipases. Plant Signal. Behav. 3 79–86. 10.4161/psb.3.2.5303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D., Colaneri A., Jones A. M. (2014). Gα modulates salt-induced cellular senescence and cell division in rice and maize. J. Exp. Bot. 65 6553–6561. 10.1093/jxb/eru372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Chong K. (2010). “Signaling and communication in plants,” in Integrated G Proteins Signaling in Plants, eds Yalovsky S., Baluska F., Jones A. (Berlin: Springer; ), 1–25. [Google Scholar]
- Wang Z., Tang Y., Zhang F., Wang H. (1999). Effect of boron and low temperature on membrane integrity of cucumber leaves. J. Plant Nutr. 22 543–550. 10.1080/01904169909365650 [DOI] [Google Scholar]
- Wilhelm K. S., Thomashow M. F. (1993). Arabidopsis thaliana cor15b, an apparent homologue of cor15a, is strongly responsive to cold and ABA, but not drought. Plant Mol. Biol. 23 1073–1077. 10.1007/BF00021822 [DOI] [PubMed] [Google Scholar]
- Wu L., Zhou M., Shen C., Liang J., Lin J. (2012). Transgenic tobacco plants over expressing cold regulated protein CbCOR15b from Capsella bursa-pastoris exhibit enhanced cold tolerance. J. Plant Physiol. 169 1408–1416. 10.1016/j.jplph.2012.05.016 [DOI] [PubMed] [Google Scholar]
- Yadav D. K., Islam S. M., Tuteja N. (2012). Rice heterotrimeric G-protein gamma subunits (RGG1 and RGG2) are differentially regulated under abiotic stress. Plant Signal. Behav. 7 733–740. 10.4161/psb.20356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ya-Nan M. A., Ming C., Dong-Bei X. U., Fang G. N., Wang E. H., Gao S. Q., et al. (2015). G-protein β subunit AGB1 positively regulates salt stress tolerance in Arabidopsis. J. Integr. Agric. 14 314–325. 10.1016/S2095-3119(14)60777-2 [DOI] [Google Scholar]
- Yuri T., David C., Ramón B. J. (2012). Diversity of heterotrimeric G-protein γ subunits in plants. BMC Res. Notes 5:608. 10.1186/1756-0500-5-608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Luo Y., Hou Y.-X., Jiang H., Chen Q., Tang H.-R. (2008). Chilling acclimation induced changes in the distribution of H2O2 and antioxidant system of strawberry leaves. Agron. J. 3 286–291. [Google Scholar]
- Zhao C., Zhang Z., Xie S., Si T., Li Y., Zhu J.-K. (2016). Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 171 2744–2759. 10.1104/pp.16.00533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Dong C. H., Zhu J. K. (2007). Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr. Opin. Plant Biol. 10 290–295. 10.1016/j.pbi.2007.04.010 [DOI] [PubMed] [Google Scholar]
- Zhu J.-K. (2016). Abiotic stress signaling and responses in plants. Cell 167 313–324. 10.1016/j.cell.2016.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.