Abstract
Our previous studies on the endogenous brassinosteroids (BRs) in Arabidopsis have provided suggestive evidence for the operation of the early C6-oxidation and the late C6-oxidation pathways, leading to brassinolide (BL) in Arabidopsis. However, to date the in vivo operation of these pathways has not been fully confirmed in this species. This paper describes metabolic studies using deuterium-labeled BRs in wild-type and BR-insensitive mutant (bri1) seedlings to establish the intermediates of the biosynthetic pathway of BL in Arabidopsis. The first evidence for the conversion of campestanol to 6-deoxocathasterone and the conversion of 6-deoxocathasterone to 6-deoxoteasterone is provided. The later biosynthetic steps (6-deoxoteasterone → 3-dehydro-6-deoxoteasterone → 6-deoxotyphasterol → 6-deoxocastasterone → 6α-hydroxycastasterone → castasterone → BL) were demonstrated by stepwise metabolic experiments. Therefore, these studies complete the documentation of the late C6-oxidation pathway. The biosynthetic sequence involved in the early C6-oxidation pathway (teasterone → 3-dehydroteasterone → typhasterol → castasterone → BL) was also demonstrated. These results show that both the early and late C6-oxidation pathways are functional in Arabidopsis. In addition we report two new observations: the presence of a new branch in the pathway, C6 oxidation of 6-deoxotyphasterol to typhasterol, and increased metabolic flow in BR-insensitive mutants.
Brassinosteroids (BRs) are now recognized as a major hormone controlling plant growth and development (Yokota, 1997; Altmann, 1998; Clouse and Feldmann, 1999). Up to now more than 40 BRs have been fully characterized (Fujioka, 1999). The biosynthetic pathways of brassinolide (BL), the most active BR, have been elucidated using cultured cells of Catharanthus roseus. Through extensive metabolic studies, parallel branched pathways for BL, namely the early C6-oxidation and late C6-oxidation pathways, have been proposed (Fujioka and Sakurai, 1997a, 1997b). The first reaction toward BL is the conversion of campesterol (CR) to campestanol (CN), and then CN is converted to castasterone (CS) through either the early C6-oxidation pathway or the late C6-oxidation pathway. Finally CS is converted to BL, the most active BR. The natural occurrence of most BR intermediates was demonstrated in cultured cells of C. roseus. In addition most of the steps were defined by feeding deuterium-labeled substrates followed by identification of the metabolites using gas chromatography-mass spectrometry (GC-MS). However, some steps remained to be validated. In fact the first proposed biosynthetic step, namely the conversion of CR to CN, was only recently refined (Noguchi et al., 1999a). The study revealed the biosynthetic scheme CR → (24R)-24-methylcholest-4-en-3β-ol → (24R)-24-methylcholest-4-en-3-one→(24R)-24-methyl-5α- cholestan-3-one → CN in cultured cells of C. roseus and seedlings of Arabidopsis. These findings were based on the identification of endogenous intermediates and demonstration of the reaction sequence by the metabolic studies. More recently the step from 6-oxocampestanol (6-OxoCN) to cathasterone (CT) has been demonstrated in cultured cells of C. roseus (Fujioka et al., 2000). This result, together with our previous studies (Fujioka and Sakurai, 1997a, 1997b) completed the documentation of the early C6-oxidation pathway. However, the sequence, CN → 6-deoxocathasterone (6-DeoxoCT) → 6-deoxoteasterone (6-DeoxoTE) in the late C6-oxidation pathway has remained hypothetical.
These biochemical studies have been supported by the recent discovery of a number of BR-deficient mutants from Arabidopsis (Clouse and Feldmann, 1999), pea (Nomura et al., 1997, 1999), and tomato (Bishop et al., 1999; Koka et al., 2000). These mutants typically exhibit dwarf phenotypes when grown in the light, and de-etiolation in the dark. The discovery of BR mutants led to wide acceptance of an essential role for BRs in plant growth and development (Yokota, 1997; Altmann, 1998; Clouse and Feldmann, 1999). Arabidopsis BR-deficient mutants, det2 (Li et al., 1996, 1997; Fujioka et al., 1997; Noguchi et al., 1999a), cpd (Szekeres et al., 1996), dwf4 (Azpiroz et al., 1998; Choe et al., 1998), dwf1/dim (Takahashi et al., 1995; Klahre et al., 1998; Choe et al., 1999a), ste1/dwf7 (Choe et al., 1999b), sax1 (Ephritikhine et al., 1999), and dwf5 (Choe et al., 2000), and the BR-insensitive mutant, bri1 (Clouse et al., 1996; Li and Chory, 1997; Noguchi et al., 1999b), were isolated and characterized. In the course of analyzing these mutants, measurements of the endogenous levels of BRs provided important information toward defining the defective steps in the BR-biosynthetic mutants and characterization of the BR-insensitive mutants. BL, CS, typhasterol (TY), teasterone (TE), 6-deoxocastasterone (6-DeoxoCS), 6-deoxotyphasterol (6-DeoxoTY), and 6-DeoxoTE were identified as endogenous BRs in Arabidopsis from various tissues such as shoots, siliques, and seeds (Fujioka et al., 1996; 1998; Noguchi et al., 1999b). All BRs identified in Arabidopsis are important components of either the early or the late C6-oxidation pathways, suggesting that both pathways are functional in this species. However, no metabolic studies in Arabidopsis have been carried out yet.
In this study we describe the metabolism of deuterium-labeled BR intermediates in seedlings of wild-type and BR-insensitive (bri1) mutants. We show that the conversion of CN to 6-DeoxoTE via 6-DeoxoCT operates in Arabidopsis. We also demonstrate the operation of the biosynthetic sequence, 6-DeoxoTE → 3-dehydro-6-deoxoteasterone (6-Deoxo3DT) → 6-DeoxoTY → 6-DeoxoCS → 6α-hydroxycastasterone (6-OHCS) → CS → BL. Furthermore, we provide evidence for the operation of the biosynthetic sequence, TE → 3-dehydroteasterone (3DT) → TY→ CS → BL in Arabidopsis, and for a new branch in the pathway: 6-DeoxoTY to TY.
RESULTS
To establish the biosynthetic pathway of BL in Arabidopsis, the metabolism of deuterium-labeled BR intermediates in Arabidopsis seedlings was investigated. Metabolites were identified by full-scan GC-MS (Table I). Identified metabolites from each substrate are summarized in Table II. The amount of each metabolite was roughly estimated from the peak area in the mass chromatogram of the GC-MS analysis (Table II).
Table I.
Representative GC-MS data used for the identification of BR metabolites
| BRs | Rt | Prominent Ion m/z |
|---|---|---|
| min | relative intensity % | |
| [2H6]6-DeoxoCT | 10.45 | 553 (M+-15, 2), 297 (3), 193 (100) |
| [2H6]6-DeoxoTE | 10.70 | 536 (M+, 40), 521 (35), 446 (10), 431 (15), 215 (100), 161 (37) |
| [2H6]6-Deoxo3DT | 10.82 | 462 (M+, 18), 283 (16), 231 (100), 161 (54) |
| [2H6]6-DeoxoTY | 10.30 | 536 (M+, 22), 521 (5), 446 (18), 431 (20), 215 (100), 161 (28) |
| [2H6]6-DeoxoCS | 10.80 | 504 (M+, 46), 489 (12), 288 (13), 273 (100), 161 (55) |
| [2H6]TE | 12.08 | 550 (M+, 21), 535 (62), 521 (100), 460 (3), 161 (19) |
| [2H6]3DT | 12.07 | 476 (M+, 14), 399 (3), 316 (14), 245 (15), 161 (100) |
| [2H6]TY | 11.33 | 550 (M+, 69), 535 (50), 521 (100), 460 (41), 161 (66) |
| [2H6]CS | 11.95 | 518 (M+, 52), 399 (8), 358 (16), 287 (25), 161 (100) |
| [2H6]BL | 12.87 | 534 (M+, 3), 374 (19), 338 (20), 177 (80), 161 (100) |
Table II.
Results of metabolic experiment of BR1-5 biosynthetic intermediates in Arabidopsis wild type and bri1-5
| Substrate | Metabolite
|
|
|---|---|---|
| Wild type (Ws-2) | bri1-5 | |
| [2H6]CN | [2H6]6-DeoxoCT (1) | [2H6]6-DeoxoCT (3) |
| [2H6]6-DeoxoCT | [2H6]6-DeoxoTE (0.5), [2H6]6-DeoxoTY (1) | [2H6]6-DeoxoTE (0.5), [2H6]6-DeoxoTY (15) |
| [2H6]6-DeoxoTE | [2H6]6-Deoxo3DT (10), [2H6]6-DeoxoTY (14), [2H6]6-DeoxoCS (0.5), [2H6]TY (0.1) | [2H6]6-Deoxo3DT (3), [2H6]6-DeoxoTY (45), [2H6]6-DeoxoCS (3), [2H6]TY (0.2), [2H6]CS (3) |
| [2H6]6-Deoxo3DT | [2H6]6-DeoxoTE (390), [2H6]6-DeoxoTY (2600), [2H6]6-deoxoCS (26), [2H6]TY (4), [2H6]CS (2) | [2H6]6-DeoxoTE (100), [2H6]6-DeoxoTY (1000), [2H6]6-DeoxoCS (4), [2H6]TY (15), [2H6]CS (15) |
| [2H6]6-DeoxoTY | [2H6]6-DeoxoTE (5), [2H6]6-Deoxo3DT (20), [2H6]6-DeoxoCS (2), [2H6]TY (0.5), [2H6]CS (2) | [2H6]6-DeoxoTE (15), [2H6]6-Deoxo3DT (26),[2H6]6-DeoxoCS (28), [2H6]TY (80), [2H6]CS (28) |
| [2H6]6-DeoxoCS | [2H6]CS (8) | [2H6]CS (10) |
| [2H6]6-OH CS | [2H6]CS (4) | [2H6]CS (20) |
| [2H6]TE | [2H6]TY (50), [2H6]CS (1) | [2H6]TY (100), [2H6]CS (3) |
| [2H6]3DT | [2H6]TE (10), [2H6]TY (500) | [2H6]TE (2), [2H6]TY (66) |
| [2H6]TY | – | [2H6]CS (1) |
| [2H6]CS | – | [2H6]BL (5) |
Values in parentheses indicate the detected amount (nanograms) of each metabolite.
The Metabolism of CN and 6-DeoxoCT in Wild-Type Seedlings
Although the natural co-occurrence of CN, 6-DeoxoCT, and 6-DeoxoTE has been demonstrated in C. roseus and tomato, metabolic conversions of CN to 6-DeoxoCT, and of 6-DeoxoCT to 6-DeoxoTE have not been observed in any plant systems so far. When [2H6]CN was fed to wild-type (Ws-2) seedlings, [2H6]6-DeoxoCT was detected together with endogenous 6-DeoxoCT by full-scan GC-MS. A mass spectrum derived from a mixture of endogenous 6-DeoxoCT and [2H6]6-DeoxoCT (as a metabolite of [2H6]CN) was obtained. Prominent ion peaks were as follows: (*, metabolite; #, endogenous) m/z 553* (0.3%), 547# (2%), 297*# (3%), 193* (8%), and 187# (100%). Although ions derived from endogenous 6-DeoxoCT were predominant in the mass spectrum, minor but distinct ions (at m/z 553 and 193) corresponding to [2H6]6-DeoxoCT trimethylsilyl derivative were detected (approximately 8% of the endogenous 6-DeoxoCT).
GC-selected ion monitoring analysis confirmed the presence of endogenous 6-DeoxoCT and [2H6]6-DeoxoCT (Rt of [2H6]6-DeoxoCT was ca 1 s earlier than that of 6-DeoxoCT). The results showed the conversion of CN to 6-DeoxoCT and natural occurrence of 6-DeoxoCT in seedlings of Arabidopsis. When [2H6]6-DeoxoCT was fed to seedlings, [2H6]6-DeoxoTE and [2H6]6-DeoxoTY were identified together with endogenous compounds. The detected amount of [2H6]6-DeoxoTE (a metabolite of [2H6]6-DeoxoCT) was approximately 20% that of endogenous 6-DeoxoTE, whereas [2H6]6-DeoxoTY (a metabolite of [2H6]6-DeoxoCT) was more abundant than endogenous 6-DeoxoTY (the ratio was 2:1). Detection of [2H6]6-DeoxoCT from the feed of [2H6]CN, and of [2H6]6-DeoxoTE from the feed of [2H6]6-DeoxoCT indicates that the conversion of CN to 6-DeoxoCT and the conversion of 6-DeoxoCT to 6-DeoxoTE occur in Arabidopsis seedlings. This is the first evidence for the conversion of CN to 6-DeoxoCT and the conversion of 6-DeoxoCT to 6-DeoxoTE. In addition the natural occurrence of 6-DeoxoCT in Arabidopsis was demonstrated for the first time. Using a different ecotype (wild type, En-2), the above experiments were repeated. The conversion of [2H6]CN to [2H6]6-DeoxoCT and the conversion of [2H6]6-DeoxoCT to [2H6]6-DeoxoTE and [2H6]6-DeoxoTY, as well as the occurrence of endogenous 6-DeoxoCT were found in En-2, confirming our findings in Ws-2.
The Metabolism of 6-DeoxoTE, 6-Deoxo3DT, and 6-DeoxoTY in Wild-Type Seedlings
The results of metabolic experiments with [2H6]6-DeoxoTE, [2H6]6-Deoxo3DT and [2H6]6-DeoxoTY are summarized in Figure 1. When [2H6]6-DeoxoTE was fed to wild-type (Ws-2) seedlings, [2H6]6-Deoxo3DT and [2H6]6-DeoxoTY were detected as major metabolites. Trace amounts of [2H6]6-DeoxoCS and [2H6]TY were also identified in this feed. When feeding [2H6]6-Deoxo3DT to seedlings, [2H6]6-DeoxoTY was detected as a major metabolite. [2H6]6-DeoxoTE, [2H6]6-DeoxoCS, [2H6]TY, and [2H6]CS were also identified as metabolites of [2H6]6-Deoxo3DT. When [2H6]6-DeoxoTY was fed to seedlings, [2H6]6-DeoxoCS, [2H6]6-DeoxoTE, [2H6]6-Deoxo3DT, [2H6]TY, and [2H6]CS were identified. From the above metabolic experiments, the conversion of 6-DeoxoTE to 6-Deoxo3DT,the conversion of 6-Deoxo3DT to 6-DeoxoTY, and the conversion of 6-DeoxoTY to 6-DeoxoCS have been definitely demonstrated in Arabidopsis. These results confirmed our previous findings (biosynthetic sequence, 6-DeoxoTE → 6-Deoxo3DT → 6-DeoxoTY → 6-DeoxoCS), which were established in cultured cells of C. roseus (Choi et al., 1997). In addition the reversible conversion between 6-DeoxoTE and 6-DeoxoTY was observed. Furthermore, [2H6]TY was clearly detected from the feeds of [2H6]6-DeoxoTE, [2H6]6-Deoxo3DT, and [2H6]6-DeoxoTY, although the amount was trace. These results show that 6-DeoxoTE, 6-Deoxo3DT, and/or 6-DeoxoTY are converted to TY in Arabidopsis. This is the first report that TY was identified as a metabolite of 6-deoxoBRs, indicating that C6 oxidation of 6-deoxoBR could occur not only in the conversion of 6-DeoxoCS to CS, but also in the conversion of 6-DeoxoTY to TY.
Figure 1.
Metabolism of 6-DeoxoBRs in wild-type seedlings of Arabidopsis. Box indicates substrate. Arrows show the observed metabolism from each substrate.
The Metabolism of 6-DeoxoCS and 6-OHCS in Wild-Type Seedlings
When [2H6]6-DeoxoCS was fed to seedlings, [2H6]CS was identified by full-scan GC-MS (Table II). After feeding [2H6]6-OHCS to seedlings, [2H6]CS was also identified. Therefore we have established that the late C6-oxidation pathway is functional in Arabidopsis seedlings (Fig. 2).
Figure 2.
Proposed biosynthetic pathways for BL in Arabidopsis.
The Metabolism of 6-OxoBR Intermediates in Wild-Type Seedlings
Metabolism of 6-oxoBR intermediates was also carried out. When [2H6]TE was fed, [2H6]TY (major) and [2H6]CS (trace) were identified and when [2H6]3DT was fed, [2H6]TY (major) and [2H6]TE (minor) were detected (ratio of the detected amounts of metabolites, [2H6]TE:[2H6]TY = 1:50). Other conversions belonging to the early C6-oxidation pathway, i.e. the conversions of CN to 6-OxoCN, of 6-OxoCN to CT, and CT to TE were not observed in wild-type seedlings.
The Metabolism of 6-DeoxoBR Intermediates in BR-Insensitive Mutants
We also performed metabolic experiments using two BR-insensitive (bri1) mutants (a null allele, bri1-4 and a weak allele, bri1-5). Recently we have shown that bri1 mutants accumulate high levels of BRs (Noguchi et al., 1999b). The results for bri1-5 are summarized in Table II.
When feeding [2H6]CN to bri1-5 seedlings, [2H6]6-DeoxoCT was detected as a metabolite. In the feed of [2H6]6-DeoxoCT to bri1-5, [2H6]6-DeoxoTE and [2H6]6-DeoxoTY were detected as metabolites. The detected amount of [2H6]6-DeoxoTY was more abundant compared with the same feeding experiment in wild-type seedlings (Table II). From the feeding of [2H6]6-DeoxoTE, [2H6]6-DeoxoTY (major), [2H6]6-Deoxo3DT, [2H6]6-DeoxoCS, [2H6]TY, and [2H6]CS were identified. When feeding [2H6]6-Deoxo3DT to bri1-5 seedlings, [2H6]6-DeoxoTY (major), [2H6]6-DeoxoTE, [2H6]6-DeoxoCS, [2H6]TY, and [2H6]CS were detected as metabolites. After [2H6]6-DeoxoTY was fed to bri1-5, [2H6]6-DeoxoCS, [2H6]6-DeoxoTE, [2H6]6-Deoxo3DT, [2H6]TY, and [2H6]CS were detected. From both the feeding of [2H6]6-DeoxoCS and [2H6]6-OHCS, [2H6]CS was detected as a metabolite. Thus the biosynthetic steps belonging to the late C6-oxidation pathway were also demonstrated in bri1-5, similar to that of wild type.
The Metabolism of 6-OxoBR Intermediates in BR-Insensitive Mutants
From the feeding of [2H6]TE, [2H6]TY, and [2H6]CS were detected as metabolites. After the feeding of [2H6]3DT, [2H6]TY (major) and [2H6]TE (minor) were detected as metabolites. The ratio of [2H6]TY and [2H6]TE was 33:1. When [2H6]TY was fed to bri1-5, [2H6]CS was detected as a metabolite, although this conversion was not observed in wild-type seedlings. Furthermore, from the feeding of [2H6]CS, [2H6]BL was identified as a metabolite. Thus, the biosynthetic sequence of TE → 3DT → TY → CS → BL was demonstrated in Arabidopsis seedlings.
When feeding [2H6]6-DeoxoCS to a null allele, bri1-4, [2H6]CS was identified as a major metabolite. It is interesting that [2H6]BL was also identified as a metabolite of [2H6]6-DeoxoCS by full-scan GC-MS.
Altogether the full biosynthetic sequence of the late C6-oxidation pathway has been demonstrated in bri1 mutants. In addition partial sequence of the early C6-oxidation pathway has been also demonstrated. The conversions of TY to CS and of CS to BL were detected in bri1 mutants, whereas the conversions were not detected in wild type. In addition the detected amounts of the metabolites in bri1 were, in most cases, higher than those in wild type. These results suggest that biosynthesis of BRs in bri1 mutants may be more active compared with wild type. The increased metabolic flow in bri1 may be associated with induction of the transcripts of biosynthetic enzymes.
DWF4 mRNA Level Is Dramatically Increased in bri1-5
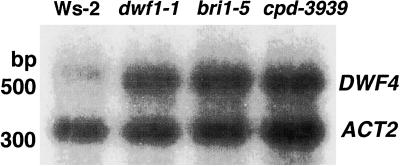
To learn whether the increased metabolic flow in bri1-5 is associated with the up-regulation of the mRNA coding for biosynthetic enzymes we examined the level of DWF4 mRNA in wild-type and BR mutants including bri1-5. It has been previously proposed that DWF4 mediates a putative rate-limiting step in the BR biosynthetic pathway, and the level of the DWF4 mRNA is extremely low (Choe et al., 1998). Thus we chose to examine DWF4 mRNA levels using reverse transcriptase (RT)-PCR techniques rather than regular northern analysis. Figure 3 displays a result of Southern analysis for DWF4 and Actin-2. The relative levels for the control Actin-2 are similar throughout wild type, dwf1-1, bri1-5, and cpd-3939, suggesting this is a reliable control for this experiment. By contrast the DWF4 mRNA level is dramatically increased in the mutants including bri1-5.
Figure 3.
DNA gel-blot analysis of DWF4 RT-PCR products of wild type (Ws-2), dwf1-1, bri1-5, and cpd-3939. DWF4 and Actin-2 cDNAs were used as probes. The level of DWF4 is dramatically increased in the mutants as compared with that of wild type.
DISCUSSION
Biosynthesis of BRs had previously been investigated mainly using cultured cells of C. roseus, and it has been proposed that BL, the most active BR, is biosynthesized from CR by two alternative pathways, namely the early C6-oxidation and the late C6-oxidation pathways (Fujioka and Sakurai, 1997a, 1997b). Many BR-deficient mutants in Arabidopsis have recently been isolated and characterized (Clouse and Feldmann, 1999). Through analyses of the mutants we have accumulated information on the endogenous BRs in this species. The BR profile in Arabidopsis suggests that the proposed pathways may operate in Arabidopsis, but no metabolic experiments had been previously carried out. In this study we have investigated the biosynthesis of BRs in seedlings of Arabidopsis by metabolic experiments with deuterium-labeled BR intermediates. These studies provide evidence for the operation of our proposed pathways for BL in Arabidopsis.
6-DeoxoTE Is Biosynthesized from CN via 6-DeoxoCT
Although 6-DeoxoCT and 6-DeoxoTE have been previously identified as endogenous BRs in several plant species (Bishop et al., 1999; Fujioka et al., 2000), their biosynthetic origin has not been verified. From their chemical structures it was expected that CN and 6-DeoxoCT were the biosynthetic precursors of 6-DeoxoCT and 6-DeoxoTE, respectively. As expected, this study revealed that the metabolite from CN was 6-DeoxoCT, and one of the metabolites from 6-DeoxoCT was 6-DeoxoTE, providing the first evidence for the biosynthetic origin of 6-DeoxoCT and 6-DeoxoTE. As a result, the biosynthetic sequence CN → 6-deoxoCT → 6-deoxoTE has been definitely established for the first time.
Early and Late C6-Oxidation Pathways Are Functional in Arabidopsis
By stepwise metabolic studies the biosynthetic sequence (6-DeoxoTE to CS) in the late C6-oxidation pathway established in C. roseus (Choi et al., 1996, 1997) has also been demonstrated in Arabidopsis seedlings. Therefore this is the first report that the full biosynthetic sequence of the late C6-oxidation pathway, CN → 6-DeoxoCT → 6-DeoxoTE → 6-Deoxo3DT → 6-DeoxoTY → 6-DeoxoCS → 6-OHCS → CS, has been established in the same plant species (Fig. 2).
The biosynthetic origin of CT was recently shown to be 6-OxoCN (Fujioka et al., 2000). That study together with our previous studies (Fujioka and Sakurai, 1997a, 1997b) completed the elucidation of the early C6-oxidation pathway in C. roseus. By contrast, this study completes the elucidation of the late C6-oxidation pathway in Arabidopsis. All combined studies substantiate our original proposed pathways for the early and late C6-oxidation pathways. In the present study the sequence of TE → 3DT → TY → CS → BL has been definitely demonstrated in Arabidopsis. Although some steps in the early C6-oxidation pathway remain to be validated in Arabidopsis, the data presented here together with BR profiles in Arabidopsis strongly suggest that both the early C6-oxidation and the late C6-oxidation pathways are functional in Arabidopsis seedlings.
Which Biosynthetic Pathway Is Important in Arabidopsis?
Our results indicate that Arabidopsis seedlings have at least two separate biosynthetic pathways for BRs. Among the native BRs in Arabidopsis, the levels of 6-DeoxoTY and 6-DeoxoCS are usually predominant in wild-type Arabidopsis, whereas the levels of 6-oxoBRs are relatively low (Choe et al., 1999b, 2000; Noguchi et al., 1999b). From a quantitative point of view it appears that the late C6-oxidation pathway is the major source of BL in light-grown seedlings of Arabidopsis. We previously reported that rescue experiments of det2 and dwf4 mutants using intermediates in each of the two pathways resulted in differential growth effects between dark- and light-grown seedlings (Fujioka et al., 1997; Choe et al., 1998). Namely, 6-deoxoBR intermediates showed stronger activity than their corresponding 6-oxoBR intermediates in the light, whereas 6-deoxoBR intermediates were slightly less active than the corresponding 6-oxidized forms in the dark. The results suggest that the late C6-oxidation pathway may play a predominant role in the light, whereas the early C6-oxidation pathway may be dominant in the dark. Under a variety of light conditions biosynthesis and metabolism of BRs could differ. Examination of BR profiles in the plants grown in the dark and other light conditions would yield information as to how the light regime is associated with differential usage of the two pathways.
Reversible Conversion between 6-DeoxoTE and 6-DeoxoTY, and TE and TY
The reversible conversion between 6-DeoxoTE and 6-DeoxoTY via 6-Deoxo3DT was observed in Arabidopsis (Fig. 1). In addition, reversible conversion between TE and TY via 3DT was also observed. GC-MS data in the feeds of [2H6]6-Deoxo3DT and [2H6]3DT revealed that the major part of the label was in 6-DeoxoTY and TY, with minor incorporation into 6-DeoxoTE and TE, respectively. Thus these reversible conversions prefer the formation of 3α-hydroxyl (6-DeoxoTY and TY) to the formation of 3β-hydroxyl (6-DeoxoTE and TE). This is very similar to the reversible conversion between TE and TY, which was found in other plant species (Abe et al., 1994; Suzuki et al., 1994, 1995a). It is most likely that pathways from 6-DeoxoTE to 6-DeoxoTY via 6-Deoxo3DT and from TE to TY via 3DT are predominant, whereas the reversible reactions from 6-DeoxoTY to 6-DeoxoTE, and from TY to TE occur as a minor event.
New Branched Pathway: 6-DeoxoTY to TY
It is interesting that TY was identified as one of the metabolites from 6-DeoxoTY in this study, although the amount was trace. So far no evidence for the conversion of 6-DeoxoTY to TY has been obtained in similar metabolic studies using cultured cells of C. roseus. It has been reported that C6 oxidation occurs to convert CN to 6-OxoCN and 6-DeoxoCS to CS in C. roseus (Suzuki et al., 1995b; Choi et al., 1996). Our present results suggest that C6 oxidation may occur in several other steps. In fact the conversion of 6-DeoxoTY to TY has been definitely shown in this study. In addition when 6-DeoxoTE or 6-Deoxo3DT was fed, TY was identified as one of their metabolites. As one possibility, 6-DeoxoTE or 6-Deoxo3DT may be converted to TE or 3DT, and then to TY in Arabidopsis. Alternatively, 6-DeoxoTE or 6-Deoxo3DT may be converted to 6-DeoxoTY, then converted to TY. However, we cannot rule out the possibility that the unexpected conversions are due to atypical enzymatic reactions possibly caused by higher concentration of fed substrates. Thus further experiments including in vitro enzyme assays will be required to confirm these new pathways. A tomato dwarf mutant has been shown recently to be defective in the conversion of 6-DeoxoCS to CS (Bishop et al., 1999). Functional expression of DWARF in yeast has revealed that DWARF catalyzes two steps, namely the conversion of 6-DeoxoCS to 6-OHCS, and the conversion of 6-OHCS to CS (Bishop et al., 1999). So it would be very interesting to know how many DWARF paralogs are present in Arabidopsis and how C6 oxidation is regulated by these C6 oxidases. If C6 oxidation occurs at every possible step in the early and late C6-oxidation pathways, it means that BR biosynthesis is composed of a metabolic grid similar to GA biosynthesis. The grid would connect members of the late C6-oxidation pathway to the corresponding members of the early C6-oxidation pathway.
CS Is Converted to BL in Arabidopsis
The conversion of CS to BL has been demonstrated only in cultured cells and seedlings of C. roseus. Several trials using different plant species such as rice and tobacco have not produced positive results (Suzuki et al., 1995a). Even in plants in which CS and BL are native, the metabolic conversion has not been shown. In wild-type seedlings of Arabidopsis the situation was the same. We have tried several times to show metabolic conversion of CS to BL, but were not successful. We have, however, succeeded in showing the conversion using BR-insensitive mutants, bri1-4 (null allele) and bri1-5 (weak allele). We reported that bri1 mutants accumulate very high levels of endogenous BRs, especially 6-oxoBRs such as BL and CS (Noguchi et al., 1999b). Therefore it could be expected that biosynthesis of BRs in bri1 mutants might be up-regulated as compared with wild type. As expected, the conversion of CS to BL was shown in bri1 mutants. In the case of bri1-4, a null allele, BL was detected even in the feed of 6-DeoxoCS. This is only the second example for the conversion of CS to BL in the plant kingdom. In addition the conversion of TY to CS, which was not observed in the wild type, was observed in this mutant. These results indicate that biosynthesis of BRs in bri1 mutant may be more active compared with wild type.
DWF4 mRNA Is Up-Regulated in bri1 Mutants
We have previously shown that the proper perception of BRs is a prerequisite to homeostasis of endogenous BR levels in Arabidopsis bri1 mutants (Noguchi et al., 1999b). Homeostasis is not feedback-regulated by an end product itself, but by successful perception of BRs. Mathur et al. (1998) suggested that the feedback-regulatory factor is translated de novo, since the regulation was abolished in the presence of the protein synthesis inhibitor cycloheximide. Accumulation of BRs could be due to many possible reasons: inactivation of a degradation pathway, increased activity of BR biosynthetic enzymes, increased stability of mRNAs, and continued transcription of the genes for BR biosynthetic enzymes. Increased steady-state levels of DWF4 mRNA in bri1-5 support both the third and fourth possibilities. In addition DWF4 mRNA level is also up-regulated in the BR-sensitive, dwf1-1, and cpd-3939 mutants, suggesting that normal BR signaling is required for transcriptional regulation of DWF4.
MATERIALS AND METHODS
Seedling Cultures
Wild-type Arabidopsis ecotypes Wassilewskija (Ws-2) and Enkheim (En-2) and BR-insensitive mutants, bri1-4 and bri1-5 (Noguchi et al., 1999b) were used in this study. Wild-type and bri1 seedlings were germinated and grown on one-half-concentrated Murashige and Skoog medium (Murashige and Skoog, 1962) containing 1% (w/v) agar and 1% (w/v) Suc in the light at 22°C. Seven days after sowing, the seedlings (wild type, 10–20 seedlings; bri1-4 and bri1-5, 40–60 seedlings) were transferred to a 200-mL flask containing 30 mL of one-half-concentrated Murashige and Skoog medium supplemented with 1% (w/v) Suc. The seedlings were incubated at 22°C in the light on a shaker (110 rpm). After 6 to 8 d in culture, deuterium-labeled substrates were added aseptically to each 200-mL flask and the seedlings were allowed to grow under the same conditions.
Deuterium-Labeled Substrates
Deuterium-labeled substrates used in this study were chemically synthesized: [2H6]BL, [2H6]CS, [2H6]TY, and [2H6]TE (Takatsuto and Ikekawa, 1986), [2H6]3DT (Suzuki et al., 1994), [2H6]CT (Fujioka et al., 1995), [2H6]6-OHCS (Fujioka et al., 2000), [2H6]6-DeoxoCS (Choi et al., 1996), [2H6]6-DeoxoTY, [2H6]6-Deoxo3DT, and [2H6]6-DeoxoTE (Choi et al., 1997), and [2H6]CN (Fujioka et al., 1997). [2H6]6-DeoxoCT was synthesized from [2H6]crinosterol (T. Watanabe, T. Noguchi, S. Fujioka, and S. Takatsuto, unpublished data).
Metabolism of Deuterium-Labeled Substrates
A MeOH solution of deuterium-labeled substrate (10 μg/10 μL or 20 μg/20 μL) was added aseptically to each flask. For the feeding of [2H6]CN, an acetone solution of the substrate (200 μg/40 μL) was added to each flask.
After incubation (2–4 d) the cultures (tissues and medium) were extracted with MeOH, and the extract was partitioned three times between CHCl3 (25 mL) and (50 mL). The CHCl3-soluble fraction was purified by a silica gel cartridge (2 g, Sep-Pak Vac Silica, Waters, Milford, MA). The column was subsequently eluted with 30 mL each of CHCl3, 2% (v/v) MeOH in CHCl3, and 7% (v/v) MeOH in CHCl3. Each eluent was subjected to ODS-HPLC (4.6 × 150 mm, Senshu Pak ODS 1151-D, Senshu Scientific Co., Ltd., Tokyo) at a flow rate 1 mL min−1. MeOH was used as a solvent for the eluate derived from the CHCl3 fraction (6-DeoxoCT was detected in retention time [Rt] of 3.5–4 min). Eighty percent (v/v) CH3CN was used as a solvent for the eluate derived from 2% (v/v) MeOH fraction (6-DeoxoTE, 6-Deoxo3DT, and 6-DeoxoTY were detected in Rt of 10–14, 14–18, and 18–22 min fractions, respectively). Sixty-five percent (v/v) CH3CN or 45% (v/v) CH3CN was used as solvent for the eluate derived from the 7% (w/v) MeOH fraction. BL, CS, TE, 3DT, TY, and 6-DeoxoCS were detected in Rt of 2- to 3-, 3- to 4-, 5- to 6-, 8- to 10-, 8- to 10-, and 11- to 14-min fractions, respectively (when 65% [v/v] CH3CN was used as the solvent), whereas BL and CS were detected in Rt of 6- to 8- and 9- to 11-min fractions, respectively (when 45% [v/v] CH3CN was used as the solvent).
GC-MS Analyses
The GC-MS analyses were carried out under the following conditions: an Automass mass spectrometer (JMS-AM150, JEOL, Tokyo) was connected to a gas chromatograph (5890A-II, Hewlett-Packard, Wilmington, DE), electron ionization (70 eV), with a source temperature of 230°C, column DB-5 (J&W, 15 m × 0.25 mm, 0.25-μm film thickness), and an injection temperature of 280°C. The column temperature program was: 80°C for 1 min, then raised to 320°C at a rate of 30°C min−1, and held at this temperature for 5 min. The interface temperature was 280°C and the carrier gas was He at a flow rate of 1 mL min−1 with splitless injection. For analyses of BL, CS, 6-DeoxoCS, 3DT, and 6-Deoxo3DT, samples were derivatized to methaneboronate, for analyses of TE, TY, 6-DeoxoTE, and 6-DeoxoTY, samples were derivatized to methaneboronate-trimethylsilyl ether, and for analyses of CT and 6-DeoxoCT, samples were derivatized to trimethylsilyl ether (Fujioka et al., 1997).
RT-PCR
The routine techniques of molecular biology were according to Sambrook et al. (1989). Five micrograms of total RNA isolated from 3-week-old Ws-2 wild type, dwf1-1, bri1-5, and cpd-3939 was subject to cDNA synthesis using a SuperScript II kit (BRL, Gaithersburg, MD) following the manufacturer's directions. cDNA samples were first treated with RNaseH, then used for PCR. For a reaction control, the Actin-2 gene (GenBank accession no. U37281) was amplified in conjunction with DWF4. The names and oligonucleotide sequences used as primers are ACT2RTF (5′-AGTGTGTCTTGTCTTATCTGGTTCG-3′), ACT2RTR (5′-AATAGCTGCATTGTCACCCGATACT-3′), D4RTF (5′-TTCTTGGTGAAACCATCGGTTATCTTAAA-3′),and D4RTR (5′-TATGATAAGCAGTTCCTGGTAGATTT-3′). The expected sizes of the PCR products are 380 bp for Actin-2 and 546 bp for DWF4. For normalization of the amplification between the two genes, 0.1 volume of the cDNA products was used for amplification of the DWF4 message, whereas 0.01 volume of the Actin-2 product was used for amplification. DWF4 and Actin-2 messages were amplified in separate tubes using the same PCR program consisting of a 3-min initial denaturation at 94°C and 35 cycles of amplification (30 s at 94°C, 30 s at 58°C, and 40 s at 72°C). The two PCR products for each genotype were combined, and the one-half of the PCR products were separated on a 1% (w/v) agarose gel. The DNA was transferred to a membrane and probed with the DWF4 full-length cDNA and 3′-untranslated region of Actin-2 cDNA.
Footnotes
This work was supported by a Grant-in-Aid for Scientific Research (B) from the Ministry of Education, Science, Sports, and Culture of Japan (grant no. 10460050 to S.F.), by the National Science Foundation (grant no. 9604439 to K.A.F.), and by the U.S. Department of Agriculture (grant no. 97–35304–4708 to F.E.T.).
LITERATURE CITED
- Abe H, Honjo C, Kyokawa Y, Asakawa S, Natsume M, Narushima M. 3-Oxoteasterone and the epimerization of teasterone: identification in lily anthers and Distylium racemosum leaves and its biotransformation into typhasterol. Biosci Biotechnol Biochem. 1994;58:986–989. [Google Scholar]
- Altmann T. Recent advances in brassinosteroid molecular genetics. Curr Opin Plant Biol. 1998;1:378–383. doi: 10.1016/s1369-5266(98)80259-8. [DOI] [PubMed] [Google Scholar]
- Azpiroz R, Wu Y, LoCascio JC, Feldmann KA. An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell. 1998;10:219–230. doi: 10.1105/tpc.10.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GJ, Nomura T, Yokota T, Harrison K, Noguchi T, Fujioka S, Takatsuto S, Jones JDG, Kamiya Y. The tomato DWARF enzyme catalyzes C-6 oxidation in brassinosteroid biosynthesis. Proc Natl Acad Sci USA. 1999;96:1761–1766. doi: 10.1073/pnas.96.4.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell. 1998;10:231–243. doi: 10.1105/tpc.10.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Gregory BD, Ross AS, Yuan H, Noguchi T, Fujioka S, Takatsuto S, Tanaka A, Yoshida S, Tax FE, Feldmann KA. The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol. 1999a;119:897–907. doi: 10.1104/pp.119.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Noguchi T, Fujioka S, Takatsuto S, Tissier CP, Gregory BD, Ross AS, Tanaka A, Yoshida S, Tax FE, Feldmann KA. The Arabidopsis dwf7/ste1 is defective in the Δ7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell. 1999b;11:207–221. [PMC free article] [PubMed] [Google Scholar]
- Choe S, Tanaka A, Noguchi T, Fujioka S, Takatsuto S, Ross AS, Tax FE, Yoshida S, Feldmann KA. Lesions in the sterol Δ7 reductase gene of Arabidopsis cause dwarfism due to a block in brassinosteroid biosynthesis. Plant J. 2000;21:431–443. doi: 10.1046/j.1365-313x.2000.00693.x. [DOI] [PubMed] [Google Scholar]
- Choi YH, Fujioka S, Harada A, Yokota T, Takatsuto S, Sakurai A. A brassinolide biosynthetic pathway via 6-deoxocastasterone. Phytochemistry. 1996;43:593–596. [Google Scholar]
- Choi YH, Fujioka S, Nomura T, Harada A, Yokota T, Takatsuto S, Sakurai A. An alternative brassinolide biosynthetic pathway via late C-6 oxidation. Phytochemistry. 1997;44:609–613. [Google Scholar]
- Clouse SD, Feldmann KA. Molecular genetics of brassinosteroid action. In: Sakurai A, Yokota T, Clouse SD, editors. Brassinosteroids: Steroidal Plant Hormones. Tokyo: Springer-Verlag; 1999. pp. 163–190. [Google Scholar]
- Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephritikhine G, Pagant S, Fujioka S, Takatsuto S, Lapous D, Caboche M, Kendrick RE, Barbier-Brygoo H. The sax1 mutation defines a new locus involved in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant J. 1999;18:315–320. doi: 10.1046/j.1365-313x.1999.00455.x. [DOI] [PubMed] [Google Scholar]
- Fujioka S. Natural occurrence of brassinosteroids in the plant kingdom. In: Sakurai A, Yokota T, Clouse SD, editors. Brassinosteroids: Steroidal Plant Hormones. Tokyo: Springer-Verlag; 1999. pp. 21–45. [Google Scholar]
- Fujioka S, Choi YH, Takatsuto S, Yokota T, Li J, Chory J, Sakurai A. Identification of castasterone, 6-deoxocastasterone, typhasterol and 6-deoxotyphasterol from the shoots of Arabidopsis thaliana. Plant Cell Physiol. 1996;37:1201–1203. doi: 10.1093/oxfordjournals.pcp.a029074. [DOI] [PubMed] [Google Scholar]
- Fujioka S, Inoue T, Takatsuto S, Yanagisawa T, Yokota T, Sakurai A. Identification of a new brassinosteroid, cathasterone, in cultured cells of Catharanthus roseus as a biosynthetic precursor of teasterone. Biosci Biotechnol Biochem. 1995;59:1543–1547. [Google Scholar]
- Fujioka S, Li J, Choi YH, Seto H, Takatsuto S, Noguchi T, Watanabe T, Kuriyama H, Yokota T, Chory J, Sakurai A. The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell. 1997;9:1951–1962. doi: 10.1105/tpc.9.11.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Noguchi T, Watanabe T, Takatsuto S, Yoshida S. Biosynthesis of brassinosteroids in cultured cells of Catharanthus roseus. Phytochemistry. 2000;53:549–553. doi: 10.1016/s0031-9422(99)00582-8. [DOI] [PubMed] [Google Scholar]
- Fujioka S, Noguchi T, Yokota T, Takatsuto S, Yoshida S. Brassinosteroids in Arabidopsis thaliana. Phytochemistry. 1998;48:595–599. doi: 10.1016/s0031-9422(98)00065-x. [DOI] [PubMed] [Google Scholar]
- Fujioka S, Sakurai A. Brassinosteroids. Nat Prod Rep. 1997a;14:1–10. doi: 10.1039/np9971400001. [DOI] [PubMed] [Google Scholar]
- Fujioka S, Sakurai A. Biosynthesis and metabolism of brassinosteroids. Physiol Plant. 1997b;100:710–715. [Google Scholar]
- Klahre U, Noguchi T, Fujioka S, Takatsuto S, Yokota T, Nomura T, Yoshida S, Chua NH. The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell. 1998;10:1677–1690. doi: 10.1105/tpc.10.10.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koka CV, Cerny RE, Gardner RG, Noguchi T, Fujioka S, Takatsuto S, Yoshida S, Clouse SD. A putative role for the tomato genes DUMPY and CURL-3 in brassinosteroid biosynthesis and response. Plant Physiol. 2000;122:85–98. doi: 10.1104/pp.122.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Biswas MG, Chao A, Russell DW, Chory J. Conservation of function between mammalian and plant steroid 5α-reductases. Proc Natl Acad Sci USA. 1997;94:3554–3559. doi: 10.1073/pnas.94.8.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J. Arole for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- Mathur J, Molnar G, Fujioka S, Takatsuto S, Sakurai A, Yokota T, Adam G, Voigt B, Nagy F, Maas C, Schell J, Koncz C, Szekeres M. Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J. 1998;14:593–602. doi: 10.1046/j.1365-313x.1998.00158.x. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 1999b;121:743–752. doi: 10.1104/pp.121.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Takatsuto S, Sakurai A, Yoshida S, Li J, Chory J. Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-en-3-one to (24R)- 24-methyl-5α-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol. 1999a;120:833–839. doi: 10.1104/pp.120.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Kitasaka Y, Takatsuto S, Reid JB, Fukami M, Yokota T. Brassinosteroid/sterol synthesis and plant growth as affected by lka and lkb mutations of pea. Plant Physiol. 1999;119:1517–1526. doi: 10.1104/pp.119.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Nakayama M, Reid JB, Takeuchi Y, Yokota T. Blockage of brassinosteroid biosynthesis and sensitivity causes dwarfism in garden pea. Plant Physiol. 1997;113:31–37. doi: 10.1104/pp.113.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai A, Fujioka S. Studies on biosynthesis of brassinosteroids. Biosci Biotechnol Biochem. 1997;61:757–762. doi: 10.1271/bbb.61.757. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Suzuki H, Fujioka S, Takatsuto S, Yokota T, Murofushi N, Sakurai A. Biosynthesis of brassinosteroids in seedlings of Catharanthus roseus, Nicotiana tabacum, and Oryza sativa. Biosci Biotechnol Biochem. 1995a;59:168–172. [Google Scholar]
- Suzuki H, Inoue T, Fujioka S, Saito T, Takatsuto S, Yokota T, Murofushi N, Yanagisawa T, Sakurai A. Conversion of 24-methylcholesterol to 6-oxo-24-methylcholestanol, a putative intermediate of the biosynthesis of brassinosteroids, in cultured cells of Catharanthus roseus. Phytochemistry. 1995b;40:1391–1397. [Google Scholar]
- Suzuki H, Inoue T, Fujioka S, Takatsuto S, Yanagisawa T, Yokota T, Murofushi N, Sakurai A. Possible involvement of 3-dehydroteasterone in the conversion of teasterone to typhasterol in cultured cells of Catharanthus roseus. Biosci Biotechnol Biochem. 1994;58:1186–1188. [Google Scholar]
- Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Gasch A, Nishizawa N, Chua NH. The DIMINUTO gene of Arabidopsis is involved in regulating cell elongation. Genes Dev. 1995;9:97–107. doi: 10.1101/gad.9.1.97. [DOI] [PubMed] [Google Scholar]
- Takatsuto S, Ikekawa N. Synthesis of deuterio-labeled brassinosteroids, [26, 28-2H6]brassinolide, [26, 28-2H6]castasterone, [26, 28-2H6]typhasterol and [26, 28-2H6]teasterone. Chem Pharm Bull. 1986;34:4045–4049. [Google Scholar]
- Yokota T. The structure, biosynthesis and function of brassinosteroids. Trends Plant Sci. 1997;2:137–143. [Google Scholar]