Abstract
Background
Esophageal squamous cell carcinoma (ESCC) is often diagnosed at an advanced and incurable stage. Information on driver genes and prognosticators in ESCC remains incomplete. The objective was to elucidate significantly mutated genes (SMGs), mutational signatures, and prognosticators in ESCC.
Patients and methods
Three MutSig algorithms (i.e. MutSigCV, MutSigCL and MutSigFN) and ‘20/20+’ ratio-metric were employed to identify SMGs. Nonnegative matrix factorization was used to decipher mutational signatures. Kaplan–Meier survival analysis, multivariate Cox and logistic regression models were applied to analyze association between mutational features and clinical parameters.
Results
We identified 26 SMGs, including 8 novel (NAV3, TENM3, PTCH1, TGFBR2, RIPK4, PBRM1, USP8 and BAP1) and 18 that have been previously reported. Three mutational signatures were identified to be prevalent in ESCC including clocklike C>T at CpG, APOBEC overactive C>T at TpCp[A/T], and a signature featured by T>C substitution. The T>C mutational signature was significantly correlated with alcohol consumption (OR: 3.59; 95% CI: 2.30–5.67; P < 0.001). This alcohol consumption signature was also observed in liver cancer and head and neck squamous cell carcinoma, and its mutational activity was substantially higher in samples with mutations in TP53. Survival analysis revealed that TENM3 mutations (HR: 5.54; CI: 2.68–11.45; P < 0.001) and TP53 hotspot mutation p.R213* (HR: 3.37; CI: 1.73–8.06; P < 0.001) were significantly associated with shortened survival outcome. The association remained statistically significant after controlling for age, gender, TNM stage and tumor grade.
Conclusions
We have uncovered several new SMGs in ESCC and defined an alcohol consumption related mutational signature. TENM3 mutations and the TP53 hotspot mutation p.R213* are independent prognosticators for poor survival in ESCC.
Keywords: esophagus, driver genes, mutational signature, prognosticator
Key Message
We carried out systematic and comprehensive analyses of 549 ESCC samples. We identified eight previously unreported driver genes and defined a mutational signature associated with alcohol consumption. Mutations in TENM3 and TP53 hotspot mutation p.R213* were significantly associated with shortened survival outcome independent of age, gender and TNM staging.
Introduction
Esophageal cancer ranks the seventh most commonly diagnosed cancer type and the sixth leading cause for cancer-related death worldwide with a 5-year survival rate as low as 13% [1]. The incidence and mortality rates of esophageal cancer are higher in China (4th in ranking) with esophageal squamous cell carcinoma (ESCC) accounting for 90% of esophageal cancer [2]. The well-established risk factors for developing ESCC include alcohol consumption and tobacco smoking [3]. Single-nucleotide polymorphisms in ALDH2 (rs671, AG/AA) and ADH1B (rs1229984, GG) were reported to associate with increase the risk of ESCC [4].
Recent next-generation sequencing studies have advanced our understanding of genetic alterations in ESCC [5–11]. Genes involved in cell cycle, RTK/PI3K/AKT circuit, chromatin remodeling, and the Notch signaling pathway are frequently altered [7]. TP53 is the most significantly mutated genes (SMGs) in ESCC with mutation frequency reaching 93% [7]. The EP300 mutation was reported to be independent prognostic factor for ESCC [7, 10]. TENM3 is a member of teneurin encoding gene family and its genomic variations have been observed in human cancers [12, 13].
The characteristic mutational signatures are the fingerprints of endogenous and exogenous factors that have acted over the course of tumorigenesis. For example, substitution of C>T at TpCpW (where W = A or T) is associated with over-activity of APOBEC RNA-editing enzyme [14]. In ESCC, the APOBEC mutational activity is significantly greater in ZNF750 mutated cancer samples as compared with those without ZNF750 mutations [15]. Prevalent C>T mutations at CpG dinucleotide via spontaneous deamination of 5-methylcytosine is associated with aging; a risk factor for cancer development.
The purposes of this study were to identify new SMGs and genetic prognosticators for patients diagnosed with ESCC, and to characterize the mutational signatures in ESCC by jointly interrogating genomic data and clinical information from published ESCC studies [5–11].
Materials and methods
Genomic data and clinical information
All somatic mutations were initially extracted from seven previous studies comprising 549 ESCC cases. Five of these seven studies have survival data. Detailed information is shown in supplementary Table S1, available at Annals of Oncology online. Clinical information is provided in supplementary Table S2, available at Annals of Oncology online. Detailed descriptions are provided in supplementary material, available at Annals of Oncology online. The genotypes of ALDH2 at rs671 and ADH1B at rs1229984 were derived from samples with bam files available. All previously called mutations were re-annotated after filtering through an in-house reference genomics database composed of a panel of 1600 healthy Chinese individuals. Previously called mutations were discarded if they present in >2 alignment bam files in the reference database.
Identification of SMGs
We used four algorithms, namely MutSigCV [16], MutSigCL [17], MutSigFN [17] and ‘20/20+’ ratio-metric [18] to identify SMGs, and applied stringent filtering criteria to eliminate false positives. We required that mutations of these genes were not only statistically significant by MutSig algorithm but also detected in ≥4 independent studies out of the seven studies included in our meta-analysis. In addition, we required that these genes were shown to be expressed in human cancer cell lines [19] and the TCGA pan-cancer dataset [20]. We also compared the mRNA expression levels of these genes in another ESCC study that only has microarray-based gene expression profiling [21]. Detailed procedures are provided in supplementary material, available at Annals of Oncology online.
Deciphering mutational signature operative in the genome
We applied the framework proposed by Kim et al. [22] to extract mutational signatures. This framework is based on Bayesian variant nonnegative matrix factorization and it can automatically determine the optimal number of mutational signatures. We also used nonnegative least approach to deconvolute the mutational portrait of ESCC against mutational signatures 1, 2, 13 and 16, which resemble signatures extracted from ESCC and was curated by the Catalogue of Somatic Mutations in Cancer (COSMIC). These COSMIC signatures were obtained from http://cancer.sanger.ac.uk/cosmic/signatures (5 September 2017, date last accessed). Detailed procedures are provided in supplementary material, available at Annals of Oncology online.
Prognostic analysis of mutated genes
Kaplan–Meier survival analysis and Cox proportional hazards model were employed to analyze the association between mutated genes and prognosis. Confounding factors that were not significant in the univariate Cox model were not included in the multivariate Cox analysis except for age and gender. Kaplan–Meier survival and Cox regression analyses were carried out with the R survival package (2.40-1). P-value < 0.05 was considered to be statistically significant. The drug treatment information for these ESCC was not available.
Results
SMGs in ESCC
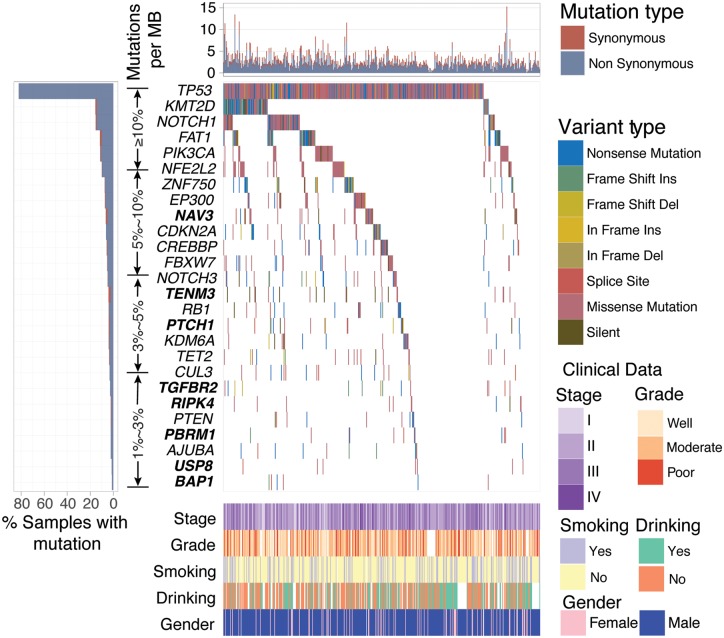
A total of 67 592 coding somatic mutations were obtained from 7 previously published studies totaling 549 ESCC cases (a median of 107 mutations per tumor). We used MutSigCV [16], MutSigCL [17], MutSigFN [17] and ‘20/20+’ ratio-metric [18] to re-annotate SMGs that met the criteria of being positively accumulated, clustered at a hotspot and of functional importance. In total, we identified 26 SMGs (Figure 1), including 18 previously reported ESCC driver genes (e.g. TP53, KMT2D and NOTCH1) and 8 novel SMGs (i.e. NAV3, TENM3, PTCH1, TGFBR2, RIPK4, PBRM1, USP8 and BAP1). According to the ‘20/20+’ ratio-metric, three newly identified SMGs, namely PTCH1, PRM1 and BAP1, were categorized as tumor suppressor genes. The mutation plots of these eight novel SMGs are shown in supplementary Figure S1, available at Annals of Oncology online. The mRNA expression level of these 26 SMGs in tumor tissues versus matched adjacent normal control tissue were examined in a separate microarray-based ESCC gene expression dataset [21]. The analyses showed that 19 SMGs were significantly upregulated or downregulated (supplementary Figure S2, available at Annals of Oncology online; Paired t-test, q < 0.1). NAV3, mutated in 6.9% of ESCC cases, was reported to be recurrently mutated in five cancer types from a previous pan-cancer study [20]; however, NAV3 function in carcinogenesis has not been well established. TENM3 was mutated in 4% of ESCC. Eleven of the 12 non-silent mutations in TENM3 were missense mutations. The ubiquitin specific protease 8 encoding gene USP8, identified as an oncogene in Cushing’s disease [23, 24], was found to harbor hotspot mutations at p.N764K (n = 3) and p.R763W (n = 2) in ESCC. Mutations of PTCH1 did not reach statistical significance in our previous ESCC study, albeit, it was suggested as a key gene implicated in ESCC [15]. RIPK4, encoding for receptor-interacting protein kinase 4, was reported to be involved in head and neck squamous cell carcinoma [25]. TGFBR2 is a major player of TGF-beta signaling pathway and its alteration has been linked to multiple human cancer types [20]. PBRM1 and BAP1 are both involved in chromatin remodeling and are frequently mutated in multiple human cancer types including renal carcinoma, HNSC, pancreatic, bladder and lung cancers [26].
Figure 1.
Mutational landscape of significantly mutated genes (SMGs) in esophageal squamous cell carcinoma (ESCC). The left panel indicates gene mutation frequency, the upper panel shows mutational prevalence with respect to synonymous and non-synonymous mutations, the middle panel depicts SMG mutation landscape across analyzed ESCC cases with different mutation types color coded differently, and the bottom panel displays clinical features such as TNM stage, tumor grade, smoking, alcohol consumption and gender. New SMGs are highlighted in bold.
To gain insights into the genetic alterations in canonical signaling pathways, we curated cancer-related signaling pathways from previous studies [20, 27, 28] and applied PathScan [29] to evaluate the mutational significance of these pathways. Our result showed that chromatin modification, DNA damage response, RAS signaling, cell cycle, genomic integrity maintenance and Notch signaling were significantly enriched for somatic mutations (Supplementary Table S3, available at Annals of Oncology online). Association of their mutation status with survival outcomes is provided in Supplementary Table S4, available at Annals of Oncology online.
Mutational signatures operative in ESCC
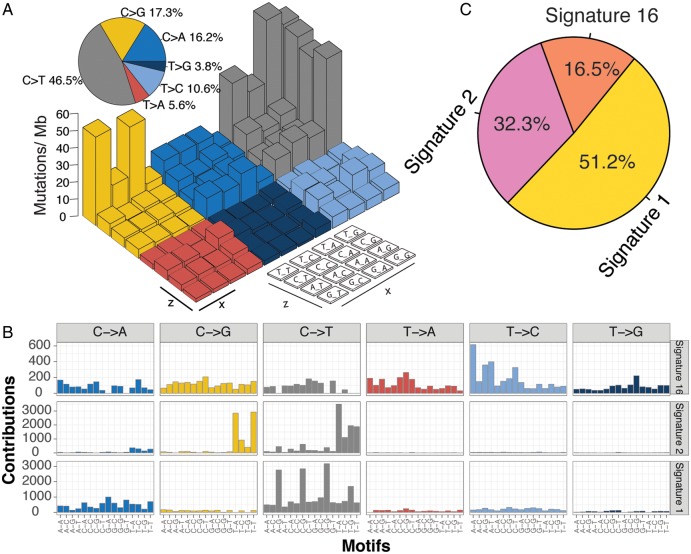
The overall mutational pattern of ESCC was dominated by C>T and C>G mutations (Figure 2A). We extracted three mutational signatures (i.e. signatures 1, 2 and 16; Figure 2B) from ESCC with varying mutational activities, which are defined by the number of mutations generated by each corresponding mutational signature (Figure 2C). These three signatures were named according to the COSMIC signature nomenclature. The clocklike signature 1, featured by C>T transitions at CpG dinucleotides, is thought to be connected with age-related accumulation of spontaneous deamination of 5-methylcytosine. Signature 2, characterized by C>T mutations at TpCpW (where W = A or T) trinucleotide sequences, is thought to result from over-activity of the APOBEC RNA-editing enzyme [14]. Signatures 1 and 2 are widespread among many human cancer types including ESCC [15]. Signature 16, contributed to 16.5% of the total mutation load and characterized by T>C at the trinucleotide, ApTpW (where W = A, G or T). To rule out the possibility that signature 16 may result from random noise, we deconvoluted somatic mutation data against four COSMIC Signatures (i.e. Signatures 1, 2, 13 and 16; see supplementary material, available at Annals of Oncology online) that closely resemble these three signatures extracted from ESCC, and we observed that signature 16 was indeed present in ESCC (supplementary Figure S3, available at Annals of Oncology online).
Figure 2.
Mutational signatures extracted from ESCC. (A) Lego plot representation of mutation patterns in 549 ESCC cases. Single-nucleotide substitutions are divided into six categories with 16 surrounding flanking bases. Inset pie chart shows the proportion of six categories of mutation patterns. (B) Three mutational signatures extracted from ESCC. (C) The mutational activities of corresponding mutational signatures.
Mutational signatures correlated with clinical features
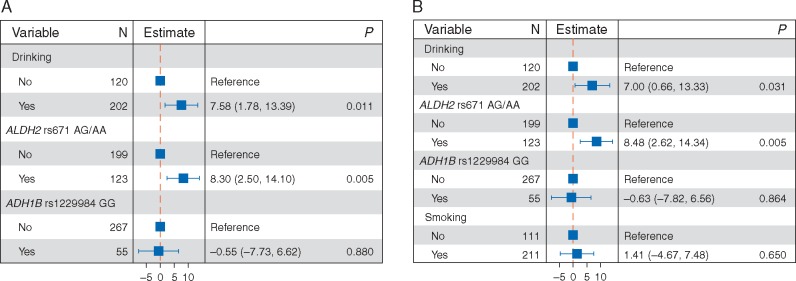
To identify mutagenic factors that are responsible for signature 16, we carried out logistic regression analysis for mutational activity of signature 16 versus alcohol consumption and risk genotypes of ALDH2 and ADH1B. Our analysis showed that increased mutational activity of signature 16 was significantly linked to alcohol consumption and the presence of the ALDH2 rs671 AG/AA polymorphism (Figure 3A). This association remained significant when tobacco smoking was taken into account (Figure 3B). We also carried out mutational signature analysis for HNSC and liver cancer, and found that signature 16 was present in these two cancer types (supplementary Figures S4 and S5, available at Annals of Oncology online, respectively). This association between alcohol consumption and signature 16 was also observed in HNSC (supplementary Figure S6, available at Annals of Oncology online). The association between alcohol consumption and signature 16 in liver cancer was not assessed due to the missing alcohol assumption information in the TCGA liver cancer dataset. In addition, unsupervised hierarchical clustering for activities of mutational signatures identified two distinctive clusters; C1/2 (supplementary Figure S7A, available at Annals of Oncology online), and their association with survival outcome was statistically significant (supplementary Figure S7B and C, available at Annals of Oncology online).
Figure 3.
The association between mutational activity of signature 16 and alcohol consumption with genotypes of ALDH2 and ADH1B (A), and tobacco smoking (B) taken into account. The confounding factors were shown on left-side of each forest plot, and the corresponding estimated odds ratio and P-value were shown on the middle and right-side panels, respectively.
SMG mutation associated with alcohol exposure
We analyzed the association between SMGs and alcohol consumption and found eight SMGs enriched in the alcohol consumption group (Fisher’s exact test, P < 0.05; supplementary Figure S8A, available at Annals of Oncology online). We further examined the difference of mutational activity of signature 16 with respect to the mutational status of these eight SMGs. We observed that increased mutational activity of signature 16 was associated with mutations in ZNF750 (median: 26.3 versus 14.0; P = 0.002), TP53 (median: 15.3 versus 11.6; P = 0.02) and EP300 (median: 23.3 versus 14.1; P = 0.01; supplementary Figure S8B, available at Annals of Oncology online). The association between TP53 mutation status and alcohol consumption signature (i.e. signature 16) was manifested by a significantly higher T>C mutation fraction of TP53 in the alcohol group versus non-alcohol group (22.2% versus 12.4%; one-sided proportion test, P = 0.006). A previous study investigating the impact of acetaldehyde (the first metabolite of alcohol) on TP53 mutations showed that acetaldehyde treatment induced T>C mutations in TP53 [30]. In HNSC, mutations in TP53 were also significantly associated with increased mutational activity of signature 16 (median: 18.3 versus 8.05; one-sided Wilcoxon test, P < 0.001). In liver cancer, this association was marginally significant (median: 33.4 versus 32.9; one-sided Wilcoxon test, P = 0.07).
Prognostic markers for ESCC
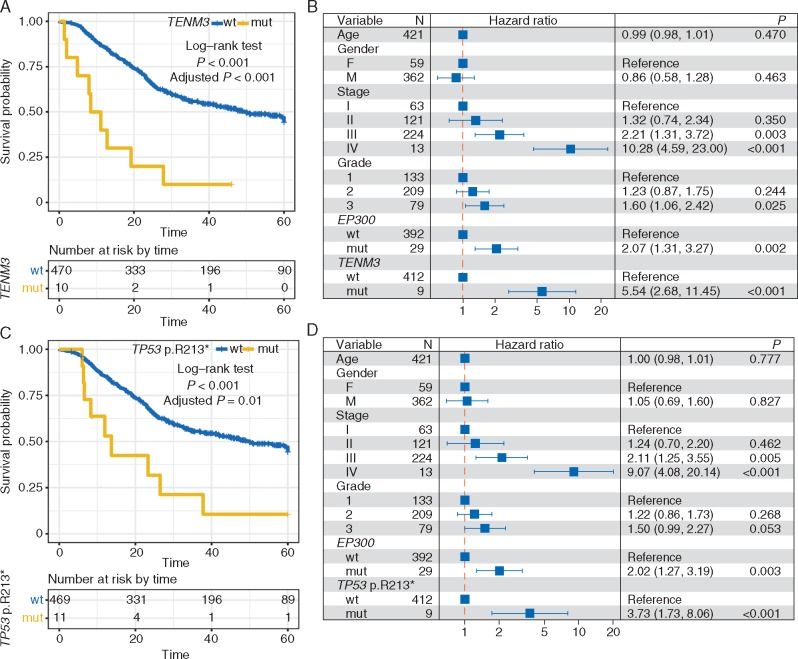
We carried out Kaplan–Meier survival analysis in each of the individual ESCC studies for the 26 identified SMGs and found that mutated TENM3 was significantly associated with the survival outcomes in 4 out of the 5 collected ESCC datasets that included survival data (supplementary Table S5, available at Annals of Oncology online; log-rank test, P < 0.05). Mutation of EP300 was significantly associated with poor survival (supplementary Figure S9, available at Annals of Oncology online). When examining the association between gene mutation and survival in the combined ESCC cohort of 549 cases, we found that mutation of TENM3 was the most significant association after controlling for multiple hypothesis tests (Figure 4A; log-rank test, adjusted P < 0.001). Moreover, mutated TENM3 remained statistically significant after taking into account age, gender, TNM staging, tumor grade and mutation of EP300 (Figure 4B). To rule out the confounding impact of geographical area, we took the geographical area as strata variable in multivariate Cox model and found that mutation of TENM3 was still significant (HR: 5.78; 95% CI: 2.78–12.05; P < 0.001). We next examined TENM3 expression and found that TENM3 was significantly overexpressed in tumor tissue versus matched normal control tissue (supplementary Figure S10A, available at Annals of Oncology online; median: 9.68 versus 7.78; Wilcoxon test, P < 0.001). ESCC patients with abnormally high expression of TENM3 (See supplementary material, available at Annals of Oncology online) were associated with poor prognosis (supplementary Figure S10B and C, available at Annals of Oncology online).
Figure 4.
Prognostic significance of TENM3 and TP53 p.R213* mutations in ESCC. (A, C) Kaplan–Meier survival analysis of TENM3 and TP53 p.R213* mutations. Log-rank test is used to evaluate statistical significance. (B, D) Multivariate Cox regression analysis of TENM3 and TP53 p.R213* mutation with age, gender, TNM stage, tumor grade and EP300 mutation taken into account.
TP53 was the most significantly mutated driver gene in the combined ESCC dataset (86.7%). In this study, we analyzed the association of TP53 hotspot mutations (n ≥ 5) and survival outcome. The result showed that TP53 p.R213* mutation (n = 12) was significantly associated with poor prognosis (Figure 4C and D). TP53 p.R213* mutation was still statistically significant (HR: 3.86; 95% CI: 1.78–8.37; P < 0.001) with geographical area taken as strata variable in a multivariate Cox model. TP53 p.R213* and TENM3 mutations remained statistically significant when they were included as confounding variables in a multivariate Cox model (supplementary Figure S11, available at Annals of Oncology online).
Discussion
In this study, we carried out a meta-analysis of 549 ESCC cases from 7 published studies and identified several less frequently mutated SMGs that were not recognized previously. We revealed a mutational signature and SMGs that are associated with alcohol consumption. We further identified mutations of TENM3 and TP53 (p.R213*) as poor prognosticators for ESCC. In addition, the existence of an alcohol consumption signature (i.e. signature 16) was also present in two recent ESCC studies [31, 32].
A major advantage for this meta-analysis is the inclusion of a large sample size for ESCC. The statistical power to detect SMGs mutated in 3% of samples is only 43%; therefore, a large sample size of ESCC samples is critical for the detection of low mutation frequency SMGs [17]. On the other hand, a potential problem is the batch effect introduced by different cohorts. To overcome this weakness, an SMG was required to be mutated in at least four independent ESCC datasets. In addition, to increase the robustness of our analysis, we used four algorithms to re-annotate mutations and identified 18 previously reported and, more importantly, eight novel SMGs. The novel SMGs include NAV3, TENM3, RIPK4, PBRM4 and USP8. Recurrent mutations of NAV3 have been reported in several cancer types [20] but not in ESCC. TENM3 was both non-silently mutated and overexpressed in tumor tissues compared to adjacent normal control tissue, suggesting TENM3 may function as an oncogene in ESCC. TENM3 was also observed to be frequently mutated in many other human cancer types (supplementary Table S6, available at Annals of Oncology online) obtained from cBioPortal [33]. The TENM3 gene encodes a protein that belongs to the teneurin family. Teneurins are highly conserved transmembrane glycoprotein receptors that have been implicated in tumor development and drug resistance [12]. Genomic aberration of TENM3 was reported in neuroblastoma, and its dysregulation was associated with survival outcomes [13].
In our analysis, mutation of TENM3 was associated with deceased survival outcomes in four datasets and ranked as the most statistically significant prognostic factor in the combined ESCC dataset. Mutations at a hotspot of TP53, p.R213*, was also shown to be an independent prognostic factor.
Another key finding from our study is identification of the alcohol consumption associated signature 16, which was not extracted from our previous study [15] likely due to limited sample size and the relatively smaller mutational activity of signature 16 in comparison with Signatures 1 and 2. In the meta-analysis, signature 16 consisted of 16.5% of total mutations in ESCC (Figure 2C). The association of signature 16 with alcohol consumption in ESCC contributed to 7.1% of total mutations in HNSC (supplementary Figure S12, available at Annals of Oncology online). This finding bridges the gap between alcohol consumption and somatic mutations in relation to ESCC tumorigenesis. The mechanisms through which alcohol and/or its metabolites (e.g. acetaldehyde) induce distinct mutations in ESCC are still elusive and require future investigation. It has been suggested that alcohol consumption might induce TP53 mutations in breast cancer [34], non-small-cell lung cancer [35] and rectal tumors [36]; probably due to accumulation of oxidative stress resulting from alcohol metabolism. Future studies will be needed to illustrate whether alcohol consumption causes TP53 mutations as a critical mechanism for ESCC development.
Supplementary Material
Acknowledgement
We thank Dr Mac Robinson at Wake Forest Baptist Comprehensive Cancer Center for editing the manuscript.
Funding
This work was partially supported by the Program for Changjiang Scholars and Innovative Research Team in University in China (IRT_14R40 to KC), and the Tianjin Municipal Education Commission (11601501-2016KJ0148 to DH). WZ is supported by a Fellowship from the National Foundation for Cancer Research, an Endowed Hanes and Willis Family Professor in Cancer at the Wake Forest Baptist Comprehensive Cancer Center, and the Cancer Center Support Grant from the National Cancer Institute to the Comprehensive Cancer Center of Wake Forest Baptist Medical Center (P30 CA012197).
Disclosure
The authors have declared no conflicts of interest.
References
- 1.GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase. No. 11. http://globocan.iarc.fr/Default.aspx (24 March 2017, date last accessed).
- 2. Chen W, Zheng R, Baade PD. et al. Cancer Statistics in China, 2015. 2016; 66(2): 115–132. [DOI] [PubMed] [Google Scholar]
- 3. Engel LS, Chow W-H, Vaughan TL. et al. Population attributable risks of esophageal and gastric cancers. J Natl Cancer Inst. 2003; 95(18): 1404–1413. [DOI] [PubMed] [Google Scholar]
- 4. Tanaka F, Yamamoto K, Suzuki S. et al. Strong interaction between the effects of alcohol consumption and smoking on oesophageal squamous cell carcinoma among individuals with ADH1B and/or ALDH2 risk alleles. Gut 2010; 59(11): 1457–1464. [DOI] [PubMed] [Google Scholar]
- 5. Cheng C, Zhou Y, Li H. et al. Whole-genome sequencing reveals diverse models of structural variations in esophageal squamous cell carcinoma. Am J Hum Genet 2016; 98(2): 256–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Qin H-D, Liao X-Y, Chen Y-B. et al. Genomic characterization of esophageal squamous cell carcinoma reveals critical genes underlying tumorigenesis and poor prognosis. Am J Hum Genet 2016; 98(4): 709–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sawada G, Niida A, Uchi R. et al. Genomic landscape of esophageal squamous cell carcinoma in a Japanese population. Gastroenterology 2016; 150(5): 1171–1182. [DOI] [PubMed] [Google Scholar]
- 8. Song Y, Li L, Ou Y. et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature 2014; 509(7498): 91–95. [DOI] [PubMed] [Google Scholar]
- 9. Lin D-C, Hao J-J, Nagata Y. et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet 2014; 46(5): 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gao Y-B, Chen Z-L, Li J-G. et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet 2014; 46(10): 1097–1102. [DOI] [PubMed] [Google Scholar]
- 11. Hao J, Lin D, Dinh HQ. et al. Spatial intratumoral heterogeneity and temporal clonal evolution in esophageal squamous cell carcinoma. Nat Genet 2016; 48(12): 1500–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ziegler A, Corvalán A, Roa I. et al. Teneurin protein family: an emerging role in human tumorigenesis and drug resistance. Cancer Lett 2012; 326(1): 1–7. [DOI] [PubMed] [Google Scholar]
- 13. Molenaar JJ, Koster J, Zwijnenburg DA. et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 2012; 483(7391): 589–593. [DOI] [PubMed] [Google Scholar]
- 14. Roberts S. a, Lawrence MS, Klimczak LJ. et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet 2013; 45(9): 970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang L, Zhou Y, Cheng C. et al. Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am J Hum Genet 2015; 96(4): 597–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lawrence MS, Stojanov P, Polak P. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013; 499(7457): 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lawrence MS, Stojanov P, Mermel CH. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014; 505(7484): 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tokheim C, Papadopoulis N, Kinzler KW. et al. Evaluating the evaluation of cancer driver genes. Proc Natl Acad Sci 2016; 113(50): 60426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Klijn C, Durinck S, Stawiski EW. et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol 2014, doi: 10.1038/nbt.3080 [DOI] [PubMed] [Google Scholar]
- 20. Kandoth C, McLellan MD, Vandin F. et al. Mutational landscape and significance across 12 major cancer types. Nature 2013; 502(7471): 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li J, Chen Z, Tian L. et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut 2014; 63(11): 1700–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim J, Mouw KW, Polak P. et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat Genet 2016; 48(6): 600–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reincke M, Sbiera S, Hayakawa A. et al. Mutations in the deubiquitinase gene USP8 cause Cushing’s disease. Nat Genet 2015; 47(1): 31–38. [DOI] [PubMed] [Google Scholar]
- 24. Ma Z-Y, Song Z-J, Chen J-H. et al. Recurrent gain-of-function USP8 mutations in Cushing’s disease. Cell Res 2015; 25(3): 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stransky N, Egloff AM, Tward AD. et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011; 333(6046): 1157–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gonzalez-Perez A, Jene-Sanz A, Lopez-Bigas N.. The mutational landscape of chromatin regulatory factors across 4, 623 tumor samples. Genome Biol 2013; 14(9): r106.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vogelstein B, Papadopoulos N, Velculescu VE. et al. Cancer genome landscapes. Science 2013; 339(6127): 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Davoli T, Xu AW, Mengwasser KE. et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 2013; 155(4): 948–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wendl MC, Wallis JW, Lin L. et al. PathScan: a tool for discerning mutational significance in groups of putative cancer genes. Bioinformatics 2011; 27(12): 1595–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Paget V, Lechevrel M, Sichel F.. Acetaldehyde-induced mutational pattern in the tumour suppressor gene TP53 analysed by use of a functional assay, the FASAY (functional analysis of separated alleles in yeast). Mutat Res 2008; 652(1): 12–19. [DOI] [PubMed] [Google Scholar]
- 31. Chen X, Zhong Q, Liu Y. et al. Genomic comparison of esophageal squamous cell carcinoma and its precursor lesions by multi-region whole-exome sequencing. Nat. Commun 2017; 8(1): 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chang J, Tan W, Ling Z. et al. Genomic analysis of oesophageal squamous-cell carcinoma identifies alcohol drinking-related mutation signature and genomic alterations. Nat. Commun 2017; 8(17): 15290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gao J, Aksoy BA, Dogrusoz U. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013; 6(269): pl1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Freudenheim JL, Bonner M, Krishnan S. et al. Diet and alcohol consumption in relation to p53 mutations in breast tumors. Carcinogenesis 2004; 25(6): 931–939. [DOI] [PubMed] [Google Scholar]
- 35. Ahrendt SA, Chow JT, Yang SC. et al. Alcohol consumption and cigarette smoking increase the frequency of p53 mutations in non-small cell lung cancer. Cancer Res 2000: 3155–3159. [PubMed]
- 36. Slattery ML, Wolff RK, Herrick JS. et al. Alcohol consumption and rectal tumor mutations and epigenetic changes. Dis Colon Rectum 2010; 53(8): 1182–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.