Abstract
Background
The genetic associations with trajectories of body fatness over the life course remain unknown.
Methods
We used a group-based modelling approach to identify trajectories of body fatness from age 5 years up to 65 for 7277 women from the Nurses’ Health Study and 4645 men from the Health Professionals Follow-up Study. We created a genetic risk score (GRS) based on 97 variants associated with adulthood body mass index (BMI) and estimated its association with trajectories using logistic regression.
Results
We identified four distinct trajectories: lean-medium, medium-medium, lean-heavy and medium-heavy. The GRS increased across the four groups in that order (P < 0.001); 47% of women and 45% of men in the first decile of the GRS were in the lean-medium group, and these proportions reduced to 26% and 28%, respectively, for the highest decile. The corresponding proportions in the medium-heavy group were 8% and 5%, increasing to 21% and 14%, respectively. For women, compared with the odds of being in the lean-medium group, a 10-allele increment in the GRS was associated with a 40% [95% confidence interval (CI), 27–54%], 43% (30–58%), and 115% (91–143%) increase in the odds of being in the medium-medium, lean-heavy and medium-heavy groups, respectively. For men, the corresponding increases in the odds were 26% (12–42%), 27% (13–43%), and 81% (53–115%), respectively.
Conclusions
Individuals with genetic variants for adulthood BMI were more likely to maintain a heavy body shape and gain weight throughout life. These findings support a persistent effect of genetic variants on body fatness across the lifespan.
Keywords: Genetic variants, trajectory analysis, obesity, life course epidemiology
Key Messages
This longitudinal study found that genetic variants for adulthood BMI were associated with the different trajectories of body fatness across the lifespan.
Individuals with more genetic BMI variants were more likely to maintain a heavy body shape and gain weight throughout life.
These findings provide evidence for a persistent effect of genetic variants on body fatness across the lifespan.
Introduction
Despite evidence for the tracking of childhood weight status into adulthood,1 there is considerable individual variation in the shape of trajectories of body fatness across the lifespan.2,3 For example, for persons who start with a lean body shape in childhood, some maintain leanness throughout adulthood whereas others gain substantial weight and become obese. These different trajectories have been found to predict the subsequent risk for chronic diseases and mortality.2–4 Therefore, understanding the determinants for the heterogeneity in the lifetime trajectories of body fatness has important biological and public health implications. It may not only shed light on the aetiology of obesity, but also facilitate identification of individuals at high risk for obesity and development of tailored strategies for prevention of obesity and its comorbidities.
Twin studies indicate that genetic factors explain 31–90% of the variation in body mass index (BMI).5 A recent meta-analysis of genome-wide association studies (GWAS) has discovered 97 common genetic variants that are associated with higher BMI for adults.6 Many of the implicated genes are highly expressed in the central nervous system, and might have a persistent effect on the regulation of body mass throughout lifetime.6–8 Therefore, studies have begun to interrogate the importance of genetic susceptibility variants across the life course. For example, some studies suggest that adult BMI alleles have a stronger association with BMI in adolescence and younger adult life relative to older adult life,9–12 possibly through accelerated growth in early childhood.13–17 However, these studies are limited by the cross-sectional design,9,10,18 the small number of assessed genetic variants10,11,19,20 and the focus on a narrow period of life course (mostly childhood).9,10,13–18 Therefore, it remains largely unknown how genetic variants influence the trajectories of body fatness across the lifespan.
To extend our knowledge, we constructed a genetic risk score (GRS) using 97 common single nucleotide polymorphisms (SNPs) associated with adult BMI, and examined its association with trajectories of body fatness from age 5 through 65 years using two large, prospective US cohorts. By classifying participants into distinct, mutually exclusive trajectory groups, we were able to directly assess the genetic contribution to the population heterogeneity in change in body fatness over the life course.
Methods
Study sample
We studied participants in two prospective cohort studies: the Nurses’ Health Study (NHS), which commenced in 1976 and has since enrolled 121 700 female registered nurses aged 30–55 years, and the Health Professionals Follow-up Study (HPFS), which was initiated in 1986 and has enrolled 51 529 male professionals aged 40–75 years. For both cohorts, follow-up questionnaires were administered biennially to collect and update medical, lifestyle and other health-related information. Between 1989 and 1990, 32 826 women from the NHS and, between 1993 and 1995, 18 225 men from the HPFS returned a blood specimen on ice packs by overnight courier. For the current study, we included 9980 women and 6406 men of European ancestry, whose genotype data were available based on previous nested case-control GWASs of various outcomes, including colorectal cancer, gout, kidney stone, type 2 diabetes, glaucoma, coronary heart disease, percent mammographic density (NHS only), endometrial cancer (NHS only), breast cancer (NHS only) and prostate cancer (HPFS only). To minimize the influence of chronic diseases on body weight, we excluded 2369 women and 1094 men who had a diagnosis of cardiovascular disease, cancer or diabetes before the age of 65 years. We also excluded 334 women and 667 men whose body shape or BMI data were not available for six or more time points from ages of 5, 10, 20, 30, 40, 45, 50, 55 and 60, to 65 years. Therefore, a total of 7277 women and 4645 men were included in the final analysis. The study protocol was approved by the institutional review boards of Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health.
Assessment of body fatness
In 1988, participants in both cohorts were asked to recall their body shape in early and middle life by choosing one of nine pictorial body diagrams (somatotypes) developed by Stunkard et al.21 which best depicted their current and previous body outlines at ages 5, 10, 20, 30 and 40. The validity of this measure for assessing adiposity in early life has been evaluated among 181 participants aged 71 to 76 years in the Third Harvard Growth Study, by comparing participants’ recalled somatotypes with their measured BMI at approximately the same ages.22 The Pearson correlation coefficients were 0.60 for age 5, 0.65 for age 10 and 0.66 for age 20 in women. The corresponding Pearson correlation coefficients in men were 0.36, 0.66 and 0.53, respectively.22
Recalled weight at age 18 years was enquired in 1980 in the NHS, and weight at age 21 years was enquired in 1986 in the HPFS. Participants in both cohorts reported their height and weight at baseline enrolment and biennially thereafter. We used these data to calculate the BMI at ages 40, 45, 50, 55, 60 and 65. To minimize random variation, we used the average BMI within 2 years for each age. To convert the BMI to the same scale as somatotypes in younger ages, we built a linear regression model in each cohort for somatotype and BMI at age 40 among participants with available data, and then used the regression coefficients and reported BMI to impute the somatotype from age 40 to 65. The imputed somatotype highly correlated with participants’ reported somatotype at age 40 (r = 0.74 in women, 0.70 in men) and in 1988 (r = 0.83 in women, 0.71 in men), indicating good performance of our rescaling method.
Trajectory modelling
We used a group-based trajectory modelling approach, separately in women and men, to identify subgroups within each cohort which shared a similar underlying trajectory of body fatness from age 5 up to 65.2,3 The model implemented by SAS Proc Traj fits longitudinal data as a discrete mixture of two or more latent trajectories via maximum likelihood.23 The model fitting and assessment were performed separately for men and women. For this study, we used a censored normal model as a polynomial function of the time scale (i.e. age). The optimal number of groups and the shapes of trajectories were selected for best fit to the data using a two-stage approach based on the change in the Bayesian Information Criterion (BIC).24 The first stage was to determine the number of groups using a quadratic form for all trajectory groups. To ensure adequate statistical power for the genetic analysis, we considered up to four groups and compared the BIC with that with 3, 2 and 1 groups, respectively. Once observing that the model with four groups fitted the data best, we then determined in the second stage the order of the polynomial function specifying the shape of each trajectory. We compared the BIC of the four group models with different functional forms, and found that the model with all groups with up to cubic order terms demonstrated the best fit to the data. Therefore, the final trajectory model was built using a cubic function of age for each of the four trajectories. We then named the trajectory groups to describe their visual patterns (i.e. lean-medium, medium-medium, lean-heavy and medium-heavy).
Based on the final model, we calculated the posterior predicted probability for each individual of being a member of each of the four trajectories. Participants were assigned into the trajectory group to which their posterior membership probability was highest. We then assessed the adequacy of our final model by calculating the average posterior probability of assignment for each group. Using ≥0.70 as the recommended criterion,24 our model demonstrated good discrimination in classifying individuals into distinctive trajectory groups: the average posterior probability for each trajectory group was 0.95, 0.92, 0.92 and 0.97 in women and 0.92, 0.92, 0.88 and 0.95 in men, respectively.
Genotyping and computation of GRS
We included the 97 SNPs that are known to be associated with BMI in adults.6 SNP genotyping and imputation have been described in detail elsewhere.25 All of the SNPs were genotyped or had a high imputation quality score (r2 ≥ 0.8), as assessed by the MACH software (version 1.0.16, Center for Statistical Genetics, University of Michigan). We created a GRS using the 97 SNPs by an established weighting method. Each SNP was recoded as 0, 1 or 2 according to the number of risk alleles (BMI increasing alleles), and weighted by its relative effect size (β coefficient) derived from the most recent meta-analysis of GWASs.6 The GRS was calculated using the equation: , where is the effect size of the on BMI. Each unit of the GRS represented one risk allele, and the GRS could theoretically range from 0 to 194, with a higher score indicating greater genetic predisposition to obesity.
Statistical analysis
All the statistical analyses were performed in each cohort separately. We first compared the GRS and BMI at different ages across the four trajectory groups by the Kruskal-Wallis test, because the normality assumption was violated for these dependent variables. We then calculated the proportion of participants in each trajectory across deciles of the GRS, and tested the statistical significance of the trend for the proportions across deciles using the multinomial Cochran-Armitage trend test.26 To assess the association between the GRS and trajectory assignment, we used the ‘lean-medium’ group as the reference and calculated the odds ratio (OR) and 95% confidence interval (CI) of being in each of the non-reference trajectories associated with 10-allele increment of GRS, by the multinomial logistic regression. We also ran the ordinal logistic regression model by using the trajectory as an ordinal response variable (0 = ’lean-medium’, 1 = ’medium-medium’, 2 = ’lean-heavy’ and 3 = ’medium-heavy’) and calculated the P-value for testing the overall trend of the GRS-trajectory association. Similarly, we assessed each of the 97 individual SNPs in relation to trajectory, using the additive genetic model. To account for multiple testing, we adjusted the P-values by estimating the false discovery rate (FDR) using the Benjamini-Hochberg method.27 SAS 9.4 was used for all analyses (SAS Institute Inc., Cary, NC, USA). All statistical tests were two-sided, with the type I error rate of α = 0.05.
Results
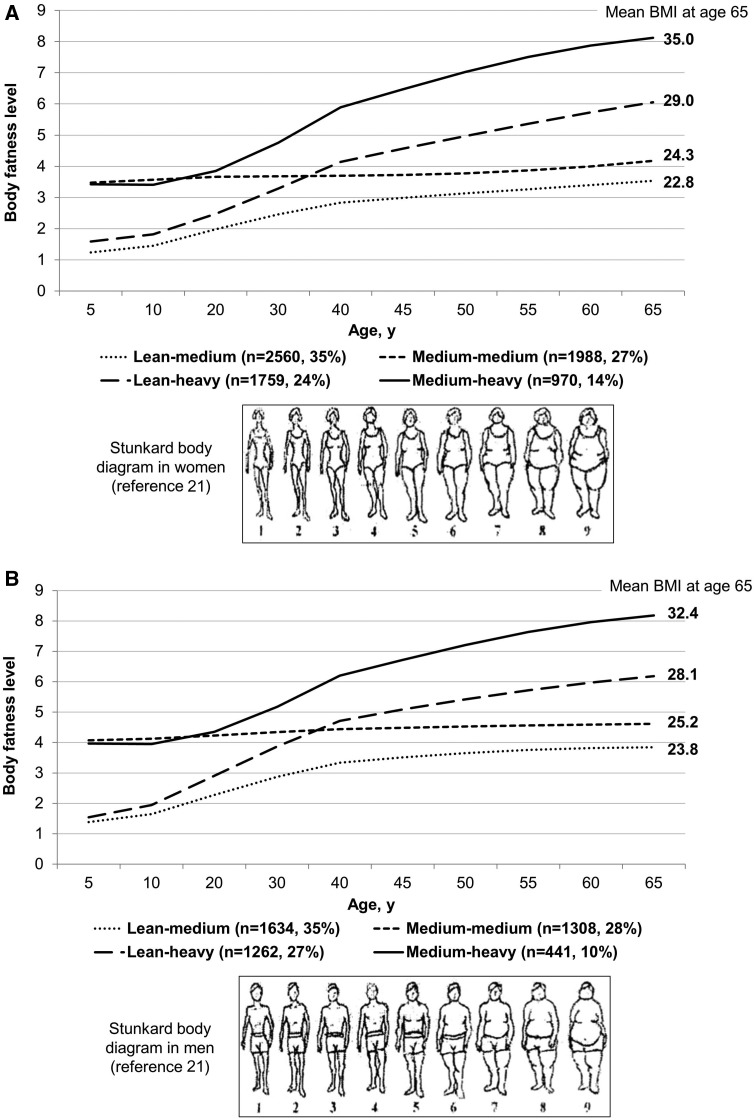
Figure 1 shows the four trajectories of body fatness from age 5 up to 65 in women and men: 35% of women and men started lean and then experienced a moderate increase in body fatness (lean-medium group); 27% of women and 28% of men maintained a medium body shape throughout life (medium-medium group); 24% of women and 27% of men started lean and then gained a substantial amount of weight (lean-heavy group); 14% of women and 10% of men started with a medium body shape and then gained more weight over age (medium-heavy group). As expected, the BMI profile throughout adulthood in each group conformed well to the patterns of the identified trajectories (Supplementary Table S1, available as Supplementary data at IJE online). For example, for women, the mean BMI in the lean-medium group increased from 19.6 to 22.8 kg/m2 between age 18 and 65, whereas in the medium-heavy group the mean BMI increased from 24.3 to 35.0 kg/m2 during this period.
Figure 1.
Trajectory of body fatness by age in women (A) and men (B).
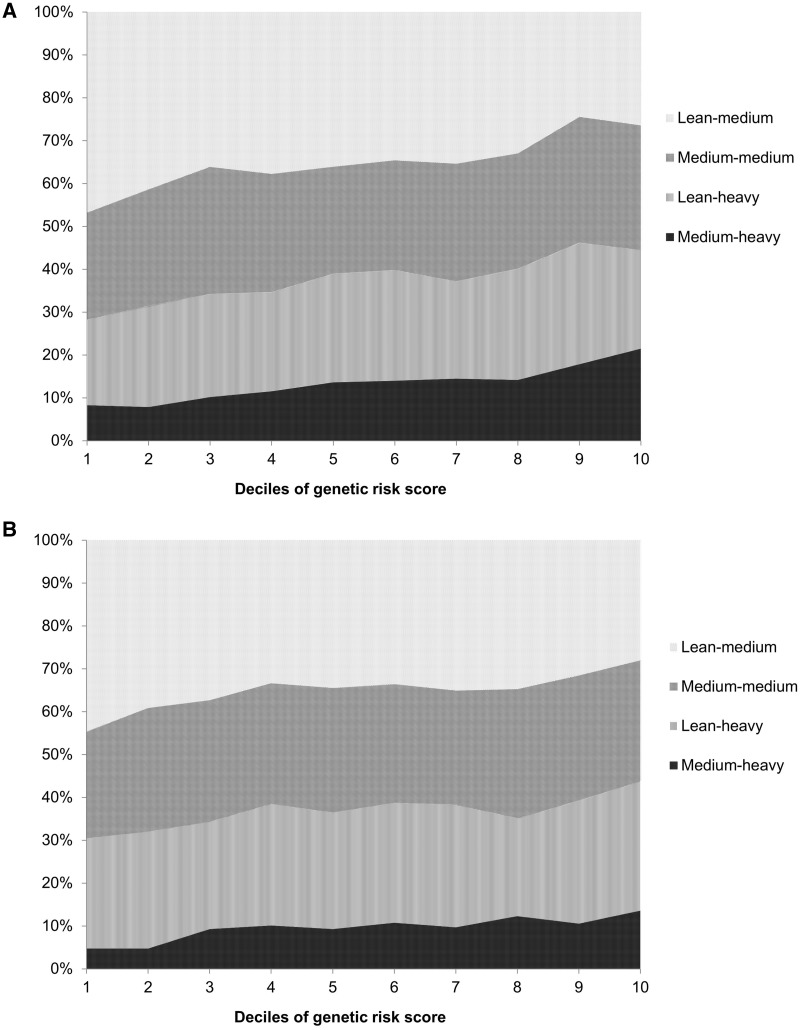
Figure 2 presents the proportion of trajectory groups according to deciles of GRS (P for trend < 0.001 for both women and men); 47% of women and 45% of men in the first decile of the GRS were in the lean-medium group, and these proportions reduced to 26% and 28%, respectively, for the highest decile. The proportions of participants in the other groups all showed an increase from the lowest to the highest deciles of the GRS: for women: 25% to 29% for the medium-medium group, 20% to 23% for the lean-heavy group and 8% to 21% for the medium-heavy group; for men: 25% to 28% for the medium-medium group, 26% to 30% for the lean-heavy group and 5% to 14% for the medium-heavy group.
Figure 2.
Proportion of trajectory groups according to deciles of genetic risk score for obesity (GRS) in women (A) and men (B).
We then assessed the association of GRS with trajectory grouping (Table 1). The GRS increased from the lean-medium, medium-medium, lean-heavy to medium-heavy groups, with the mean of 87.0, 88.3, 88.4 and 89.9 in women, and 87.1, 88.0, 88.0 and 89.4 in men, respectively (P < 0.001). For women, compared with the odds of being in the lean-medium group, a 10-allele increment in the GRS was associated with a 40% (95% CI, 27–54%), 43% (30–58%), and 115% (91–143%) increase in the odds of being in the medium-medium, lean-heavy and medium-heavy groups, respectively (P for trend < 0.001). For men, the corresponding increases in the odds were 26% (12–42%), 27% (13–43%),= and 81% (53–115%), respectively (P for trend < 0.001).
Table 1.
Associations of trajectory of body fatness with genetic risk score (GRS) in women and men
| Lean-medium | Medium-medium | Lean-heavy | Medium-heavy | P for trend | |
|---|---|---|---|---|---|
| Women | |||||
| Mean GRS (SD) | 87.0 (6.3) | 88.3 (6.2) | 88.4 (6.0) | 89.9 (6.2) | <0.001 |
| OR (95% CI) per 10-allele increment in GRSa | 1 (reference) | 1.40 (1.27–1.54) | 1.43 (1.30–1.58) | 2.15 (1.91–2.43) | <0.001 |
| Men | |||||
| Mean GRS (SD) | 87.1 (6.4) | 88.0 (6.3) | 88.0 (6.3) | 89.4 (5.9) | <0.001 |
| OR (95% CI) per 10-allele increment in GRSa | 1 (reference) | 1.26 (1.12–1.42) | 1.27 (1.13–1.43) | 1.81 (1.53–2.15) | <0.001 |
SD, standard deviation.
aMultinomial logistic regression was used to derive ORs and 95% CIs. Ordinal logistic regression was used to calculate the P-value for trend test.
Among the 97 SNPs in the GRS, 24 SNPs in women and 12 in men were nominally associated with the trajectories of body fatness (P for trend < 0.05; Supplementary Tables S2 and S3, available as Supplementary data at IJE online). After correction for multiple testing, eight SNPs in women and one SNP in men remained to be associated with the trajectories with an FDR P-value of < 0.05 (Table 2). Among these SNPs, rs1558902 in FTO showed the strongest association with the trajectories: relative to being in the lean-medium trajectory, one risk-allele increment in women was associated with a 12% (95% CI, 3–22%), 23% (12–34%) and 35% (22–50%) increase in the odds of being in the medium-medium, lean-heavy and medium-heavy groups, respectively (FDR P-value < 0.001). For men, the corresponding increases in the odds were 12% (1–25%), 8% (−3–20%), and 46% (25–69%), respectively (FDR P-value = 0.02). In addition, we found a modest association with the trajectories in women for the variants in other genes, including PMS2L11, OLFM4, GNPDA2, RALYL, ADCY3, LMX1B and MTCH2. The odds ratio for comparing the extreme trajectories ranged from 1.10 to 1.28.
Table 2.
Odds ratio (95% confidence interval) of trajectory of body fatness per one allele increment in individual SNPs in women and men (only SNPs with FDR < 0.05 are shown)a
| SNP | Nearest gene | Lean-medium | Medium-medium | Lean-heavy | Medium-heavy | P for trend | FDR |
|---|---|---|---|---|---|---|---|
| Women | |||||||
| rs1558902 | FTO | 1 (reference) | 1.12 (1.03–1.22) | 1.23 (1.12–1.34) | 1.35 (1.22–1.50) | <0.001 | <0.001 |
| rs2245368 | PMS2L11 | 1 (reference) | 1.17 (1.04–1.31) | 1.15 (1.02–1.30) | 1.28 (1.11–1.47) | <0.001 | 0.02 |
| rs12429545 | OLFM4 | 1 (reference) | 1.05 (0.93–1.19) | 1.12 (0.98–1.27) | 1.26 (1.09–1.47) | 0.003 | 0.04 |
| rs10938397 | GNPDA2 | 1 (reference) | 1.13 (1.04–1.23) | 1.08 (0.99–1.18) | 1.24 (1.12–1.38) | <0.001 | 0.02 |
| rs2033732 | RALYL | 1 (reference) | 1.10 (1.00–1.21) | 1.09 (0.99–1.20) | 1.21 (1.07–1.37) | 0.003 | 0.04 |
| rs10182181 | ADCY3 | 1 (reference) | 1.10 (1.01–1.19) | 1.14 (1.05–1.25) | 1.14 (1.02–1.26) | 0.001 | 0.03 |
| rs10733682 | LMX1B | 1 (reference) | 1.09 (1.00–1.18) | 1.13 (1.03–1.23) | 1.14 (1.03–1.27) | 0.002 | 0.03 |
| rs3817334 | MTCH2 | 1 (reference) | 1.10 (1.01–1.19) | 1.16 (1.06–1.26) | 1.10 (0.99–1.22) | 0.004 | 0.04 |
| Men | |||||||
| rs1558902 | FTO | 1 (reference) | 1.12 (1.01–1.25) | 1.08 (0.97–1.20) | 1.46 (1.25–1.69) | <0.001 | 0.02 |
a‘Lean-medium’ group was used as the reference group. Multinomial logistic regression was used to derive the odds ratio and 95% confidence interval. Ordinal logistic regression was used to calculate the P-value for trend test.
To test the robustness of our findings with participants with modest accuracy of trajectory assignment, we excluded 925 women and 832 men whose trajectory assignment probability was below 0.80. The results remained essentially unchanged (Supplementary Table S4, available as Supplementary data at IJE online). To examine whether participants who were diagnosed with chronic diseases may be more likely to be influenced by genetic BMI variants, we also performed a sensitivity analysis by restricting to 5055 women and 2837 men who were used as controls in the previous GWAS studies for various outcomes. Our results also did not materially change (Supplementary Table S4). Finally, given the possibility that a different set of SNPs may influence adiposity across the life course, we performed the trajectory analysis by starting at age 30. As shown in Supplementary Figure S1 (available as Supplementary data at IJE online), similar trajectory patterns were identified, with good concordance in the trajectory group assignments with those in our primary analysis [weighted Kappa, 0.65 (95% CI, 0.64–0.66, P = 0.006) in women and 0.42 (95% CI, 0.39–0.44, P = 0.01) in men]. Similar associations between the GRS and trajectories were also observed (Supplementary Table S4).
Discussion
To our knowledge, this is the first study that examines the effect of BMI-associated genetic variants on trajectories of body fatness over the life course. Our results indicate that individuals with more BMI risk alleles were more likely to have a heavy body shape in early life and also to gain further weight as they aged. Participants who carried 10 more risk alleles had about a doubled likelihood of having a medium-heavy trajectory relative to having a lean-medium trajectory. These findings suggest a persistent effect of adulthood BMI-related genetic variants on the trajectories of body fatness across the lifespan. Therefore, these genetic variants may be used for early identification of individuals who will likely maintain a heavy body shape throughout life, to facilitate targeted intervention.
Previous studies on the genetic associations with adiposity have largely focused on growth and BMI changes during childhood and adolescence. The findings suggest that individuals who carry more adulthood BMI-related genetic variants demonstrate accelerated weight gain in early infancy and childhood,17,28,29 show earlier adiposity rebound13,14 and maintain higher BMI throughout early life.13 However, evidence is limited regarding how genetic variants influence body weight variations over the life course. So far the only data available are from the British National Survey of Health and Development (NSHD), which included 2537 participants with repeated measurements of BMI from age 2 to 53 years.15 Using 11 adult BMI-associated SNPs to create the GRS, the study found a positive association between the GRS and BMI at all ages, which reached its maximum at ages 11 and 20 years and then stabilized. These results differ from our findings, because despite similar levels of body fatness at age 20 years (Figure 1), the medium-heavy group in our study demonstrated a much stronger association with the GRS than the medium-medium group, suggesting that the genetic effect on adiposity may persist beyond early adulthood. It is possible that population differences between the two studies, at least partly explain the discrepant results. In contrast to the participants in our cohorts who were mostly born in the USA between 1925 and 1940, the NSHD participants were all born in 1946 and had very low prevalence of childhood obesity due to postwar food rationing in the UK from birth to age 8 years. Given the recent data suggesting that the GRS was more strongly associated with BMI among persons born in later cohorts,30 possibly due to modification by obesogenic environment, it is possible that delayed exposure to the obesogenic conditions in our participants contributes to the extended penetrance of the genetic influence into middle adulthood. It is also possible that standardization of diet achieved by food rationing in the NSHD cohort leads to stronger observed genetic effects on childhood versus later weight gain.15
Consistent with our observation for a persistent effect of genetic variants on adiposity from age 5 to 65 years, several studies have linked genetic BMI variants to adulthood weight change. A Swedish study assessed a GRS comprising 32 BMI-associated SNPs in relation to BMI change from age 25 through late adulthood.31 A higher GRS was associated with a steeper increase in BMI until the age of 65 years, but after age of 65 years no consistent pattern was found,31 possibly due to the influence of late life diseases and ageing-related body composition changes. Similarly, a more recent study showed that a 31-SNP-based GRS for obesity was strongly associated with increased annual weight gain and substantial weight gain from age 20 to middle age (mean: 58 years).32 However, paradoxically the study found that a higher number of BMI-increasing risk alleles were associated with decreased weight gain during and after middle age.32 Although this observation is difficult to explain, given the possibility that early life weight gain may predispose to later weight loss due to either physiological regulations or obesity-related morbidities, it highlights the unique advantage of our trajectory approach in assessing the risk factors for changes in body fatness over time. Instead of dividing the life course into arbitrary age periods, the trajectory approach respects the continuity of body growth throughout life and classifies individuals into distinct, mutually exclusive groups, thus allowing us to directly test the genetic contributions to between-person differences in the dynamics of body fatness over the life course.
Among all of the known genetic variants, the SNP in FTO (rs1558902) showed the strongest associations with trajectories of body fatness in our study, with an increasing OR from the medium-medium, lean-heavy to medium-heavy groups. Previous investigations have consistently established a predominant effect of FTO on obesity, likely through the hypothalamic regulation of appetite, energy homeostasis and metabolic rate.33–35 However, data about the life course influence of FTO loci on adiposity remain inconclusive. Whereas several meta-analyses of cross-sectional studies observed a stronger BMI-raising effect of genetic variations at FTO in younger adults relative to older adults,10,12 others did not find any heterogeneity between age groups.36 Because participants in the same age group may be born in different periods with distinct exposure experiences throughout life, these studies were unable to disentangle age, birth cohort and period effects. A longitudinal study in the NSHD found that the association of FTO variant with BMI strengthened during childhood and adolescence, peaked at age 20 years and then weakened during adulthood.11 In contrast, our previous analysis in the NHS and HPFS cohorts indicated that the FTO-BMI association did not decline until age 50–55.20 As discussed above, differences in population experiences over the life course may have contributed to the discrepant results between the two studies. For other genetic variants, in agreement with our findings, previous studies have identified positive associations with early life adiposity for adult BMI-related variants in several genes, such as ADCY3,18,37OLFM4,18,37GNPDA2,9,37,38LMX1B37 and MTCH2,10 although the underlying biological mechanisms remain to be elucidated.
Despite the similar patterns in the two sexes, we found that the magnitude of the relationship of GRS with trajectories appeared to be stronger in women than in men. However, strict comparison between the two sexes is difficult because the trajectories were created within each cohort separately and sex-specific reference groups were used in the analysis. It is possible that better performance of the pictograms in assessing body fatness in women (see Methods) may have contributed to the stronger results than in men. Previous studies did not provide strong evidence for the sex heterogeneity in the genetic influence on BMI,12 although a few SNPs showed stronger associations with BMI in women than in men.6
Our study takes advantage of the rich genetic and anthropometric data and long-term follow-up of two well-established cohorts. Compared with previous studies, we examined a much larger number of genetic variants that have been associated with adult BMI. Furthermore, because our study participants are all health professionals born within a narrow window of time, the influence of macro-environmental or birth cohort factors was minimized. Additionally, the trajectory method we used also provides an attractive alternative to traditional analysis by directly examining the between-person heterogeneity. Taken together with our previous findings, that individuals with distinct trajectories were at strikingly different risk of chronic diseases (e.g. cancer and cardiometabolic diseases) and early death in later life,2–4 these investigations provide novel data for the causes and consequences of adiposity across the lifespan.
We acknowledge the limitations of our study. First, the trajectories were constructed up to age 65 years, thus precluding assessment for late adulthood. However, our choice for the end age was based on the data suggesting that BMI after age 65 may not be a good indicator for adiposity, due to ageing-related muscle loss.39 Furthermore, the incidence of chronic diseases started to increase rapidly after age 65, thus making it difficult, if not impossible, to distinguish the biologically and behaviourally conferred variations of body weight from those resulting from obesity-related morbidities.40 Second, body shape assessed by recalled somatotype in early life and self-reported BMI in adulthood are subject to measurement error. However, the good performance of the instrument as indicated by previous validation studies suggests that measurement error is unlikely to have a substantial influence on trajectory assignment.22 Third, for the sake of statistical power, a limited number of trajectories were derived that may not accurately reflect each person’s profile of body fatness. However, the good discrimination of our trajectory model and well-tracked change in BMI across the trajectories indicate that these trajectories can provide a parsimonious summary of the predominant features of lifetime body fatness in our population, without a significant loss of information. This is further supported by the similar results obtained after excluding participants with suboptimal trajectory assignment.
In conclusion, we found that genetic variants for adulthood BMI were associated with the different trajectories of body fatness across the lifespan. Individuals with more risk alleles were more likely to have a heavier body shape throughout life. Our results support a persistent effect of genetic variants on body fatness across the lifespan. Future studies are warranted to examine whether this persistence is modifiable by early life exposures and to determine the critical window during which genetic risk can be most alleviated by early intervention.
Supplementary Data
Supplementary data are available at IJE online.
Funding
This work was supported by the National Institutes of Health (UM1 CA186107, R01 CA49449, UM1 CA167552, and 1U54CA155626). M.S. is supported by the 2017 AACR-AstraZeneca Fellowship in Immuno-oncology Research (Grant Number 17-40-12-SONG). The funders had no role in design and conduct of the study; collection, management, analysis and interpretation of the data; preparation, review and approval of the manuscript; or the decision to submit the manuscript for publication.
Supplementary Material
Acknowledgements
We would like to thank the participants and staff of the Nurses’ Health Study and the Health Professionals Follow-up Study for their valuable contributions, as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY.
Author Contributions
M.S. researched data and wrote the manuscript. Y.Z. researched data and reviewed/edited the manuscript. L.Q. researched data and reviewed/edited the manuscript. F.B.H. reviewed/edited the manuscript. A.T.C. reviewed/edited the manuscript. E.L.G. reviewed/edited the manuscript and supervised the study.
Conflict of interest: The authors have no conflict of interest to disclose.
References
- 1. Singh AS, Mulder C, Twisk JW, van Mechelen W, Chinapaw MJ. Tracking of childhood overweight into adulthood: a systematic review of the literature. Obes Rev 2008;9:474–88. [DOI] [PubMed] [Google Scholar]
- 2. Song M, Willett WC, Hu FB, et al. Trajectory of body shape across the lifespan and cancer risk. Int J Cancer 2016;138:2383–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Song M, Hu FB, Wu K, et al. Trajectory of body shape in early and middle life and all cause and cause specific mortality: results from two prospective US cohort studies. BMJ 2016;353:i2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zheng Y, Song M, Manson JE, Giovannucci EL, Hu FB. Group-based trajectory of body shape from age 5 to 55 years and cardio-metabolic disease risk in two US cohorts. Am J Epidemiol 2017, June 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Min J, Chiu DT, Wang Y. Variation in the heritability of body mass index based on diverse twin studies: a systematic review. Obes Rev 2013;14:871–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Locke AE, Kahali B, Berndt SI, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015;518:197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu Q, Clark MS, Palmiter RD. Deciphering a neuronal circuit that mediates appetite. Nature 2012;483:594–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Llewellyn CH, Trzaskowski M, van Jaarsveld CH, Plomin R, Wardle J. Satiety mechanisms in genetic risk of obesity. JAMA Pediatr 2014;168:338–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Graff M, Ngwa JS, Workalemahu T, et al. Genome-wide analysis of BMI in adolescents and young adults reveals additional insight into the effects of genetic loci over the life course. Hum Mol Genet 2013;22:3597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Graff M, Gordon-Larsen P, Lim U, et al. The influence of obesity-related single nucleotide polymorphisms on BMI across the life course: the PAGE study. Diabetes 2013;62:1763–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hardy R, Wills AK, Wong A, et al. Life course variations in the associations between FTO and MC4R gene variants and body size. Hum Mol Genet 2010;19:545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Winkler TW, Justice AE, Graff M, et al. The influence of age and sex on genetic associations with adult body size and shape: a large-scale genome-wide interaction study. PLoS Genet 2015;11:e1005378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Belsky DW, Moffitt TE, Houts R, et al. Polygenic risk, rapid childhood growth, and the development of obesity: evidence from a 4-decade longitudinal study. Arch Pediatr Adolesc Med 2012;166:515–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Warrington NM, Howe LD, Wu YY, et al. Association of a body mass index genetic risk score with growth throughout childhood and adolescence. PloS One 2013;8:e79547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elks CE, Loos RJ, Hardy R, et al. Adult obesity susceptibility variants are associated with greater childhood weight gain and a faster tempo of growth: the 1946 British Birth Cohort Study. Am J Clin Nutr 2012;95:1150–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Warrington NM, Wu YY, Pennell CE, et al. Modelling BMI trajectories in children for genetic association studies. PloS One 2013;8:e53897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Elks CE, Heude B, de Zegher F, et al. Associations between genetic obesity susceptibility and early postnatal fat and lean mass: an individual participant meta-analysis. JAMA Pediatr 2014;168:1122–30. [DOI] [PubMed] [Google Scholar]
- 18. Warrington NM, Howe LD, Paternoster L, et al. A genome-wide association study of body mass index across early life and childhood. Int J Epidemiol 2015;44:700–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qi L, Kraft P, Hunter DJ, Hu FB. The common obesity variant near MC4R gene is associated with higher intakes of total energy and dietary fat, weight change and diabetes risk in women. Hum Mol Genet 2008;17:3502–08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qi L, Kang K, Zhang C, et al. Fat mass- and obesity-associated (FTO) gene variant is associated with obesity: longitudinal analyses in two cohort studies and functional test. Diabetes 2008;57:3145–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stunkard AJ, Sorensen T, Schulsinger F. Use of the Danish Adoption Register for the study of obesity and thinness. Res Publ Assoc Res Nerv Ment Dis 1983;60:115–20. [PubMed] [Google Scholar]
- 22. Must A, Willett WC, Dietz WH. Remote recall of childhood height, weight, and body build by elderly subjects. Am J Epidemiol 1993;138:56–64. [DOI] [PubMed] [Google Scholar]
- 23. Jones BL, Nagin DS. Advances in group-based trajectory modeling and an SAS procedure for estimating them. Sociol Method Res 2007;35:542–71. [Google Scholar]
- 24. Nagin DS. Group-based Modeling of Development. Cambridge, MA: Harvard University Press, 2005. [Google Scholar]
- 25. Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genets 2007;39:870–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szabo A. Testing for trend with a nominal outcome. Eastern North American Meeting (ENAR) Spring Meeting, 6–9 March 2016. Austin, TX, 2016.
- 27. Benjamini Y, Hochberg Y. Controlling the false discovery rate:a practical and powerful approach to multiple testing. J R Stat Soc B (Methodol) 1995;57:289–300. [Google Scholar]
- 28. Elks CE, Loos RJ, Sharp SJ, et al. Genetic markers of adult obesity risk are associated with greater early infancy weight gain and growth. PLoS Med 2010;7:e1000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Steinsbekk S, Belsky D, Guzey IC, Wardle J, Wichstrom L. Polygenic risk, appetite traits, and weight gain in middle childhood: a longitudinal study. JAMA Pediatr 2016;170:e154472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Walter S, Mejia-Guevara I, Estrada K, Liu SY, Glymour MM. Association of a genetic risk score with body mass index across different birth cohorts. JAMA 2016;316:63–69. [DOI] [PubMed] [Google Scholar]
- 31. Dahl AK, Reynolds CA, Fall T, Magnusson PK, Pedersen NL. Multifactorial analysis of changes in body mass index across the adult life course: a study with 65 years of follow-up. Int J Obes (Lond) 2014;38:1133–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rukh G, Ahmad S, Ericson U, et al. Inverse relationship between a genetic risk score of 31 BMI loci and weight change before and after reaching middle age. Int J Obes (Lond) 2016;40:252–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cecil JE, Tavendale R, Watt P, Hetherington MM, Palmer CN. An obesity-associated FTO gene variant and increased energy intake in children. N Engl J Med 2008;359:2558–66. [DOI] [PubMed] [Google Scholar]
- 34. Fischer J, Koch L, Emmerling C, et al. Inactivation of the Fto gene protects from obesity. Nature 2009;458:894–98. [DOI] [PubMed] [Google Scholar]
- 35. Church C, Lee S, Bagg EA, et al. A mouse model for the metabolic effects of the human fat mass and obesity associated FTO gene. PLoS Genet 2009;5:e1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hertel JK, Johansson S, Sonestedt E, et al. FTO, type 2 diabetes, and weight gain throughout adult life: a meta-analysis of 41 504 subjects from the Scandinavian HUNT, MDC, and MPP studies. Diabetes 2011;60:1637–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Felix JF, Bradfield JP, Monnereau C, et al. Genome-wide association analysis identifies three new susceptibility loci for childhood body mass index. Hum Mol Genet 2016;25:389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao J, Bradfield JP, Zhang H, et al. Role of BMI-associated loci identified in GWAS meta-analyses in the context of common childhood obesity in European Americans. Obesity (Silver Spring) 2011;19:2436–39. [DOI] [PubMed] [Google Scholar]
- 39. Kyle UG, Genton L, Hans D, Karsegard L, Slosman DO, Pichard C. Age-related differences in fat-free mass, skeletal muscle, body cell mass and fat mass between 18 and 94 years. Eur J Clin Nutr 2001;55:663–72. [DOI] [PubMed] [Google Scholar]
- 40. Kalyani RR, Corriere M, Ferrucci L. Age-related and disease-related muscle loss: the effect of diabetes, obesity, and other diseases. Lancet Diabetes Endocrinol 2014;2:819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.