This work reveals the conserved function of D14 from dicots and monocots, and defines rice D14 as an unconventional hormone receptor that can restore strigolactone signaling in the Arabidopsis d14 mutant
Keywords: Arabidopsis, DWARF14, phytohormone, receptor, rice, strigolactone
Abstract
Strigolactones (SLs) act as an important class of phytohormones to regulate plant shoot branching, and also serve as rhizosphere signals to mediate interactions of host plants with soil microbes and parasitic weeds. SL receptors in dicots, such as DWARF14 in Arabidopsis (AtD14), RMS3 in pea, and ShHTL7 in Striga, serve as unconventional receptors that hydrolyze SLs into a D-ring-derived intermediate CLIM and irreversibly bind CLIM to trigger SL signal transduction. Here, we show that D14 from the monocot rice can complement Arabidopsis d14 mutant and interact with the SL signaling components in Arabidopsis. Our results further reveal that rice D14, similar to SL receptors in dicots, also serves as an unconventional hormone receptor that generates and irreversibly binds the active form of SLs. These findings uncover the conserved functions of D14 proteins in monocots and dicots.
Introduction
Strigolactones (SLs) are a family of carotenoid-derived phytohormones (Gomez-Roldan et al., 2008; Umehara et al., 2008) that play a vital role in the control of plant shoot branching, a key agricultural trait that determines plant architecture and affects crop yield (Song et al., 2017; Zhang et al., 2017). SLs also act as rhizosphere signals to establish symbiotic interactions between plants and soil microbes (Akiyama et al., 2005; Kretzschmar et al., 2012; Gutjahr et al., 2015), and regulate parasitic interactions by stimulating germination and growth of parasitic weeds such as Striga (Cook et al., 1966; Cardoso et al., 2014; Conn et al., 2015; Toh et al., 2015; Tsuchiya et al., 2015; Gobena et al., 2017; Lumba et al., 2017a). Moreover, SLs also regulate hypocotyl elongation, root growth, leaf development and senescence (Snowden et al., 2005; Kapulnik et al., 2011; Ruyter-Spira et al., 2011; Waters et al., 2012b; Waters and Smith, 2013; Yamada et al., 2014; Soundappan et al., 2015; Ueda and Kusaba, 2015; Wang et al., 2015; Li et al., 2016). Recently, SLs were revealed to regulate various plant stress responses including drought tolerance and disease resistance (Umehara et al., 2010; Dor et al., 2011; Kohlen et al., 2011; Bu et al., 2014; Ha et al., 2014; Decker et al., 2017).
Genetic and molecular characterization of highly branched mutants in various plant species, such as rice dwarf14 (d14), d3, d10, d17, d27, and d53 (Ishikawa et al., 2005; Arite et al., 2007, 2009; Gao et al., 2009; Lin et al., 2009; Liu et al., 2009; Jiang et al., 2013; Zhou et al., 2013), Arabidopsis more axillary growth1 (max1), max2, max3, max4, and d14 (Woo et al., 2001; Stirnberg et al., 2002; Sorefan et al., 2003; Booker et al., 2004, 2005; Shen et al., 2007; Nelson et al., 2011; Waters et al., 2012a, b; Abe et al., 2014; Chevalier et al., 2014; Yao et al., 2016), pea ramosus1 (rms1), rms2, rms3, rms4, and rms5 (Beveridge et al., 1996; Morris et al., 2001; Foo et al., 2005; Johnson et al., 2006; de Saint Germain et al., 2016), and petunia decreased apical dominance 1 (dad1), dad2, and dad3 (Snowden et al., 2005; Simons et al., 2007; Hamiaux et al., 2012), suggests that SL biosynthesis and signaling pathway are largely conserved in diverse plant species.
Rice D3 or its ortholog in Arabidopsis (MAX2), petunia (PhMAX2A), or pea (RMS4) encodes an F-box protein (Johnson et al., 2006; Shen et al., 2007; Stirnberg et al., 2007; Drummond et al., 2011; Hamiaux et al., 2012), a subunit of SCF (Skp1–Cullin1–F-box protein) ubiquitin ligase complex that functions in substrate recognition for proteasome-mediated proteolysis. Rice D14 or its orthologs, such as Arabidopsis D14, petunia DAD2, or pea RMS3, encodes an α/β hydrolase that hydrolyzes SLs (Hamiaux et al., 2012; Nakamura et al., 2013; Zhao et al., 2013; de Saint Germain et al., 2016; Yao et al., 2016) and interacts with its respective F-box protein in an SL-dependent manner to recruit various repressors, such as D53 in rice and SMXL6/7/8 (SUPPRESSOR OF MAX2 1-LIKE6/7/8) proteins in Arabidopsis, for ubiquitination and degradation (Hamiaux et al., 2012; Jiang et al., 2013; Zhou et al., 2013; Soundappan et al., 2015; Wang et al., 2015; Liang et al., 2016; Yao et al., 2016). Degradation of these repressors will subsequently de-repress their directly targeted transcription factors, such as IPA1 (Ideal Plant Architecture 1) in rice (Song et al., 2017), to activate downstream genes essential for various SL-regulated plant responses (Lumba et al., 2017b; Waters et al., 2017). It is intriguing that the receptors AtD14 and D14 are also degraded in an SL-induced and MAX2/D3-dependent manner (Chevalier et al., 2014; Hu et al., 2017).
Recently, AtD14 was defined as a non-canonical hormone receptor that possesses dual functions: AtD14 acts as an enzyme to hydrolyze SLs and generate the active form of the hormone molecule CLIM (the covalently linked intermediate molecule), and also serves as a receptor of SL to bind CLIM irreversibly and undergo significant conformational change for interacting with downstream components and triggering SL signal transduction (Snowden and Janssen, 2016; Yao et al., 2016; Fang and Chen, 2017; Yao et al., 2017). Similarly to AtD14, RMS3 in pea and ShHTL7 in Striga hermonthica also hydrolyze strigolactone into the D-ring-derived intermediate CLIM and covalently bind CLIM in an irreversible manner to trigger SL signaling (de Saint Germain et al., 2016; Yao et al., 2017).
Rice is an important crop and also serves as a model plant for the study of monocots. It is known that rice, but not Arabidopsis, is the host plant for both symbiotic arbuscular mycorrhizal fungi (AMF) and parasitic Striga (Yoshida and Shirasu, 2009; Waters et al., 2017). In this study, we employ bioinformatics, genetic, and biochemical approaches to investigate functional conservations between rice D14 and Arabidopsis AtD14, and examine whether the rice D14 is similar to AtD14 in generating and perceiving the active form of SLs.
Materials and methods
Generation of transgenic plants
The modified binary vector pCAMBIA1300-cFlag (Yao et al., 2016) carrying the full coding sequence of Arabidopsis thaliana D14 (AtD14), Oryza sativa D14, or N-terminus (amino acids 1–51) truncated O. sativa D14 (D14ΔN) under the control of the 35S promoter was introduced into the Atd14-5 mutant (Yao et al., 2016) by using the Agrobacterium-mediated floral dip method. Similarly, the pCAMBIA1300-cFlag vector carrying D14ΔN under the control of the AtD14 promoter was introduced into the Atd14-1 mutant (Waters et al., 2012b) to generate transgenic plants, AtD14pro:D14ΔN. The primary rosette branching numbers were counted for 7-week-old plants which were germinated on plates and grown in soil under a light/dark photoperiod of 16 h/8 h at 22 °C.
Real-time PCR (RT-PCR) analysis
Rosette leaves of plants were collected for RNA extraction. Total RNA was prepared with a TransZol Kit (TransGen) and used in the reverse transcription reaction with the reagent TransScript® RT/RI Enzyme Mix (TransGen). The first-strand cDNA was used as the template for the subsequent RT-PCR, which was performed to amplify AtD14 (primers 5'-ATGAGTCAACACAACATCTTAG-3' and 5'-GATGATTCCGATCATAGCG-3'), and D14 (primers 5'-TGACCTCTTCGCCAAGCTTG-3' and 5'-TCTTGAAGACG GTCTGGCAGAC-3') in the plants with the indicated genotypes. The A. thaliana ACTIN1 was employed as a control (primers 5'-TGTTGAGAAGAACTACGAGC-3' and 5'- AAGCACTTCCTGTGAACAAT-3').
Hypocotyl measurements
The Arabidopsis seeds were sterilized and germinated on Murashige and Skoog (MS) medium with or without 3 μM rac-GR24 (an SL analog, Chiralix) under continuous low light at 22 °C for 7 d. Hypocotyl length was measured by Digimizer software.
Leaf morphology analysis
Plants were grown in soil under a light/dark photoperiod of 16 h/8 h at 22 °C for an additional 3 weeks after germination and growth on MS medium for 1 week. For each genotype, 20 plants were used for observation of leaf morphology. Whole plants and their sixth leaves were photographed and harvested for further measurement. The leaf length (the distance between the leaf tip and the base of petiole) and leaf width (the greatest distance across the leaf lamina perpendicular to the proximal/distal axis of the leaf) were measured manually using a ruler.
Protein preparation
N-terminus-truncated O. sativa D14ΔN or full-length A. thaliana D14 (AtD14) was expressed in Escherichia coli strain BL21 (DE3) (Novagen) as an N-terminal glutathione S-transferase (GST) tag-fusion protein. After being purified by glutathione Sepharose 4B (GE Healthcare) affinity chromatography, GST–D14ΔN or GST–AtD14 protein was released by 10 mM glutathione (GSH) elution or on-column cleavage to remove the GST tag, then further purified by HiTrap Q (GE Healthcare) followed by Superdex 200 10/300 (GE Healthcare) in a buffer containing 10 mM Tris–HCl, pH 8.0, 150 mM NaCl, and 5 mM DTT.
Full-length A. thaliana SMXL6 was expressed in sf9 insect cells with an N-terminal Flag tag and purified by anti-Flag beads (Sigma, A2220) according to the manufacturer’s manual.
The full-length O. sativa D3 or A. thaliana MAX2 was fused with His6 and co-expressed with ASK1, which stabilizes F-box proteins (Yan et al., 2013; Yao et al., 2016; Li et al., 2017), in sf9 insect cells. After purification by Ni-NTA (Novagen) affinity chromatography, the His-D3–ASK1 or His-MAX2–ASK1 complex was eluted and further purified by HiTrap Q followed by Superdex 200 10/300 in a buffer containing 20 mM MES, pH 6.5, 150 mM NaCl, and 5 mM DTT.
Pull-down assay
For the interaction between D14 and MAX2, ~20 μg of His-MAX2–ASK1 was used as the bait and ~12 μg of GST–D14 was used as the prey in the presence of 20 μM rac-GR24 or its solvent DMSO as the control. The reaction mixtures were incubated with Ni-NTA beads (Qiagen) at 4 °C for 1 h in the reaction buffer [50 mM Tris–HCl, pH 6.8, 100 mM NaCl, 25 mM imidazole, 10% (v/v) glycerol, 0.1% Tween-20, 20 mM 2-mercaptoethanol]. After washing six times with the reaction buffer, the protein complexes on the beads were released and then subjected to western blot analysis. The pull-down assay of AtD14 with MAX2 was performed in a similar way to serve as a positive control.
For the interaction between D14 and SMXL6, ~20 μg of Flag-SMXL6 protein was used as the bait and ~12 μg of GST–D14 as the prey in the presence of 20 μM rac-GR24 or its solvent DMSO as the control. The reaction mixtures were incubated with anti-flag beads at 4 °C for 1 h in the reaction buffer (50 mM Tris–HCl, pH 7.0, 150 mM NaCl, 0.5% Tween-20). After washing six times with the reaction buffer, the protein complexes on the beads were released and then subjected to western blot analysis. The pull-down assay of AtD14 with SMXL6 was performed in a similar way to serve as a positive control.
GST-fused proteins were detected by a monoclonal anti-GST antibody (Abmart). The polyvinylidene difluoride (PVDF) membrane was stained with Memstain (Applygen) to show equal loading.
Co-immunoprecipitation (Co-IP) assay in protoplasts
Protoplasts prepared from the smxl6 smxl7 smxl8 triple mutant or the wild type were transformed with transient expression plasmids as described (Wang et al., 2015). After transformation with the hemagglutinin (HA)-AtD14 and green fluorescent protein (GFP)–MAX2 plasmids and incubation at 21 °C for 11 h, protoplasts were pre-treated with 100 µM rac-GR24 for 1 h in W5 solution. Cells were then collected and broken in the protein extraction buffer [50 mM Tris–HCl, 150 mM NaCl, 10% (v/v) glycerol, 0.1% Nonidet P-40, and 1×complete protease inhibitor cocktail], and immunoprecipitation (IP) with agarose-conjugated anti-GFP monoclonal antibody (MBL) was subsequently carried out in the presence or absence of 100 µM rac-GR24 at 4 °C. The HA-AtD14 recombinant proteins were detected with the anti-HA monoclonal antibody (Millipore), and the GFP–MAX2 fusion proteins and GFP were detected with the anti-GFP monoclonal antibody (Sigma). The total proteins extracted from protoplasts before IP were used as inputs.
Size exclusion chromatography (SEC) assay
Purified D14 (~10 μM) and D3–ASK1 proteins (~5 μM) were incubated with 200 μM 5-deoxystrigol (5DS; OlChemIm Ltd) or an equal amount of DMSO as the solvent control at 25 °C for 1 h in buffer containing 20 mM MES, pH 6.5, 150 mM NaCl, 5 mM DTT. The reaction mixture was then injected onto a Superdex 200 10/300 column for analysis at a flow rate of 0.3 ml min–1. The fractions (0.5 ml per fraction) were analyzed by SDS–PAGE and visualized by Coomassie Brilliant Blue staining.
Mass spectrometric analysis of covalent modification
The gel bands of D14 from the SEC-separated D14–D3–ASK1 complex induced by 5DS were excised for mass spectrometric analysis as previously described (Yao et al., 2016). Briefly, tandem mass spectrometry (MS/MS) spectra from each LC-MS/MS run were searched against the D14 protein database using the Proteome Discoverer (Version 1.4) searching algorithm. The search criteria were as follows: full enzymatic specificity for trypsin was required, two missed cleavages were allowed, carbamidomethylation was set as a fixed modification (on the cysteine residue), oxidation (on the methionine residue) was set as a variable modification, precursor ion mass tolerance was 10 ppm for all mass spectra acquired in the Orbitrap mass analyzer, and fragment ion mass tolerance was 0.02 Da for all MS/MS spectra acquired in the ion trap. A high confidence score filter [false discovery rate (FDR) <1%) was used to select the ‘hit’ peptides, and their corresponding MS/MS spectra were manually inspected.
Results
Phylogenetic analysis and sequence alignment of D14 orthologs from monocots and dicots
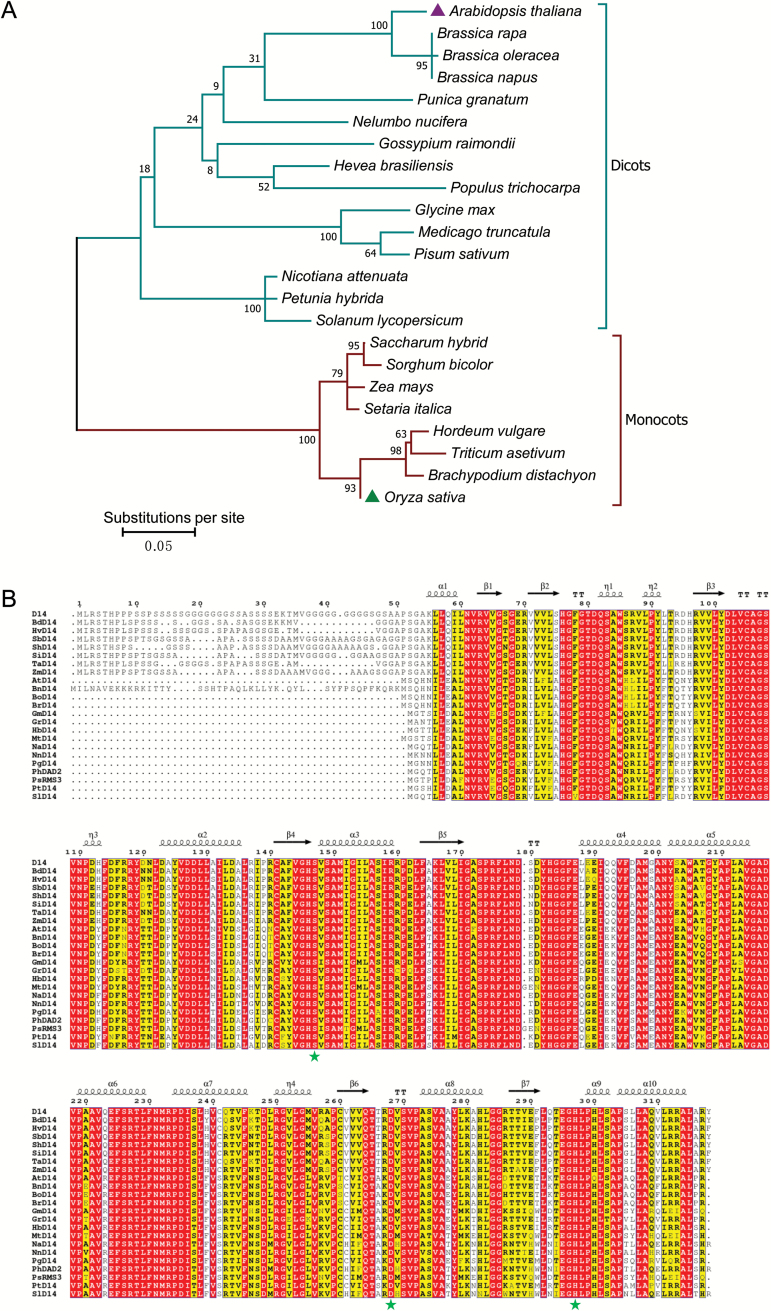
To investigate the evolutionary relationships among D14 orthologs in monocots and dicots, we searched public sequence databases using BLAST with the rice (O. sativa) D14 protein sequence as a query to obtain the predicted sequences of D14 orthologs from important monocots and dicots (Waters et al., 2012b; Conn et al., 2015; Bythell-Douglas et al., 2017) (Fig. 1A). The phylogenetic analysis showed that D14 proteins from the monocots such as O. sativa, Brachypodium distachyon, Triticum asetivum, Hordeum vulgare, Setaria italica, Zea mays, Sorghum bicolor, and Saccharum hybrid have closer phylogenic relationships, while D14 orthologs from all the tested dicots exhibit closer relationships (Fig. 1A).
Fig. 1.
Phylogenetic analysis and sequence alignment of D14 orthologs from monocots and dicots. (A) Phylogenetic analysis of D14 orthologs from monocots and dicots. The phylogenetic tree was generated with 23 full-length amino acid sequences of D14 orthologs using the Maximum Likelihood method based on the WAG model (100 replicates) in MEGA7 (Whelan and Goldman, 2001; Kumar et al., 2016). The percentage of trees in which the associated taxa clustered together is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The GenBank accession numbers of D14 orthologs in the presented species are, from top to bottom: Arabidopsis thaliana D14 (NP_566220), Brassica rapa D14 (XP_009130408), Brassica oleracea D14 (XP_013638430), Brassica napus D14 (CDY42894), Punica granatum D14 (OWM70752), Nelumbo nucifera D14 (XP_010248100), Gossypium raimondii D14 (XP_012451974), Hevea brasiliensis D14 (XP_021646820), Populus trichocarpa D14 (XP_002302409), Glycine max D14 (XP_003557012), Medicago truncatula D14 (XP_003589086), Pisum sativum RMS3 (AMB61024), Nicotiana attenuata D14 (XP_019258478), Petunia hybrida DAD2 (AFR68698), Solanum lycopersicum D14 (XP_004238093), Saccharum hybrid D14 (AJY78078), Sorghum bicolor D14 (XP_002468316), Zea mays D14 (NP_001150635), Setaria italica D14 (XP_004985292), Hordeum vulgare D14 (AJP07999), Triticum asetivum D14 (AK332360), Brachypodium distachyon D14 (XP_003558555), and Oryza sativa D14 (XP_015631400). (B) Sequence alignment and structural annotation of D14 orthologs. ESPript was used to analyze the multiple sequence alignments generated by Clustal Omega (Sievers et al., 2011; Robert and Gouet, 2014) with the D14 orthologs listed in (A). Secondary structure elements of the rice D14 crystal structure (PDB code: 4IH9) are displayed on top of the alignments. Identical and conserved residues are highlighted by red and yellow backgrounds, respectively. The three catalytic residues, Ser, Asp, and His, are indicated by green stars.
Further sequence alignment and structural annotation showed that the examined D14 orthologs from various plant species all exhibit considerable identities at the amino acid level and have the same catalytic triad Ser–His–Asp and α/β hydrolase fold (Fig. 1B; Supplementary Table S1 at JXB online), suggesting the conserved physiological functions for different D14 proteins. Consistent with phylogenetic analysis (Fig. 1A), protein sequences of D14 orthologs from monocots or dicots, respectively, are more conserved. For example, the rice D14 exhibits no less than 80% identity with its orthologs from all the tested monocots, but displays only ~50% identity with those from the examined dicots (Supplementary Table S1). Interestingly, all the examined monocot D14 proteins (including rice D14) have an additional glycine- and serine-rich N-terminus (at least 7 glycine and 10 serine residues among the N-terminal 55 residues), which is absent in all the examined dicot D14 proteins (Fig. 1B). It is unclear whether full-length D14 proteins in monocots and dicots might have some divergent physiological functions due to their discrepant N-termini.
Rice D14 rescues phenotypes of the Arabidopsis d14 mutant and interacts with Arabidopsis MAX2 and SMXL6 proteins
We further investigated whether the physiological function of D14 proteins is conserved in the monocot rice and the dicot Arabidopsis. We generated the transgenic Arabidopsis 35Spro:D14 and 35Spro:D14ΔN by introducing full-length rice D14 or N-terminus (residues 1–51) truncated rice D14 (D14ΔN) under the control of the Cauliflower mosaic virus (CaMV) 35S promoter into the Arabidopsis Atd14-5 mutant, a weak allele of the Atd14 mutant (Yao et al., 2016). We also introduced the 35S promoter-driven AtD14 into Atd14-5 to generate the 35Spro:AtD14 plants for comparison. As shown in Supplementary Table S2, 66.1% of 35Spro:AtD14 plants (39 out of 59 transgenic lines) and 33.9% of 35Spro:D14ΔN plants (21 out of 62 lines) display a similar branching phenotype to the wild-type Col-0 (with ≤3 branches) while only 3 out of 52 (5.8%) 35Spro:D14 transgenic lines rescues the branching phenotype of Atd14-5 well. These genetic complementation results demonstrate that the highly branched phenotype of the Arabidopsis Atd14-5 mutant can be rescued by both the full-length rice D14 and the N-terminus-truncated rice D14 (D14ΔN), but the complementation ratio is very low for the case of full-length rice D14.
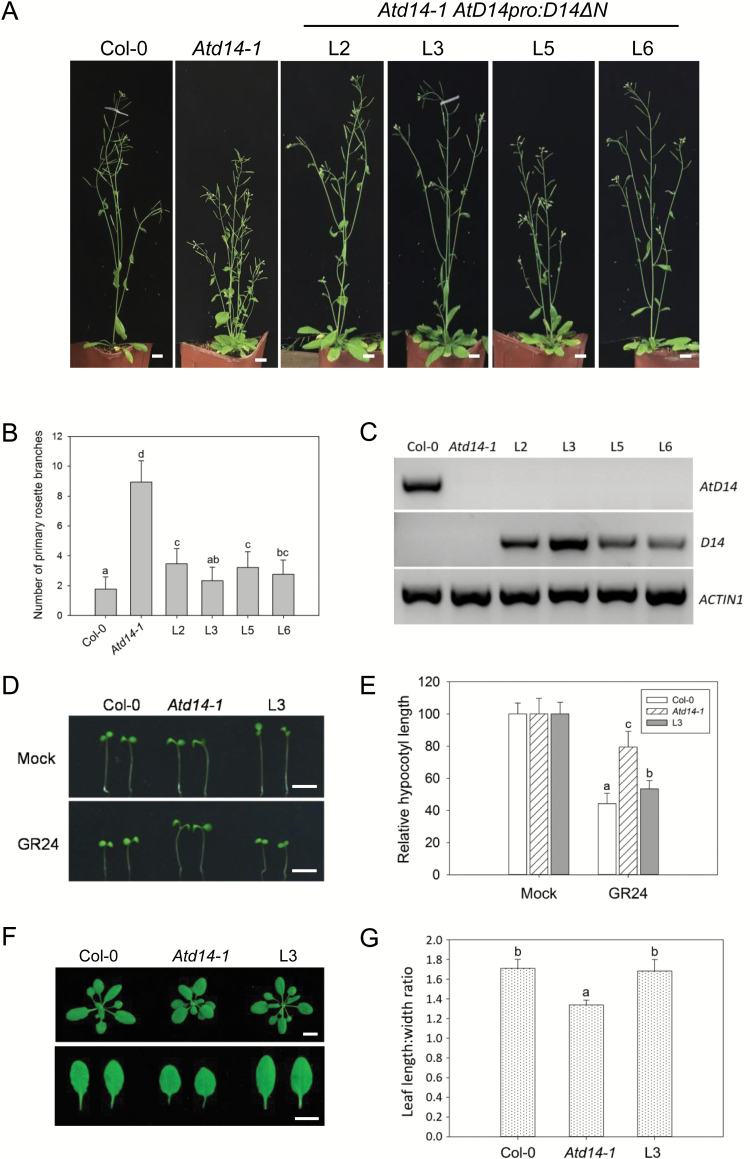
We further introduced D14ΔN under the control of the native AtD14 promoter into the T-DNA insertion knockout mutant Atd14-1 (Waters et al., 2012b) to generate the AtD14pro:D14ΔN plants. The results showed that 50% of the AtD14pro:D14ΔN plants exhibit a similar branching phenotype to the wild-type Col-0 (Fig. 2A, B; Supplementary Table S2), demonstrating that the highly branched phenotype of the Arabidopsis Atd14-1 mutant can be well rescued by rice D14ΔN (Fig. 2A–C).
Fig. 2.
Rice D14 rescues the Arabidopsis d14 mutant. (A and B) Rice D14 rescued the branching phenotype of Atd14-1. (A) Representative branching phenotypes of 7-week-old Col-0, Atd14-1, and four independent T3 transgenic lines AtD14pro:D14ΔN L2 (line 2), L3, L5, and L6 in the Atd14-1 background; scale bars=1 cm. (B) Quantitative analysis of primary rosette branches of the indicated plants; data are means ±SD (n=20). Error bars indicate the SD; bars with the same letter are not significantly different from one another (ANOVA+Tukey HSD, P<0.01). (C) RT–PCR analysis of the AtD14 or D14 transcript levels in the indicated plants described in (A). The Arabidopsis ACTIN1 was used as an internal control. (D and E) Rice D14 rescued the hypocotyl phenotype of Atd14-1. (D) Representative hypocotyl phenotypes of 7-day-old Col-0, Atd14-1, and AtD14pro:D14ΔN L3 (T3) seedlings; scale bars=5 mm. (E) Relative hypocotyl lengths of the indicated seedlings; data are means ±SD (n=30). Error bars indicate the SD; bars with the same letter are not significantly different from one another (ANOVA+Tukey HSD, P<0.01). (F and G) Rice D14 rescued the leaf phenotype of Atd14-1. (F) Representative leaf phenotypes of 4-week-old Col-0, Atd14-1, and AtD14pro:D14ΔN L3 (T3); scale bars=1 cm. (G) Quantitative analysis on the leaf length/leaf width ratio for the sixth leaves of the indicated plants; data are means ±SD (n=20). Error bars indicate the SD; bars with the same letter are not significantly different from one another (ANOVA+Tukey HSD, P<0.01).
Moreover, we explored whether rice D14ΔN is able to complement Atd14-1 in other SL-regulated physiological phenotypes including hypocotyl elongation (Scaffidi et al., 2014; Umehara et al., 2015) and leaf morphology (Waters et al., 2012b). As shown in Fig. 2D–G, the Atd14-1 mutant shows impaired sensitivity to rac-GR24 treatment on hypocotyl inhibition and has rounder and broader leaves than the wild-type Col-0, which is consistent with previous observations (Waters et al., 2012b; Scaffidi et al., 2014; Umehara et al., 2015). However, the hypocotyl elongation of AtD14pro:D14ΔN plants (L3) was obviously inhibited when treated with rac-GR24, and the leaf phenotypes of AtD14pro:D14ΔN plants (L3) were also similar to those of Col-0 (Fig. 2D–G). These results demonstrate that rice D14 can rescue many SL-regulated physiological phenotypes of the Arabidopsis d14 mutant well.
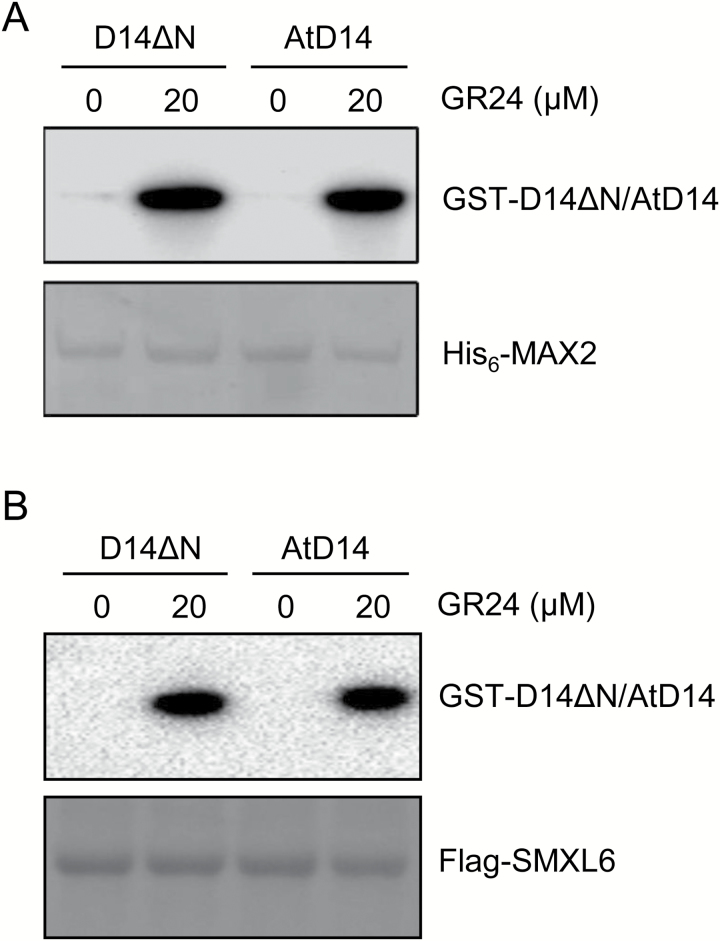
Consistent with the functional complementation of Atd14 by rice D14 and D14ΔN, biochemical pull-down assays and structural analysis showed that, similar to AtD14, both rice D14 and D14ΔN are able to interact efficiently with Arabidopsis MAX2 and SMXL6 proteins in an SL-dependent manner (Fig. 3; Supplementary Fig. S1). These results reveal the molecular basis of the functional complementation by showing the conserved functions of D14 proteins at the protein–protein interaction level.
Fig. 3.
Rice D14 physically interacts with the Arabidopsis SL signaling components. (A) Rice D14 efficiently bound Arabidopsis MAX2 in the presence of rac-GR24. Pull-down assay using recombinant His6-MAX2 and GST–D14 or GST–AtD14 in the absence or presence of rac-GR24. GST-fused proteins were detected by anti-GST antibody and the PVDF membrane was stained with MemStain to show equal loading. (B) Rice D14 efficiently bound Arabidopsis SMXL6 in the presence of rac-GR24. Pull-down assay using recombinant Flag-SMXL6 and GST–D14 or GST–AtD14 in the absence or presence of rac-GR24. GST-fused proteins were detected by anti-GST antibody and the PVDF membrane was stained with MemStain to show equal loading.
Taken together, our results demonstrate that rice D14 can rescue the phenotype of the Arabidopsis d14 mutant well, which is probably attributed to the conserved function of rice D14 to interact with Arabidopsis MAX2 and SMXL6 proteins in the presence of SLs.
The in vivo interaction of the SL receptor with its F-box protein does not require repressors
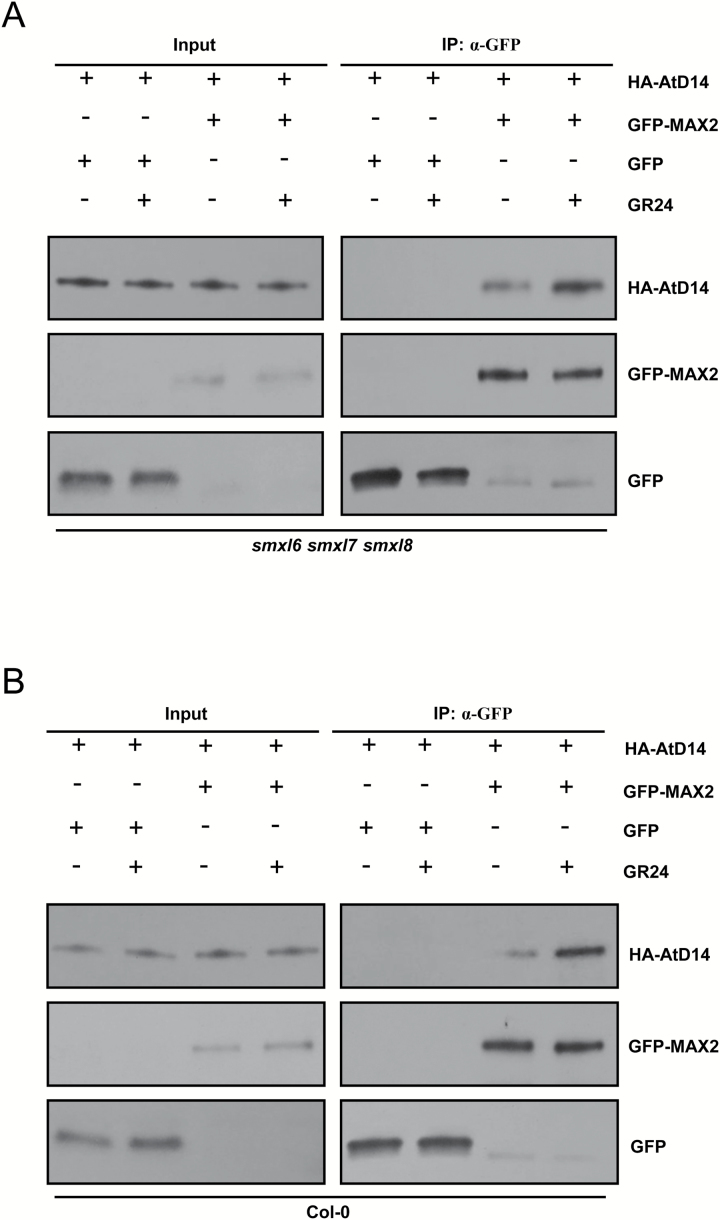
The interaction dynamics or sequential binding among SL signaling components remains an open question (Wang and Smith, 2016). It is known that the SL-induced interaction of receptor with repressor does not depend on the F-box protein MAX2 in vitro or in vivo (Wang et al., 2015). However, it is unclear whether or not the SL-induced in vivo interaction of the SL receptor with its F-box protein is independent of repressors. To answer this question, we employed our well-established Arabidopsis protoplasts transformation and Co-IP system to investigate the interaction between AtD14 and MAX2 in the wild-type Col-0 and the smxl6 smxl7 smxl8 triple mutant. We found that AtD14 was able to interact weakly with MAX2 in both the smxl6 smxl7 smxl8 triple mutant (Fig. 4A) and the wild-type Col-0 (Fig. 4B), and that such interactions in both the triple mutant and Col-0 were obviously enhanced by the addition of exogenous rac-GR24 (Fig. 4). These data demonstrate that the SL-induced in vivo interaction of AtD14 with MAX2 is independent of SMXLs. Together with previous studies (Wang et al., 2015), these results imply that the in vivo interactions between any two components among the receptor (AtD14 or D14), the F-box protein (MAX2 or D3), and the repressor (SMXLs or D53) do not require the presence of the third one.
Fig. 4.
AtD14 interacts with MAX2 independently of SMXL6/7/8. The in vivo interactions between HA-AtD14 and GFP–MAX2 revealed by co-immunoprecipitation (Co-IP) assay in protoplasts prepared from the smxl6 smxl7 smxl8 triple mutant (A) and the wild-type Col-0 (B). After transformation and incubation for 11 h, protoplasts were pre-treated with rac-GR24 for 1 h, cells were broken, and then immunoprecipitation (IP) using agarose-conjugated anti-GFP monoclonal antibody was carried out in the presence or absence of 100 µM rac-GR24. The HA-AtD14 recombinant protein was detected by anti-HA monoclonal antibody, and the GFP–MAX2 fusion protein and GFP were detected by anti-GFP monoclonal antibody. Input means extracted crude proteins without immunoprecipitation.
Rice D14 is an unconventional hormone receptor for SLs
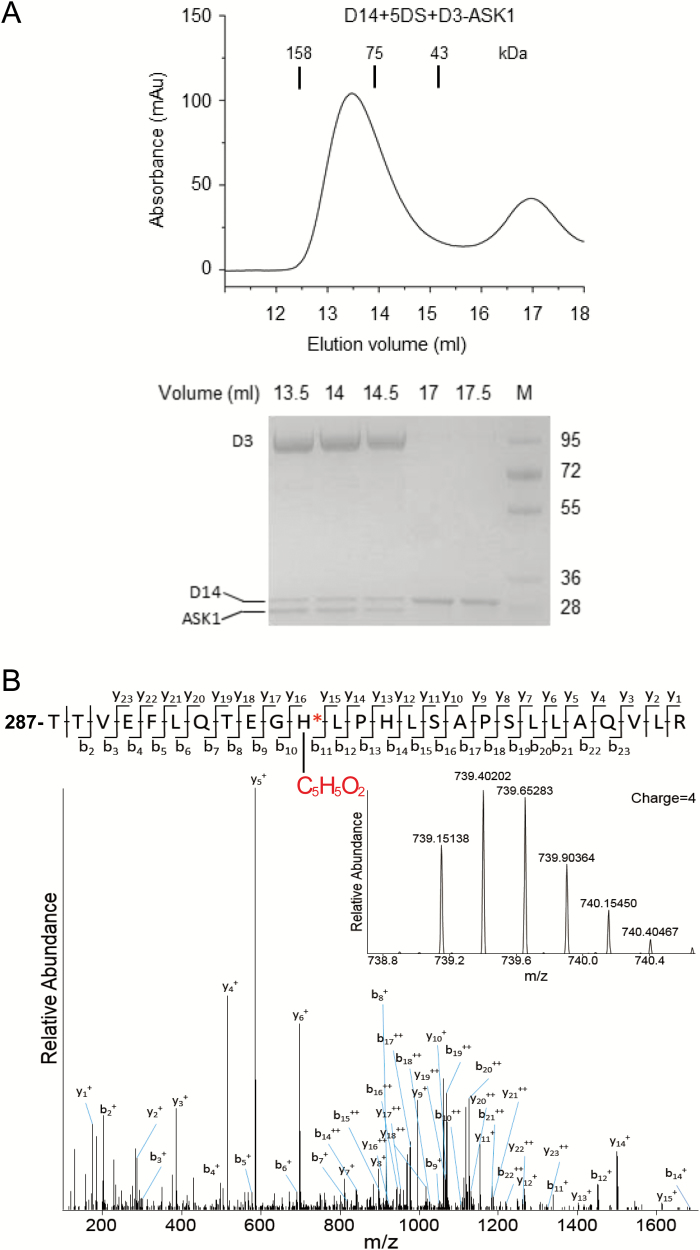
To investigate further whether rice D14, similar to AtD14, generates and covalently binds the active SL molecule CLIM, we employed the SEC approach to prepare the 5DS-induced D14–D3–ASK1 complex for MS/MS analysis. The D14–D3–ASK1 complex was eluted earlier (fraction peak ~13.4 ml) (Fig. 5A, upper panel), and then subjected to SDS–PAGE to separate D14 protein (Fig. 5A, lower panel) for further trypsin digestion followed by MS/MS analysis. Peptide matching from MS/MS spectra identified a chemically modified peptide (287-TTVEFLQTEGHLPHLSAPSLLAQVLR-312) of D14 with a molecular weight shift of 96.0211 Da on the catalytic residue H297 (Fig. 5B), which is identical to the accessional molecular weight on the corresponding histidine residue (H247) of AtD14 (Yao et al., 2016). As the control, no modified peptide was identified when D14 without 5DS treatment was subjected to MS/MS analysis (Supplementary Fig. S2).
Fig. 5.
Rice D14 generates and covalently binds the active form of SLs. (A) 5DS induced the interaction of rice D14 and D3 in the SEC assay. Upper panel: SEC analysis of the interaction between D14 and D3–ASK1 in the presence of 5DS; the elution volumes of the molecular weight markers are indicated above the peaks. Lower panel: SDS–PAGE analysis of peak fractions from the upper panel; M, molecular weight ruler (kDa). (B) Rice D14 hydrolyzed 5DS and generated the C5H5O2 modification on the catalytic residue H297. A quadruply charged peptide (287-TTVEFLQTEGHLPHLSAPSLLAQVLR-312) of D14 with the 5DS-derived C5H5O2 modification on H297 was identified by MS/MS (m/z=739.40202). The modified peptide was isolated from the trypsin digestion products of D14 in the 5DS-induced D14–D3–ASK1 complex collected in SEC (A). Labeled peaks correspond to masses of y and b ions of the peptide displayed on the top, respectively. The asterisked ‘H’ indicates the modified H297.
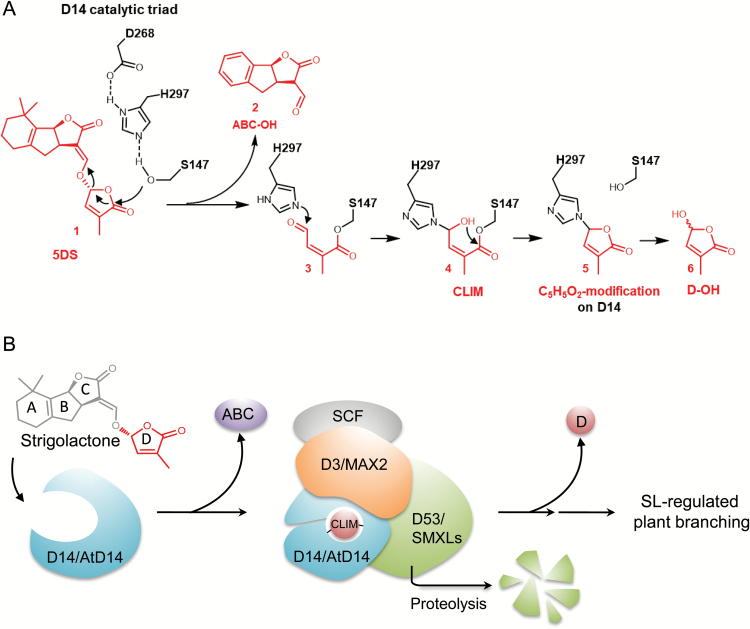
Taking advantage of the comprehensive analyses on D14-mediated SL perception in our recent work (Yao et al., 2016), we are able to deduce that this molecular weight shift of 96.0211 Da (Fig. 5B) corresponds to the chemical formula C5H4O2, which indicates covalent C5H5O2 modification on H297 of D14 (compound 5, Fig. 6A) and denotes the presence of D-ring-derived intermediate compound 4 (CLIM; Fig. 6A) as the active form of SLs in the SL-induced D14–D3 complex. Moreover, the same C5H5O2 modification on rice D14 was also detected in planta when 35Spro:D14ΔN plants were treated with 5DS (Supplementary Fig. S3).
Fig. 6.
Proposed mechanism of SL perception by rice D14. (A) Schematic diagram of a proposed rice D14-mediated hydrolysis process of 5DS. The hydrolysis of 5DS (1) is proposed to involve a nucleophilic attack by S147, which produces ABC-OH (2) and compound 3, and the generation of D-OH (6). The Nε2 atom of H297 attacks the aldehyde carbon atom of the S147-linked compound 3 to form the H297- and S147-linked linear compound 4, referred to as the covalently linked intermediate molecule (CLIM). Compound 4 initiates an intramolecular nucleophilic attack to generate the H297-linked circular compound 5, which appears as a C5H5O2 modification on H297 detected by MS/MS and denotes the existence of CLIM inside the D3-bound D14. Compound 5 would be further hydrolyzed from H297 to produce D-OH (6). A similar deduction of the AtD14-mediated (+)-GR24 hydrolysis process can be found in detail in our recent study (Yao et al., 2016). (B) A simplified model of SL perception. D14/AtD14 docks SL in the catalytic cavity, hydrolyzes SL into a D-ring-derived intermediate (CLIM), which is covalently sealed inside the catalytic center of D14/AtD14 to promote the interaction with the D3/MAX2-based SCF complex and the repressor D53/SMXLs for triggering SL-regulated plant branching (Yao et al., 2016).
Together with previous studies about D14 function in rice (Arite et al., 2009; Gao et al., 2009; Liu et al., 2009; Jiang et al., 2013; Zhou et al., 2013), our results collectively uncover the conserved function of D14 proteins in the monocot rice and the dicot Arabidopsis, and suggest that rice D14 acts as an unconventional hormone receptor to generate and perceive the active form of SLs.
Discussion
Understanding of hormone perception is central to comprehending hormone action. Biologists over the past century have established a general perception mechanism for phytohormones: receptors specifically and reversibly bind their ligands with high affinity to initiate hormone signaling, and eventually release the unchanged ligands for the next round of perception. However, recent works on SL perception in dicots (de Saint Germain et al., 2016; Yao et al., 2016, 2017) have defined Arabidopsis D14, pea RMS3, and Striga HTL7 as unconventional receptors that hydrolyze SLs into the active form of hormone (CLIM), covalently bind CLIM to trigger SL signaling, and ultimately release an inactive hydrolysis product D-OH. Here, our data suggest that D14 in the monocot rice possesses the same physiological functions as AtD14, and also acts as an unconventional hormone receptor to generate and perceive CLIM, and expectedly undergo conformational changes (Hamiaux et al., 2012; Zhao et al., 2015; Yao et al., 2016) for recruitment of signaling components (such as D3 and D53) (Jiang et al., 2013; Zhou et al., 2013), thereby triggering SL signal transduction (Fig. 6B).
It is intriguing that a glycine- and serine-rich N-terminus is present in all the examined monocot D14s but absent in those of all the tested dicots (Fig. 1), and the N-terminus-truncated rice D14 showed a much higher complementation ratio than the full-length rice D14 when expressed in the Arabidopsis mutant Atd14-5 (Supplementary Table S2). However, the underlying molecular mechanism remains to be investigated in the future. Such a glycine- and serine-rich N-terminus does not affect the D14 interaction with MAX2 or SMXL6 (Supplementary Fig. S1), and is possibly structurally flexible (Kagiyama et al., 2013). Given that D14 can be transported via phloem in rice (Kameoka et al., 2016), it would be interesting to investigate whether the additional glycine- and serine-rich N-terminal sequence of rice D14 functions as a signal peptide for D14 localization and/or transport in vivo.
Divergent features of the SL signaling pathway in monocotyledonous and dicotyledonous species are also found in the downstream signal transduction process. In the dicots Arabidopsis and pea, the gene BRANCHED1 (BRC1), which encodes a TCP transcription factor, has been demonstrated to be a key SL-responsive gene in the downstream SL signaling pathway (Aguilar-Martínez et al., 2007; Póza-Carrion et al., 2007; Mashiguchi et al., 2009; González-Grandío et al., 2013; Wang et al., 2015). However, in the monocots rice and maize, the BRC1 ortholog TEOSINTE BRANCHED1 (TB1) was not up-regulated by GR24 treatment (Minakuchi et al., 2010; Guan et al., 2012). Moreover, a recent study identified rice IPA1 (OsSPL14), a member of the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factor family (Jiao et al., 2010; Miura et al., 2010), as a direct target of the repressor D53 to participate in the SL-mediated regulation of rice tillering (Song et al., 2017). The loss-of-function mutant of IPA1 is insensitive to GR24 treatment and shows more tillers than wild-type rice plants (Song et al., 2017). Similar regulation of Triticum aestivum (Ta)D53 on TaSPL13/17 from bread wheat was also observed recently (Liu et al., 2017). However, the Arabidopsis mutant containing loss-of-function mutations in both SPL9 and SPL15 (the orthologs of the IPA1/OsSPL14 gene) still responds to the SL analog GR24 and shows reduced branching to a level similar to that of the wild type (Bennett et al., 2016). These phenomena suggest that the downstream SL signaling pathway seems not to be fully conserved between monocots and dicots. Further study is needed to better understand the elusive downstream SL signaling pathway in various plant species.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. The N-terminus of rice D14 does not affect the interaction with Arabidopsis components MAX2 or SMXL6.
Fig. S2. No modified peptide was identified when D14 without 5DS treatment was subjected to MS/MS analysis.
Fig. S3. SL can generate the C5H5O2 modification of rice D14 in planta.
Table S1. Protein identities between D14 orthologs and D14 or AtD14.
Table S2. Branch numbers of Atd14 transgenic plants with full-length or truncated rice D14.
Acknowledgements
We thank Drs Steven M. Smith (University of Tasmania) and Mark T. Waters (The University of Western Australia) for providing the Arabidopsis mutant Atd14-1, and Dr Haiteng Deng and Mr Wenhao Zhang (Tsinghua University) for help with LC-MS/MS analysis. This work was supported by the National Key R&D Program of China (grant 2016YFA0500501), the National Natural Science Foundation of China (grants 31421001 and 91635301), and grants for RY from the China Association for Science and Technology, the Postdoctoral Fellowship of Tsinghua-Peking Joint Center for Life Sciences, and the China Postdoctoral Science Foundation.
Glossary
Abbreviations:
- BRC
BRANCHED
- CLIM
covalently linked intermediate molecule
- D
DWARF
- DAD
DECREASED APICAL DOMINANCE
- GFP
green fluorescent protein
- IP
immunoprecipitation
- IPA
Ideal Plant Architecture
- MAX
MORE AXILLARY GROWTH
- MS/MS
tandem mass spectrometry
- RMS
RAMOSUS
- SCF
Skp–Cullin–F-box
- SEC
size exclusion chromatography
- SL
strigolactone
- SMXL
SUPPRESSOR OF MAX2 1-LIKE
- SPL
SQUAMOSA PROMOTER BINDING PROTEIN-LIKE
- TB
TEOSINTE BRANCHED.
References
- Abe S, Sado A, Tanaka K et al. . 2014. Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proceedings of the National Academy of Sciences, USA 111, 18084–18089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Martínez JA, Poza-Carrión C, Cubas P. 2007. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. The Plant Cell 19, 458–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K, Matsuzaki K, Hayashi H. 2005. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827. [DOI] [PubMed] [Google Scholar]
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J. 2007. DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. The Plant Journal 51, 1019–1029. [DOI] [PubMed] [Google Scholar]
- Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J. 2009. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant and Cell Physiology 50, 1416–1424. [DOI] [PubMed] [Google Scholar]
- Bennett T, Liang Y, Seale M, Ward S, Müller D, Leyser O. 2016. Strigolactone regulates shoot development through a core signalling pathway. Biology Open 5, 1806–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge CA, Ross JJ, Murfet IC. 1996. Branching in pea (Action of genes Rms3 and Rms4). Plant Physiology 110, 859–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O. 2004. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Current Biology 14, 1232–1238. [DOI] [PubMed] [Google Scholar]
- Booker J, Sieberer T, Wright W, Williamson L, Willett B, Stirnberg P, Turnbull C, Srinivasan M, Goddard P, Leyser O. 2005. MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Developmental Cell 8, 443–449. [DOI] [PubMed] [Google Scholar]
- Bu Q, Lv T, Shen H et al. . 2014. Regulation of drought tolerance by the F-box protein MAX2 in Arabidopsis. Plant Physiology 164, 424–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bythell-Douglas R, Rothfels CJ, Stevenson DWD, Graham SW, Wong GK, Nelson DC, Bennett T. 2017. Evolution of strigolactone receptors by gradual neo-functionalization of KAI2 paralogues. BMC Biology 15, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso C, Zhang Y, Jamil M et al. . 2014. Natural variation of rice strigolactone biosynthesis is associated with the deletion of two MAX1 orthologs. Proceedings of the National Academy of Sciences, USA 111, 2379–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier F, Nieminen K, Sánchez-Ferrero JC, Rodríguez ML, Chagoyen M, Hardtke CS, Cubas P. 2014. Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis. The Plant Cell 26, 1134–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn CE, Bythell-Douglas R, Neumann D, Yoshida S, Whittington B, Westwood JH, Shirasu K, Bond CS, Dyer KA, Nelson DC. 2015. Plant evolution. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science 349, 540–543. [DOI] [PubMed] [Google Scholar]
- Cook CE, Whichard LP, Turner B, Wall ME, Egley GH. 1966. Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154, 1189–1190. [DOI] [PubMed] [Google Scholar]
- Decker EL, Alder A, Hunn S et al. . 2017. Strigolactone biosynthesis is evolutionarily conserved, regulated by phosphate starvation and contributes to resistance against phytopathogenic fungi in a moss, Physcomitrella patens. New Phytologist 216, 455–468. [DOI] [PubMed] [Google Scholar]
- de Saint Germain A, Clavé G, Badet-Denisot MA et al. . 2016. An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nature Chemical Biology 12, 787–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dor E, Joel DM, Kapulnik Y, Koltai H, Hershenhorn J. 2011. The synthetic strigolactone GR24 influences the growth pattern of phytopathogenic fungi. Planta 234, 419–427. [DOI] [PubMed] [Google Scholar]
- Drummond RS, Sheehan H, Simons JL, Martínez-Sánchez NM, Turner RM, Putterill J, Snowden KC. 2011. The expression of petunia strigolactone pathway genes is altered as part of the endogenous developmental program. Frontiers in Plant Science 2, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Chen XY. 2017. Branching out. Science China Life Sciences 60, 108–110. [DOI] [PubMed] [Google Scholar]
- Foo E, Bullier E, Goussot M, Foucher F, Rameau C, Beveridge CA. 2005. The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea. The Plant Cell 17, 464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Qian Q, Liu X, Yan M, Feng Q, Dong G, Liu J, Han B. 2009. Dwarf 88, a novel putative esterase gene affecting architecture of rice plant. Plant Molecular Biology 71, 265–276. [DOI] [PubMed] [Google Scholar]
- Gobena D, Shimels M, Rich PJ, Ruyter-Spira C, Bouwmeester H, Kanuganti S, Mengiste T, and Ejeta G. 2017. Mutation in sorghum LOW GERMINATION STIMULANT 1 alters strigolactones and causes Striga resistance. Proceedings of the National Academy of Sciences, USA 114, 4471–4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB et al. . 2008. Strigolactone inhibition of shoot branching. Nature 455, 189–194. [DOI] [PubMed] [Google Scholar]
- González-Grandío E, Poza-Carrión C, Sorzano CO, Cubas P. 2013. BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. The Plant Cell 25, 834–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JC, Koch KE, Suzuki M, Wu S, Latshaw S, Petruff T, Goulet C, Klee HJ, McCarty DR. 2012. Diverse roles of strigolactone signaling in maize architecture and the uncoupling of a branching-specific subnetwork. Plant Physiology 160, 1303–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C, Gobbato E, Choi J et al. . 2015. Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science 350, 1521–1524. [DOI] [PubMed] [Google Scholar]
- Ha CV, Leyva-Gonzalez MA, Osakabe Y et al. . 2014. Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proceedings of the National Academy of Sciences, USA 111, 851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiaux C, Drummond RS, Janssen BJ, Ledger SE, Cooney JM, Newcomb RD, Snowden KC. 2012. DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Current Biology 22, 2032–2036. [DOI] [PubMed] [Google Scholar]
- Hu Q, He Y, Wang L, et al. 2017. DWARF14, a receptor covalently linked with the active form of strigolactones, undergoes strigolactone-dependent degradation in rice. Frontiers in Plant Science 8, 1935. doi:10.3389/fpls.2017.01935 [DOI] [PMC free article] [PubMed]
- Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J. 2005. Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant and Cell Physiology 46, 79–86. [DOI] [PubMed] [Google Scholar]
- Jiang L, Liu X, Xiong G et al. . 2013. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504, 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D et al. . 2010. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nature Genetics 42, 541–544. [DOI] [PubMed] [Google Scholar]
- Johnson X, Brcich T, Dun EA, Goussot M, Haurogné K, Beveridge CA, Rameau C. 2006. Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiology 142, 1014–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiyama M, Hirano Y, Mori T, Kim SY, Kyozuka J, Seto Y, Yamaguchi S, Hakoshima T. 2013. Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes to Cells 18, 147–160. [DOI] [PubMed] [Google Scholar]
- Kameoka H, Dun EA, Lopez-Obando M, Brewer PB, de Saint Germain A, Rameau C, Beveridge CA, Kyozuka J. 2016. Phloem transport of the receptor DWARF14 protein is required for full function of strigolactones. Plant Physiology 172, 1844–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapulnik Y, Delaux PM, Resnick N et al. . 2011. Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 233, 209–216. [DOI] [PubMed] [Google Scholar]
- Kohlen W, Charnikhova T, Liu Q, Bours R, Domagalska MA, Beguerie S, Verstappen F, Leyser O, Bouwmeester H, Ruyter-Spira C. 2011. Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiology 155, 974–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar T, Kohlen W, Sasse J, Borghi L, Schlegel M, Bachelier JB, Reinhardt D, Bours R, Bouwmeester HJ, Martinoia E. 2012. A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 483, 341–344. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Yao R, Ma S, Hus S, Li S, Wang Y, Yan C, Xie D, Yan J.. 2017. Efficient ASK-assisted system for expression and purification of plant F-box proteins. Plant Journal 92, 736–743. [DOI] [PubMed] [Google Scholar]
- Li S, Chen L, Li Y, Yao R, Wang F, Yang M, Gu M, Nan F, Xie D, Yan J. 2016. Effect of GR24 stereoisomers on plant development in Arabidopsis. Molecular Plant 9, 1432–1435. [DOI] [PubMed] [Google Scholar]
- Liang Y, Ward S, Li P, Bennett T, Leyser O. 2016. SMAX1-LIKE7 signals from the nucleus to regulate shoot development in Arabidopsis via partially EAR motif-independent mechanisms. The Plant Cell 28, 1581–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Wang R, Qian Q et al. . 2009. DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. The Plant Cell 21, 1512–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Cheng X, Liu P, Sun J. 2017. miR156-targeted SBP-box transcription factors interact with DWARF53 to regulate TEOSINTE BRANCHED1 and BARREN STALK1 expression in bread wheat. Plant Physiology 174, 1931–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Wu C, Fu Y, Hu G, Si H, Zhu L, Luan W, He Z, Sun Z. 2009. Identification and characterization of HTD2: a novel gene negatively regulating tiller bud outgrowth in rice. Planta 230, 649–658. [DOI] [PubMed] [Google Scholar]
- Lumba S, Holbrook-Smith D, McCourt P. 2017a The perception of strigolactones in vascular plants. Nature Chemical Biology 13, 599–606. [DOI] [PubMed] [Google Scholar]
- Lumba S, Subha A, McCourt P. 2017b Found in translation: applying lessons from model systems to strigolactone signaling in parasitic plants. Trends in Biochemical Sciences 42, 556–565. [DOI] [PubMed] [Google Scholar]
- Mashiguchi K, Sasaki E, Shimada Y, Nagae M, Ueno K, Nakano T, Yoneyama K, Suzuki Y, Asami T. 2009. Feedback-regulation of strigolactone biosynthetic genes and strigolactone-regulated genes in Arabidopsis. Bioscience, Biotechnology, and Biochemistry 73, 2460–2465. [DOI] [PubMed] [Google Scholar]
- Minakuchi K, Kameoka H, Yasuno N et al. . 2010. FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant and Cell Physiology 51, 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, Asano K, Matsuoka M, Kitano H, Ashikari M. 2010. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nature Genetics 42, 545–549. [DOI] [PubMed] [Google Scholar]
- Morris SE, Turnbull CG, Murfet IC, Beveridge CA. 2001. Mutational analysis of branching in pea. Evidence that Rms1 and Rms5 regulate the same novel signal. Plant Physiology 126, 1205–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Xue YL, Miyakawa T et al. . 2013. Molecular mechanism of strigolactone perception by DWARF14. Nature Communications 4, 2613. [DOI] [PubMed] [Google Scholar]
- Nelson DC, Scaffidi A, Dun EA, Waters MT, Flematti GR, Dixon KW, Beveridge CA, Ghisalberti EL, and Smith SM. 2011. F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 108, 8897–8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poza-Carrión C, Aguilar-Martínez JA, Cubas P. 2007. Role of TCP gene BRANCHED1 in the control of shoot branching in Arabidopsis. Plant Signaling and Behavior 2, 551–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Research 42, W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyter-Spira C, Kohlen W, Charnikhova T et al. . 2011. Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones?Plant Physiology 155, 721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi A, Waters MT, Sun YK, Skelton BW, Dixon KW, Ghisalberti EL, Flematti GR, Smith SM. 2014. Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiology 165, 1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Luong P, Huq E. 2007. The F-box protein MAX2 functions as a positive regulator of photomorphogenesis in Arabidopsis. Plant Physiology 145, 1471–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D et al. . 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons JL, Napoli CA, Janssen BJ, Plummer KM, Snowden KC. 2007. Analysis of the DECREASED APICAL DOMINANCE genes of petunia in the control of axillary branching. Plant Physiology 143, 697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden KC, Janssen BJ. 2016. Structural biology: signal locked in. Nature 536, 402–404. [DOI] [PubMed] [Google Scholar]
- Snowden KC, Simkin AJ, Janssen BJ, Templeton KR, Loucas HM, Simons JL, Karunairetnam S, Gleave AP, Clark DG, Klee HJ. 2005. The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. The Plant Cell 17, 746–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Lu Z, Yu H et al. . 2017. IPA1 functions as a downstream transcription factor repressed by D53 in strigolactone signaling in rice. Cell Research 27, 1128–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K, Booker J, Haurogné K et al. . 2003. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes and Development 17, 1469–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundappan I, Bennett T, Morffy N, Liang Y, Stanga JP, Abbas A, Leyser O, Nelson DC. 2015. SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. The Plant Cell 27, 3143–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P, Furner IJ, Ottoline Leyser HM. 2007. MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. The Plant Journal 50, 80–94. [DOI] [PubMed] [Google Scholar]
- Stirnberg P, van De Sande K, Leyser HM. 2002. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129, 1131–1141. [DOI] [PubMed] [Google Scholar]
- Toh S, Holbrook-Smith D, Stogios PJ, Onopriyenko O, Lumba S, Tsuchiya Y, Savchenko A, McCourt P. 2015. Structure–function analysis identifies highly sensitive strigolactone receptors in Striga. Science 350, 203–207. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Yoshimura M, Sato Y et al. . 2015. Parasitic plants. Probing strigolactone receptors in Striga hermonthica with fluorescence. Science 349, 864–868. [DOI] [PubMed] [Google Scholar]
- Ueda H, Kusaba M. 2015. Strigolactone regulates leaf senescence in concert with ethylene in Arabidopsis. Plant Physiology 169, 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M, Cao M, Akiyama K, Akatsu T, Seto Y, Hanada A, Li W, Takeda-Kamiya N, Morimoto Y, Yamaguchi S. 2015. Structural requirements of strigolactones for shoot branching inhibition in rice and Arabidopsis. Plant and Cell Physiology 56, 1059–1072. [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Magome H, Takeda-Kamiya N, Yamaguchi S. 2010. Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant and Cell Physiology 51, 1118–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S et al. . 2008. Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200. [DOI] [PubMed] [Google Scholar]
- Wang L, Smith SM. 2016. Strigolactones redefine plant hormones. Science China Life Sciences 59, 1083–1085. [DOI] [PubMed] [Google Scholar]
- Wang L, Wang B, Jiang L, Liu X, Li X, Lu Z, Meng X, Wang Y, Smith SM, Li J. 2015. Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-Like SMXL repressor proteins for ubiquitination and degradation. The Plant Cell 27, 3128–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Brewer PB, Bussell JD, Smith SM, Beveridge CA. 2012a The Arabidopsis ortholog of rice DWARF27 acts upstream of MAX1 in the control of plant development by strigolactones. Plant Physiology 159, 1073–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Gutjahr C, Bennett T, Nelson DC. 2017. Strigolactone signaling and evolution. Annual Review of Plant Biology 68, 291–322. [DOI] [PubMed] [Google Scholar]
- Waters MT, Nelson DC, Scaffidi A, Flematti GR, Sun YK, Dixon KW, Smith SM. 2012b Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139, 1285–1295. [DOI] [PubMed] [Google Scholar]
- Waters MT, Smith SM. 2013. KAI2- and MAX2-mediated responses to karrikins and strigolactones are largely independent of HY5 in Arabidopsis seedlings. Molecular Plant 6, 63–75. [DOI] [PubMed] [Google Scholar]
- Whelan S, Goldman N. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Molecular Biology and Evolution 18, 691–699. [DOI] [PubMed] [Google Scholar]
- Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG. 2001. ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. The Plant Cell 13, 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Furusawa S, Nagasaka S, Shimomura K, Yamaguchi S, Umehara M. 2014. Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta 240, 399–408. [DOI] [PubMed] [Google Scholar]
- Yan J, Li H, Li S, Yao R, Deng H, Xie Q, Xie D. 2013. The Arabidopsis F-box protein CORONATINE INSENSITIVE1 is stabilized by SCFCOI1 and degraded via the 26S proteasome pathway. The Plant Cell 25, 486–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R, Li J, Xie D. 2017. Recent advances in molecular basis for strigolactone action. Science China Life Sciences. doi:10.1007/s11427-017-9195-x. [DOI] [PubMed] [Google Scholar]
- Yao R, Ming Z, Yan L et al. . 2016. DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536, 469–473. [DOI] [PubMed] [Google Scholar]
- Yao R, Wang F, Ming Z, Du X, Chen L, Wang Y, Zhang W, Deng H, Xie D. 2017. ShHTL7 is a non-canonical receptor for strigolactones in root parasitic weeds. Cell Research 27, 838–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Shirasu K. 2009. Multiple layers of incompatibility to the parasitic witchweed, Striga hermonthica. New Phytologist 183, 180–189. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yu H, Ma B et al. . 2017. A natural tandem array alleviates epigenetic repression of IPA1 and leads to superior yielding rice. Nature Communications 8, 14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LH, Zhou XE, Wu ZS et al. . 2013. Crystal structures of two phytohormone signal-transducing α/β hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Research 23, 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LH, Zhou XE, Yi W et al. . 2015. Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3. Cell Research 25, 1219–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Lin Q, Zhu L et al. . 2013. D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504, 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.