A critical survey of strigolactone receptor–ligand structures available in the literature shows that the models frequently contain features not supported by the X-ray data.
Keywords: Crystal structure, DWARF14, ligand binding, Oryza sativa, strigolactone, strigolactone receptor
Abstract
Strigolactones, a group of terpenoid lactones, control many aspects of plant growth and development, but the active forms of these plant hormones and their mode of action at the molecular level are still unknown. The strigolactone protein receptor is unusual because it has been shown to cleave the hormone and supposedly forms a covalent bond with the cleaved hormone fragment. This interaction is suggested to induce a conformational change in the receptor that primes it for subsequent interaction with partners in the signalling pathway. Substantial efforts have been invested into describing the interaction of synthetic strigolactone analogues with the receptor, resulting in a number of crystal structures. This investigation combines a re-evaluation of models in the Protein Data Bank with a search for new conditions that may permit the capture of a receptor–ligand complex. While weak difference density is frequently observed in the binding cavity, possibly due to a low-occupancy compound, the models often contain features not supported by the X-ray data. Thus, at this stage, we do not believe that any detailed deductions about the nature, conformation, or binding mode of the ligand can be made with any confidence.
Introduction
Strigolactones (SLs) are phytohormones that control many aspects of plant growth. SLs are produced mainly in the roots, which they shape morphologically, especially in relation to nutrient deprivation. They are also transported to and, to a limited extent, synthesized in the shoots, where they exert pleiotropic functions in development and stress responses, most prominently repression of axillary bud outgrowth (Gomez-Roldan et al., 2008, Umehara et al., 2008). As exogenous signals, SLs promote the symbiotic association between land plants and arbuscular mycorrhizal fungi that improve nutrient uptake (Akiyama et al., 2005; Bouwmeester et al., 2007; Al-Babili and Bouwmeester, 2015). Secreted into the soil, SLs can also function as germination signals for parasitic plants and may cause considerable crop failure. For instance, parasitic weeds such as witchweed (Striga sp.) and broomrape (Orobanche sp.) have become completely dependent on this interaction for germination of their seeds (Cook et al., 1966; Cardoso et al., 2011).
Despite their relatively recent discovery as plant hormones (Gomez-Roldan et al., 2008; Umehara et al., 2008), rapid progress has been made in understanding SL function, including the identification of a signalling pathway for SLs. This pathway involves the SL receptors of the DWARF14 (D14) class of α/β-hydrolase enzymes, the MORE AXILLARY GROWTH2 (MAX2) F-box proteins, and the SUPPRESSOR OF MAX2 1 (SMAX1) and SMAX1-LIKE (SMXL) family of chaperonin-like proteins. In Oryza sativa (rice), MAX2 proteins interact with D14 in an SL-dependent manner (Zhou et al., 2013) and act as part of a Skp, Cullin, F-box-containing complex (or SCF complex) that is assumed to regulate ubiquitination and degradation of target proteins. The SMXL proteins are targets of MAX2/D14-dependent protein degradation. This pathway appears to be evolutionarily conserved; homologues of these proteins have also been identified in, among others, Arabidopsis (Arabidopsis thaliana), pea (Pisum sativum), and petunia (Petunia hybrida) (Arite et al, 2009; Waters et al., 2012; Hamiaux et al., 2012; reviewed in Bennett and Leyser, 2014).
There are notable similarities between SLs and their receptors and the perception and signalling mechanisms of several other plant hormones, including auxins (Hayward et al., 2009) and gibberellins. The main analogies include response triggering by proteasome-mediated destruction of ubiquitinated repressors, and the central role of F-box protein adaptors in this process. Similarities extend to the involvement of members in the α/β-hydrolase protein family as receptors for hormonal ligands, such as in the case of the Gibberellin Insensitive Dwarf1 (GID1) protein for bioactive gibberellins (Sun, 2010). However, unlike the gibberellin receptor GID1, which is enzymatically inactive and stably interacts with its target hormone, SL receptors destroy their target.
In rice, the SL signal is transmitted through interaction with a D14 receptor, the DWARF3 (D3, orthologue of MAX2 in Arabidopsis) F-box protein and the DWARF53 (D53) protein, which has been identified as a target protein (Jiang et al., 2013; Zhou et al., 2013). The D14 α/β-hydrolase enzyme uses a Ser–His–Asp catalytic triad to perceive and hydrolyse SLs simultaneously. Recent results (de Saint Germain et al., 2016; Yao et al., 2016) indicate that a covalent bond with histidine may form upon hydrolysis of the active hormone at the receptor.
A major challenge is to identify the active form of SLs and their molecular mode of action. On interaction with D14, the synthetic SL analogue GR24 is hydrolysed to two compounds (Hamiaux et al., 2012; Nakamura et al., 2013), a tricyclic lactone (the ABC ring moiety), and 5-hydroxy-3-methylbutenolide (the D-OH ring), or possibly some other degradation intermediate such as 2,4,4-trihydroxy-3-methyl-3-butenal (TMB; Zhao et al, 2013). This hydrolysis is relatively slow (Hamiaux et al., 2012; de Saint Germain et al., 2016), raising hopes that a complex between the receptor and the hydrolysed product may be captured in a crystal structure. However, the challenges involved in such studies are numerous and include issues such as the low contribution to the total scattering of the ligand compared with the protein partner, the high concentration of components used for the purification, crystallization, and cryo-cooling that may often outcompete the desired ligand, and importantly the incompatibility of ligand binding with the packing of the protein in the crystal lattice. These questions will be elaborated further below.
We searched for conditions that may allow the crystallization of a complex between rice D14 and SL analogues, such as GR24, and have compared the resulting structures with the structures of putative complexes deposited in the databases. The results from our survey may serve as an illustration of critical issues in protein–ligand crystallization efforts for the D14–SL complex.
Materials and methods
DNA cloning
The OsD14 cDNA fragment lacking the sequence encoding the first 51 N-terminal amino acids was amplified from a cDNA pool obtained from roots of Oryza sativa (Japonica subgroup) by PCR using the Phusion® High-Fidelity PCR Master Mix (Thermo Fisher Scientific) with forward primer 5'-GGATCCAGCGGGGCGAAGCTGCT-3' (BamHI site underlined) and reverse primer 5'-GGTACCTTAGT ACCGGGCGAGAG-3' (KpnI site underlined). The PCR product was analysed by agarose gel electrophoresis. The eluted D14 PCR fragment was prepared for TA cloning by adding 3'-adenine ends using PCR Master Mix (Thermo Fisher Scientific) and cloned into pGEM-T (Promega). After confirming the correct D14-short coding sequence by Sanger sequencing, the fragment was cloned into the protein expression plasmid pHUE (Catanzariti et al., 2004) by using the endonuclease restriction sites BamHI and KpnI. The resulting construct (OsD14-short) consists of OsD14 lacking the first 51 amino acids, fused at the N-terminus to ubiquitin carrying an N-terminal His-tag and a C-terminal cleavage site for a ubiquitin-specific peptidase 2 (Usp-2). The reactive serine in the active site of OsD14 was substituted by an alanine (S147A). Mutation was achieved by using the Quick Change II Site-Directed Mutagenis Kit (Agilent Technologies) and reactions were performed as described in the user’s manual with oligos 5'-CAC TCC GTC GCC GCC ATG ATC GGC ATC-3' (sense) and 5'-GAT GCC GAT CAT GGC GGC GAC GGA GTG-3' (antisense). The expression vector pHUE-OsD14-short was used as template for the PCR. Successful introduction of the mutation was confirmed by DNA sequencing.
Protein expression
OsD14 (short, tagged version as described above) was heterologously expressed in Escherichia coli strain BL21 DE3 (Life Technologies). After transformation with OsD14-short, E. coli was grown at 37 °C in 2× YT medium (1.6% tryptone, 1% yeast extract, 0.5% NaCl) containing 1 mM betaine and 100 μg ml−1 ampicillin until OD600=0.6 was reached. Cells were then cooled to 20 °C, induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and cultivated overnight at 20 °C.
Protein purification
The cells were harvested by centrifugation and re-suspended in 10% glycerol, 20 mM Bis-Tris propane pH 8, 0.5 M NaCl (Buffer A). After incubation with DNase, the cells were lysed using a cell disruptor CF Range (Constant Systems Ltd) at 35 kpsi.
The cell lysate was applied to a gravity flow Ni-NTA column (Bio-Rad), washed with 50 mM imidazole in Buffer A, and eluted with 500 mM imidazole in Buffer A. After lowering the salt concentration by diluting the eluate with 20 mM Bis-Tris propane pH 8 followed by a protein concentrating step using a Vivaspin centrifugal concentrator (Sartorius), the protein was incubated at 37 °C with 0.1 mg ml–1 Usp-2 (Catanzariti et al., 2004) to remove the His-ubiquitin tag. In the subsequent step, the His-ubiquitin tag and the His-tagged Usp-2 protease were removed from OsD14 by applying the solution a second time to an Ni-NTA column. Prior to this step, imidazole was removed using a PD10 column (GE Healthcare) equilibrated with Buffer A. The eluate containing the cleaved OsD14 was collected, diluted 30 times in buffer MQ (5% glycerol, 20 mM Bis-Tris propane pH 9.0), and applied to a 1 ml MonoQ column equilibrated with MQ buffer. OsD14 was eluted with a gradient of 0–0.5 M NaCl in MQ buffer. Fractions containing OsD14 eluted at ~0.1 M NaCl were pooled and concentrated to 2–4 mg ml−1. The mutant S147A was purified in the same manner as the native OsD14.
Crystallization and data collection
OsD14 (lacking the N-terminal 51 amino acids) was crystallized at room temperature (293 K). Crystallization conditions were screened using commercial screens and a mosquito® Crystal robot (TTP Labtech) with 0.2 μl drop size. Initial crystals were obtained in 10% 2-methylpentanediol (MPD), 0.1 M HEPES pH 7.0 (pHClear Suite, Qiagen). These crystals belong to space group P212121 with unit cell parameters a=48.1, b=88.6, c=118.9 Å, α, β, γ=90°, two molecules in the asymmetric unit, and 42% solvent content. Data were collected to 1.35 Å resolution at the ESRF beam line ID23-2. In the following, to exclude potential ligands (MPD and HEPES), extensive screening was made with alternative buffers and precipitating agents at 293 K using the hanging drop vapour diffusion method. Crystals of apo OsD14 were obtained using polyethylene glycol (PEG) 6000 at concentrations of 5–30% and in the pH range 6.0–9.5. These conditions were subsequently tried for crystallization of complexes of native OsD14 and a S147A mutant with SL analogues by soaking and co-crystallization. We used a number of compounds of various sizes (Prandi et al., 2011; Artuso et al., 2015, Supplementary Fig. S1 at JXB online), including GR24 [a racemic mixture of (+)-GR24 and (–)-GR24]. The compounds were dissolved in acetone to a concentration of 0.2 M, and either added to a drop containing a crystal for soaking experiments or to the solution of OsD14 prior to crystallization for co-crystallization experiments. Incubation times were 20–60 min. Prior to data collection, crystals were soaked in a reservoir solution containing cryo-protectant (30% glycerol and any analogue present in the crystallization droplet), transferred into a nylon loop (Hampton Research), flash-cooled in liquid nitrogen, and maintained at 100 K for data collection. Data collection statistics are presented in Table 1.
Table 1.
Data collection and refinement statistics
| OsD14 MPD complex | |
|---|---|
| Protein Data Bank id | 6ELX |
| Data collection | |
| Beam line | ESRF ID23-2 |
| Wavelength (Å) | 0.9919 |
| Space group | P212121 |
| Wavelength (Å) | 0.9919 |
| Unit cell parameters (Å) | a=48.1 b=88.6 c=118.9; α, β, γ,=90° |
| Solvent content (%) | 42.0 |
| Resolution (Å) | 44.61–1.35 (1.40–1.35) |
| No. of observations | 516 416 (36044) |
| No. of unique reflections | 107 535 (8569) |
| Completeness (%) | 95.72 (77.32) |
| Multiplicity | 4.8 (4.2) |
| R merge | 0.0510 (0.4184) |
| R meas a | 0.0572 (0.4757) |
| <I/(σI)> | 16.15 (2.91) |
| CC1/2 | 0.999 (0.813) |
| Refinement | |
| Resolution range (Å) | 44.61–1.35 (1.40–1.35) |
| No. of reflections | 107 538 (8569) |
| R work b | 0.159 (0.217) |
| R free c | 0.177 (0.229) |
| No. of atoms | |
| Protein | 4388 |
| Ligands | 58 |
| Waters | 607 |
| Average B-values (Å2) | 16.58 |
| Estimated from Wilson plot | 11.96 |
| Protein | 14.76 |
| Ligands | 32.73 |
| Solvent | 27.74 |
| Rms deviations from ideal values | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.19 |
| Ramachandran analysisd | |
| Favoured (%) | 98.1 |
| Allowed (%) | 1.9 |
| Outliers (%) | 0.0 |
Values in parentheses are for the outer resolution shell.
a R meas=∑h ∑l (nh/nh–1)1/2 | Ihl–<Ih> |/∑h ∑l <Ih> (Evans, 2006; Evans and Murshudov, 2013).
b R work=∑hkl| |Fo|–|Fc| |/∑hkl |Fo| where Fo and Fc are the observed and calculated structure factor amplitudes, respectively.
c R free calculated from a randomly chosen 5% of all unique reflections.
d From ‘MolProbity’ (Chen et al., 2010).
Structure determination and refinement
The X-ray diffraction data were processed, scaled, and merged with the XDS suite (Kabsch, 2010) with 5% of the data set aside for Rfree calculations. Phases were obtained by molecular replacement in Phenix (Adams et al. 2010) using the structure of RbsQ (PDB id 1wom; Kaneko et al., 2005) as a search model. Refinement of the initial model was performed with Phenix alternated with inspection and manual rebuilding in Coot (Emsley et al., 2010). Solvent molecules were identified using the water insertion command in Phenix and assigned based on peak heights of residual electron density, hydrogen-bonding patterns, and B-factors. Refinement statistics are presented in Table 1. The initial model was then used as a search model to solve all subsequent structures.
Analysis of deposited structures
A structural superposition of OsD14 with deposited structures in the PDB was performed with the least-squares superposition function in O (Jones et al., 1991). The default distance cut-off limit of 3.8 Å was used.
Initial σA-derived maximum-likelihood electron density maps, 2mFo–DFc and mFo–DFc (Read, 1986, Pannu and Read, 1996), where m is the figure of merit and D is the Luzzati factor (Luzzati, 1953), were calculated using the deposited models and diffraction data downloaded from the PDB. The 2mFo–DFc maps were coloured blue (or white) and contoured at 1.0 sigma (i.e. 1.0 SD above the mean electron density) and show where the model is expected to be, whereas the mFo–DFc maps were coloured green (3.0 sigma) and red (–3.0 sigma); the green mesh shows where atoms are missing in the current model, while the red mesh shows where atoms are present in the model but not in the crystal. The initial maps were calculated without any prior manipulation or refinement of the deposited co-ordinates. In a second step, omit maps were calculated in the same manner after excluding the ligand from the calculation. Thirdly, after inspection of native and omit maps, in the relevant cases a suitable ligand was built into the positive difference density appearing at the 3.0 sigma level followed by 10 cycles of maximum likelihood restrained refinement in REFMAC5 (Murshudov et al., 2011) or Phenix (Adams et al., 2010). During this operation, care was taken to use the programs and scripts of the originators, as described in the respective PDB header.
The figures were created with ‘PyMOL’ (Version 1.6.0.0, Schrödinger, LLC).
Results and Discussion
Structure solution and overall structure
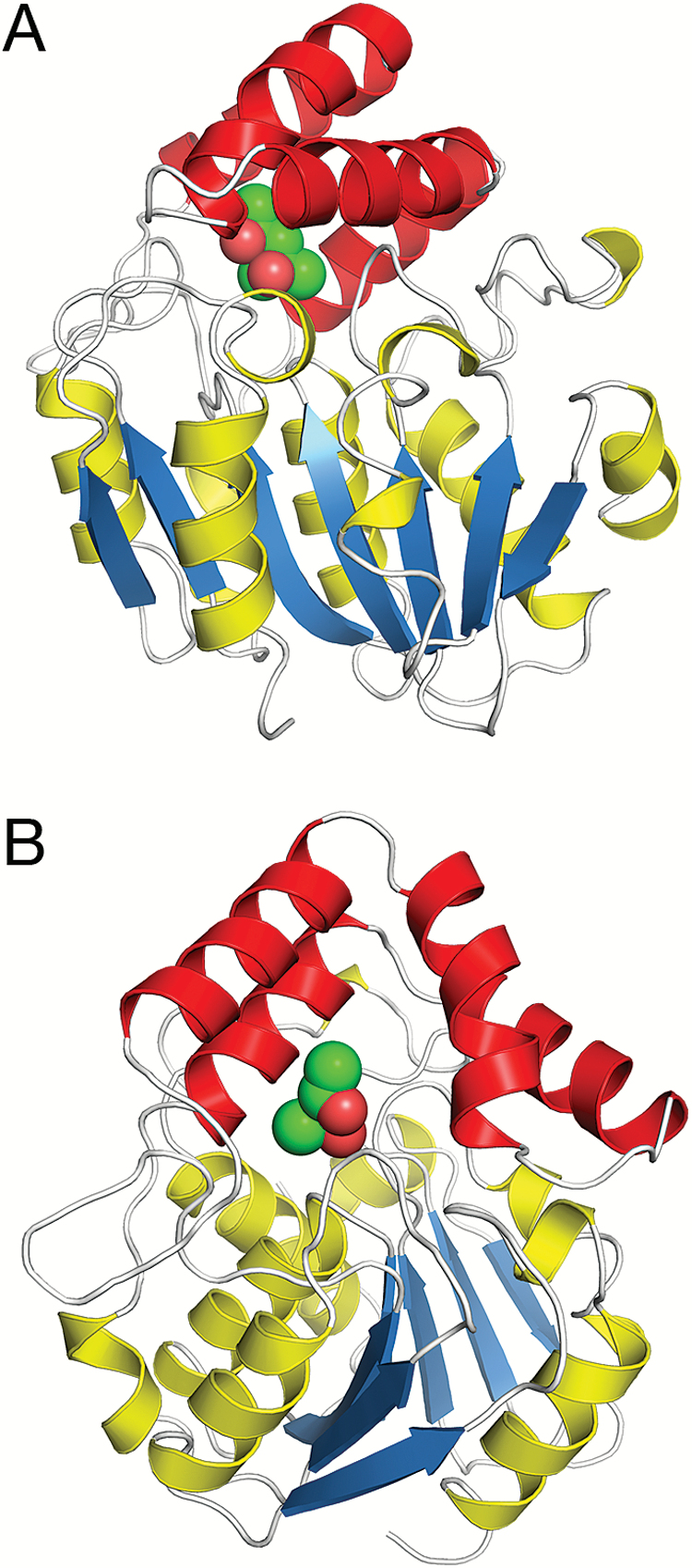
The structure of O. sativa Dwarf14 (OsD14) was determined by X-ray crystallography to a resolution of 1.35 Å from a construct lacking the first 51 residues. The N-terminal 51 residues comprise an unusual combination of repeated glycine and serine residues, suggesting that this part may be flexible in the mature protein and/or that it may be glycosylated. OsD14 has a α/β-hydrolase fold (Gaiji et al., 2012) with a seven-stranded β-sheet core domain flanked by seven α-helices, of which two are shorter 310-helices. The short version of the rice D14, lacking the first 51 residues, starts with a helix, which is also present in the α/β-hydrolase structure RbsQ (PDB id 1wom) that was used as a search molecule for the structure solution. The central seven-stranded sheet of OsD14 is built of six parallel strands with the first N-terminal strand running antiparallel to the core (Fig. 1A). Two of the flanking helices are located on one side of the sheet and the remaining five reside on the other side. Inserted between strands five and six of the central sheet are four helices connected to the core by a β-hairpin loop on its N-terminal side and a shorter loop on the other side. This domain, commonly named the helical cap, forms a V-shaped lid over a deep cavity, which is assumed to bind the SL hormone (Fig. 1B). The cavity is relatively large and lined by hydrophobic residues. At the bottom of the cavity is a canonical catalytic triad consisting of Ser147, His297, and Asp268.
Fig. 1.
(A) Overall structure of OsD14. β-Strands of the core are coloured blue, associated helices yellow, and helices in the helical cap red. MPD is depicted as green spheres. (B) Binding of MPD in the ligand-binding cavity showing the opening of the cavity towards the solution. The two views in (A) and (B) are related by a 90° rotation with respect to the vertical axis.
Binding cavity
After model building of the protein, residual density was observed in the ligand-binding cavity. We identified this as originating from MPD, which was used as a precipitating agent during crystallization. MPD was added as a racemic mixture of (4S)-2-methyl-2,4-pentanediol and (4R)-2-methyl-2,4-pentanediol, and it appears that both stereoisomers were able to bind in the cavity. Judging from the electron density, there was a preference for binding the 4S stereoisomer in one subunit (arbitrarily named molecule B in the PDB co-ordinate file). MPD is located some distance away from the catalytic triad, forms several hydrophobic interactions with residues lining the cavity (Phe78, Phe176, Phe186, Val194, Val244, and Phe245), and hydrogen-bonds to the hydroxyl oxygen of Tyr209 and to Oγ of Ser147 and Nε2 of His297 via a water molecule (Fig. 2A). In the second subunit (named molecule A), the binding of the 4R stereoisomer appears to be preferred, and the direct interaction with the Tyr209 hydroxyl group is replaced by a solvent-mediated contact. However, most probably there was a (unequal) mixture of the two stereoisomers binding at both sites. At this data resolution, estimating the contribution of the individual stereoisomers to each site is not feasible. The helical cap covers MPD like a dome, but is open to solvent on one side in this conformational state of the receptor (Fig. 1B). The undesired binding of MPD in the ligand-binding cavity of OsD14 illustrates a common problem in crystallographic investigations of ligand binding to proteins, as elaborated below.
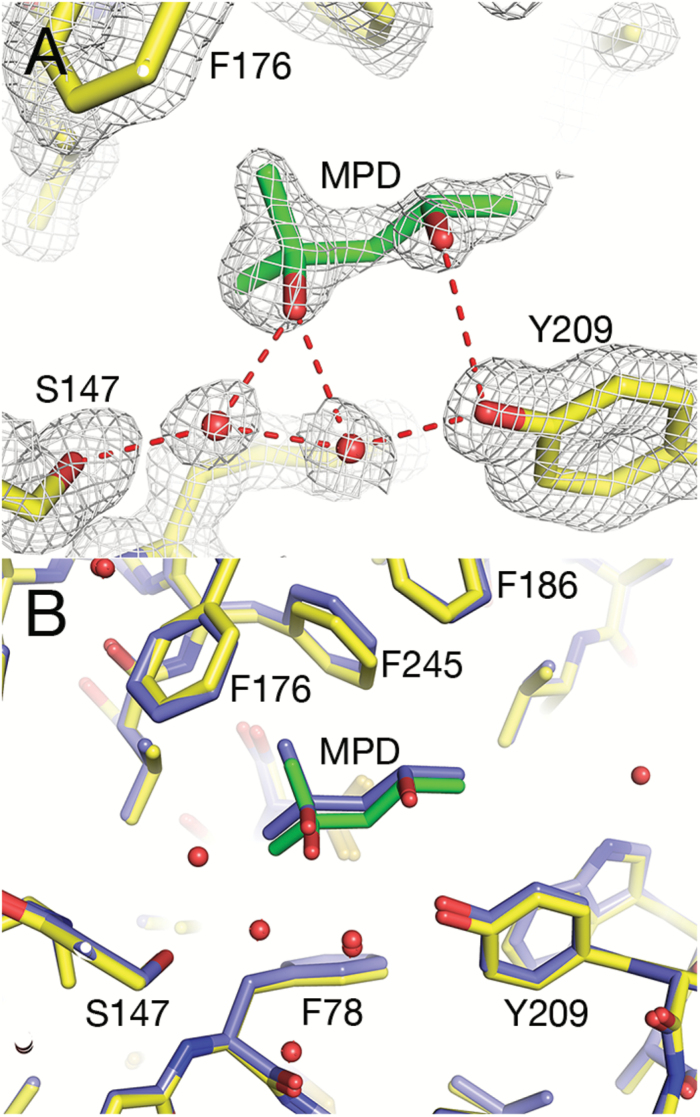
Fig. 2.
Binding cavity of OsD14. (A) Electron density around the ligand-binding site of OsD14, showing MPD built into the density. 2mFo–DFc map contoured at 1.0 sigma (white) (B) Superposition of OsD14 with the structure of O. sativa D14 in PDB id 3wo4 (coloured blue).
Difficulties in the crystallographic validation of structures of protein–ligand complexes
The interpretation of structures from electron density maps involves some level of subjectivity (Brändén and Jones, 1990), the extent of which depends on, for example, the quality of the X-ray data, the resolution, inherent disorder, and the experience of the interpreter. The now common practice of depositing the co-ordinates and the structure factor amplitudes in the PDB, which are then subjected to curation and analysis, has helped to raise the awareness of such errors. To aid this process, there are a number of programs available for validation and analysis of macromolecular structures (Read, 2011), which have helped to reduce significantly the number of erroneous protein structure models.
Validation of structures of protein–ligand complexes remains more problematic because of the infinitely variable chemical character of ligands that bind to protein, such as cofactors, substrate analogues, products, inhibitors, crystallization additives, etc., compared with the limited repertoire of the amino acids that constitute the protein structure. Common indicators of quality such as geometry and R factors work less well for small ligands; the geometry of small ligands is often poorly determined, and the low contribution of the ligand to the total scattering compared with that of the protein partner makes R factors a blunt indicator in this case. In addition, ligand occupancy depends on ligand binding affinity and concentration, and is most often incomplete. The high concentration of buffer molecules, crystallization cocktail components, etc. means that these molecules may often outcompete the desired ligand, especially if the affinity of the protein for the ligand in question is not very high. In the unfortunate situation when the binding of a desired ligand leads to conformational changes that are incompatible with the packing of the protein in the crystal lattice, crystallization may be hindered even when the ligand affinity is high. In this case, a less likely complex with common buffer molecules may crystallize instead. As a final hurdle, data collection at cryogenic temperatures requires the addition at high concentration of cryo-protectant molecules that often resemble, and therefore outcompete, the native ligand. Although clearly very important, issues on ligand binding to proteins have only recently come into focus (see, for example, Pozharski et al., 2013; Weichenberger et al., 2013; Adams et al., 2016).
Thus, the binding of MPD in the ligand-binding cavity of D14 is not surprising; the concentration of MPD is high (10%, v/v) and this compound, commonly used in crystallization experiments, may bind in an open cavity poised for binding a ligand—it may even outcompete a ligand initially bound. To avoid MPD binding in the ligand-binding cavity, extensive crystallization screening was carried out with alternative precipitating agents. Well diffracting crystals of apo OsD14 were obtained using PEG 6000 as a precipitating agent at concentrations of ~8–20% and in the pH range 6.5–9.5. Because of the slower spontaneous hydrolysis of SL analogue compounds at lower pH, crystallization of ligand complexes of OsD14 was generally performed at pH 6.5. A range of compounds were tried: a racemic mixture of GR24 and four other analogues of various sizes (see Supplementary Fig. S1). A mutation of the catalytic Ser147 was introduced and crystallization of the mutant S147A was performed in the same manner. This resulted in a number of diffraction data sets and corresponding structures; a selection is presented in Supplementary Table S1. Analysis of the potential ligand structures showed only minor structural changes: the overall structures were very similar and there were no major conformational transitions [root-mean-square deviations (rmsds) to the MPD-bound structure (PDB id 6elx) 0.15–0.56 Å for 267 Cα atoms of one molecule, Supplementary Table S1]. There was spurious additional density in the ligand-binding cavity indicating the binding of a compound (or a mixture of compounds) at low occupancy. This is shown for structures 5–18 and the mutant S147A (Supplementary Table S1), which were soaked with Compound C and co-crystallized with Compound B, respectively (Supplementary Fig. S1). In 5–18, the extra density extends from Ser147 into the binding cavity and defines a binding site lined by residues Phe78, Phe176, Phe186, Trp205, Tyr209, Cys241, Val244, Phe245, Ser270, and His297 (Supplementary Fig. S2A). The density partly overlaps with the binding site for MPD in the deposited structure (6elx) and we suspect the partial binding of the cryoprotectant glycerol in this structure. However, the contrast to the well-defined density of MDP, which binds at high occupancy, although it is held mainly by solvent-mediated contacts, is striking. As in the MPD-bound structure, Ser270 and Cys241 have double conformations but, in contrast to the MPD-bound structure, the side chain of Ser147 also has two conformations in the C-soaked complex. There is extended extra electron density on the Oγ atom, indicating the binding of a compound to one of the conformers. It is not possible to determine the nature of the bond, covalent or non-covalent, given the low occupancy. As expected, S147A lacks extra density at residue 147, and the density in the cavity appears more continuous (Supplementary Fig. S2B).
Failure to detect distinct binding of SL analogues (or hydrolysis products) to D14 may be explained in several ways. The low occupancy may be explained by low concentration of the SL analogue because of its low solubility in water. Also, if crystal lattice forces do not support the conformational transition required for SL binding, the compound may not be able to bind, or crystallization components may bind in its stead. Finally, if our construct is able to (slowly) hydrolyse the compounds (as already reported for identical constructs, e.g. Zhao et al., 2015), there may have been a mixed binding of hydrolysed and non-hydrolysed compounds in the cavity as well as glycerol and other compounds from the crystallization mixture. We can only speculate on this possibility, because the activity of our construct was not monitored. In spite of this caveat, detailed re-analysis of SL structures deposited in the PDB (see below) shows that these may be common problems, also when structures are obtained from proteins whose activity in solution had been experimentally confirmed before crystallization.
Comparison with other SL receptor structures
The interaction of MPD with our OsD14 is similar to that of MPD in the model of O. sativa D14 presented by Kagiyama et al. (2013; PDB id 3w04) (Fig. 2B). The two models superimpose with an rmsd of 0.337 Å for 262 Cα atoms (rmsd 0.680 Å for 526 Cα atoms of the two molecules in the asymmetric unit). Electron density for MPD in crystal structure 3w04 is visible in both molecules (A and B) and the binding of the ligand is stabilized by contacts of O4 with the hydroxyl group of Tyr209 and by solvent-mediated hydrogen bonds between O2 of MPD and Oγ of Ser147 and Nε2 of His297.
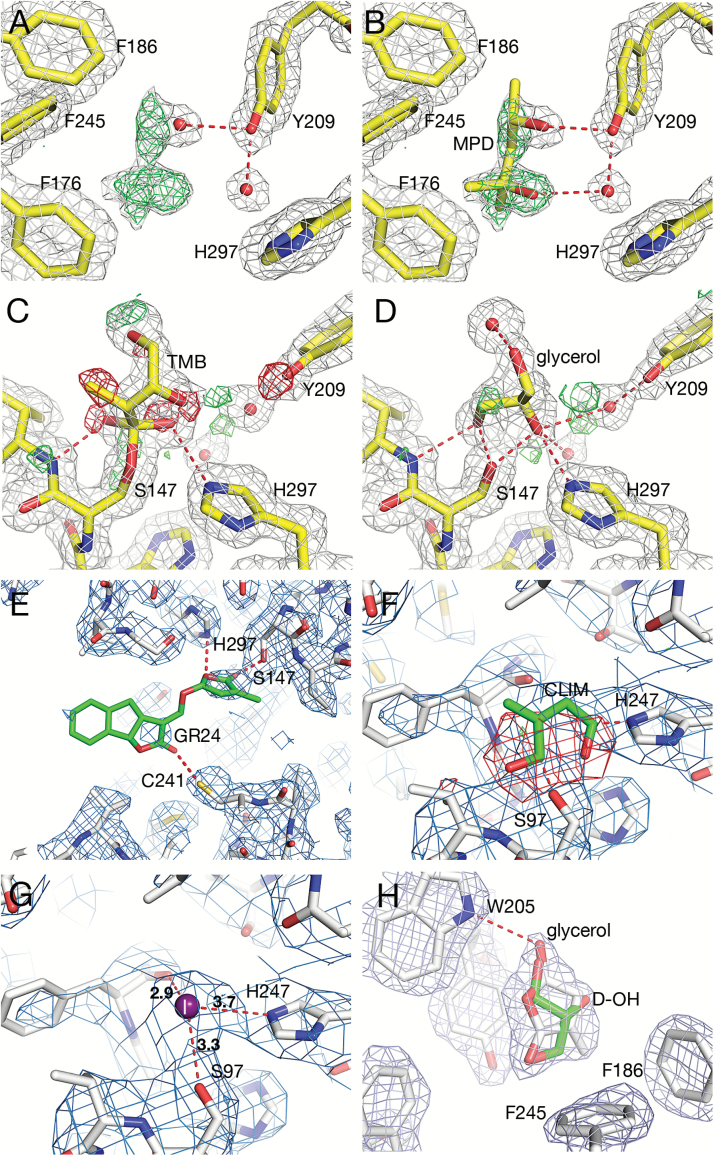
MPD seems poised to bind in the ligand-binding cavity of D14. In the model of the O. sativa apo D14 (PDB id 4ih9; Zhao et al., 2013), extensive positive electron density was detected in the mFo–DFc map in one of the monomers (named B) in the asymmetric unit (Fig. 3A). The density, which extends from a solvent molecule (water B463) into the ligand-binding pocket, may be interpreted as MPD [present at 5% (v/v) during crystallization]. MPD modelled (Fig. 3B) at this site (replacing water B463) would be held in place by hydrogen bonds from O4 to the hydroxyl of Tyr209(159) (residue numbers refer to the numbering of the full-length rice sequence, whereas numbers in parentheses refer to the number in the PDB co-ordinate file) and by a solvent-mediated hydrogen bond between MPD O2 and the hydroxyl of Tyr209(159).
Fig. 3.
Comparison of ligand binding in diverse D14 structures. Numbering of residues in (A–E) and (H) refers to the full-length O. sativa sequence, whereas (F) and (G) show the A. thaliana D14 with a shorter sequence. Dotted lines indicate atoms within hydrogen-bonding distance. Bonding distances are in Å. (A) Positive density was detected in the ligand-binding cavity of O. sativa D14 (PDB id 4ih9) close to a water molecule. 2mFo–DFc map contoured at 1.0 sigma (white), and mFo–DFc map contoured at 3.0 sigma (green). (B) Same as (A) with MPD built into the vacant density, replacing water. (C) O. sativa D14 (PDB id 4iha; Zhao et al., 2013) with its degradation intermediate, 2,4,4,-trihydroxy-3-methyl-3-butenal (TMB). 2mFo–DFc map contoured at 1.0 sigma (white) and mFo–DFc contoured at 3.0 sigma (green) and –3.00 sigma (red) calculated using submitted co-ordinates. Note the red mesh indicating that these modelled atoms are not present in the crystal. (D) Glycerol built into the TMB site. 2mFo–DFc map contoured at 1.0 sigma (white) and 2mFo–DFc contoured at 3.0 sigma (green) and –3.0 sigma (red) calculated after 20 cycles of refinement in Refmac5. This shows a better fit to the electron density than that in (C). (E) O. sativa D14 (PDB id 5dj5, Zhao et al., 2015) with GR24 (green) modelled into the ligand-binding cavity. Omit 2mFo–DFc map (blue) calculated after removing the ligand. The map was contoured at 1.0 sigma. Note the weak density for the ligand. (F) A. thaliana D14 receptor in a complex with the proteins D3 from O. sativa and ASK1 from A. thaliana (PDB id 5hzg; Yao et al., 2016). The GR24 hydrolysis intermediate, CLIM (green), modelled into the ligand-binding cavity. 2mFo–DFc map contoured at 1.0 sigma (blue) and mFo–DFc map contoured at –3.0 sigma (red) calculated from submitted co-ordinates. Note the red mesh indicating that these modelled atoms are not present in the crystal. (G) Iodide ion (purple sphere) modelled into the CLIM site. 2mFo–DFc map contoured at 1.0 sigma (blue). This shows a reasonable fit to the electron density, and no peaks were found in the mFo–DFc maps contoured at ±3.0 sigma. (H) O. sativa L. cv. Shiokari D14 (PDB id 3wio; Nakamura et al., 2013). 2mFo–DFc map contoured at 1.0 sigma calculated from submitted co-ordinates. Glycerol (green) was modelled into the density at the opening of the cavity and superposed on the D-OH co-ordinates (in white). This shows that glycerol fits equally well as D-OH to the density.
In a related structure (PDB id 4iha; Zhao et al., 2013), covalent binding of a putative degradation intermediate, 2,4,4-trihydroxy-3-methyl-3-butenal (TMB), to O. sativa D14 was described. However, analysis of electron density maps (2mFo–DFc and mFo–DFc) calculated from deposited data indicates a less than satisfactory fit of the intermediate to the electron density. In one subunit (named A), electron density is lacking (negative electron density indicating parts of the model that are not supported by the data) for O2, O4, and C5 of TMB (red mesh in Fig. 3C). In the second subunit (named B), there is negative electron density for O2 and O4 of the intermediate and there is extra density (positive density indicating features present in the data that are not accounted for by the model) close to C4 (not shown). TMB is the hydrate and open form of D-OH and is likely to co-exist with D-OH. Our analysis of the electron density maps indicates that a mixture of TMB and D-OH may fit better in the density. Alternatively, glycerol [present at 20% (v/v) during cryo-cooling] may also bind at the same site. Glycerol (Fig. 3D) would be held in place by an extensive network of hydrogen bonds: between O1 and one water molecule, between O2 and Oγ of Ser147(97), Nε2 of His297(247), and two water molecules, and between O3 and Oγ of Ser147(97) and the backbone N atoms of Val148(98) and Phe78(28). The conclusion is that glycerol is a more likely ligand than TMB in the crystal structure deposited as 4iha, but the presence of some small positive peaks (green) in the difference density map indicates that a mixture of the two ligands may bind at the site.
Glycerol was also modelled in the internal cavity of petunia DAD2 (PDB id 4dnp) by Hamiaux et al. (2012). Inspection of electron density maps calculated from the deposited model reveals that glycerol is only partially supported by electron density; O1 is supported by electron density, but lacks interaction with the protein, whereas O2 is supported by electron density and hydrogen-bonds to Nε2 of the catalytic His246 (not shown). There is no density for the O3 hydroxyl, which in the model would be hydrogen-bonded to the catalytic Ser96 and to two solvent molecules, but apparently the placing of the O3 hydroxyl was inferred from another crystal structure (see the supplementary experimental procedures in Hamiaux et al., 2012).
Binding of the SL analogue GR24 to rice D14 was detected in solution by Zhao et al. (2015). To capture the complex in the crystal, GR24 was added at a high molar ratio to a dilute solution of D14 that was subsequently concentrated for crystallization. Electron density maps calculated from the structure show only very weak density in the ligand-binding cavity (PDB id 5dj5) and there are no significant conformational differences between the crystal structures with or without GR24, as would be expected if a ligand was bound. The electron density map calculated after removing GR24 from the structure shows some faint electron density close to the hydroxyl group of the catalytic Ser147(97), whereas there is no density indicating the presence of the ABC moiety (Fig. 3E). The very weak residual density in the omit maps indicates only partial occupancy of a (small) potential ligand, which in this case would be impossible to identify. Because of the low solubility of GR24 in water, it may fall out of solution during concentration/crystallization. It may also be that the crystal packing does not support the conformational changes required to bind GR24 and that therefore apo D14 was crystallized, without the ligand being able to bind.
A conformational transition was detected by Yao et al. (2016) in the crystal structure (PDB id 5hzg) of a part of the SCF degradation complex involved in SL signal propagation, where D14 is shown to undergo a shape change that closes its large open ligand-binding cavity. The crystallized complex consists of the proteins A. thaliana D14, O. sativa D3, and A. thaliana SCF-component ASK1 (Arabidopsis Skp1-like). The conformational transition consists of rotational movements, coiling, unfolding, and extending of the four α-helices of the helical cap, resulting in the shrinking of the ligand-binding cavity and the formation of a lid that closes the cavity. This finding contrasts with the results from a crystal structure of a TIR–ASK1 complex active in auxin perception, where auxin does not induce conformational changes, but instead acts as a molecular glue in mediating ligand–receptor interactions (Tan et al., 2007).
A small ligand appears to be trapped in the cavity of D14 and was identified as a GR24 hydrolysis intermediate, named CLIM (covalently linked intermediate molecule), covalently linked to His247, Ser97, or both (Yao et al., 2016). Analysis of electron density maps calculated from co-ordinates of the complex (Fig. 3F) shows that the size of the density peak around the intermediate is too small to accommodate CLIM, leaving a major part (atoms C4, C5, O1, and O2) of the putative intermediate outside of the electron density (blue mesh in Fig. 3F). In addition, a large negative peak in the mFo–DFc difference map (red mesh in Fig. 3F) indicates the presence of atoms in the model which are not present in the crystal. The crystallization solution contained 200 mM KI and 10 mM Pr(III) acetate. We conclude that the most likely ligand is an iodide ion. Insertion of iodide into density followed by 10 cycles of restrained refinement (according to the protocol described by the authors) shows this to be consistent with the shape and position of the density peak (Fig. 3G). However, the modelling of a small compound at the relatively low resolution of the data (3.3 Å) is not straightforward. In addition, refinement appears to be underdetermined; that is, the number of parameters to be refined (62 704) exceeds the number of unique reflections (49 694). Refinement of grouped B-factors (one per residue instead of one per atom) would be recommended in this case (see, for example, p. 20 of Wlodawer et al., 2008). Acetate was present at 10 mM concentration in the crystallization solution; therefore, acetylation of the (reactive) catalytic serine may also be considered. If CLIM was not bound in the active site cavity, then what triggered the conformational transition? One possibility is that GR24 may be initially bound and fragmented, thereby triggering the shape change that allows the ternary complex D14–D3–ASK1 to form. The intermediate fragment may subsequently be replaced by iodide ion, which was present at high concentration in the crystallization cocktail.
From all of the above, the fate of an SL or an SL analogue binding in the cavity of the receptor is still to be considered unknown. What is the exact structure of the putative intermediate and how does it interact with the receptor? Based on a structure of rice D14 (PDB id 3wio; Nakamura et al., 2013) a mechanism was suggested where D-OH binds not at the bottom of the ligand-binding cavity near the catalytic triad but at the opening of the cavity. D-OH is suggested to act as a plug for the catalytic cavity and to induce a hydrophobic patch to facilitate the interaction of D14 and its target proteins. Electron density maps calculated from the submitted co-ordinates show a reasonable fit to the electron density of a compound, H3M, that mimics D-OH but lacks the required planarity of the five-membered ring (Fig. 3H). H3M was modelled in one of the molecules in 3wio, whereas the ligand-binding cavity is empty in both molecules A and B in the apo structure (PDB id 3vxk). There is no conformational change detected in comparing the structures of the apo enzyme and the H3M ligand complex. However, modelling of glycerol [present at 10% (v/v) during purification] shows that this compound also fits the electron density at the opening of the cavity (Fig. 3H). There is also the possibility that a mixture of compounds may bind. Thus, also in this case, the nature of the bound compound may be considered unknown.
Conclusions
To identify the active form(s) of SL in plants and their molecular mode of action has long been a major technical challenge and a hotspot in SL biology. However, the unusual nature of the SL hormone receptor, simultaneously able to perceive and hydrolyse (destroy) the hormone, has made this a formidable task. There is evidence from studies in solution suggesting that the receptor may cleave the SL and form a covalent bond to the hormone fragment (de Saint Germain et al., 2016; Yao et al., 2016). This in turn may trigger a dramatic change in shape of the receptor, thereby forming surfaces that can interact with signalling partners. The rate of cleavage appears to be extremely slow, and may in fact stall at the first turnover (Hamiaux et al., 2012; de Saint Germain et al., 2016). The chemical nature of the hormone fragment, supposedly a small molecule, ~5 carbon atoms in size, is another obstacle in the way of determining a structure of a complex between the receptor and its hormone. Results from our attempts at capturing a complex of D14 with SL analogues and our analysis of available structures of SL receptor complexes indicate that in most cases the modelled compound may either be outcompeted by common buffer and crystallization compounds, or may bind at very low occupancy, making modelling extremely uncertain. It is also possible that the conformational transitions accompanying SL binding may be incompatible with the packing of the protein in the crystal lattice, leading to the apo protein being titrated out of solution (meaning that the subpopulation of the apo enzyme was preferentially crystallized, leaving behind any complex that incorporated the ligand). We conclude that the active forms of SL and their interaction with the SL receptor are yet to be identified.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Data collection and refinement statistics for D14–ligand complex searches.
Fig. S1. Strigolactone analogues used in crystallization experiments.
Fig. S2. Electron densities in the ligand-binding cavity of OsD14 native and S147A mutant proteins.
Acknowledgements
We wish to acknowledge the European Synchrotron Radiation Facility, Grenoble, for providing beam time and data collection facilities, and thank the staff at beam line ID23-2 for excellent support. This work was supported by a COST grant from the European Union (STREAM. Action no. FA1206) and by the STRItools project from the Compagnia di San Paolo (‘Addressing Horizon 2020’, 2014) to CP and FC.
Glossary
Abbreviations:
- CLIM
covalently linked intermediate molecule
- D14
DWARF14
- GID1
Gibberellin Insensitive Dwarf1
- MAX
MORE AXILLARY GROWTH
- MPD
2-methylpentanediol
- rmsd
root-mean-square deviation
- SCF
Skp, Cullin, F-box containing complex
- SL
strigolactone; SMAX1, SUPPRESSOR OF MAX2 1
- SMXL
SMAX1-LIKE
- TMB
2,4,4-trihydroxy-3-methyl-3-butenal.
References
- Adams PD, Aertgeerts K, Bauer C et al. 2016. Outcome of the first wwPDB/CCDC/D3R ligand validation workshop. Structure 24, 502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica. Section D, Biological Crystallography 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K, Matsuzaki K, Hayashi H. 2005. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827. [DOI] [PubMed] [Google Scholar]
- Al-Babili S, Bouwmeester HJ. 2015. Strigolactones, a novel carotenoid-derived plant hormone. Annual Review of Plant Biology 66, 161–186. [DOI] [PubMed] [Google Scholar]
- Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J. 2009. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant and Cell Physiology 50, 1416–1424. [DOI] [PubMed] [Google Scholar]
- Artuso E, Ghibaudi E, Lace B et al. 2015. Stereochemical assignment of strigolactone analogues confirms their selective biological activity. Journal of Natural Products 78, 2624–2633. [DOI] [PubMed] [Google Scholar]
- Bennett T, Leyser O. 2014. Strigolactone signalling: standing on the shoulders of DWARFs. Current Opinion in Plant Biology 22, 7–13. [DOI] [PubMed] [Google Scholar]
- Bouwmeester HJ, Roux C, Lopez-Raez JA, Bécard G. 2007. Rhizosphere communication of plants, parasitic plants and AM fungi. Trends in Plant Science 12, 224–230. [DOI] [PubMed] [Google Scholar]
- Brändén C-I, Jones TA. 1990. Between objectivity and subjectivity. Nature 343, 687–689. [Google Scholar]
- Cardoso C, Ruyter-Spira C, Bouwmeester HJ. 2011. Strigolactones and root infestation by plant-parasitic Striga, Orobanche and Phelipanche spp. Plant Science 180, 414–420. [DOI] [PubMed] [Google Scholar]
- Catanzariti AM, Soboleva TA, Jans DA, Board PG, Baker RT. 2004. An efficient system for high-level expression and easy purification of authentic recombinant proteins. Protein Science 13, 1331–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica. Section D, Biological Crystallography 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook CE, Whichard LP, Turner B, Wall ME, Egley GH. 1966. Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154, 1189–1190. [DOI] [PubMed] [Google Scholar]
- de Saint Germain A, Clavé G, Badet-Denisot MA et al. 2016. An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nature Chemical Biology 12, 787–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallographica. Section D, Biological Crystallography 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. 2006. Scaling and assessment of data quality. Acta Crystallographica. Section D, Biological Crystallography 62, 72–82. [DOI] [PubMed] [Google Scholar]
- Evans PR, Murshudov GN. 2013. How good are my data and what is the resolution?Acta Crystallographica. Section D, Biological Crystallography 69, 1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiji N, Cardinale F, Prandi C, Bonfante P, Ranghino G. 2012. The computational-based structure of Dwarf14 provides evidence for its role as potential strigolactone receptor in plants. BMC Research Notes 5, 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB et al. 2008. Strigolactone inhibition of shoot branching. Nature 455, 189–194. [DOI] [PubMed] [Google Scholar]
- Hamiaux C, Drummond RS, Janssen BJ, Ledger SE, Cooney JM, Newcomb RD, Snowden KC. 2012. DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Current Biology 22, 2032–2036. [DOI] [PubMed] [Google Scholar]
- Hayward A, Stirnberg P, Beveridge C, Leyser O. 2009. Interactions between auxin and strigolactone in shoot branching control. Plant Physiology 151, 400–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Liu X, Xiong G et al. 2013. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504, 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallographica. Section A, Foundations of Crystallography 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch W. 2010. XDS. Acta Crystallographica. Section D, Biological Crystallography 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiyama M, Hirano Y, Mori T, Kim SY, Kyozuka J, Seto Y, Yamaguchi S, Hakoshima T. 2013. Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes to Cells 18, 147–160. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Tanaka N, Kumasaka T. 2005. Crystal structures of RsbQ, a stress-response regulator in Bacillus subtilis. Protein Science 14, 558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzzati V. 1953. Resolution d’une structure cristalline lorsque les positions d’une partie des atomes sont connues—traitement statistique. Acta Crystallographica 6, 142–152. [Google Scholar]
- Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallographica. Section D, Biological Crystallography 67, 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Xue YL, Miyakawa T et al. 2013. Molecular mechanism of strigolactone perception by DWARF14. Nature Communications 4, 2613. [DOI] [PubMed] [Google Scholar]
- Pannu NS, Read RJ. 1996. Improved structure refinement through maximum likelihood. Acta Crystallographica. Section A, Foundations of Crystallography 52, 659–668. [Google Scholar]
- Pozharski E, Weichenberger CX, Rupp B. 2013. Techniques, tools and best practices for ligand electron-density analysis and results from their application to deposited crystal structures. Acta Crystallographica. Section D, Biological Crystallography 69, 150–167. [DOI] [PubMed] [Google Scholar]
- Prandi C, Occhiato EG, Tabasso S, Bonfante P, Novero M, Scarpi D, Bova ME, Miletto I. 2011. New potent fluorescent analogues of strigolactones: synthesis and biological activity in parasitic weed germination and fungal branching. European Journal of Organic Chemistry 2011, 3781–3793. [Google Scholar]
- Read RJ. 1986. Improved fourier coefficients for maps using phases from partial structures with errors. Acta Crystallographica. Section A, Foundations of Crystallography 42, 140–149. [Google Scholar]
- Read RJ, Adams PD, Arendall WB 3rd et al. 2011. A new generation of crystallographic validation tools for the protein data bank. Structure 19, 1395–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP. 2010. Gibberellin–GID1–DELLA: a pivotal regulatory module for plant growth and development. Plant Physiology 154, 567–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. 2007. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645. [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S et al. 2008. Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200. [DOI] [PubMed] [Google Scholar]
- Waters MT, Nelson DC, Scaffidi A, Flematti GR, Sun YK, Dixon KW, Smith SM. 2012. Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139, 1285–1295. [DOI] [PubMed] [Google Scholar]
- Weichenberger CX, Pozharski E, Rupp B. 2013. Visualizing ligand molecules in Twilight electron density. Acta Crystallographica. Section F, Structural Biology and Crystallization Communications 69, 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodawer A, Minor W, Dauter Z, Jaskolski M. 2008. Protein crystallography for non-crystallographers, or how to get the best (but not more) from published macromolecular structures. FEBS Journal 275, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R, Ming Z, Yan L et al. 2016. DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536, 469–473. [DOI] [PubMed] [Google Scholar]
- Zhao LH, Zhou XE, Wu ZS et al. 2013. Crystal structures of two phytohormone signal-transducing α/β hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Research 23, 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LH, Zhou XE, Yi W et al. 2015. Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3. Cell Research 25, 1219–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Lin Q, Zhu L et al. 2013. D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504, 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.