This is the first case of wild-type roots being unable to suppress branching in a strigolactone-deficient scion. MdCCD7 RNAi primary shoots also exhibited an increased growth rate relative to the wild type.
Keywords: CAROTENOID CLEAVAGE DIOXYGENASE (CCD), Malus×domestica, shoot growth rate, strigolactone, sylleptic branching
Abstract
Branching has a major influence on the overall shape and productivity of a plant. Strigolactones (SLs) have been identified as plant hormones that have a key role in suppressing the outgrowth of axillary meristems. CAROTENOID CLEAVAGE DIOXYGENASE (CCD) genes are integral to the biosynthesis of SLs and are well characterized in annual plants, but their role in woody perennials is relatively unknown. We identified CCD7 and CCD8 orthologues from apple and demonstrated that MdCCD7 and MdCCD8 are able to complement the Arabidopsis branching mutants max3 and max4 respectively, indicating conserved function. RNAi lines of MdCCD7 show reduced gene expression and increased branching in apple. We performed reciprocal grafting experiments with combinations of MdCCD7 RNAi and wild-type ‘Royal Gala’ as rootstocks and scion. Unexpectedly, wild-type roots were unable to suppress branching in MdCCD7 RNAi scions. Another key finding was that MdCCD7 RNAi scions initiated phytomers at an increased rate relative to the wild type, resulting in a greater node number and primary shoot length. We suggest that localized SL biosynthesis in the shoot, rather than roots, controls axillary bud outgrowth and shoot growth rate in apple.
Introduction
Plant architecture is largely dictated by the activities of meristems (Hallé et al., 1978; Steeves and Sussex, 1989). The shoot apical meristem (SAM) forms the primary axis by initiating new phytomers: repeating units of leaf, axillary meristem, node, and internode. The activity of axillary meristems determines the pattern of branching, which contributes greatly to the overall shape of a plant. In annual plants, axillary meristems form and some of these grow out into secondary shoots, a process that is highly plastic and influenced by genetic, developmental, environmental, and nutritional factors, and interactions with surrounding plants (Khayat and Zieslin, 1982; Kim et al., 2010; Huché-Thélier et al., 2011; Pierik and Testerink, 2014; Drummond et al., 2015). In perennial species, branching is even more complex because axillary meristems can have multiple fates: extending into a shoot without a dormant period (sylleptic shoot), developing into a vegetative shoot after a dormant period (proleptic shoot), developing into a floral bud that opens the following spring, or remaining dormant indefinitely (Costes et al., 2014). In addition, environmental signals such as temperature and day length, planting density, internal competition for carbon between buds, and the effects of pruning and plant manipulation all integrate to regulate the outgrowth of axillary meristems and buds (Girault et al., 2008, 2010; Henry et al., 2011; Rabot et al., 2012; Djennane et al., 2014; Fanwoua et al., 2014).
Plant hormones regulate the outgrowth of axillary meristems through a series of complex interactions (reviewed by Domagalska and Leyser, 2011; Müller and Leyser, 2011; Janssen et al., 2014; Rameau et al., 2015). Auxin has long been implicated in the mechanism of apical dominance over axillary meristems (Thimann and Skoog, 1933; Snow, 1937; Sachs and Thimann, 1964). More recently, genetic mutants that show increased branching have led to the discovery of the newest class of plant hormones, strigolactones (SLs) (Gomez-Roldan et al., 2008; Umehara et al., 2008). These mutants include the more axillary growth (max) mutants of Arabidopsis (Arabidopsis thaliana) (Sorefan et al., 2003; Stirnberg et al., 2002; Turnbull et al., 2002), the ramosus (rms) mutants of pea (Pisum sativum) (Beveridge, 2000; Morris et al., 2001; Rameau et al., 2002), the decreased apical dominance (dad) mutants of petunia (Petunia hybrida) (Napoli, 1996; Napoli and Ruehle, 1996), and the dwarf (d) and high-tillering dwarf1 (htd1) mutants of rice (Oryza sativa) (Zou et al., 2006; Arite et al., 2007).
The biosynthesis of SLs is still not completely understood, but includes the isomerization of a carotenoid precursor by DWARF27, followed by cleavage and cyclization to give carlactone by two CAROTENOID CLEAVAGE DIOXYGENASE (CCD) genes, CCD7 and CCD8 (Alder et al., 2012). Later steps of biosynthesis may vary by species and final products made, but include a cytochrome P450 [e.g. MAX1 in Arabidopsis and Os01g077900 and Os01g0701400 in rice (Zhang et al., 2014)], an oxidoreductase-like enzyme [LATERAL BRANCHING OXIDASE (Brewer et al., 2016], and a sulphotransferase [LOW GERMINATION STIMULANT1 (Gobena et al., 2017)].
CCD7 and CCD8 genes have been identified and corresponding mutant phenotypes have been well characterized in a number of species. MAX3, RMS5, DAD3, and D17/HTD1 are orthologous genes encoding CCD7 (Booker et al., 2004; Johnson et al., 2006; Zou et al., 2006; Drummond et al., 2009), and MAX4, RMS1, DAD1, and D10 are orthologous genes encoding CCD8 (Sorefan et al., 2003; Snowden et al., 2005; Arite et al., 2007). Grafting experiments have demonstrated that a wild-type (WT) rootstock can restore normal branching patterns in max3, max4, rms5, rms1, dad3, and dad1 mutants that are deficient in CCD7 or CCD8, indicating that these genes are necessary to synthesize a long-range signal that inhibits branching (Beveridge et al., 1994; Napoli, 1996; Foo et al., 2001; Morris et al., 2001; Turnbull et al., 2002; Sorefan et al., 2003; Simons et al., 2007). This SL signal can originate in roots, but more complex grafting experiments also indicate that sufficient signal to suppress branching can also be stem derived and only moves acropetally (Napoli, 1996; Foo et al., 2001; Turnbull et al., 2002; Simons et al., 2007).
The phenotypes resulting from SL deficiency are well characterized in annual plants, but less so in woody perennials. Grafting experiments involving combinations of SL-deficient scion and roots are highly informative, but have never been performed in any perennial fruit or nut crop. Given that many of these crops are routinely grafted onto clonal rootstocks, it is important to understand the effects of SL deficiency in scion and rootstocks. Knockdown of the kiwifruit (Actinidia chinensis) CCD8 orthologue resulted in an increased number of branches (Ledger et al., 2010). SL pathway genes have been identified in poplar (Populus), and knockdown of the poplar CCD8 orthologues resulted in short, highly branched trees (Czarnecki et al., 2014; Muhr et al., 2016). In willow (Salix spp.), an allelic variant of SxMAX4 that does not fully complement the max4 mutant was associated with increased branching after coppicing (Salmon et al., 2014). The identification of such allelic variants can be useful in developing new germplasm.
Apple (Malus×domestica) is a deciduous woody perennial fruit tree grown in commercial orchards, home gardens, and in the wild. In commercial orchards, tree growth is managed to provide optimal light interception essential for high yields. Maintaining optimal shoot architecture is time consuming and costly; therefore, knowledge about the genes regulating bud outgrowth are of importance to breeding programmes. To this end, we identified apple orthologues of CCD7 and CCD8, and demonstrated that they are able to complement max3 and max4, respectively, and that RNAi knockdown of MdCCD7 in apple resulted in increased sylleptic branching. Reciprocal grafting experiments with RNAi MdCCD7 and WT scions and rootstocks gave an unexpected result: WT roots were unable to rescue normal branching patterns in RNAi MdCCD7 scions. Our results suggest that localized SL biosynthesis in the shoot, but not in roots, controls sylleptic branching in apple trees.
Materials and methods
Identification and cloning of MdCCD7 and MdCCD8 genes
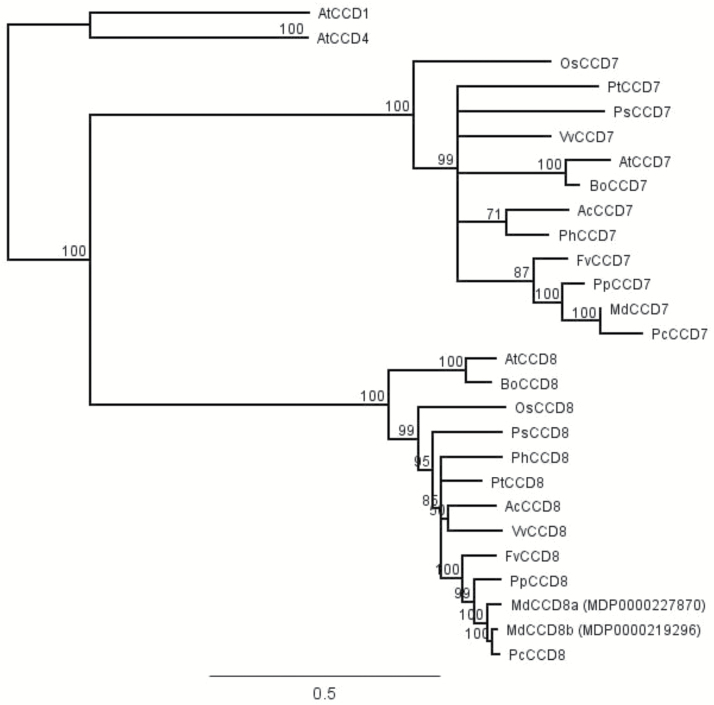
The apple CCD7 and CCD8a genes were isolated using a combination of searching an in-house EST data base, degenerate primer PCR, gene walking, and 5'/3' RACE as described previously (Ledger et al., 2010). Full-length cDNAs of MdCCD7 and MdCCD8a were amplified from ‘Royal Gala’ (RG) root mRNA. A second MdCCD8 gene, hereafter referred to as MdCCD8b, was identified later by whole-genome sequencing. All the primers used in this study are listed in Supplementary Table S1 at JXB online. Amino acid sequences of MdCCD7, MdCCD8a/b, and other full-length CCD proteins were aligned using ClustalW. The phylogenetic tree shown in Figure 1 was constructed using the maximum-likelihood principle and bootstrap values of 500 replicates using Geneious Pro software version 10 (Auckland, New Zealand).
Fig. 1.
Unrooted phylogenetic tree of the CAROTENOID CLEAVAGE DIOXYGENASE (CCD) gene family. Only full-length members of the family are included. The predicted sequences were aligned using ClustalW in Geneious (version 10). Phylogenetic relationships were calculated using the maximum-likelihood principle, and bootstrap values with 500 replicates were determined using Geneious. The scale bar is the number of substitutions per site. Accession numbers for the sequences are as follows: Actinidia chinensis AcCCD7 (ADP37985.1), AcCCD8 (ADP37984.1); Arabidopsis thaliana AtCCD1 (AT3G63520), AtCCD4 (AT4G19170), AtCCD7 (AT2G44990.1), AtCCD8 (AT4G32810); Brassica oleracea BoCCD7 (XP_013635602.1), BoCCD8 (XP_013589279.1); Fragaria vesca FvCCD7 (XP_004306976.2), FvCCD8 (XP_011458989.1); Malus×domestica MdCCD7 (MF034498), MdCCD8a (XP_008378214.1), MdCCD8b (XP_008352014.1); Oryza sativa OsCCD7 (LOC_Os04g46470.1), OsCCD8 (XP_015642760.1); Petunia×hybrida PhCCD7 (ACY01408.1), PhCCD8 (AAW33596.1); Pisum sativum PsCCD7 (ABD67496.2), PsCCD8 (AAS66906.1); Populus trichocarpa PtCCD7 (XP_006375244.1), PtCCD8 (XP_002324797.1); Prunus persica PpCCD7 (XP_007221108.1), PpCCD8 (XP_007222386.2); Pyrus communis PcCCD7 (PCP005718), PcCCD8 (PCP000841); Vitis vinifera VvCCD7 (XP_002274198.1), VvCCD8 (XP_002281239.2).
Complementation of Arabidopsis max3 and max4 mutants
Full-length cDNA clones of MdCCD7 and MdCCD8a were cloned into pHEX2 (Hellens et al., 2005), both under the control of the Cauliflower mosaic virus (CaMV) 35S promoter. The overexpression constructs were transformed into Agrobacterium tumefaciens strain LBA4404. The MdCCD7 overexpression construct was transformed into the Arabidopsis max3-1 mutant, and the MdCCD8a overexpression construct was transformed into the Arabidopsis max4-2 mutant using the floral dip method (Clough and Bent, 1998). Seven independent lines of MdCCD7/max3 and eight independent lines of MdCCD8a/max4 were generated. T2 seeds were selected on kanamycin in tissue culture then transferred to the glasshouse and grown under long days (16 h light:8 h dark). Control plants containing pHEX2 (an empty vector control) were germinated on kanamycin, and max3-1 and max4-2 seeds were germinated in tissue culture without antibiotic selection. The number of rosette branches and primary shoot length were recorded 10 weeks after planting, once flowering was complete. Between 5 and 16 biological replicates of each independent transformation line were phenotyped. Mean rosette branch number was compared by ANOVA followed by Fisher’s protected least significant difference (LSD) multiple comparison test to determine if the MdCCD7 and MdCCD8a lines were able to complement the max3-1 and max4-2 mutant phenotypes, respectively (GenStat 17th edition, VSN International, Hemel Hempstead, UK).
Transgenic RNAi knockdown of MdCCD7 and MdCCD8 in apple and phenotype measurements
To generate RNAi knockdown lines of MdCCD7 and MdCCD8 in apple, a 424 bp fragment from the RG cDNA sequence of MdCCD7 and a 482 bp fragment from the RG cDNA sequence of MdCCD8a were both cloned as inverted repeats into the vector pTKO2 (Snowden et al., 2005) to produce RNAi constructs for each gene (Supplementary Table S2). It was assumed that the MdCCD8a construct could knock down both MdCCD8a and MdCCD8b. These constructs were transformed into A. tumefaciens strain LBA4404 and then used to generate nine independent lines of MdCCD7 RNAi and 15 of MdCCD8 RNAi using the protocol described by Yao et al. (1995). Regenerated shoots were subcultured and maintained on selective media for 5 months. Eight untransformed RG plants that were regenerated in tissue culture were used as controls. In late 2008 to early 2009 (New Zealand summer), plants were transferred into potting mix and grown in a glasshouse under natural light and ambient temperature.
In the spring of 2010, all trees were cut back to 20 cm and one shoot was allowed to regenerate into a primary axis. After growth had terminated, the length and node number of the primary axis and any sylleptic shoots were recorded for each RNAi line and seven RG trees. Average internode length was determined by dividing primary axis length by node number. Some trees flowered and set fruit in 2013, and examination of the fruit suggested a possible increase in number of locules and seeds in the RNAi lines. In spring of 2014, flowers were hand pollinated to increase fruit set. In autumn of 2015, a total of 5–25 fruit from each line and from eight RG trees were cut in half, photographed, and locule and seed number recorded. Leaf blade length and width and petiole length were measured on six fully expanded leaves taken from at least three different sylleptic shoots. Graphing of data was performed in Excel® 2013 (Microsoft Corporation, Redman, WA, USA).
Quantitative real-time PCR
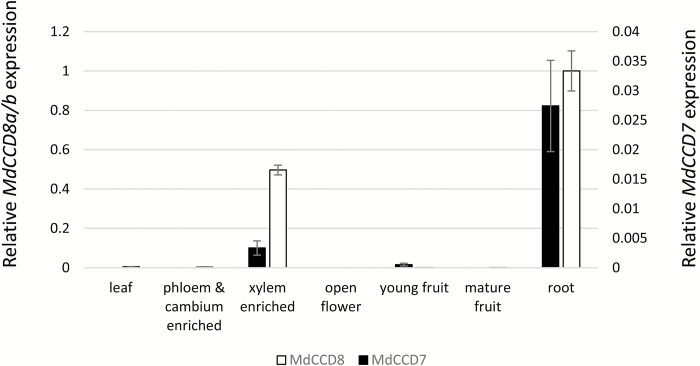
To verify reduced expression of MdCCD7 and MdCCD8 in the RNAi lines, root tissue was collected in early summer from transgenic and control plants 3 months after transfer of the plants to the glasshouse. Only 7 of the 15 MdCCD8 lines had enough roots to remove some for RNA isolation without jeopardizing the health of the plants. The MdCCD8 primers detected the combined expression of both MdCCD8a and MdCCD8b. RNA was also extracted from WT RG leaves, roots, open flowers, young fruits [14 days after full bloom (DAFB)], mature fruit (146 DAFB), and stem tissue to determine expression levels of MdCCD7 and MdCCD8 in these tissues. To collect stem tissue, a section of bark was removed from a field-grown RG tree, the inside of the bark was sampled for phloem- and cambium-enriched tissue, and the tree side was sampled for xylem-enriched tissue (Fig. 3; all values are relative to MdCCD8 expression in roots). To measure MdCCD7 expression in roots of grafted trees, root tissue was collected from 4–5 biological replicates of each scion–rootstock combination from trees grown in 2014–2015. All tissue was snap-frozen in liquid nitrogen and stored at –80 °C until RNA extraction. Total RNA was extracted using a cetyltrimethylammonium bromide (CTAB) method as modified by Ledger et al. (2010). The extracted total RNA was treated with DNase I and the quality and concentration of RNA were determined using either an Agilent 2100 Bioanalyser (Agilent Technologies, Palo Alto, CA, USA) or a Fragment Analyser (Advanced Analytical, Ankey, USA). Only RNAs with an RNA integrity number of ≥8.0 were made into cDNA. First-strand cDNA was synthesized from 1.0 µg of total RNA using poly(dT) primer and Primescript Reverse Transcriptase (TaKaRa, Kusatsu, Japan).
Fig. 3.
Expression of CAROTENOID CLEAVAGE DIOXYGENASE (CCD) genes in ‘Royal Gala’ apple. qRT-PCR expression of MdCCD7 and MdCCD8 in wild-type tissues. Values are means of three technical replicates ±SE, normalized to internal control genes and relative to MdCCD8 expression in roots. Tissues without bars had expression levels below the threshold of detection. The scale for MdCCD7 is on the right axis and for MdCCD8 is on the left.
Quantitative real-time PCR (qRT-PCR) was performed with KAPA sybr fast qRT-PCR mastermix (Wilmington, MA, USA) on a Roche 480 LightCycler® (Basel, Switzerland). cDNAs were diluted 1:20 and run in three technical replicates, with 1.0 µl of template in a reaction volume of 7.5 µl. PCRs were set up in a 384-well plate by a Biomek liquid handling robot (Waltham, MA, USA) to minimize pipetting errors. PCR cycles were as follows: initial denaturation at 95 °C for 5 min, followed by 45 cycles of 94 °C 10 s, 55 °C 15 s, 72 °C 10 s, and a final melt curve analysis to determine if a single product was amplified. Primers were designed by Primer 3 (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) to amplify products of 100–120 bp (Supplementary Table S1). For each analysis, a no-cDNA template was included as a negative control and actin, glyceraldehyde phosphate dehydrogenase (GAPDH), and EST 32701 were used as reference genes. Primer efficiencies and relative expression for three technical replicates per sample were calculated using Roche 480 Light Cycling software version SW1.5 (Basel, Switzerland). One-way ANOVA and graphing were performed with OriginLab 8.5 (Northampton, MA, USA).
Clonal propagation of scion and rootstock material
The MdCCD7 RNAi line AS3354 was selected for the reciprocal grafting experiments. The AS3354 tree (hereafter referred to as ccd7 for simplicity) and WT RG stems were clonally propagated by cutting into segments, grafting onto donor rootstocks, and inducing the ccd7 and RG scions to root using an aerial layering technique. In late winter (August), segments of ccd7 and WT stems were cleft grafted onto donor rootstocks and 2–3 shoots grew out per plant. In January, the lower leaves of the shoots (~20 cm) were removed, the stem base was girdled, and the wound was treated with indole-3-butyric acid (1000 mg l-1) to induce rooting. Wet sphagnum moss was placed over the girdle and wrapped securely within plastic. Holes were punctured in the plastic to allow aeration before aerial layers were covered with aluminium foil to prevent overheating. By autumn, roots had formed around the base of the ccd7 and WT scions. These rooted shoots were excised from the donor rootstock while dormant, and roots were bedded into moist sawdust until winter grafting.
Grafting experiments and data analysis
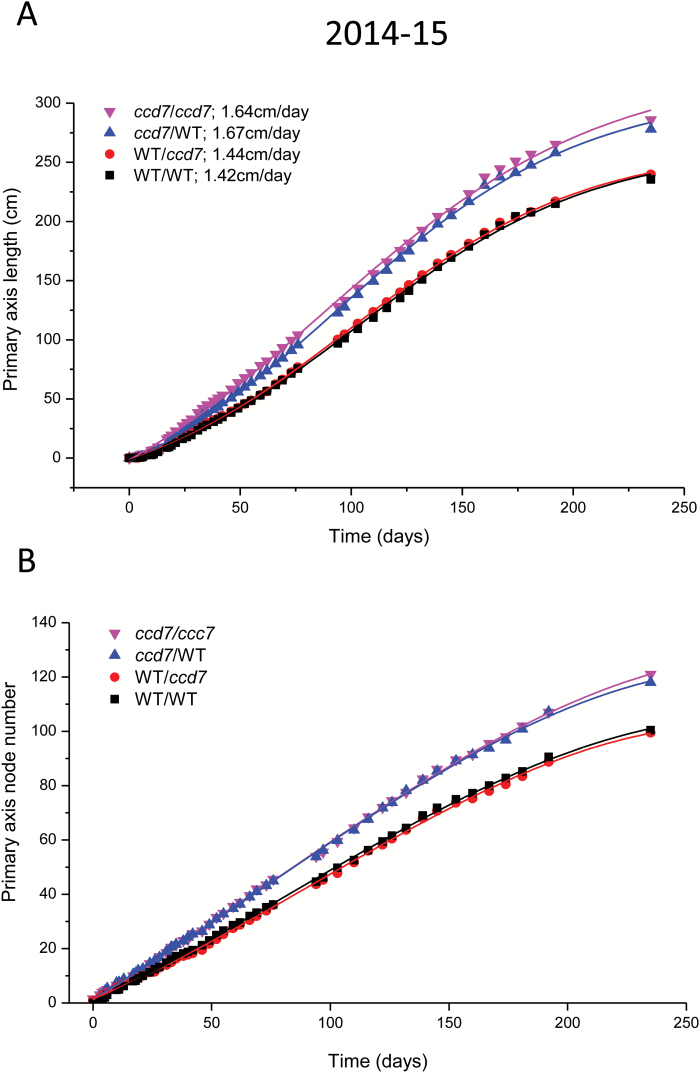
Scions and rootstocks of uniform diameter (8–12 mm) were grafted in August 2013, 2014, and 2016 using standard bench grafting techniques (Seleznyova et al., 2003). Root restriction is known to limit sylleptic branching (van Hooijdonk, 2009), therefore trees were grown in 50 litre black polythene planter bags spaced at least 1 m apart, watered by an automated system, under natural light in a glasshouse. The amount of ccd7 material limited the number of grafted trees that could be generated in any year. Any ccd7 material not used in the first experiment was propagated to generate more scions and rootstocks for the second and third experiments. Throughout this paper, the scion genotype is given before that of the rootstock (scion/rootstock). In the first experiment (2013–2014), we generated three ccd7/ccd7, four ccd7/WT, three WT/ccd7, and four WT/WT control trees. In the second experiment (2014–2015), we generated three ccd7/ccd7, five ccd7/WT, four WT/ccd7, and five WT/WT control trees. For the first two experiments, phenotyping began once the scion bud opened and the primary shoot began to extend. Primary shoot length and node number, sylleptic shoot node number, and the nodal position on the primary axis were recorded every 4 weeks in 2013–2014 and every 2 d in 2014–2015 until shoot growth terminated. In the third experiment (2016–2017), five trees each of WT/WT and ccd7/WT were generated. These were measured for final primary shoot length, node number, and position of sylleptic shoots along the primary axis. Graphing and one-way ANOVA were performed in OriginLab 8.5. Lines on the graphs shown in Fig. 7 and Supplementary figures are connected with B-spline. Maximum primary shoot growth rates were estimated by fitting a Boltzmann growth function to smoothed growth data for each treatment, and the maximum growth rates were calculated from the fitted functions. Unlike shoot length, node appearance is a discrete integer, which complicates calculation of the growth rate in terms of nodes d–1.
Fig. 7.
Growth of the primary axis for each ‘Royal Gala’ apple graft combination (scion/rootstock) in 2014–2015. Growth was measured in terms of increase in (A) primary axis length and (B) node number over the growing season. The maximum growth rate (cm d–1) was estimated by fitting a Boltzmann function to the smoothed growth data and is listed after each symbol in the key. WT, wild-type ‘Royal Gala’.
Results
Identification of MdCCD7 and MdCCD8a/b
A partial clone of a CCD8 gene was identified in an in-house apple EST database, but no sequences with homology to CCD7 genes were found. The full-length apple MdCCD7 and MdCCD8a genes were identified using a combination of degenerate primer PCR, genome walking, and 5'/3' RACE. The full-length cDNA clone of MdCCD7 has an ORF of 1842 bp (MF034498) and corresponds to two truncated gene models on linkage group (LG) 2 (MDP0000197409 and MDP0000139334). We identified one MdCCD8 gene (identical to XP_008378214.1), and whole-genome sequencing revealed a second gene (XP_008352014.1), consistent with Malus being an allopolyploid (Evans and Campbell, 2002; Xiang et al., 2017). The two MdCCD8 genes are 94.5% homologous to one another at the amino acid level, suggesting that they are homeologous genes rather than different alleles of the same gene, which tend to be 98–99% homologous (Foster et al., 2007; Chagné et al., 2012). Both MdCCD8 cDNAs have an ORF of 1692 bp. MdCCD8a (XP_008378214.1) corresponds to MDP0000227870 on LG8, and MdCCD8b (XP_008352014.1) corresponds to MDP0000219296 on LG15. The chromosomal regions containing MDP0000227870 and MDP0000219296 have been duplicated, providing further support that these are homeologous genes (Velasco et al., 2010).
The predicted MdCCD7 and MdCCD8a/b amino acid sequences were used in a phylogenetic analysis of the CCD family (Fig. 1). This included full-length CCD7 and CCD8 protein sequences from other plants and AtCCD1 and AtCCD4 as outgroups. The MdCCD7 gene is in the clade including the Arabidopsis, pea, petunia, and rice CCD7 genes identified by the branching mutants max3, rms5, dad3, and d17/htd1, and the predicted orthologues from other species. The MdCCD8 gene is in the clade that includes Arabidopsis, pea, petunia, and rice CCD8 genes identified by the max4, rms1, dad1, and d10 branching mutants and the predicted orthologues from other species. Both MdCCD7 and MdCCD8a/b cluster closely with the respective CCD proteins from the Rosaceae family.
MdCCD7 and MdCCD8 complement the Arabidopsis branching mutants max3 and max4, respectively
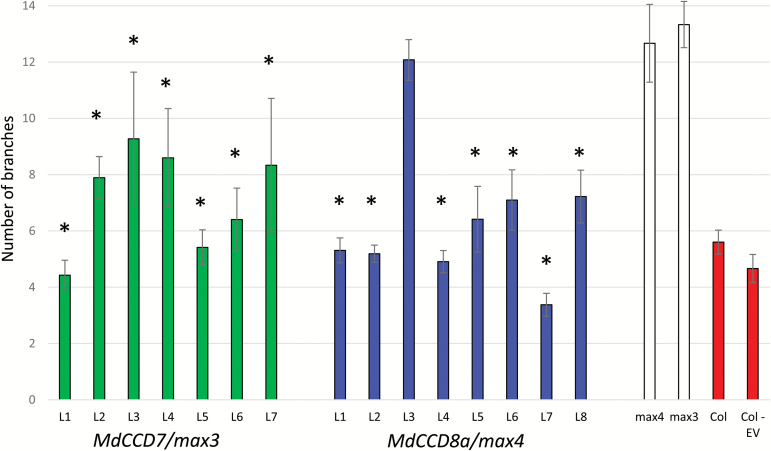
To determine if the CCD protein function is conserved between apple and Arabidopsis, we transformed MdCCD7 into the Arabidopsis max3 mutant and MdCCD8a into max4. The apple genes were introduced into the Arabidopsis mutants under the control of the CaMV 35S promoter. Mean rosette branch number was compared by ANOVA followed by Fisher’s protected LSD multiple comparison test. All seven of the MdCCD7 lines and seven of the eight MdCCD8a independent lines had significantly fewer rosette branches than either max3 or max4 (Fig. 2). The apple genes were able to complement the Arabidopsis branching mutants functionally, indicating conserved CCD7 and CCD8 function.
Fig. 2.
Complementation of Arabidopsis branching mutants with MdCCD7 and MdCCD8a. Number of rosette branches in seven independent transgenic lines of 35S:MdCCD7 in max3-1 and eight lines of 35S:MdCCD8a in max4-2 compared with max3-1, max4-2, wild-type ‘Columbia’ (Col) Arabidopsis, and plants containing an empty pHEX vector (Col-EV). Values are means ±SE, n=5–16 for each independent line. Means were compared using ANOVA, followed by Fisher’s protected LSD multiple comparison test; an asterisk above the bar indicates a significant difference from max3-1 and max4-2 at P≤0.05.
MdCCD7 and MdCCD8 transcripts are most abundant in roots
We measured MdCCD7 and MdCCD8 transcript abundance in apple using qRT-PCR. RNA was isolated from a range of WT apple tissues and developmental stages. The highest level of both MdCCD7 and MdCCD8 expression was in roots (Fig. 3). Expression of both genes was also detected in xylem-enriched stem tissue, but was negligible in leaf, flowers, fruit, and phloem/cambium-enriched stem tissue. The abundance of the MdCCD8 transcript was >13- and 20-fold that of MdCCD7 in roots and xylem-enriched stem tissue, respectively.
Knockdown of MdCCD7 expression in apple results in increased sylleptic branching
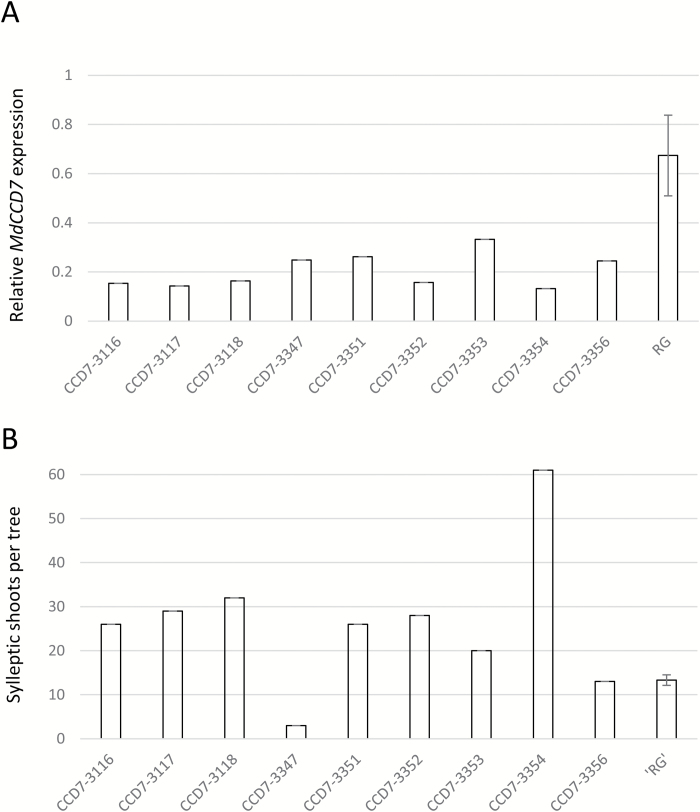
Transgenic RNAi lines of MdCCD7 and MdCCD8 were generated in RG apple to determine the effect of reduced CCD expression on sylleptic branching. Nine independent lines of RNAi MdCCD7, 15 of RNAi MdCCD8, and eight control RG trees were generated. Because of the time required to obtain progeny or even propagate apple trees clonally, each line consisted of the original transformed tree. The transcript abundance of MdCCD7 and MdCCD8 in the roots of transgenic RNAi lines was determined 3 months after transfer to the glasshouse. Transcript abundance of MdCCD7 in individual RNAi lines was 2- to 5-fold lower than in RG controls and very consistent between RNAi lines (Fig. 4A). The transcript abundance of MdCCD8 was reduced in only three of the eight RNAi lines tested, suggesting that the RNAi construct was not very effective in reducing MdCCD8 expression (Supplementary Fig. S1A). The hairpin sequence was identical to that of MdCCD8a, but slightly more divergent to the MdCCD8b gene model (Supplementary Table S2).
Fig. 4.
Expression of MdCCD7 and sylleptic shoot number of MdCCD7 RNAi ‘Royal Gala’ (RG) apple lines. qRT-PCR expression of (A) MdCCD7 in the roots of nine individual MdCCD7 RNAi lines and (B) total number of sylleptic shoots per tree in nine individual MdCCD7 RNAi lines relative to RG controls. For the controls, values are the means of eight biological replicates of RG ±SE.
In the spring of 2010, trees were cut back to 20 cm and a single primary shoot was allowed to develop from an axillary bud. Final shoot length and node number were recorded for primary and sylleptic shoots once growth had terminated. With the exception of two lines, all of the MdCCD7 RNAi lines had more sylleptic shoots than the controls (Fig. 4B). In contrast, only 4 of the 15 MdCCD8 RNAi lines had more sylleptic shoots than the controls (Supplementary Fig. S1B). Both MdCCD7 and MdCCD8 RNAi tended to have slightly shorter primary axes and had fewer nodes than controls (Supplementary Fig. S2). The MdCCD8 RNAi lines did not show greatly reduced MdCCD8 expression or increased branching, and therefore we focused on more detailed analysis of sylleptic branching in the MdCCD7 RNAi lines.
Fruit and leaf phenotypes of RNAi lines
The petunia dad1 and dad3 mutants (mutations in CCD7 and CCD8, respectively) produce about half the seed yield per capsule as the WT (Simons et al., 2007). Some of the RNAi trees first fruited in 2013, and a preliminary investigation indicated that several lines had fruit with an abnormal number of locules and seeds (Supplementary Fig. S3A). A more detailed analysis of fruit in 2015 indicated that locule numbers in most of the RNAi lines were similar to those of RG, but the number of seeds per fruit were reduced in about half the MdCCD7 and MdCCD8 lines (Supplementary Fig. S3B). Previous studies have found that SL-deficient mutants have altered leaf morphology (Beveridge et al., 1997; Scaffidi et al., 2013; Lauressergues et al., 2015). There was much more variability in leaf dimensions in about half the RNAi lines relative to that in RG, but no consistent trend was observed (Supplementary Fig. S4).
Grafting experiments demonstrate that MdCCD7 expression in the scion, but not the rootstock, supresses branching
Grafting experiments were performed to determine whether reduced MdCCD7 expression in roots or shoots altered sylleptic shoot outgrowth. Sylleptic branching is strongly influenced by tree spacing, so these experiments required a large containment glasshouse. Initiating roots on woody material is technically difficult, which also limited the number of trees that could be grown each year. MdCCD7 RNAi line AS3354 had the lowest transcript abundance of MdCCD7 and the highest number of sylleptic shoots. This line, hereafter referred to as ccd7, was selected for clonal propagation and subsequent grafting experiments. Four combinations of scion and rootstock were created between ccd7 and WT RG, including ccd7 and WT homografts (ccd7/ccd7 and WT/WT, respectively). During the 2013–2014 season, primary and sylleptic shoot growth was measured monthly until the cessation of growth.
At the beginning of the season when the lower nodes of the primary shoot were extending, the ccd7/ccd7 trees had the highest cumulative sum of sylleptic shoots per tree (Supplementary Fig. S5A). There was a slight effect of WT rootstocks at the base of ccd7 scions, but, as the uppermost nodes developed towards the end of the season, the ccd7/WT (scion/rootstock) trees developed the greatest number of sylleptic shoots per tree; that is, WT rootstocks failed to supress branching in ccd7 scions. By the end of the season, WT/ccd7 trees developed the same number of sylleptic shoots as did the WT/WT trees, both of which were significantly fewer than on the ccd7/WT and ccd7/ccd7 trees (Supplementary Fig. S5A). Grouping the data by scion or rootstock genotype, it was clear that trees with ccd7 scions had a significantly higher number of sylleptic shoots than those with WT scions (Supplementary Fig. S5B). Moreover, rootstock genotype had no significant effect on final sylleptic shoot number (Supplementary Fig. S5C). Interestingly, the scion genotype effect became greater towards the top of the primary axis. Sylleptic shoots on ccd7 scions had twice as many nodes as sylleptic shoots on WT scions (Supplementary Fig. S6A).
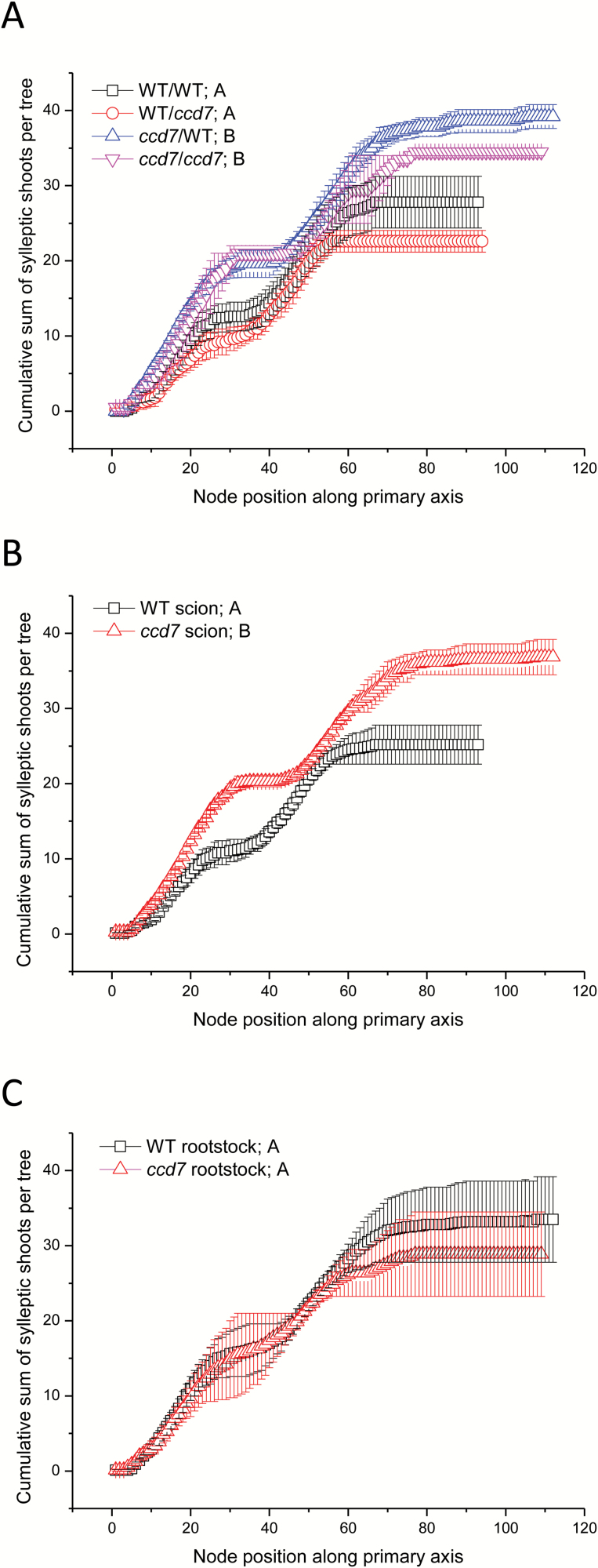
The experiment was repeated in 2014 with a new set of grafted trees to confirm the consistency of our results. Similar trends were observed in the second experiment. WT rootstocks were unable to suppress sylleptic branching in ccd7 scions, even at the lowest nodes (Fig. 5A). Regardless of rootstock genotype, the ccd7 scions developed significantly more sylleptic shoots, especially at the upper nodes of the primary axis (Fig. 5A). Once again, the genotype of the scion, not the rootstock, determined the cumulative sum of sylleptic shoots (Fig. 5B, C). All trees showed two distinct flushes of sylleptic shoot development; however, the first flush was much more pronounced in the ccd7 scions (Fig. 5B). Sylleptic branches had twice as many nodes and occurred higher on the primary axes of ccd7 scions relative to WT scions (Supplementary Fig. S6B).
Fig. 5.
The cumulative sum of sylleptic shoots per ‘Royal Gala’ apple tree for each graft combination in 2014–2015. For each tree, the presence or absence of a sylleptic shoot was recorded at each node along the primary axis from the base to the tip. The cumulative sum at each node for (A) all four graft combinations (scion/rootstock), (B) trees grouped by scion genotype, and (C) rootstock genotype. Symbols are means of 3–5 biological replicates ±SE. Means were compared by one-way ANOVA; different letters after each symbol in the key represent a significant difference at P≤0.05. WT, wild-type ‘Royal Gala’.
Only WT/WT and ccd7/WT trees were generated in 2016. Sylleptic shoot position and final primary axis length were recorded at the end of the growing season. Consistent with data from the previous 2 years, ccd7/WT trees developed significantly more sylleptic shoots than WT/WT trees, especially at the upper nodes of the primary axis (Fig. 6; and Supplementary Fig. S7).
Fig. 6.
WT/WT and ccd7/WT ‘Royal Gala’ apple trees grown in 2016–2017. Photographs of a representative (A) WT/WT tree and (B, C) ccd7/WT trees (scion/rootstock); the red bar represents 1 m and red arrows show graft junctions. WT, wild-type ‘Royal Gala’.
Reduced expression of MdCCD7 in the scion increases primary axis growth rate
In all three grafting experiments, we found that ccd7 scions had a significantly larger final node number and primary shoot length than wild-type scions (Supplementary Fig. S8). To determine if this resulted from a longer period of growth or a higher growth rate, we recorded the node number and length of the primary shoot throughout the growing season for the first two grafting experiments. Primary shoot growth rate, in terms of both increase in length and node initiation, showed a similar trend in both years. The ccd7/ccd7 and ccd7/WT trees had a higher primary shoot growth rate than WT/ccd7 and WT/WT trees (Fig. 7; Supplementary Fig. S9). The maximum growth rate of ccd7 scions was ~15% greater than that of the WT scions (Fig. 7). All trees terminated growth at about the same time. Average internode length was similar between all graft combinations in all three experiments (data not shown).
No graft-transmissible knockdown of MdCCD7 in wild-type roots
Our findings differ from many previous reports where CCD expression in the roots is able to restore wild-type branching in CCD mutant scions. One possible explanation for our data was that the hairpin used in the MdCCD7 RNAi lines moved from the scion and knocked down expression of MdCCD7 in wild-type roots. To test this, we measured MdCCD7 transcript abundance in roots from the four graft combinations. MdCCD7 expression was 5- to7-fold higher in WT roots than in ccd7 roots regardless of scion genotype (Supplementary Fig. S10). This result indicates that the failure of WT scions to repress branching in ccd7 scions is not caused by a graft-transmissible knockdown in MdCCD7 expression in WT roots.
Discussion
In this study, we identified the apple orthologues of CCD7 and CCD8 and demonstrated that they were able to complement the Arabidopsis branching mutants max3 and max4, respectively. This indicates that CCD7 and CCD8 function is conserved between the woody perennial apple and the annual Arabidopsis. Reduced expression of MdCCD7 in apple by RNAi resulted in an increased number of sylleptic shoots in the majority of lines. In contrast, only 4 of the 15 MdCCD8 RNAi lines showed slightly increased sylleptic shoot number. The hairpin constructs were designed before whole-genome sequencing identified two copies of CCD8 in apple. The MdCCD8 hairpin construct included a region that was identical to MdCCD8a, but slightly more divergent from MdCCD8b; therefore, it is possible that only MdCCD8a was effectively silenced. Loss of either CCD7 or CCD8 has resulted in blocked SL biosynthesis and highly branched and dwarfed phenotypes in a range of plants (Beveridge et al., 1997; Morris et al., 2001; Turnbull et al., 2002; Booker et al., 2004; Ishikawa et al., 2005; Snowden et al., 2005; Arite et al., 2007; Drummond et al., 2009).
An important caveat for this work is that we were unable to generate complete knockouts of MdCCD7 and MdCCD8, so we selected the RNAi line with the lowest MdCCD7 expression for grafting experiments and detailed architectural measurements. This line showed consistent results in three independent grafting experiments, and showed an increase in sylleptic branching with a concomitant reduction in MdCCD7 transcripts. However, the increase in nodes produced on the primary axis of ccd7 scions (Supplementary Fig. S8) contrasted with the result obtained in the initial phenotyping of the complete set of RNAi lines (Supplementary Fig. S2). The reason for this difference is unknown; however, it is worth noting that the trees were grown in different conditions for these analyses. In particular, the complete set of RNAi lines were grown in much smaller pots with less spacing between trees than the grafted trees (5 litres versus 50 litres). In addition, the age of the trees at phenotyping, and time since removal from tissue culture differed between the experiments. Future studies to examine the role of SLs in apple tree growth would benefit from the use of a technique such as genome editing to knock out MdCCD7 and MdCCD8a/b expression completely and using multiple lines in grafting experiments.
MdCCD7 genotype of the scion, not rootstock, determines the extent of branching in apple
Grafting experiments with CCD mutants or RNAi lines have consistently shown that WT roots are able to restore normal branching in scions that lack either CCD7 or CCD8 (Beveridge et al., 1997; Morris et al., 2001; Turnbull et al., 2002; Booker et al., 2004; Simons et al., 2007; Muhr et al., 2016). In contrast, our results demonstrated that WT roots were unable to suppress branching in ccd7/WT compared with the amounts of branching in WT/WT. It is worth noting that the difference in cumulative sylleptic shoot number between ccd7 and WT scions became greater at the upper nodes of the primary axis. Researchers studying CCD genes in poplar have suggested that local SL biosynthesis in the shoot may be more important for bud inhibition in trees because they are much larger than annual plants (Czarnecki et al., 2014; Muhr et al., 2016). Indeed, WT scions grafted to CCD7- or CCD8-deficient roots all show a WT branching pattern in Arabidopsis, pea, petunia, and poplar, supporting the idea that scion expression of CCD7 or CCD8 is sufficient, but not necessary to suppress branching. Our results suggest that MdCCD7 expression in the scion is necessary to inhibit sylleptic branching in apple.
To our knowledge, this is the first study to present a dynamic picture of sylleptic shoot outgrowth. Final branch number does not give any information about where on the primary axis branching occurs. By following the cumulative sylleptic shoot number, we were able to identify that the increase in sylleptic branching became more pronounced at the upper nodes of ccd7 scions. We suggest that localized SL biosynthesis in the stem becomes more important at nodes further from the root system. Previous studies have shown that the propensity for sylleptic branching in both apple and pear is determined by the scion genotype (Costes and Guédon, 2002; Seleznyova et al., 2012). Perhaps this reflects the input of SL signalling pathways.
Extent of sylleptic branching may reflect differential sensitivity to SLs
Sylleptic branching is a highly plastic trait that is controlled by genetic, environmental, and developmental factors (Hallé et al., 1978; Crabbé, 1987; Génard et al., 1994; Wu and Hinckley, 2001; Cline and Dong-Il, 2002; Marron et al., 2006;). The SL pathway is thought to integrate multiple environmental and internal signals such as light quality, nutrient availability, and input from other hormone signalling pathways to modulate growth (Domagalska and Leyser, 2011; Djennane et al, 2014; Janssen et al, 2014; Drummond et al, 2015; Rameau et al., 2015). A study comparing poplar clones with high and low sylleptic branching found that the former had an increased sensitivity to auxin and a decreased sensitivity to cytokinin, whereas the latter showed the opposite (Cline and Dong-Il, 2002). Even within one species, there appears to be a range of sensitivity to hormones that regulate branching. RNAi lines of MAX genes in poplar have been shown to increase sylleptic branching greatly (Muhr et al., 2016) or have no effect at all (Brunner et al., 2011).
The function of CCD proteins in SL biosynthesis is conserved between poplar and apple, although there were some interesting phenotypic differences between the amiMAX4 and MdCCD7 RNAi trees. The amiMAX4 poplar trees had 5–15 times more sylleptic shoots than WT poplar (Muhr et al., 2016). Even the most highly branched MdCCD7 RNAi line had <5-fold more sylleptic shoots than WT trees. The amiMAX4 homografted trees had 8–10 times more sylleptic shoots than WT/WT trees, whereas our ccd7/ccd7 trees had roughly twice the number of sylleptic branches as the WT/WT trees.
The differences between the CCD knockdown phenotypes in poplar versus apple could be due to the effectiveness of the knockdown of gene expression or could reflect inherent differences in branching behaviour. The amiMAX4 construct was transformed into the hybrid Populus×canescens, which is a low syllepsis clone. Depending on the scion genotype and growth conditions, apple trees can have several flushes of sylleptic branching and produce up to 30 sylleptic shoots in the first year of growth. It is possible that apple trees have a higher threshold of SLs required for repression of bud outgrowth.
Reduced MdCCD7 expression associated with higher growth rate of the primary shoot
In addition to increased branching, CCD mutants or knockdown lines generally have a shorter primary axis and shorter internode lengths (Beveridge et al., 1997; Morris et al., 2001; Booker et al., 2004; Simons et al., 2007; Guan et al., 2012; Muhr et al., 2016). In contrast, we found that the primary axes of MdCCD7 RNAi scions were longer, with more internodes than WT scions. Average internode length was unaffected by reduced MdCCD7 expression, but the rate of growth, in terms of both length and node number, was increased relative to that of the WT. The rate of node initiation or plastochron index reflects the activity of the SAM, whereas internode elongation is due to a combination of cell division and elongation proximal to the shoot apex. de Saint Germain and co-workers have demonstrated that the artificial SL GR24 was able to restore internode length in the pea rms1 mutant by increasing cell number, not cell length (de Saint Germain et al., 2013). These authors proposed that SLs normally repress cell division in axillary meristems, but promote cell division in internodes. Based on our findings, we suggest that the reduction of SLs in ccd7 scions resulted in more active apical and axillary meristems, possibly because of increased cell division. Whether these effects are independent or causally related remains to be investigated. Unlike annual plants, perennial plants must balance growth during multiple seasons with maintaining reserves for homeostasis during winter and early spring growth. Given the different survival goals and the role of SLs in integrating signals controlling growth, perhaps it is not surprising that some effects of SLs could be different in annual and perennial plants.
Conclusions
We have identified CCD7 and CCD8 genes from apple and shown that their function is conserved. RNAi lines of MdCCD7 had reduced MdCCD7 expression and increased sylleptic branching in apple. Reciprocal grafting experiments with one RNAi line provided evidence that MdCCD7 expression in the scion was necessary to restore WT sylleptic branch numbers. WT roots were unable to repress sylleptic branching in scions of this RNAi line. The growth rate of the primary axis was higher in this line than in the WT. Based on our findings, we propose that localized SL synthesis in the shoot may have a predominant role in suppressing sylleptic shoot outgrowth and primary shoot growth rate in apple.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Expression of MdCCD8 and and sylleptic shoot number of MdCCD8 RNAi ‘Royal Gala’ (RG) apple lines.
Fig. S2. Primary axis length and node number for MdCCD7 and MdCCD8 RNAi ‘Royal Gala’ (RG) apple lines.
Fig. S3. Fruit characteristics of RNAi and ‘Royal Gala’ (RG) apple fruit.
Fig. S4. Dimensions of RNAi and ‘Royal Gala’ (RG) apple leaves.
Fig. S5. The cumulative sum of sylleptic shoots per ‘Royal Gala’ apple tree for each graft combination in 2013–2014.
Fig. S6. Total node number of sylleptic shoots.
Fig. S7. The cumulative sum of sylleptic shoots per ‘Royal Gala’ apple tree for ccd7/WT and WT/WT trees in 2016–2017.
Fig. S8. Final primary axis length and node number of grafted ‘Royal Gala’ apple trees (scion/rootstock).
Fig. S9. Growth of the primary axis for each ‘Royal Gala’ apple graft combination (scion/rootstock) in 2013–2014.
Fig. S10. Expression of MdCCD7 in roots of grafted ‘Royal Gala’ apple trees.
Table S1. PCR primers used in this study.
Table S2. Sequences for RNAi constructs and homology to the targeted gene.
Supplementary Material
Acknowledgements
We thank Monica Dragulescu and Ian King (Plant and Food Research) for maintaining the trees, The Foundation for Research, Science and Technology and Plant and Food research for funding, and Drs Donald Hunter and Erika Varkonyi-Gasic for helpful comments on the manuscript.
References
- Alder A, Jamil M, Marzorati M et al. 2012. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335, 1348–1351. [DOI] [PubMed] [Google Scholar]
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J. 2007. DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. The Plant Journal 51, 1019–1029. [DOI] [PubMed] [Google Scholar]
- Beveridge CA. 2000. Long-distance signalling and a mutational analysis of branching in pea. Plant Growth Regulation 32, 193–203. [Google Scholar]
- Beveridge CA, Ross JJ, Murfet IC. 1994. Branching mutant rms-2 in Pisum sativum (grafting studies and endogenous indole-3-acetic acid levels). Plant Physiology 104, 953–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge CA, Symons GM, Murfet IC, Ross JJ, Rameau C. 1997. The rms1 mutant of pea has elevated indole-3-acetic acid levels and reduced root-sap zeatin riboside content but increased branching controlled by graft-transmissible signal(s). Plant Physiology 115, 1251–1258. [Google Scholar]
- Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O. 2004. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Current Biology 14, 1232–1238. [DOI] [PubMed] [Google Scholar]
- Brewer PB, Yoneyama K, Filardo F et al. 2016. LATERAL BRANCHING OXIDOREDUCTASE acts in the final stages of strigolactone biosynthesis in Arabidopsis. Proceedings of the National Academy of Sciences, USA 113, 6301–6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner A, Sheng X, Edwards J, Fujino T, Wang C-T, DiFazio S. 2011. Regulation of shoot-system development in Populus. BMC Proceedings 5, I11. [Google Scholar]
- Chagné D, Crowhurst RN, Troggio M et al. 2012. Genome-wide SNP detection, validation, and development of an 8K SNP array for apple. PLoS One 7, e31745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MG, Dong-IL K. 2002. A preliminary investigation of the role of auxin and cytokinin in sylleptic branching of three hybrid poplar clones exhibiting contrasting degrees of sylleptic branching. Annals of Botany 90, 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method forAgrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Costes E, Crespel L, Denoyes B, Morel P, Demene MN, Lauri PE, Wenden B. 2014. Bud structure, position and fate generate various branching patterns along shoots of closely related Rosaceae species: a review. Frontiers in Plant Science 5, 666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costes E, Guédon Y. 2002. Modelling branching patterns on 1-year-old trunks of six apple cultivars. Annals of Botany 89, 513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbé J. 1987. Aspects particuliers de la morphogenese caulinaire des vegetaux ligneux et introduction a leur etude quantitative. Bruxelles: IRSIA. [Google Scholar]
- Czarnecki O, Yang J, Wang X, Wang S, Muchero W, Tuskan GA, Chen JG. 2014. Characterization of MORE AXILLARY GROWTH genes in Populus. PLoS One 9, e102757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Germain A, Ligerot Y, Dun EA, Pillot JP, Ross JJ, Beveridge CA, Rameau C. 2013. Strigolactones stimulate internode elongation independently of gibberellins. Plant Physiology 163, 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djennane S, Hibrand-Saint Oyant L, Kawamura K et al. 2014. Impacts of light and temperature on shoot branching gradient and expression of strigolactone synthesis and signalling genes in rose. Plant, Cell and Environment 37, 742–757. [DOI] [PubMed] [Google Scholar]
- Domagalska MA, Leyser O. 2011. Signal integration in the control of shoot branching. Nature Reviews. Molecular Cell Biology 12, 211–221. [DOI] [PubMed] [Google Scholar]
- Drummond RS, Janssen BJ, Luo Z, Oplaat C, Ledger SE, Wohlers MW, Snowden KC. 2015. Environmental control of branching in petunia. Plant Physiology 168, 735–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond RS, Martínez-Sánchez NM, Janssen BJ, Templeton KR, Simons JL, Quinn BD, Karunairetnam S, Snowden KC. 2009. Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE7 is involved in the production of negative and positive branching signals in petunia. Plant Physiology 151, 1867–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RC, Campbell CS. 2002. The origin of the apple subfamily (Maloideae; Rosaceae) is clarified by DNA sequence data from duplicated GBSSI genes. American Journal of Botany 89, 1478–1484. [DOI] [PubMed] [Google Scholar]
- Fanwoua J, Bairam E, Delaire M, Buck-Sorlin G. 2014. The role of branch architecture in assimilate production and partitioning: the example of apple (Malus domestica). Frontiers in Plant Science 5, 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Turnbull CG, Beveridge CA. 2001. Long-distance signaling and the control of branching in the rms1 mutant of pea. Plant Physiology 126, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T, Kirk C, Jones WT, Allan AC, Espley R, Karunairetnam S, Rakonjac J. 2007. Characterisation of the DELLA subfamily in apple (Malus × domestica Borkh.). Tree Genetics and Genomes 3, 187–197. [Google Scholar]
- Génard M, Pagès L, Kervella J. 1994. Relationship between sylleptic branching and components of parent shoot development in the peach tree. Annals of Botany 74, 467–470. [Google Scholar]
- Girault T, Abidi F, Sigogne M, Pelleschi-Travier S, Boumaza R, Sakr S, Leduc N. 2010. Sugars are under light control during bud burst in Rosa sp. Plant, Cell and Environment 33, 1339–1350. [DOI] [PubMed] [Google Scholar]
- Girault T, Bergougnoux V, Combes D, Viemont JD, Leduc N. 2008. Light controls shoot meristem organogenic activity and leaf primordia growth during bud burst in Rosa sp. Plant, Cell and Environment 31, 1534–1544. [DOI] [PubMed] [Google Scholar]
- Gobena D, Shimels M, Rich PJ, Ruyter-Spira C, Bouwmeester H, Kanuganti S, Mengiste T, Ejeta G. 2017. Mutation in sorghum LOW GERMINATION STIMULANT 1 alters strigolactones and causes Striga resistance. Proceedings of the National Academy of Sciences, USA 114, 4471–4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB et al. 2008. Strigolactone inhibition of shoot branching. Nature 455, 189–194. [DOI] [PubMed] [Google Scholar]
- Guan JC, Koch KE, Suzuki M, Wu S, Latshaw S, Petruff T, Goulet C, Klee HJ, McCarty DR. 2012. Diverse roles of strigolactone signaling in maize architecture and the uncoupling of a branching-specific subnetwork. Plant Physiology 160, 1303–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallé F, Oldeman RAA, Tomlinson PB. 1978. Tropical trees and forests: an architectural analysis. Berlin: Springer. [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. 2005. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry C, Rabot A, Laloi M et al. 2011. Regulation of RhSUC2, a sucrose transporter, is correlated with the light control of bud bursta in Rosa sp. Plant, Cell and Environment 34, 1776–1789. [DOI] [PubMed] [Google Scholar]
- Huché-Thélier L, Boumaza R, Demotes-Mainard S, Canet A, Symoneaux R, Douillet O, Guérin V. 2011. Nitrogen deficiency increases basal branching and modifies visual quality of the rose bushes. Scientia Horticulturae 130, 325–334. [Google Scholar]
- Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J. 2005. Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant and Cell Physiology 46, 79–86. [DOI] [PubMed] [Google Scholar]
- Janssen BJ, Drummond RS, Snowden KC. 2014. Regulation of axillary shoot development. Current Opinion in Plant Biology 17, 28–35. [DOI] [PubMed] [Google Scholar]
- Johnson X, Brcich T, Dun EA, Goussot M, Haurogné K, Beveridge CA, Rameau C. 2006. Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiology 142, 1014–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khayat E, Zieslin N. 1982. Environmental factors involved in the regulation of sprouting of basal buds in rose plants. Journal of Experimental Botany 33, 1286–1292. [Google Scholar]
- Kim HK, van Oosterom E, Dingkuhn M, Luquet D, Hammer G. 2010. Regulation of tillering in sorghum: environmental effects. Annals of Botany 106, 57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauressergues D, André O, Peng J, Wen J, Chen R, Ratet P, Tadege M, Mysore KS, Rochange SF. 2015. Strigolactones contribute to shoot elongation and to the formation of leaf margin serrations in Medicago truncatula R108. Journal of Experimental Botany 66, 1237–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledger SE, Janssen BJ, Karunairetnam S, Wang T, Snowden KC. 2010. Modified CAROTENOID CLEAVAGE DIOXYGENASE8 expression correlates with altered branching in kiwifruit (Actinidia chinensis). New Phytologist 188, 803–813. [DOI] [PubMed] [Google Scholar]
- Marron N, Bastien C, Sabatti M, Taylor G, Ceulemans R. 2006. Plasticity of growth and sylleptic branchiness in two poplar families grown at three sites across Europe. Tree Physiology 26, 935–946. [DOI] [PubMed] [Google Scholar]
- Morris SE, Turnbull CG, Murfet IC, Beveridge CA. 2001. Mutational analysis of branching in pea. Evidence that Rms1 and Rms5 regulate the same novel signal. Plant Physiology 126, 1205–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhr M, Prüfer N, Paulat M, Teichmann T. 2016. Knockdown of strigolactone biosynthesis genes in Populus affects BRANCHED1 expression and shoot architecture. New Phytologist 212, 613–626. [DOI] [PubMed] [Google Scholar]
- Müller D, Leyser O. 2011. Auxin, cytokinin and the control of shoot branching. Annals of Botany 107, 1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli C. 1996. Highly branched phenotype of the petunia dad1-1 mutant is reversed by grafting. Plant Physiology 111, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli CA, Ruehle J. 1996. New mutations affecting meristem growth and potential in Petunia hybrida Vilm. Journal of Heredity 87, 371–377. [Google Scholar]
- Pierik R, Testerink C. 2014. The art of being flexible: how to escape from shade, salt, and drought. Plant Physiology 166, 5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabot A, Henry C, Ben Baaziz K et al. 2012. Insight into the role of sugars in bud burst under light in the rose. Plant and Cell Physiology 53, 1068–1082. [DOI] [PubMed] [Google Scholar]
- Rameau C, Bertheloot J, Leduc N, Andrieu B, Foucher F, Sakr S. 2015. Multiple pathways regulate shoot branching. Frontiers in Plant Science 5, 741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rameau C, Murfet IC, Laucou V, Floyd RS, Morris SE, Beveridge CA. 2002. Pea rms6 mutants exhibit increased basal branching. Physiologia Plantarum 115, 458–467. [DOI] [PubMed] [Google Scholar]
- Sachs T, Thimann KV. 1964. Release of lateral buds from apical dominance. Nature 201, 939–940. [Google Scholar]
- Salmon J, Ward SP, Hanley SJ, Leyser O, Karp A. 2014. Functional screening of willow alleles in Arabidopsis combined with QTL mapping in willow (Salix) identifies SxMAX4 as a coppicing response gene. Plant Biotechnology Journal 12, 480–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi A, Waters MT, Ghisalberti EL, Dixon KW, Flematti GR, Smith SM. 2013. Carlactone-independent seedling morphogenesis in Arabidopsis. The Plant Journal 76, 1–9. [DOI] [PubMed] [Google Scholar]
- Seleznyova AN, Dayatilake GA, Watson AE, Tustin DS. 2012. After initial invigoration by heading, young pear trees show reduction in axis vigour and increased propensity to flower. Functional Plant Biology 40, 34–43. [DOI] [PubMed] [Google Scholar]
- Seleznyova AN, Thorp TG, White M, Tustin S, Costes E. 2003. Application of architectural analysis and AMAPmod methodology to study dwarfing phenomenon: the branch structure of ‘Royal Gala’ apple grafted on dwarfing and non-dwarfing rootstock/interstock combinations. Annals of Botany 91, 665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons JL, Napoli CA, Janssen BJ, Plummer KM, Snowden KC. 2007. Analysis of the DECREASED APICAL DOMINANCE genes of petunia in the control of axillary branching. Plant Physiology 143, 697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow R. 1937. On the nature of correlative inhibition. New Phytologist 36, 283–300. [Google Scholar]
- Snowden KC, Simkin AJ, Janssen BJ et al. 2005. The decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. The Plant Cell 17, 746–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K, Booker J, Haurogné K et al. 2003. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes and Development 17, 1469–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves TA, Sussex IM. 1989. Patterns in plant development. New York: Cambridge University Press. [Google Scholar]
- Stirnberg P, van de Sande K, Leyser HMO. 2002. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129, 1131–1141. [DOI] [PubMed] [Google Scholar]
- Thimann KV, Skoog F. 1933. Studies on the growth hormone of plants: III. The inhibiting action of the growth substance on bud development. Proceedings of the National Academy of Sciences, USA 19, 714–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull CGN, Booker JP, Leyser HMO. 2002. Micrografting techniques for testing long-distance signalling in Arabidopsis. The Plant Journal 32, 255–262. [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S et al. 2008. Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200. [DOI] [PubMed] [Google Scholar]
- van Hooijdonk BM. 2009. The physiological basis of vigour control by apple rootstocks—an unresolved paradigm. Palmerston North, New Zealand: Massey University. [Google Scholar]
- Velasco R, Zharkikh A, Affourtit J et al. 2010. The genome of the domesticated apple (Malus × domestica Borkh.). Nature Genetics 42, 833–839. [DOI] [PubMed] [Google Scholar]
- Wu RL, Hinckley TM. 2001. Phenotypic plasticity of sylleptic branching: genetic design of tree architecture. Critical Reviews in Plant Sciences 20, 467–485. [Google Scholar]
- Xiang Y, Huang CH, Hu Y, Wen J, Li S, Yi T, Chen H, Xiang J, Ma H. 2017. Evolution of Rosaceae fruit types based on nuclear phylogeny in the context of geological times and genome duplication. Molecular Biology and Evolution 34, 262–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao JL, Cohen D, Atkinson R, Richardson K, Morris B. 1995. Regeneration of transgenic plants from the commercial apple cultivar Royal Gala. Plant Cell Reports 14, 407–412. [DOI] [PubMed] [Google Scholar]
- Zhang Y, van Dijk AD, Scaffidi A et al. 2014. Rice cytochrome P450 MAX1 homologs catalyze distinct steps in strigolactone biosynthesis. Nature Chemical Biology 10, 1028–1033. [DOI] [PubMed] [Google Scholar]
- Zou J, Zhang S, Zhang W et al. 2006. The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. The Plant Journal 48, 687–698. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.