Why was the cohort set up?
Colorectal cancer has long been one of the most frequently diagnosed cancers in the world, with an estimated 1.4 million new cases diagnosed each year (9.8% of worldwide cancer diagnoses) and the cause of 694 000 deaths (8.5% of all worldwide cancer deaths) in 2012.1 In 1996, as a commitment to reduce morbidity and mortality from this disease, the National Cancer Institute (NCI) of the U.S. National Institutes of Health invited investigators to apply for funding to establish a ‘Cooperative Family Registry for Colorectal Cancer Studies’ (RFA: CA-96-011). The main NIH stated aims were: to collect pedigree information, epidemiological data and related biological specimens from participants with and without colorectal cancer and with and without a family history of the disease, as a resource for interdisciplinary studies on the aetiology of colorectal cancer; and to identify a population at high risk of colorectal cancer that could benefit from preventive strategies. This cohort profile provides an update of the Colon Cancer Family Registry, described in detail in Newcomb et al.2
The basic premise of this initiative is that family-based designs across the spectrum of risk, in which cases, controls and their relatives are all recruited into a single research infrastructure, would enable the study of genetic aetiology, gene penetrance, gene-gene interaction and interaction with lifestyle factors. Thus, in 1997, the Colon Cancer Family Registry was established with funding support from the NCI. For Phase I (1998–2002), 5 years of funding was awarded to six Colon Cancer Family Registry sites:
Cancer Care Ontario (Toronto, ON, Canada);
Fred Hutchinson Cancer Research Center (Seattle, WA, USA);
Mayo Clinic (Rochester, MN, USA);
University of Hawaii (Honolulu, Hawaii, USA);
University of Southern California Consortium (comprising the Universities of Southern California, Minnesota, North Carolina, Colorado and Arizona, Dartmouth University and the Cleveland Clinic Foundation, USA);
University of Queensland (Brisbane, QLD, Australia).
The Colon Cancer Family Registry received funding renewals for Phase II (2003–07) and Phase III (2008–12) with the addition of:
University of Melbourne (Melbourne, VIC, Australia) substituting for the University of Queensland;
Memorial University (Newfoundland, Canada) as a collaborative site within the Cancer Care Ontario site.
In 2004–11, the ethnic/racial minority component of the Colon Cancer Family Registry was expanded through the recruitment of additional African American and Japanese American families with a separate NCI grant that included the University of Hawaii, the University of Southern California, the University of North Carolina, the Fred Hutchinson Cancer Research Center and the Cancer Prevention Institute of California.
Phase IV (2013–18) of the Colon Cancer Family Registry was funded by the NCI as a Cancer Epidemiology Cohort, and consequently renamed the Colon Cancer Family Registry Cohort (CCFRC). This phase saw the addition of:
Stanford University (CA, USA) as the administering site for the Colon Cancer Family Registry, and
Mayo Clinic (Scottsdale, AZ, USA) as the administering site for the Mayo Clinic CCFRC site.
Who is in the cohort?
Recruitment sampling schemes and inclusion and exclusion criteria varied by CCFRC site and funding phase. Details of the recruitment methods at each CCFRC site have been published previously.2 Briefly, recruitment protocols fall broadly into two main categories: population-based and clinic-based. Population-based probands were either people with a diagnosis of recently diagnosed colorectal cancer (case-probands) identified from cancer registries, or people without a prior diagnosis of colorectal cancer (control-probands) randomly sampled from the general population living in the relevant recruitment area using Medicare and Driver’s License files, telephone subscribers lists or electoral rolls, who were frequency-matched for age to the case-probands. Clinic-based probands were people with or without colorectal cancer who were attendees at a family cancer clinic or genetics clinic. Cases with known familial adenomatous polyposis were excluded. Once recruited, probands were asked for permission to contact their relatives for recruitment. The CCFRC recruited 42 489 participants—from 15 049 families—who completed a baseline questionnaire between 1998 and 2012 (Table 1). Recruitment numbers within clinic-based families was, on average, twice that for population-based families (5.3 vs. 2.6 relatives per family, respectively). The majority of participants self-reported as Caucasian/White followed by Asian ethnicities and African American/Black (Table 2).
Table 1.
Number of families and participants of the Colon Cancer Family Registry Cohort by sex and colorectal cancer (CRC) status at baseline recruitment
| Males | Females | Total | |
|---|---|---|---|
| N (%) | N (%) | N (%) | |
| Population-based familiesa | 13,190 | ||
| Probands with CRC (case-probands) | 4321 (29.2) | 4419 (24.6) | 8740 (26.7) |
| Relatives with CRC | 332 (22) | 372 (21) | 704 (21) |
| Relatives without CRC | 7769 (52.5) | 10516 (58.5) | 18285 (55.8) |
| Probands without CRC (control-probands) | 2071 (14.0) | 2205 (12.3) | 4276 (13.0) |
| Relatives of control-probandsb | 310 (21) | 467 (26) | 777 (24) |
| Total population-based individuals | 14803 | 17979 | 32782 |
| Clinic-based familiesc | 1859 | ||
| Probands and relatives with CRC | 1139 (26.1) | 1108 (20.7) | 2247 (23.1) |
| Probands and relatives without CRC | 3221 (73.9) | 4239 (79.3) | 7460 (76.9) |
| Total clinic-based individuals | 4360 | 5347 | 9707 |
| Total families | 15049 | ||
| Total participants | 19163 | 23326 | 42489 |
Probands recruited from a population-based source.
Only the University of Melbourne recruited relatives of control-probands.
Probands recruited from a family cancer clinic source.
Table 2.
Distribution of participants by race
| Racea | Proportion of participants (%) |
|---|---|
| Native American | 0.9 |
| Asian | 5.5 |
| Pacific Islander | 0.3 |
| African American/Black | 4.8 |
| Caucasian/White | 86.0 |
| More than one race | 1.0 |
| Not reported | 1.5 |
Self-reported by questionnaire.
How often have they been followed up?
We have used both active and passive follow-up methods to update the cohort, where active follow-up includes direct contact with participants and passive follow-up includes indirect methods; details as follows.
Active follow-up
Approximately every 4–5 years after completing their baseline questionnaire, all participants of population-based case-families (but not control-families) and clinic-based families were asked, either by telephone interview or by self-completed questionnaire (mailed or online), for updates on their personal and family history of cancer as well as history of surgery, cancer screening and some risk factors.
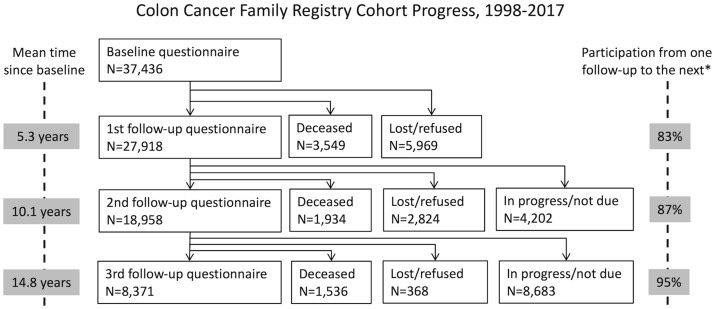
Of the 37 436 participants who completed baseline questionnaires and were approached for follow-up, 27 918 completed the first follow-up questionnaire [response proportion (or response ‘rate’ of those alive) 83%], 3549 died before being approached for the first follow-up and 5969 could not be contacted or refused follow-up. Of the 27 918 participants who had completed the first follow-up, 18 958 completed their second follow-up questionnaire (response rate 87%), 1934 had died, 2824 were either uncontactable or refused, and 4202 are still in process. Of the 18 958 participants who had completed the second follow-up, 8371 had completed their third follow-up questionnaire (response rate 95%), 1536 had died, 368 were either uncontactable or refused and 8683 are still in process (Figure 1).
Figure 1.
Progress of follow-up of participants of the Colon Cancer Family Registry Cohort (as of June 2017). Participation is defined as the percentage of those who were alive at contact attempt who completed the questionnaire.
The total number of person-years of follow-up by participants who completed a follow-up questionnaire is 276 762 person-years. As this is a family study, the vital status and cancer diagnoses of participants were also ascertained, even if they did not participate in the follow-up themselves, based on interviews of any relatives who were also participants. Including the reports by relatives, the total number of person-years of follow-up of all participants who completed a baseline questionnaire was 338 970 person-years, an average of 9.1 years per participant. These comprise approximately: 49 000 person-years for those recruited within 2 years after colorectal cancer diagnosis (thus relevant for studies of colorectal cancer survival and risk of metachronous cancer); 39 000 person-years for relatives with CRC and probands recruited more than 2 years after colorectal cancer diagnosis (thus relevant for studies of survivors of colorectal cancer); and 251 000 person-years for those with no previous diagnosis of colorectal cancer (thus relevant for studies of colorectal cancer risk and aetiology)—Table 3.
Table 3.
Numbers of incident colorectal cancer diagnosis and deaths occurring in study participants (except controls) of the Colon Cancer Family Registry since baseline recruitment by different cohort types, as of June 2017
| Number of participants | Number of incident colorectal cancer at any age (%) | Number of incident colorectal cancer under age 50 years (%) | Number of deaths (%) | Average follow-up (years)d | |
|---|---|---|---|---|---|
| Colorectal cancer within 2 years preceding recruitmenta | 6765 | 144 (2.1) | 31 (0.5) | 2694 (39.8) | 7.5 |
| Colorectal cancer >2 years preceding recruitmentb | 4623 | 202 (4.4) | 25 (0.5) | 1411 (30.5) | 8.8 |
| No history of colorectal cancer prior to recruitmentc | 26048 | 478 (1.8) | 114 (0.4) | 2914 (11.2) | 10.0 |
| Total | 37436 | 824 (2.2) | 170 (0.5) | 7019 (18.7) | 9.4 |
Cohort useful for studies of colorectal cancer survival and risk of metachronous cancer.
Cohort useful for studies of survivors of colorectal cancer.
Cohort useful for studies of colorectal cancer risk and aetiology.
Based on follow-up interview or report from participating relative.
Passive follow-up
One or more of the following passive follow-up activities have been conducted at each site of the CCFRC: data linkage with local and national death files, population-based cancer registries and electoral rolls; annual newsletters; reviews by genetic counsellors; and other mailings to participants. Passive follow-up was regularly conducted on all participants—at intervals that varied by site, type of follow-up activity and cost—to obtain information on new cancers, vital status and cause of death, and to update contact information.
Incident cancers and deaths during follow-up
During active and passive follow-up, all new reports of colorectal polyps and all cancers were recorded. Attempts were made to verify cancers using medical records, cancer registry data and confirmatory reports from relatives. To date, 824 (2.2%) participants have been diagnosed with an incident colorectal cancer since baseline; of those, 170 were diagnosed before the age of 50 years (Table 3); and 3582 (9.5%) participants have been diagnosed with an incident non-colorectal cancer since baseline. The total 4164 incident non-colorectal cancers were as follows: 772 skin, 568 breast, 599 prostate, 97 gastric, 52 small bowel, 103 hepatobiliary, 102 pancreas, 147 renal, 40 ureteric, 150 urinary bladder, 76 brain, 355 lung, 27 bone, 219 blood, 163 endometrial, 73 ovarian and 35 cervical cancers, and 586 in other organs. A total of 7019 (19%) participants (including those with and without colorectal cancer at baseline) are known to have died since baseline (Figure 1).
What has been measured?
At the baseline recruitment, CCFRC participants were asked to complete a detailed family history of cancer, a risk factor questionnaire, permission to access medical records pertaining to any colorectal cancer diagnoses, permission to access colorectal cancer tumours and, depending on the degree of relationship to the proband, to provide a blood (or buccal wash) sample—Table 4.
Table 4.
Resources available from the Colon Cancer Family Registry Cohort, as of June 2017
| Males | Females | Total | |||
|---|---|---|---|---|---|
| N (%)f | N (%)f | N (%)f | |||
| Population-based case familiesa | Probands | Baseline questionnaire | 4321 (22.5) | 4419 (18.9) | 8740 (20.6) |
| Food frequency questionnaire | 2096 (22.7) | 2409 (20.5) | 4505 (21.5) | ||
| Blood/buccal samples | 3759 (27.3) | 3886 (22.8) | 7645 (24.8) | ||
| Polyp material | 14 (7.9) | 8 (4.0) | 22 (5.8) | ||
| Cancer material | 3561 (73.6) | 3447 (70.2) | 7008 (71.9) | ||
| Diagnosis and treatment | 1563 (84.2) | 1526 (85.7) | 3089 (85.0) | ||
| Relativesb | Baseline questionnaire | 7740 (40.4) | 10328 (44.3) | 18068 (42.5) | |
| Food frequency questionnaire | 3323 (36.0) | 4700 (40.0) | 8023 (38.3) | ||
| Blood/buccal samples | 4751 (34.4) | 6731 (39.5) | 11482 (37.2) | ||
| Polyp material | 7 (3.9) | 8 (4.0) | 15 (3.9) | ||
| Cancer material | 265 (5.5) | 325 (6.6) | 590 (6.1) | ||
| Diagnosis and treatment | 40 (2.2) | 44 (2.5) | 84 (2.3) | ||
| Spouse controlsc | Baseline questionnaire | 361 (1.9) | 560 (2.4) | 921 (2.2) | |
| Food frequency questionnaire | 135 (1.5) | 197 (1.7) | 332 (1.6) | ||
| Blood/buccal samples | 149 (1.1) | 225 (1.3) | 374 (1.2) | ||
| Population-based control familiesd | Probands | Baseline questionnaire | 2071 (10.8) | 2205 (9.5) | 4276 (10.1) |
| Food frequency questionnaire | 1142 (12.4) | 1023 (8.7) | 2165 (10.3) | ||
| Blood/buccal samples | 1399 (10.1) | 1497 (8.8) | 2896 (9.4) | ||
| Relativesb | Baseline epi data | 310 (1.6) | 467 (2.0) | 777 (1.8) | |
| Food frequency questionnaire | 260 (2.8) | 383 (3.3) | 643 (3.1) | ||
| Blood/buccal samples | 6 (0.0) | 5 (0.0) | 11 (0.0) | ||
| Clinic-based Colon Cancer familiese | Probands with CRCa | Baseline questionnaire | 699 (3.6) | 644 (2.8) | 1343 (3.2) |
| Food frequency questionnaire | 247 (2.7) | 270 (2.3) | 517 (2.5) | ||
| Blood/buccal samples | 645 (4.7) | 625 (3.7) | 1270 (4.1) | ||
| Polyp material | 24 (13.5) | 29 (14.4) | 53 (13.9) | ||
| Cancer material | 561 (11.6) | 526 (10.7) | 1087 (11.2) | ||
| Diagnosis and treatment | 239 (12.9) | 204 (11.5) | 443 (12.2) | ||
| Probands no CRCc | Baseline questionnaire | 137 (0.7) | 304 (1.3) | 441 (1.0) | |
| Food frequency questionnaire | 62 (0.7) | 164 (1.4) | 226 (1.1) | ||
| Blood/buccal samples | 101 (0.7) | 250 (1.5) | 351 (1.1) | ||
| Polyp material | 13 (7.3) | 27 (13.4) | 40 (10.5) | ||
| Cancer material | 20 (0.4) | 67 (1.4) | 87 (0.9) | ||
| Diagnosis and treatment | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Relativesb | Baseline questionnaire | 3524 (18.4) | 4399 (18.9) | 7923 (18.6) | |
| Food frequency questionnaire | 1953 (21.2) | 2596 (22.1) | 4549 (21.7) | ||
| Blood/buccal samples | 2983 (21.6) | 3814 (22.4) | 6797 (22.0) | ||
| Polyp material | 120 (67.4) | 130 (64.4) | 250 (65.8) | ||
| Cancer material | 429 (8.9) | 542 (11.0) | 971 (10.0) | ||
| Diagnosis and treatment | 14 (0.8) | 6 (0.3) | 20 (0.6) | ||
| Total | All population- and clinic-based probands and relatives | Baseline questionnaire | 19163 | 23326 | 42489 |
| Food frequency questionnaire | 9218 | 11742 | 20960 | ||
| Blood/buccal samples | 13793 | 17033 | 30826 | ||
| Polyp material | 178 | 202 | 380 | ||
| Cancer material | 4836 | 4907 | 9743 | ||
| Diagnosis and treatment | 1856 | 1780 | 3636 |
Proband had a history of colorectal cancer (CRC) at baseline interview.
Affected or unaffected with colorectal cancer at baseline interview.
Spouse of proband, had no history of colorectal cancer at baseline interview.
Proband had no history of colorectal cancer at baseline interview.
Proband was recruited from a family cancer clinic.
% of total items obtained. For example 20.6% of all baseline questionnaires completed by probands of population-based case families.
Baseline risk factor questionnaires
All participants (probands and their participating relatives) were asked to complete the same detailed baseline risk factor survey using standardized questionnaires via personal or telephone interviews or mailed questionnaires. Items included demography, lifestyle factors, screening, medication and family history.2 Four CCFRC sites also asked participants to complete a self-administered food frequency dietary questionnaire. Three CCFRC sites (University of Hawaii, Cancer Care Ontario and University of Southern California consortium) used the questionnaire developed by the Multiethnic Cohort study in Hawaii and California.3 The University of Melbourne used the questionnaire developed by the Melbourne Collaborative Cohort Study.4
Follow-up risk factor questionnaires
At each follow-up, participants were asked for the following events that might have occurred since the previous contact: cancer diagnoses; bowel and gynaecological surgery; screening for colorectal cancer; polyps; and cancer diagnoses and deaths in relatives. Some CCFRC sites opted to include additional questions pertaining to colorectal cancer risk factors. All baseline and follow-up questionnaires used by each CCFRC site can be accessed at: [http://www.coloncfr.org/questionnaires].
Family history
One or more participants from each family was asked to provide their family history of cancer by answering a standard set of questions for each of their relatives (irrespective of cancer history) including: sex and date of birth; cancer sites (except non-melanoma skin cancer), and ages or dates at diagnoses (for those with a cancer history); vital status and, if deceased, date of death. All CCFRC sites recorded detailed family history information for each first- and second-degree relative, and some sites expanded to third-degree relatives, depending on site-specific protocols (detail in Newcomb et al.2). Attempts were made to verify the anatomical site, extent of disease, age at diagnosis and pathology of tumours. Sources of verification used included pathology reports, medical and surgical records, cancer registry information and death certificates.
Blood/mouthwash samples
Participants were asked to provide a blood or mouthwash sample. Of those who agreed, 93% provided a blood sample (Table 4).5 DNA was extracted from blood and mouthwash samples under CCFRC quality-control protocols, to maximize target DNA concentration and fragment size. To provide an unlimited supply of DNA and RNA for probands and selected relatives, lymphoblastoid cell lines of case-probands were immortalized using Epstein-Barr virus.6
Tumours and pathology
Paraffin-embedded colorectal cancer tumours, as well as diagnostic pathology reports, were obtained from treating facilities with the consent of the participant or the next-of-kin if the participant was deceased. In addition, some sites also obtained polyps and non-colorectal tumours, especially cancers commonly identified as part of Lynch syndrome. Multiple sections were cut from each tumour and normal-tissue block, stained with haematoxylin and eosin (H&E) and reviewed by pathologists. For each colorectal cancer, a pathology review was completed (either by examination of the H&E slides or extraction of relevant data from available pathology reports) to obtain the following standardized set of tumour features: grade, histological type, stage (depth of infiltration in large bowel wall and spread to regional lymph nodes), lymphovascular invasion and perineural invasion. Sections were stored for future research at each CCFRC site. Two sites (Ontario and University of Southern California consortium) have made tumour microarrays (TMAs) from colorectal cancers (n 1278).
Virtual tissue repository
CCFRC has created a digitized library of pathology slides (electronic representations of traditional glass slides). A total of 4510 H&E stained slides of histological sections of colorectal tumours from the probands were scanned using either the NanoZoomer Digital Pathology scanner (Hamamatsu Corp.) or the Aperio ScanScope digital slide scanner. Each image is stored as a series of 752 x 480 pixel jpeg image tiles that are reconstructed with relevant software. Typical size of these images is between 200mb and 1.5gb per slide. All images were archived on five image servers: one for short-term storage and four for long-term storage.
Clinical records
Clinical treatment and outcome records were requested from 3830 case-probands and for 111 relatives with an incident colorectal cancer diagnosed since baseline, and have been abstracted into standardized items for analysis.
Molecular characterization of tumours
Probands’ colorectal cancers were characterized for DNA mismatch repair (MMR) deficiency by polymerase chain reaction (PCR)-based microsatellite instability (MSI) tests and/or by immunohistochemistry (IHC) for the four DNA MMR proteins.7 Colorectal cancer tumour DNA was tested for the BRAF V600E somatic point mutation and somatic mutations in codons 12 and 13 of KRAS.8,9 Tumours were also tested for methylation of the MLH1 gene promoter (an epigenetic phenotype used to indicate that MMR deficiency is more likely to have been caused by a somatic epigenetic event in MLH1 than by a germline mutation in MLH1).10 Characterization of the CpG Island Methylator Phenotype (CIMP) was also performed by assessing quantitative methylation across five gene promoters (CACNA1G, IGF2, NEUROG1, RUNX3 and SOCS1)11—Table 5.
Table 5.
Molecular characterizations of participants (probands and relatives) at the Colon Cancer Family Registry Cohort, as of June 2017
| Molecular test | Number of participants tested | Number of colorectal cancer tumours tested | Test results |
|---|---|---|---|
| Tumour | |||
| DNA MSI | 5147 | 5305 |
|
| IHC for MMR proteins | 8036 | 8338 |
|
| CIMPa | 3855 | 3888 |
|
| MLH1 methylation | 3041 | 3412 |
|
| BRAF V600E mutation | 7080 | 7322 |
|
| KRAS mutation | 4014 | 4154 |
|
| Blood | |||
| MMR geneb |
|
|
|
| MUTYHb |
|
|
|
CIMP, CpG island methylator phenotype.
Tumours were classified as CIMP-positive if more than three of five genes gave percentage of methylated reference value ≥ 10.
Sequencing (probands) or predictive testing (relatives).
Molecular characterization of germline DNA
Screening for germline mutations in MLH1, MSH2, MSH6, PMS2 and EPCAM was performed for all population-based probands who had a colorectal tumour displaying an MSI-High phenotype or a loss of expression of one or more of the MMR proteins expression by IHC, and for the youngest-onset colorectal cancer case participant from each clinic-based family, regardless of MSI or MMR-protein expression status. All case-probands were genotyped for 12 previously identified MUTYH variants: c.536A>G p.(Tyr179Cys), c.1187G>A p.(Gly396Asp), c.312C>A p.(Tyr104Ter), c.821G>A p.(Arg274Gln), c.1438G>T p.(Glu480Ter), c.1171C>T p.(Gln391Ter), c.1147delC p.(Ala385ProfsTer23), c.933 + 3A>C p.(Gly264TrpfsX7), c.1437_1439delGGA p.(Glu480del), c.721C>T, p.(Arg241Trp), c.1227_1228dup p.(Glu410GlyfsX43) and c.1187-2A>G p.(Leu397CysfsX89) (detail in Cleary et al.12). Where available, blood samples from the relatives of probands with a pathogenic mutation were tested for the specific mutation (MMR gene or MUTYH) identified in the proband (predictive testing). Of the CCFRC participants: 2118 were identified as carrying a mutation in one of the MMR genes (761 in MLH1, 976 in MSH2, 243 in MSH6, 109 in PMS2 and 29 in EPCAM); and 451 were identified as carrying either a monoallelic (n = 392) or biallelic mutation (n = 59) in MUTYH. Since baseline, these mutation carriers have contributed a total of 18 514 MMR-mutation person-years and 3732 MUTYH-mutation person-years. In addition, targeted sequencing was conducted of 36 known or putative colorectal cancer susceptibility genes (including the MMR genes) for 1231 cases including cases with familial colorectal cancer type X;13 early-onset (age < 50 years at colorectal cancer diagnosis), or suspected Lynch syndrome.
CCFRC has genome-wide single nucleotide polymorphism (SNP) genotyping data for 10 716 participants (6732 cases and 2435 controls) by various platforms (including OncoArray), all imputed to the 1000 Genomes Project.14 We excluded known carriers of germline mutations MMR genes and MUTYH.
What has it found? Key findings and publications
The CCFRC resource has been used for more than 400 original peer-reviewed publications—see [http://coloncfr.org/publications]. Here, we highlight a few findings that illustrate the power of this cohort to understand genetic and environmental risk factors for colorectal cancer.
Lynch syndrome (cancer caused by inherited mutations in DNA MMR genes MLH1, MSH2, MSH6 and PMS2 or EPCAM) has been a major research focus of the CCFRC. These studies have included analyses of the identification of Lynch syndrome15–18 and associated rare variant classification,19–21 age-specific risk of cancer (penetrance),22–25 effect of extent of colon resection on the risk of metachronous cancer,26 prevalence of Lynch syndrome in colorectal cancer cases27 and in the general population,28 pathology of Lynch syndrome tumours,29 acceptability and impact of genetic testing,30–33 and modifiers of penetrance for Lynch syndrome-associated cancers.34–42 A prime example is the prospective cohort analysis of MMR gene mutation carriers to estimate cancer risks, i.e. penetrance of Lynch syndrome.22 A total of 446 MMR gene mutation carriers with an average age of 40 years and who had no cancer diagnosis were followed up for 5–10 years —the largest prospective study ever of Lynch syndrome in which participants completed a risk factor questionnaire. The 5- and 10-year risks of several cancer types were calculated, with the highest risk observed for colorectal cancer (8% 10-year risk 20 times population risk), endometrial cancer (10% 10-year risk 30 times population risk) and ovarian cancer (3% 10-year risk 19 times population risk). A novel finding of this study was that these mutation carriers also appear to have an increased risk of breast cancer. A total of 1029 of their relatives who were not mutation carriers were also assessed for cancer risk. They were found to be at the same risk as the general population and therefore can be screened as someone at average risk, despite often having a strong family history of cancer.
The CCFRC also makes major contributions to the search for new genetic risk factors, primarily due to its large sample size and because of its family-based design. This allows for the collection of DNA samples from case-probands as well as their relatives, and the verification of reports of cancer by relatives rather than relying on self-report only. This research includes the investigation of SNP associations,43–50 genome panel testing55 and statistical modelling of risk.28,51,52 Many of these studies stemmed from the research within the CCFRC on syndromic classification of familial colorectal cancer, primarily the work leading to the description of ‘familial colorectal cancer type X’, the phenotype of MMR-proficient colorectal cancer cases who fulfilled the Amsterdam Criteria for hereditary non-polyposis colon cancer,13 which has become an accepted category of familial colorectal cancer.53
What are the main strengths and weaknesses?
The unique strength of the CCFRC is its prospective, observational design, with familial enrichment and molecular characterization. Participants have deliberately been over-sampled for familial risk, and therefore the CCFRC differs from the usual cancer research cohort in novel ways that allow inferences not otherwise possible.54 This facilitates a deeper and broader research agenda that covers aetiological factors (both genetic and environmental), molecular characterization, behavioural issues and clinical research relevant to people at increased familial risk.
Participants can be categorized on their underlying familial risk profile based on their genotype, family history and risk factor data, which allows the effects of environmental risk factors to be investigated for varying levels of putative genetic risk: i.e. for studies of gene-environment interactions. The availability of genotype data allows prospective studies of the risk-modifying effects of genetic and non-genetic factors, and the effectiveness of targeted screening/surveillance by genetic sub-groups. The CCFRC can be, and has been, used for a range of gene discovery research including classic linkage studies, genome-wide association studies and whole-exome and whole-genome studies.55 Furthermore, because a large proportion of CCFRC participants were diagnosed with colorectal cancer just before recruitment and have risk factor data as well as blood samples, powerful studies of prognostic factors can be undertaken. The CCFRC also facilitates novel behavioural, psychosocial and health utilisation research for clinical translation.
From a practical perspective, conducting family studies can be challenging because of the often complex nature of familial relationships, as well as the additional layers of protocol that need to be incorporated to protect privacy within families (for example, procedures to ensure that sensitive information is not inadvertently passed to other family members). We have demonstrated that these issues, however, can be managed through carefully designed study protocols and training. We strongly believe that the benefits of a family cohort far outweigh its limitations, and that more epidemiologists should consider this design when conducting aetiological research focused on environmental risk factors across the risk spectrum.
Can I use the data? Where can I find out more?
From its inception, the CCFRC has functioned under the principle that it is a resource for research on the aetiology, risk and prognosis of colorectal cancer for all researchers, including those not affiliated with CCFRC. To this end, CCFRC welcomes collaborative applications to access and analyse both electronic data (questionnaire, genotypes, medical records, family history etc.) and biospecimens (DNA, blood, serum, and tumour specimens). Of the total 294 approved applications to use CCFRC resources, 157 (53%) have come from external investigators.
The CCFRC provides internal and external researchers fair and equitable access to this unique resource. Collaborating investigators have established numerous funded projects. For information on how to collaborate and access data for the CCFRC, including cohort data described here, please see [http://coloncfr.org/].
Profile in a nutshell
The Colon Cancer Family Registry Cohort (CCFRC) was established for the purpose of research on the genetic and environmental aetiology of colorectal cancer.
The 42 489 study participants from 15 049 families were recruited between 1998 and 2012 in the USA, Canada, Australia and New Zealand. They include: recently diagnosed colorectal cancer cases from population-based cancer registries; controls from population-based sources; patients from family cancer clinics with a strong family history of colorectal cancer; and their relatives.
Every 4–5 years after baseline, all population-based case-families and clinic-based families were followed up and re-surveyed. The total follow-up of 37 436 participants covers approximately 339 000 person-years (mean follow-up 9.1 years). Since baseline, 824 (2.2%) participants were diagnosed with a colorectal cancer and 3582 (9.5%) were diagnosed with a non-colorectal cancer.
At baseline, all participants completed the same risk factor questionnaire for a detailed personal and family history of cancer, and a wide range of risk factors. At each follow-up, participants were asked for updates on their personal and family history of cancer, screening, surgery, death and some risk factors. Blood samples and tumour specimens have been collected and used for extensive genetic and molecular characterization including Lynch syndrome.
CCFRC resources are available for collaborative research [http://www.coloncfr.org/].
Acknowledgements
The authors thank all study participants of the CCFRC and staff for their contributions to this project. The authors also acknowledge the major contributions to the CCFRC since its inception in 1997 made by the following investigators: Dennis Ahnen, Kristen Anton, Julie Arnold, Melyssa Aronson, Kelly Aujard, Bharati Bapat, John Baron, Melissa Barker, Adrian Bickerstaffe, Terrilea Burnett, Iona Cheng, James Church, Timothy Church, Mark Clendenning, Darshana Daftary, Melissa DeRycke, Elizabeth Dicks, Anh Diep, Dave Duggan, Mary Jane Esplen, Douglass Fisher, Samantha Fox, Amy French, Graham Giles, Karen Glanz, Jack Goldblatt, Richard Goldberg, Ellen Goode, William Grady, Cary Greenberg, Jane Green, Roger Green, John Grove, Robert Gryfe, Patricia Harmon, Eric Holowaty, Spring Holter, John Hopper, Louise Keogh, Hyeja Kim, Judy Kirk, Peter Lance, Mercy Laurino, Barbara Leggett, A. Joan Levine, Paul Limburg, Jan Lowery, Laurie Lydum, Finlay Macrae, Lisa Madlensky, Karen Makar, Rachel Malen, Judi Maskiell, Pamela McAllister, Ellen McGannon, Gail McKeown-Eyssen, John McLaughlin, Heide Miller-Pakvasa, Gabriela Moslein, Nathalie Nguyen, Sandy Nigon, Patrick Parafrey, Susan Parry, Susan Peterson, Amanda Phipps, Aaron Pollett, Mark Redston, Scott Rogers, Robert Sandler, Sheri Schully, Teresa Selander, Daniella Seminara, Stacey Shiovitz, Kim Siegmund, Thomas Smyrk, Douglas Snazel, Melissa Southey, John Stubbs, Graeme Suthers, Duncan Thomas, Kathy Tucker, Dee West, Michael Woods, Ban Younghusband and Joanne Young.
Funding
This work was supported by grant UM1 CA167551 from the National Cancer Institute and through cooperative agreements with the following CCFRC sites: Australasian Colorectal Cancer Family Registry (U01 CA074778 and U01/U24 CA097735); Mayo Clinic Cooperative Family Registry for Colon Cancer Studies (U01/U24 CA074800); Ontario Familial Colorectal Cancer Registry (U01/U24 CA074783); Seattle Colorectal Cancer Family Registry (U01/U24 CA074794); University of Hawaii Colorectal Cancer Family Registry (U01/U24 CA074806); and USC Consortium Colorectal Cancer Family Registry U01/U24 CA074799). The targeted minority recruitment was supported by grant R01 CA104132. The genome-wide association studies (GWAS) were supported by grants U01 CA 122839, R01 CA143237 and U19 CA148107. The CIMP and KRAS mutation testing was supported by R01 CA118699.
Additional support for case ascertainment was provided from the Surveillance, Epidemiology and End Results (SEER) Program of the National Cancer Institute to the Fred Hutchinson Cancer Research Center (Control Nos. N01-CN-67009 and N01-PC-35142, and Contract No. HHSN2612013000121), the Hawai‘i Department of Health (Control Nos. N01-PC-67001 and N01-PC-35137, and Contract No. HHSN26120100037C), and the California Department of Public Health (contracts HHSN261201000035C awarded to the University of Southern California and HHSN261201000140C awarded to the Cancer Prevention Institute of California), the following US state cancer registries: AZ, CO, MN, NC and NH, and by the Victorian Cancer Registry, Australia and the Ontario Cancer Registry, Canada. A.K.W. is an NHMRC Early Career Fellow. M.A.J. is an NHMRC Senior Research Fellow. J.L.H. is a NHMRC Senior Principal Research Fellow. D.D.B. is a University of Melbourne Research at Melbourne Accelerator Program (R@MAP) Senior Research Fellow and NHMRC R.D. Wright Career Development Fellow. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating sites in the CCFRC, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government or the CCFRC. Authors had full responsibility for the design of the study, the collection of the data, the analysis and interpretation of the data, the decision to submit the manuscript for publication and the writing of the manuscript.
Conflict of interest: The authors have no conflict of interest to declare with respect to this manuscript.
Contributor Information
Colon Cancer Family Registry Cohort Investigators:
Dennis Ahnen, Kristen Anton, Julie Arnold, Melyssa Aronson, Kelly Aujard, Bharati Bapat, John Baron, Melissa Barker, Adrian Bickerstaffe, Terrilea Burnett, Iona Cheng, James Church, Timothy Church, Mark Clendenning, Darshana Daftary, Melissa DeRycke, Elizabeth Dicks, Anh Diep, Dave Duggan, Mary Jane Esplen, Douglass Fisher, Samantha Fox, Amy French, Graham Giles, Karen Glanz, Jack Goldblatt, Richard Goldberg, Ellen Goode, William Grady, Cary Greenberg, Jane Green, Roger Green, John Grove, Robert Gryfe, Patricia Harmon, Eric Holowaty, Spring Holter, John Hopper, Louise Keogh, Hyeja Kim, Judy Kirk, Peter Lance, Mercy Laurino, Barbara Leggett, A Joan Levine, Paul Limburg, Jan Lowery, Laurie Lydum, Finlay Macrae, Lisa Madlensky, Karen Makar, Rachel Malen, Judi Maskiell, Pamela McAllister, Ellen McGannon, Gail McKeown-Eyssen, John McLaughlin, Heide Miller-Pakvasa, Gabriela Moslein, Nathalie Nguyen, Sandy Nigon, Patrick Parafrey, Susan Parry, Susan Peterson, Amanda Phipps, Aaron Pollett, Mark Redston, Scott Rogers, Robert Sandler, Sheri Schully, Teresa Selander, Daniella Seminara, Stacey Shiovitz, Kim Siegmund, Thomas Smyrk, Douglas Snazel, Melissa Southey, John Stubbs, Graeme Suthers, Duncan Thomas, Kathy Tucker, Dee West, Michael Woods, Ban Younghusband, and Joanne Young
References
- 1. Ferlay J, Soerjomataram I, Dikshit R. et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359–86. [DOI] [PubMed] [Google Scholar]
- 2. Newcomb PA, Baron J, Cotterchio M. et al. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev 2007;16:2331–43. [DOI] [PubMed] [Google Scholar]
- 3. Kolonel LN, Henderson BE, Hankin JH. et al. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am J Epidemiol 2000;151:346–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ireland P, Jolley D, Giles G. et al. Development of the Melbourne FFQ: a food frequency questionnaire for use in an Australian prospective study involving an ethnically diverse cohort. Asia Pac J Clin Nutr 1994;3:19–31. [PubMed] [Google Scholar]
- 5. Lum A, Le Marchand L.. A simple mouthwash method for obtaining genomic DNA in molecular epidemiological studies. Cancer Epidemiol Biomarkers Prev 1998;7:719–24. [PubMed] [Google Scholar]
- 6. Miller G. Immortalization of human lymphocytes by Epstein-Barr virus. Yale J Biol Med 1982;55:305–10. [PMC free article] [PubMed] [Google Scholar]
- 7. Lindor NM, Burgart LJ, Leontovich O. et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol 2002;20:1043–48. [DOI] [PubMed] [Google Scholar]
- 8. Buchanan DD, Sweet K, Drini M. et al. Risk factors for colorectal cancer in patients with multiple serrated polyps: a cross-sectional case series from genetics clinics. PLoS One 2010;5:e11636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Buchanan DD, Win AK, Walsh MD. et al. Family history of colorectal cancer in BRAF p.V600E-mutated colorectal cancer cases. Cancer Epidemiol Biomarkers Prev 2013;22:917–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Levine AJ, Win AK, Buchanan DD. et al. Cancer risks for the relatives of colorectal cancer cases with a methylated MLH1 promoter region: data from the Colorectal Cancer Family Registry. Cancer Prev Res (Phila) 2012;5:328–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weisenberger DJ, Levine AJ, Long TI. et al. Association of the colorectal CpG island methylator phenotype with molecular features, risk factors, and family history. Cancer Epidemiol Biomarkers Prev 2015;24:512–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cleary SP, Cotterchio M, Jenkins MA. et al. Germline MutY human homologue mutations and colorectal cancer: a multisite case-control study. Gastroenterology 2009;136:1251–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lindor NM, Rabe K, Petersen GM. et al. Lower cancer incidence in Amsterdam-I Criteria families without mismatch repair deficiency: familial colorectal cancer type X. JAMA 2005;293:1979–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 2012;491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clendenning M, Buchanan DD, Walsh MD. et al. Mutation deep within an intron of MSH2 causes Lynch syndrome. Fam Cancer 2011;10:297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Clendenning M, Walsh MD, Gelpi JB. et al. Detection of large scale 3' deletions in the PMS2 gene amongst Colon-CFR participants: have we been missing anything? Fam Cancer 2013;12:563–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clendenning M, Macrae FA, Walsh MD. et al. Absence of PMS2 mutations in colon-CFR participants whose colorectal cancers demonstrate unexplained loss of MLH1 expression. Clin Genet 2013;83:591–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Toon CW, Walsh MD, Chou A. et al. BRAFV600E Immunohistochemistry facilitates universal screening of colorectal cancers for Lynch Syndrome. Am J Surg Pathol 2013;37:1592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thompson BA, Goldgar DE, Paterson C. et al. A multifactorial likelihood model for MMR gene variant classification incorporating probabilities based on sequence bioinformatics and tumor characteristics: a report from the Colon Cancer Family Registry. Hum Mutat 2013;34:200–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thompson BA, Greenblatt MS, Vallee MP. et al. Calibration of multiple in silico tools for predicting pathogenicity of mismatch repair gene missense substitutions. Hum Mutat 2013;34:255–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thompson BA, Spurdle AB, Plazzer JP. et al. Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat Genet 2013;46:107–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Win AK, Young JP, Lindor NM. et al. Colorectal and other cancer risks for carriers and noncarriers from families with a DNA mismatch repair gene mutation: a prospective cohort study. J Clin Oncol 2012;30:958–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baglietto L, Lindor NM, Dowty JG. et al. Risks of Lynch syndrome cancers for MSH6 mutation carriers. J Natl Cancer Inst 2010;102:193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dowty JG, Win AK, Buchanan DD. et al. Cancer risks for MLH1 and MSH2 mutation carriers. Hum Mutat 2013;34:490–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Senter L, Clendenning M, Sotamaa K. et al. The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology 2008;135:419–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Parry S, Win AK, Parry B. et al. Metachronous colorectal cancer risk for mismatch repair gene mutation carriers: the advantage of more extensive colon surgery. Gut 2011;60:950–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Buchanan DD, Clendenning M, Rosty C. et al. Tumor testing to identify lynch syndrome in two Australian colorectal cancer cohorts. J Gastroenterol Hepatol 2017;32:427–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Win AK, Jenkins MA, Dowty JG. et al. Prevalence and penetrance of major genes and polygenes for colorectal cancer. Cancer Epidemiol Biomarkers Prev 2017;26:404–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jenkins MA, Hayashi S, O'Shea AM. et al. Pathology features in Bethesda guidelines predict colorectal cancer microsatellite instability: a population-based study. Gastroenterology 2007;133:48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim JS, Coyte PC, Cotterchio M. et al. The impact of receiving predictive genetic information about lynch syndrome on individual colonoscopy and smoking behaviors. Cancer Epidemiol Biomarkers Prev 2016;25:1524–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Graves KD, Sinicrope PS, Esplen MJ. et al. Communication of genetic test results to family and health-care providers following disclosure of research results. Genet Med 2014;16:294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Keogh LA, Fisher D, Sheinfeld Gorin S. et al. How do researchers manage genetic results in practice? The experience of the multinational Colon Cancer Family Registry. J Community Genet 2014;5:99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Flander L, Speirs-Bridge A, Rutstein A. et al. Perceived versus predicted risks of colorectal cancer and self-reported colonoscopies by members of mismatch repair gene mutation-carrying families who have declined genetic testing. J Genet Couns 2014;23:79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Win AK, Dowty JG, English DR. et al. Body mass index in early adulthood and colorectal cancer risk for carriers and non-carriers of germline mutations in DNA mismatch repair genes. Br J Cancer 2011;105:162–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Win AK, Dowty JG, Antill YC. et al. Body mass index in early adulthood and endometrial cancer risk for mismatch repair gene mutation carriers. Obstet Gynecol 2011;117:899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chau R, Dashti SG, Ait Ouakrim D. et al. Multivitamin, calcium and folic acid supplements and the risk of colorectal cancer in Lynch syndrome. Int J Epidemiol 2016;45:940–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dashti SG, Buchanan DD, Jayasekara H. et al. Alcohol consumption and the risk of colorectal cancer for mismatch repair gene mutation carriers. Cancer Epidemiol Biomarkers Prev 2017;26:366–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dashti SG, Chau R, Ouakrim DA. et al. Female hormonal factors and the risk of endometrial cancer in lynch syndrome. JAMA 2015;314:61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ait Ouakrim D, Dashti SG, Chau R. et al. Aspirin, Ibuprofen, and the risk of colorectal cancer in Lynch Syndrome. J Natl Cancer Inst 2015;107:djv170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pande M, Lynch PM, Hopper JL. et al. Smoking and colorectal cancer in Lynch Syndrome: Results from the Colon Cancer Family Registry and The University of Texas M.D. Anderson Cancer Center. Clin Cancer Res 2010;16:1331–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Win AK, Hopper JL, Buchanan DD. et al. Are the common genetic variants associated with colorectal cancer risk for DNA mismatch repair gene mutation carriers? Eur J Cancer 2013;49:1578–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Win AK, Clendenning M, Crawford W. et al. Genetic variants within the hTERT gene and the risk of colorectal cancer in Lynch syndrome. Genes Cancer 2015;6:445–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kuroiwa-Trzmielina J, Wang F, Rapkins RW. et al. SNP rs16906252C>T is an expression and methylation quantitative trait locus associated with an increased risk of developing MGMT-methylated colorectal cancer. Clin Cancer Res 2016;22:6266–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Al-Tassan NA, Whiffin N, Hosking FJ. et al. A new GWAS and meta-analysis with 1000Genomes imputation identifies novel risk variants for colorectal cancer. Sci Rep 2015;5:10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Salomon MP, Li WL, Edlund CK. et al. GWASeq:targeted re-sequencing follow up to GWAS. BMC Genomics 2016;17:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Carvajal-Carmona LG, Cazier JB, Jones AM. et al. Fine-mapping of colorectal cancer susceptibility loci at 8q23.3, 16q22.1 and 19q13.11: refinement of association signals and use of in silico analysis to suggest functional variation and unexpected candidate target genes. Hum Mol Genet 2011;20:2879–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cicek MS, Cunningham JM, Fridley BL. et al. Colorectal cancer linkage on chromosomes 4q21, 8q13, 12q24, and 15q22. PLoS One 2012;7:e38175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Poynter JN, Figueiredo JC, Conti DV. et al. Variants on 9p24 and 8q24 are associated with risk of colorectal cancer: results from the Colon Cancer Family Registry. Cancer Res 2007;67:11128–32. [DOI] [PubMed] [Google Scholar]
- 49. Figueiredo JC, Hsu L, Hutter CM. et al. Genome-wide diet-gene interaction analyses for risk of colorectal cancer. PLoS Genet 2014;10:e1004228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Garcia-Albeniz X, Rudolph A, Hutter C. et al. CYP24A1 variant modifies the association between use of oestrogen plus progestogen therapy and colorectal cancer risk. Br J Cancer 2016;114:221–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jenkins MA, Baglietto L, Dite GS. et al. After hMSH2 and hMLH1 - what next? Analysis of three-generational, population-based, early-onset colorectal cancer families. Int J Cancer 2002;102:16671. [DOI] [PubMed] [Google Scholar]
- 52. Lindor NM, Rabe KG, Petersen GM. et al. Parent of origin effects on age at colorectal cancer diagnosis. Int J Cancer 2010;127:361–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shiovitz S, Copeland WK, Passarelli MN. et al. Characterisation of familial colorectal cancer type X, Lynch syndrome, and non-familial colorectal cancer. Br J Cancer 2014;111:598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hopper JL. Disease-specific prospective family study cohorts enriched for familial risk. Epidemiol Perspect Innov 2011;8:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. DeRycke MS, Gunawardena SR, Middha S. et al. Identification of novel variants in colorectal cancer families by high-throughput exome sequencing. Cancer Epidemiol Biomarkers Prev 2013;22:1239–51. [DOI] [PMC free article] [PubMed] [Google Scholar]