Abstract
Background
Genomic analysis of plasma cell-free DNA is transforming lung cancer care; however, available assays are limited by cost, turnaround time, and imperfect accuracy. Here, we study amplicon-based plasma next-generation sequencing (NGS), rather than hybrid-capture-based plasma NGS, hypothesizing this would allow sensitive detection and monitoring of driver and resistance mutations in advanced non-small cell lung cancer (NSCLC).
Patients and methods
Plasma samples from patients with NSCLC and a known targetable genotype (EGFR, ALK/ROS1, and other rare genotypes) were collected while on therapy and analyzed blinded to tumor genotype. Plasma NGS was carried out using enhanced tagged amplicon sequencing of hotspots and coding regions from 36 genes, as well as intronic coverage for detection of ALK/ROS1 fusions. Diagnostic accuracy was compared with plasma droplet digital PCR (ddPCR) and tumor genotype.
Results
A total of 168 specimens from 46 patients were studied. Matched plasma NGS and ddPCR across 120 variants from 80 samples revealed high concordance of allelic fraction (R2 = 0.95). Pretreatment, sensitivity of plasma NGS for the detection of EGFR driver mutations was 100% (30/30), compared with 87% for ddPCR (26/30). A full spectrum of rare driver oncogenic mutations could be detected including sensitive detection of ALK/ROS1 fusions (8/9 detected, 89%). Studying 25 patients positive for EGFR T790M that developed resistance to osimertinib, 15 resistance mechanisms could be detected including tertiary EGFR mutations (C797S, Q791P) and mutations or amplifications of non-EGFR genes, some of which could be detected pretreatment or months before progression.
Conclusions
This blinded analysis demonstrates the ability of amplicon-based plasma NGS to detect a full range of targetable genotypes in NSCLC, including fusion genes, with high accuracy. The ability of plasma NGS to detect a range of preexisting and acquired resistance mechanisms highlights its potential value as an alternative to single mutation digital PCR-based plasma assays for personalizing treatment of TKI resistance in lung cancer.
Keywords: amplicon-based next generation sequencing, plasma genotyping, circulating tumor DNA, resistance mechanisms, fusion genes, EGFR
Key Message
We report compelling accuracy using tagged amplicon sequencing for the detection of a full range of genotypes, including fusion genes, in cell-free DNA from advanced non-small-cell lung cancer patients. This alternative to hybrid-capture approaches has the ability for early detection of diverse mechanisms of resistance to EGFR-TKIs, including acquired resistance mutations and amplifications.
Introduction
Genotype-directed treatment of non-small cell lung cancer (NSCLC) has led to dramatic improvement in the management of selected patients harboring a targetable oncogenic driver [1]. The limited availability of tissue to test an increasing number of potentially actionable genotypes and a better understanding of druggable mechanisms of resistance [2] have created a need for a rapid, repeatable and noninvasive access to the tumor biology throughout treatment. Genotyping of plasma cell-free DNA (cfDNA) is already an established diagnostic tool that can guide rapid initiation of TKI therapy in EGFR-mutant NSCLC [3, 4], avoiding some invasive biopsies. However, the most established assays are digital PCR-based, detecting mutations at only a single site in a predefined gene.
Unlike digital PCR plasma genotyping, next-generation sequencing (NGS) of cfDNA has the potential to more broadly assess the molecular profile of the tumor. Hybrid capture-based NGS of plasma cfDNA has already been well evaluated in NSCLC [5–7]. While this technical approach permits sequencing of dozens of genes and detection of complex variants, including rearrangements, concordance with matched tumor genotyping has been suboptimal in some series [5, 6]. Amplicon-based NGS is a well-established alternate technology, which uses target gene enrichment by PCR with a set of primers for exons or hotspots of selected genes [8], and is the basis for the tumor NGS assay that recently received approval by the US FDA (Oncomine™ Dx Target, ThermoFisher). While this technology is less well studied for NGS of cfDNA, where the levels of input DNA (and tumor fraction) are usually very low, we hypothesized barcoded amplicon-based NGS would provide excellent sensitivity with limited sequencing artifact, and would represent a compelling alternative to hybrid capture-based plasma NGS.
Methods
Patients were identified with stage IIIB/IV, progressive NSCLC harboring a known tumor genotype and consented for plasma collection as part of two ongoing correlative studies at our institution. Plasma was collected for analysis before receipt of targeted therapy; when feasible, plasma was also collected at the initial toxicity evaluation and with restaging scans until development of resistance. We first studied a cohort of 30 patients with EGFR-mutant NSCLC and T790M-positive resistance receiving osimertinib; upon successful proof of principle, we then additionally studied a cohort of 16 patients harboring other rare targetable genotypes, totaling 168 time points from 46 subjects. Plasma analyses were carried out blinded to clinical information such as tumor genotype. Sensitivity and specificity for plasma droplet digital PCR (ddPCR) and plasma NGS assays were calculated using clinically carried out tumor genotyping as reference standard; tumor genotyping was carried out using hybrid-capture NGS whenever possible [9]. Concordance of variant allelic fraction (AF) between plasma ddPCR and NGS was calculated using Kendall concordance coefficient.
Droplet digital PCR
Plasma genotyping using ddPCR was carried out for all cases with EGFR-mutant NSCLC, as well as to validate selected non-EGFR hotspot mutations based on assay availability (e.g. KRAS, PIK3CA, BRAF mutations). ddPCR was carried out at the Belfer Center for Applied Cancer Science as described previously [3]. Remaining aliquots of plasma were allocated for plasma NGS, requiring a minimum of 1–2 mL of plasma or a corresponding quantity of extracted cfDNA.
Plasma NGS
Amplicon-based plasma NGS was carried out by Inivata (Morrisville, NC), using InVisionTM, an enhanced version of TAm-Seq technology, based on methods previously described [10–12]. Thirty-six cancer-related genes were sequenced using gene-specific primers designed to hotspots and entire coding regions of interest (supplementary Figure S1, available at Annals of Oncology online). Extracted cfDNA is first quantified by digital PCR targeting a 108 bp region of the ribonuclease P/MRP subunit p30 (RPP30) gene [13]. Next generation sequencing libraries are then prepared from 2000 to 16 000 amplifiable copies of the genome (∼6.6 to 53 ng of amplifiable DNA) using a two-step PCR amplification process incorporating replicate and patient-specific barcodes and Illumina sequencing adaptors. In the first step PCR reaction, amplicons ranging from 73 to 155 bp are generated which were designed and optimized for the DNA fragment size found in circulation. Each sample is analyzed multiple times allowing the identification of false-positive and true-positive calls [10, 12]. After further clean-up using SPRI beads, samples are quantified and pooled to generate a normalized library of 12 nM. About 1.8 pM libraries are sequenced on the Illumina NextSeq with 5% PhiX added to monitor sequencing performance. A minimum Phred quality score of 30 for each base was required for inclusion in the analytics. Sequencing files were analyzed using Inivata’s proprietary Somatic Mutation Analysis (ISoMA) pipeline.
In a subset of cases known to harbor oncogenic fusions, a separate aliquot of plasma cfDNA was tested using a novel technology designed to identify ALK and ROS1 breakpoints. The novel PCR-based assay has been designed to capture all major EML4-ALK variants in NSCLC, encompassing 95% of variants found in COSMIC (version 78). The panel also captures 90% of ROS1 fusions in NSCLC as described in the COSMIC database and identifies the breakpoints occurring between CD74-ROS1, SLC34A2-ROS1, SDC4-ROS1 and EZR-ROS1. The ALK and ROS1 assay covers ∼50 kb of intronic and exonic sequences, allowing the identification of precise DNA breakpoints in regions that are frequently re-arranged. Libraries were prepared and sequenced on the Illumina NextSeq 500 as described above.
Sequencing files were analyzed using Inivata’s proprietary FUSP pipeline, which identifies specific DNA sequences brought together creating the fusions outlined above.
Results
Detecting known EGFR mutations
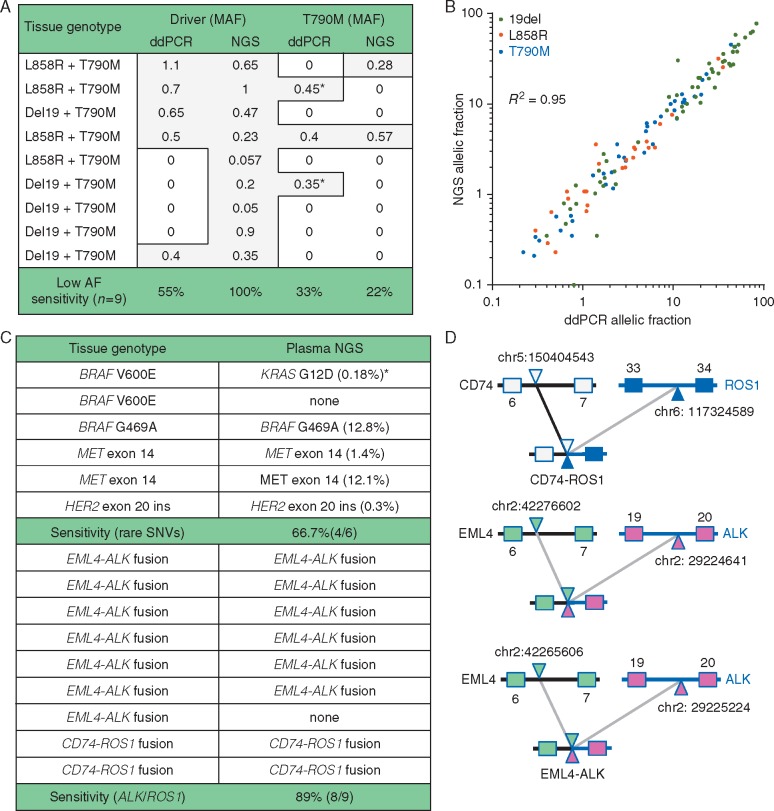
Using tissue genotyping as a reference standard, and ddPCR for orthogonal validation, sensitivity of plasma NGS was analyzed across 30 cases with EGFR-mutant NSCLC and acquired T790M. Sensitivity for detection of the driver EGFR mutation was 100% (30/30) with plasma NGS and 87% (26/30) for plasma ddPCR (P = 0.11; 10/11 L858R, 16/19 exon 19 deletion). Sensitivity for the detection of T790M was 77% (23/30) for plasma NGS and 80% (24/30) for ddPCR. Discordance was only seen at low AF, below 1.1% (Figure 1A), where plasma NGS detected four driver mutations missed with ddPCR. Two T790M mutations detected with ddPCR and not with NGS were, retrospectively, below our ddPCR threshold for clinical reporting and could have been false positives (supplementary Figure S2, available at Annals of Oncology online). Quantitative concordance of AF between NGS and ddPCR was excellent across 120 variants (from 80 specimens) positive for an EGFR mutation with both assays (R2 = 0.95, Figure 1B).
Figure 1.
(A) Focusing on nine cases of EGFR-mutant non-small cell lung cancer (NSCLC) with the lowest tumor DNA shed (< 1.1% AF), sensitivity appears better with amplicon-based plasma NGS compared with plasma ddPCR. *Two T790M mutations were detected with ddPCR but not NGS, though the ddPCR signal was below the level for clinical reporting and may have been a false positive (supplementary Figure S2, available at Annals of Oncology online). AF, allelic fraction. (B) Quantitative concordance was high (R2 = 0.95) across 120 EGFR variants from 80 specimens detected both with plasma NGS and plasma ddPCR. (C) Detection of a range of fusions and rare genotypes using amplicon-based plasma NGS. One apparent false positive (*) secondarily tested negative by ddPCR for both KRAS and BRAF mutations. (D) Three examples where fusion detection permits the determination of fusion partner and breakpoint.
Detection of rare variants and fusion genes
Studying nine cases with known ALK or ROS1 fusions, sensitivity of plasma NGS was 89% (6/7 EML4-ALK, 2/2 CD74-ROS1; Figure 1C and D); the one missed ALK case was the single patient studied with stage IIIB disease. Studying six cases with other mutations in the kinase domain (two MET splice mutation, three BRAF mutations and one HER2 exon 20 insertion), four were correctly identified (Figure 1C). Two cases of BRAF V600E in stage IV patients were undetectable with plasma NGS, and additionally were found to be undetectable on ddPCR. Interestingly one patient with a BRAF V600E mutation on tumor genotyping (RT-PCR) instead had a KRAS G12D mutation detected on plasma NGS (0.2% AF); this patient was a heavy smoker who did not respond to BRAF inhibitor therapy.
Specificity across other non-driver variants
To study specificity, we studied 19 cases with tumor NGS available (8 pre-osimertinib, 5 post-osimertinib and 6 pre-treatment specimens with other rare genotypes). For this analysis, we excluded each patient’s driver oncogene, to avoid acquired resistance mutations, and limited our analysis to genes covered by both tissue and plasma NGS panels. Composite specificity-by-gene was 99.5% across 665 genes sequenced, with 3 false positives (Table 1). First, one PIK3CA E545K mutation was found on plasma NGS at low AF (0.6%) but not in tumor; the mutation was confirmed in cfDNA by plasma ddPCR at the same AF (0.6%). Second, three point mutations in CTNNB1 were found at low AF (S37F 0.3%, S45C 1% and S45F 0.7%) on plasma NGS following osimertinib; corresponding tissue NGS did not reveal these, though tumor content may have been suboptimal (TP53 H193R mutation was found at 10% in tissue versus 30% in plasma). Third, an IDH1 R132H mutation was detected at low AF (0.3%) on plasma NGS but not in the corresponding tumor NGS; this variant recurred at multiple timepoints at a stable AF in this patient’s cfDNA, suspicious for clonal hematopoiesis (supplementary Figure S3, available at Annals of Oncology online) [14].
Table 1.
Comparison of 19 cases with matched pretreatment (n = 14) or post-treatment (n = 5) tumor NGS
| Driver | Pre-TKI tissue NGS | Pre-TKI plasma NGS |
|---|---|---|
| EGFR | TP53 A161T (55%) | None |
| EGFR | TP53 R110L (56%) | TP53 R110L (7.5%) |
| EGFR | TP53 166_167GA>A (64%) | TP53 frameshift (2.3%) |
| EGFR | None | None |
| EGFR | TP53 F134L (37%) | TP53 F134L (6.56%) |
| EGFR | None | PIK3CA E545K (0.6%) |
| EGFR | TP53 L43* (32%) | TP53 L43 (7.97%) |
| EGFR | TP53 C242F (76%) | TP53 C242F (7.78%) |
| BRAF | STK11 D343N (40%) | STK11 D343N (50%) |
| MET | TP53 R248W | TP53 R248W (0.6%) |
| ROS1 | None | None |
| ROS1 | None | None |
| ALK | None | None |
| ALK | TP53 R337C | TP53 R337C (1.2%) |
| Driver | Post-TKI Tissue NGS | Post-TKI Plasma NGS |
| EGFR | TP53 R282W (44%) | TP53 R282W (27%) |
| EGFR | KRAS Q61K (30%) | KRAS Q61K (2.8%) |
| TP53 Q104* (53%) | TP53 Q104* (12%) | |
| EGFR | None | None |
| EGFR | TP53 H193R (MAF 10%) | TP53 H193R (27%) |
| CTNNB1 (S37F 0.3%, S45C 1%, S45F 0.7%) | ||
| EGFR | TP53 del (MAF 53%) | TP53 frameshift (MAF 1.21%) |
| IDH1 R132H (0.33%) | ||
Limited to non-driver variants covered by the NGS panel (supplementary Figure S1, available at Annals of Oncology online), 3 plasma NGS-positive/tissue NGS-negative discordant results were seen (indicated in bold, 3/684 genes sequenced, specificity 99.6%).
Detection of resistance mechanisms using plasma NGS
We studied resistance in 25 osimertinib-treated subjects with detectable EGFR driver mutations with plasma NGS at the time of resistance. Fifteen patients (60%) lost T790M at resistance (Table 2), and four of them had a non-EGFR resistance mutation identified: one PIK3CA E545K mutation (0.13% AF), two BRAF V600E mutations (12.5% AF, 0.4% AF) and one KRAS G12S mutation (0.2% AF). Gene amplifications were also detected at resistance in four patients (Figure 2): two HER2 amplifications (one occurring concomitantly with BRAF V600E), one MET amplification and one FGFR1 amplification. Ten patients maintained EGFR T790M at resistance, eight of whom acquired a tertiary EGFR C797S resistance mutation. In three patients, two C797S variants (c.2889T>A and c.2390G>C) were present at resistance at different AF (Table 2). One of these three patients additionally acquired a novel EGFR Q791P mutation (2.6% AF; supplementary Figure S4, available at Annals of Oncology online). This mutation was confirmed in cfDNA using another amplicon-based NGS approach (QIAseq DNA Targeted Lung Panel, Qiagen; 2% AF). Interestingly, two patients with maintained T790M additionally acquired canonical KRAS mutations (KRAS G13D 0.2% AF; KRAS Q61K 2.8% AF).
Table 2.
Detection of acquired resistance mechanisms to osimertinib at resistance [limited to patients with a resistance mechanism identified (15/25)]
| Driver at resistance | T790M at resistance | Resistance, post-osimertinib NGS (beside EGFR driver and T790M) |
|---|---|---|
| Del19 63.4% | Lost | BRAF V600E 12.4% |
| HER2 amp | ||
| Del19 2.4% | Lost | BRAF V600E 0.4% |
| Del19 19% | Lost | PIK3CA E545K 1.2% |
| Del19 12% | Lost | MET amp |
| Del19 37.9% | Lost | HER2 amp |
| Del19 1.8% | Lost | FGFR1 amp |
| L858R 0.21% | Lost | KRAS G12S 0.2% |
| Driver at resistance | T790M at resistance | Resistance, post-osimertinib NGS (beside EGFR driver and T790M) |
| Del19 43% | 46.4% | EGFR C797S 26.6% |
| Del19 15.2% | 11% | EGFR C797S 7.9% |
| KRAS Q61K 2.8% | ||
| Del19 27.4% | 11.2% | EGFR C797S 0.4% |
| PTEN Y27C 21.9% | ||
| KRAS G13D 0.25% | ||
| L858R 7.6% | 7.3% | EGFR C797S T_A 1.3% |
| EGFR C797S G_C 1.1% | ||
| L858R 3.2% | 3.6% | EGFR C797S T_A 2.7% |
| EGFR C797S G_C 1% | ||
| EGFR Q791P A_C 2.6% | ||
| del19 37.7% | 18.3% | BRAF V600E 0.4% |
| EGFR C797S T_A 13.5% | ||
| del19 12% | 7% | EGFR C797S T_A 2.9% |
| del19 31.6% | 9.2% | EGFR C797S T_A 6.2% |
| EGFR C797S G_C 0.9% | ||
Top: 7/15 patients with acquired resistance to osimertinib and loss of T790M had a detectable mechanism of resistance. Bottom: 8/10 patients with acquired resistance to osimertinib and maintained T790M had one or multiple mechanisms of resistance to osimertinib, all of them having one or multiple tertiary EGFR mutation(s).
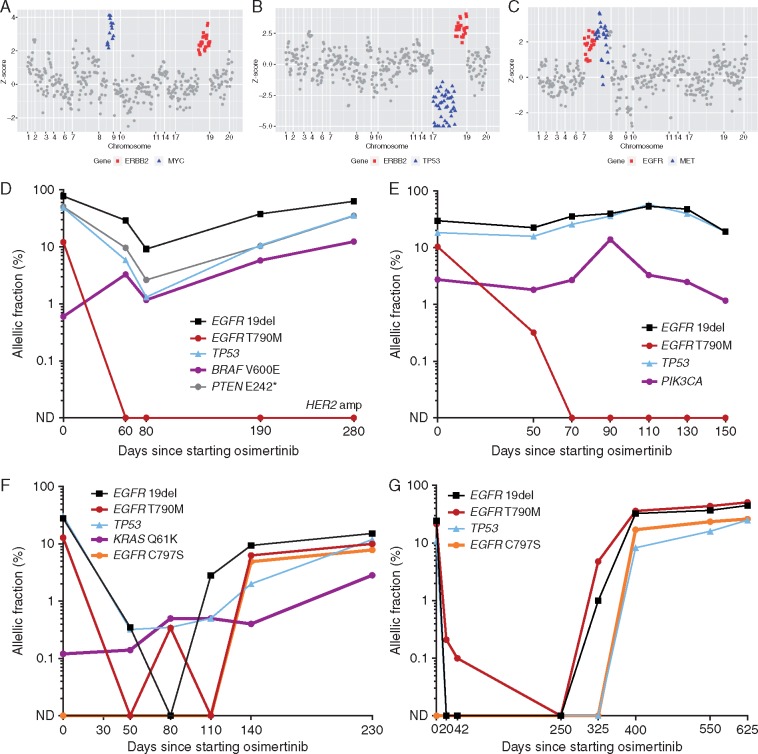
Figure 2.
(A and B) Acquired gene amplifications detected in plasma cfDNA at the time of resistance to osimertinib. (A) Acquired ERBB2 and MYC amplification (driver AF 63%). (B) Acquired ERBB2 amplification and TP53 deletion (driver AF 38%). (C) EGFR amplification and acquired MET amplification (driver AF 22%). (D–G) Early detection of mechanisms of resistance to osimertinib through serial NGS of cfDNA in four patients. (D and E) Two cases with mixed response in plasma (complete clearance of T790M, incomplete response of the driver) had loss of T790M at the time of resistance. Possible competing resistance mutations (D: BRAF V600E; E: PIK3CA E545K) coexisting with T790M at baseline can be detected pretreatment. (F and G) In two patients with maintained T790M at resistance, serial plasma NGS can detect acquired tertiary EGFR mutations several months before clinical progression (F: 3 months; G: 7.5 months). In one patient (F), plasma NGS could also detect a competing KRAS Q61K mutation at low AF at baseline that with increased AF at resistance.
Resistance genotyping was also piloted in seven specimens from patients with rare genotypes treated with various TKIs (supplementary Table S1, available at Annals of Oncology online). A mechanism of resistance could only be detected in one ALK-positive case (ALK C1156Y, 0.55% AF) after treatment with crizotinib. One ROS1 G2032R acquired resistance mutation, detected in tissue NGS in a ROS1 case after crizotinib, was not detected in plasma.
Early detection of resistance through serial plasma NGS
At least 3 serial plasma specimens (up to 8) were studied for 25 subjects treated with osimertinib, to pilot the early detection of resistance mechanisms. In four cases, a competing resistance mutation (KRAS Q61K, 2 BRAF V600E, PIK3CA E545K) could be detected pretreatment by plasma NGS. In these four cases, a complete and rapid clearance of the T790M clone was seen without immediate plasma clearance of the driver EGFR mutation (Figure 2). No cases of EGFR C797S could be detected pre-osimertinib, though monitoring multiple timepoints on therapy revealed this mutation can be seen multiple months before clinical progression (Figure 2).
Discussion
In this blinded clinical validation, we demonstrate the ability of amplicon-based plasma NGS to sensitively detect a wide range of molecular alterations, including chromosomal rearrangements, in advanced NSCLC. Few prior studies have aimed to clinically validate amplicon-based NGS of cfDNA. Couraud et al. tested a 12-gene panel on the IonTorrent platform and found an overall sensitivity of 58% [15]. The first generation of the TamSeq technology was developed by Forshew et al. on a dilution series of circulating DNA containing increasing frequencies of a rare allele, using a 48-primer set covering coding regions and hotspots in 6 genes. It was then validated using plasma samples from metastatic ovarian cancer patents. This foundational work reported high sensitivity, specificity and quantitative concordance with digital PCR, and also the ability to follow the subclonal evolution of tumors in a limited number of patients [10]. We confirm in an NSCLC population the high sensitivity of the InVisionTM assay, matching (and in some cases exceeding) the sensitivity of plasma ddPCR. This high sensitivity, combined with accurate quantification and an ability to detect a full spectrum of genomic variants (something difficult to do with PCR assays), makes this NGS-based approach a compelling alternative to ddPCR for detection of T790M and other actionable mutations.
We also report for the first time the ability of a novel amplicon-sequencing assay to detect gene fusions, with high sensitivity in cfDNA; the one missed ALK case was a patient with stage IIIB disease. In contrast, existing data on hybrid-capture NGS have suggested suboptimal sensitivity (54%) for the detection of EML4-ALK fusions [16]. Our team has also previously reported the detection of fusions genes (2/3 ALK, 2/3 RET, 2/2 ROS1) using a bias-corrected, targeted cfDNA NGS, detecting breakpoints and fusions partners using a single assay [7]. Further prospective evaluation is needed to clarify the potential sensitivity advantages of the different plasma NGS approaches for low AF mutations and the full spectrum of targetable fusions.
Though NGS of tumor tissue may be an imperfect reference standard in patients with drug resistance, our data nonetheless suggest compelling specificity with the InVision assay, which is reassuring given the plasma-positive, tumor-negative discordance seen at times with hybrid-capture NGS [6]. We identified four potential false positives—one we confirmed with plasma ddPCR, one is consistent with CHIP, and one we believe is likely due to subclonal resistance heterogeneity [14]. The only concerning false positive was a KRAS G12D mutation found at low AF (0.2%), which was not detectable with plasma ddPCR, but consistent with the patient’s clinical presentation, and this case highlights the challenges of validating assays which are potentially more sensitive than established validated assays. Still, the lack of any false positives for EGFR driver mutations highlights that targetable mutations, even when found at a very low AF, can be considered actionable when found with a well validated assay.
Our data highlight the potential value of amplicon-based plasma NGS for characterizing treatment resistance in NSCLC. In patients treated with osimertinib, we could detect common mechanisms of resistance to EGFR-TKIs [2], including expected point mutations, expected gene amplifications (MET, HER2), and a novel tertiary EGFR mutation (Q791P), which was cross-validated by another sequencing approach. Surprisingly, we found three low level KRAS mutations (one which was confirmed in tumor and has been reported previously), suggesting this may be a recurring mechanism of resistance to osimertinib. Serial plasma NGS also offers potential insights—for example in some cases with an incomplete response in the driver EGFR mutation (which has been suggested previously to indicate poorer treatment outcomes [17]), we could detect early presence of a coexistent resistance mutation. The ability of plasma NGS to detect resistance mutations coexistent with T790M as well as detecting a range of tertiary EGFR mutations makes it a potentially powerful alternative to established PCR-based assays for resistance genotyping. Furthermore, the detection in a few cases (Figure 2) of resistance mutations at low AF that overgrow the T790M on osimertinib therapy supports the potential clinical value of such low AF resistance mutations, and deserves further study.
Conclusion
In conclusion, we demonstrate herein the ability of amplicon-based NGS to detect with a wide range of targetable genotypes in advanced NSCLC, including high accuracy for point mutations and indels, and compelling initial data for gene fusions and CNVs. This approach also permits early detection of resistance mechanisms during treatment, making it a potentially valuable tool to guide early modifications of targeted therapies. Amplicon-based plasma NGS has attractive sensitivity and specificity and deserves further study as an alternative to hybrid-capture approaches.
Funding
Supported in part by the Expert Miracles Foundation (no grant number applies), the Damon Runyon Cancer Research Foundation (no grant number applies), the National Cancer Institute at the National Institutes of Health (R01–CA135257), the U.S. Department of Defense (LCRP Career Development Award) (LC130385), the Anna Fuller Fund (no grant number applies), the Stading Younger Cancer Research Foundation Thoracic Oncology Fund (no grant number applies), and NCI Cancer Clinical Investigator Team Leadership Award 591 (P30 CA006516 supplement). NG received an award from Fond de Dotation pour la Recherche en Santé Respiratoire and Fondation du Souffle contre les Maladies Respiratoires (no grant number applies).
Disclosure
CPP received honoraria form Astra Zeneca, Bio-Rad and Clovis and is part of the Advisory Board of Dropworks. GRO received consulting fees from AstraZeneca, Ariad/Takada and Inivata, and honoraria from Bio-Rad, Chugai, Guardant and Sysmex. GJ, VP, KH and JFB are employees and share-holders of Inivata Ltd. Inivata Ltd commercializes assays based on the technology described in this paper. NG received an award from Fond de Dotation pour la Recherche en Santé Respiratoire and Fondation du Souffle contre les Maladies Respiratoires. All remaining authors have declared no conflicts of interest.
Supplementary Material
References
- 1. Barlesi F, Mazieres J, Merlio J-P. et al. Routine molecular profiling of patients with advanced non-small cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet 2016; 387(10026): 1415–1426. [DOI] [PubMed] [Google Scholar]
- 2. Yu HA, Arcila ME, Rekhtman N. et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013; 19(8): 2240–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sacher AG, Paweletz C, Dahlberg SE. et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol 2016; 2(8): 1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Oxnard GR, Thress KS, Alden RS. et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small cell lung cancer. J Clin Oncol 2016; 34(28): 3375–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thompson JC, Yee SS, Troxel AB. et al. Detection of therapeutically targetable driver and resistance mutations in lung cancer patients by next-generation sequencing of cell-free circulating tumor DNA. Clin Cancer Res 2016; 22(23): 5772–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schwaederlé MC, Patel SP, Husain H. et al. Utility of genomic assessment of blood-derived circulating tumor DNA (ctDNA) in patients with advanced lung adenocarcinoma. Clin Cancer Res 2017; 23(17): 5101–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Paweletz CP, Sacher AG, Raymond CK. et al. Bias-corrected targeted next-generation sequencing for rapid, multiplexed detection of actionable alterations in cell-free DNA from advanced lung cancer patients. Clin Cancer Res 2016; 22(4): 915–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chang F, Li MM.. Clinical application of amplicon-based next-generation sequencing in cancer. Cancer Genet 2013; 206(12): 413–419. [DOI] [PubMed] [Google Scholar]
- 9. Sholl LM, Do K, Shivdasani P. et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight 2016; 1(19): e87062.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Forshew T, Murtaza M, Parkinson C. et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 2012; 4(136): 136ra68.. [DOI] [PubMed] [Google Scholar]
- 11. Gale D, Plagnol V, Lawson A. et al. Abstract 3639: Analytical performance and validation of an enhanced TAm-Seq circulating tumor DNA sequencing assay. AACR Cancer Res 2016. doi:10.1158/1538-7445.AM2016-3639. [Google Scholar]
- 12. Rosenfeld N, Forshew T, Marass F, Murtaza M. A method for detecting a genetic variant. World Intellectual Property Organization patent WO2016009224A1 (2016).
- 13. Dawson S-J, Tsui DWY, Murtaza M. et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013; 368(13): 1199–1209. [DOI] [PubMed] [Google Scholar]
- 14. Zhang BM, Aleshin A, Lin CY. et al. IDH2 mutation in a patient with metastatic colon cancer. N Engl J Med 2017; 376(20): 1991–1992. [DOI] [PubMed] [Google Scholar]
- 15. Couraud S, Vaca-Paniagua F, Villar S. et al. Noninvasive diagnosis of actionable mutations by deep sequencing of circulating free DNA in lung cancer from never-smokers: a proof-of-concept study from BioCAST/IFCT-1002. Clin Cancer Res 2014; 20(17): 4613–4624. [DOI] [PubMed] [Google Scholar]
- 16. Cui S, Zhang W, Xiong L. et al. Use of capture-based next-generation sequencing to detect ALK fusion in plasma cell-free DNA of patients with non-small cell lung cancer. Oncotarget 2017; 8(2): 2771–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oxnard GR, Hu Y, Tracy P. et al. Overgrowth of competing resistance mechanisms, such as an acquired KRAS mutation, underlies a poor prognosis subtype of acquired resistance to osimertinib in T790M-positive NSCLC. AACR; Cancer Res 2017; 77(13 Suppl): Abstract nr 4112. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.