Abstract
Land plants produce specialized low molecular weight metabolites to adapt to various environmental stressors, such as UV radiation, pathogen infection, wounding and animal feeding damage. Due to the large variety of stresses, plants produce various chemicals, particularly plant species-specific alkaloids, through specialized biosynthetic pathways. In this study, using a draft genome sequence and querying known biosynthetic cytochrome P450 (P450) enzyme-encoding genes, we characterized the P450 genes involved in benzylisoquinoline alkaloid (BIA) biosynthesis in California poppy (Eschscholzia californica), as P450s are key enzymes involved in the diversification of specialized metabolism. Our in silico studies showed that all identified enzyme-encoding genes involved in BIA biosynthesis were found in the draft genome sequence of approximately 489 Mb, which covered approximately 97% of the whole genome (502 Mb). Further analyses showed that some P450 families involved in BIA biosynthesis, i.e. the CYP80, CYP82 and CYP719 families, were more enriched in the genome of E. californica than in the genome of Arabidopsis thaliana, a plant that does not produce BIAs. CYP82 family genes were highly abundant, so we measured the expression of CYP82 genes with respect to alkaloid accumulation in different plant tissues and two cell lines whose BIA production differs to estimate the functions of the genes. Further characterization revealed two highly homologous P450s (CYP82P2 and CYP82P3) that exhibited 10-hydroxylase activities with different substrate specificities. Here, we discuss the evolution of the P450 genes and the potential for further genome mining of the genes encoding the enzymes involved in BIA biosynthesis.
Keywords: Benzylisoquinoline alkaloid, Cytochrome P450, Draft genome sequence, Eschscholzia californica, Genome mining, Macarpine
Introduction
Land plants produce specialized low molecular weight metabolites to adapt to various environmental stresses, such as UV radiation, pathogen infection, wounding and damage caused by animal feeding (Sato 2014, Wurtzel and Kutchan 2016). Due to the variations in stresses, plants produce diverse chemicals, especially plant species-specific alkaloids, through specialized biosynthesis pathways (Facchini 2001, Facchini et al. 2004, Ziegler and Facchini 2008). For example, isoquinoline alkaloids (IQAs) are specifically found in the Ranunculaceae, Papaveraceae, Berberidaceae and Rutaceae families, among others, and often consist of chemically diverse structures that are specific to plant species, including morphinans (e.g. narcotic morphine in opium poppy), emetines (e.g. anti-amoebic emetine in ipecac) and galantamines (e.g. acetylcholinesterase-inhibiting galantamine in snowdrop), as well as more generally distributed protoberberines (e.g. antibiotic berberine in the Berberidaceae, Papaveraceae, Ranunculaceae and Rutaceae families), benzophenanthridines (e.g. antibiotic sanguinarine in Papaveraceae), aporphines (e.g. the potent anti-HIV agent magnoflorine in Ranunculaceae) and pavines (e.g. eschscholizine in Papaveraceae), among others (Hagel and Facchini 2013). Biosynthetic pathways involved in specialized metabolism have thus far been characterized using metabolite identification and tracer experiments. Biosynthetic enzymes have been characterized and purified from metabolite-producing plant organs and/or in vitro cultured cells to confirm the pathways, and cDNAs for biosynthetic enzymes have been isolated (Sato 2013, Sato 2014). For example, in the 1970s, Zenk’s group characterized many IQA biosynthetic pathways, after which several groups purified their main enzymes and isolated the cDNA of those enzymes using the sequences of purified proteins involved in IQA biosynthesis (Tanahashi and Zenk 1990, Stadler and Zenk 1993, Kraus and Kutchan 1995, Takeshita et al. 1995, Morishige et al. 2000, Choi et al. 2002, Liscombe et al. 2009). Due to advances in molecular technologies, especially DNA sequencing, expressed sequence tag (EST) libraries [later involving transcriptome data produced by using next-generation sequencing (NGS)] were used to select several candidate enzyme-encoding genes based on the signature sequences of O-methyltransferase (OMT) and cytochrome P450 (P450) (Morishige et al. 2002, Ikezawa et al. 2007, Ikezawa et al. 2009), the relative abundances of transcripts or the results of correlation analyses between the accumulation of metabolites and transcripts (Liscombe et al. 2009, Takemura et al. 2013). Integrated transcriptome and metabolome analyses have been especially powerful tools for selecting candidates, particularly when plant clones and/or cell culture lines with different levels of metabolite productivity are used (Liscombe et al. 2009, Takemura et al. 2013); however, preparing adequate cell and plant collections is difficult. Therefore, we attempted to isolate candidate genes directly from the genome sequence because tomato and potato genome information has been successfully used both to analyze the steroidal glycoalkaloid biosynthesis enzyme-encoding gene cluster and to isolate candidate genes in those plant species (Itkin et al. 2013).
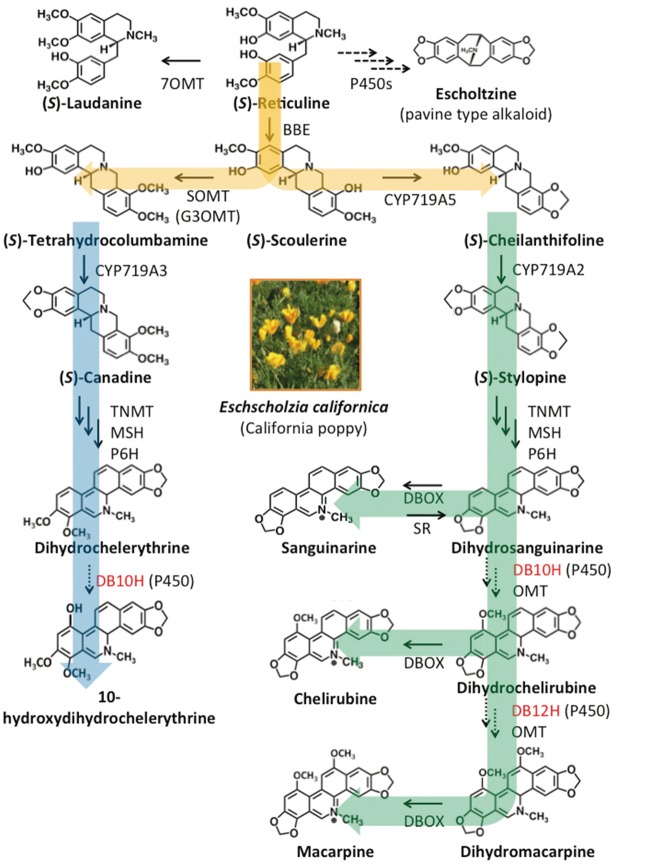
In this study, we used Eschscholzia californica cv. Hitoezaki (California poppy), a member of the Papaveraceae family that produces various types of benzylisoquinoline alkaloids (BIAs), i.e. aporphine-, pavine-, protoberberine-, protopine- and benzophenanthridine-type alkaloids (Fabre et al. 2000). The major alkaloid biosynthesis pathways and biosynthetic enzymes of these BIAs have been characterized at the molecular level (Hagel and Facchini 2013, Sato 2013). In fact, benzophenanthridine alkaloid biosynthesis from (S)-reticuline to dihydrosanguinarine has been characterized at the genetic level (Dittrich and Kutchan 1991, Ikezawa et al. 2007, Liscombe and Facchini 2007, Ikezawa et al. 2009, Beaudoin and Facchini 2013, Takemura et al. 2013), and this information has been used to reconstruct its biosynthetic pathway (Fossati et al. 2014, Hori et al. 2016). However, the genes involved in the biosynthesis of macarpine have not been characterized, although two P450s and two OMTs (Fig. 1), i.e. dihydrobenzophenanthridine alkaloid 10-hydroxylase (DB10H), dihydrobenzophenanthridine alkaloid 12-hydroxylase (DB12H), 10-hydroxydihydrobenzophenanthridine alkaloid 10-O-methyltransferase (10OMT) and 12-hydroxydihydrobenzophenanthridine alkaloid 12-O-methyltransferase (12OMT), involved in macarpine biosynthesis have been identified (De-Eknamkul et al. 1992, Kammerer et al. 1994). Macarpine biosynthesis is especially interesting because its biosynthetic pathway from reticuline involves six P450s: cheilanthiforine synthase (CYP719A5), stylopine synthase/canadine synthase (CYP719A2/A3), N-methylstylopine hydroxylase (CYP82N5), protopine 6-hydroxylase (CYP82N2v2), DB10H and DB12H. Additionally, the anti-proliferative effects of macarpine on several cancer cell lines have received increased attention (Slaninová et al. 2007).
Fig. 1.
Benzylisoquinoline alkaloid biosynthetic pathway in California poppy. Solid arrows indicate pathways in which biosynthetic enzyme genes have been identified. Dotted arrows indicate proposed pathways. BBE, berberine bridge enzyme; CYP719A2, stylopine synthase; CYP719A3, stylopine/canadine synthase; CYP719A5, cheilanthifoline synthase; DB10H, dihydrobenzophenanthridine alkaloid 10-hydroxylase; DB12H, dihydrobenzophenanthridine alkaloid 12-hydroxylase; DBOX, dihydrobenzophenanthridine alkaloid oxidase; MSH, N-methylstylopine hydroxylase; OMT, O-methyltransferase; 7OMT, reticuline 7-O-methyltransferase; P450, cytochrome P450; P6H, protopine 6-hydroxylase; SOMT, scoulerine 9-O-methyltransferase; TNMT, tetrahydroprotoberberine N-methyltransferase; SR, sanguinarine reductase.
Because the evolution of specialized metabolism is enhanced by the diversification of key enzymes such as P450 (Mizutani and Sato 2011, Nelson and Werck-Reichhart 2011), we also focused on the diversification of the P450 family in our draft genome sequence research. Our results suggest that draft genome sequencing can be a useful resource both for mining the biosynthetic enzyme genes involved in specialized metabolism and for developing tools for bioengineering. In particular, the identification of many silenced genes suggests that plant cells have the potential to expand their specialized metabolism, as reported in microbial cell systems (Ozaki et al. 2013, Rutledge and Challis 2015).
The full and/or comparative sequencing of related genomes is usually used in these types of studies. By using Illumina sequencing and genome assembly, we obtained an E. californica draft genome sequence that covered >95% of the whole genome (502 Mb). After the assembly, we searched the genome sequence and its gene clusters for sequences related to BIA biosynthesis using known biosynthetic enzyme genes. Our search results indicated that all characterized biosynthetic enzyme genes were found in the draft genome, indicating that the draft sequence sufficiently covered the genome; however, a gene cluster for BIA biosynthesis was not found. Further characterization of the identified P450 genes indicated that the California poppy genome was enriched with genes belonging to the CYP80, CYP82 and CYP719 families; these genes are involved in BIA biosynthesis in E. californica but not in Arabidopsis thaliana. Notably, CYP82 genes were clustered in some scaffolds, and the results of additional expression analyses suggested that these genes functioned differently in plant tissues and cultured cells with unique alkaloid profiles. A functional analysis using transgenic yeast revealed that CYP82P2 and CYP82P3 function as 10-hydroxylases in dihydrobenzophenanthridine alkaloid biosynthesis and exhibit different substrate specificities and expression profiles. The molecular evolution of P450 and metabolic diversification of BIA biosynthesis are also discussed.
Results
Draft genome sequencing and assembly
After the genomic DNA was digested, 150 bp of several paired-end (PE) and mate-pair (MP) libraries (PE300, PE500, MP3k, MP5k, MP8k, MP10k, MP15k and MP20k) containing average insert sizes of 342–15,125 bp were sequenced via Illumina sequencing 610 million, 510 million, 220 million, 250 million, 120 million, 360 million, 130 million and 170 million times, respectively, producing a coverage value 790 times the genome size (Supplementary Table S1). The sequences were then assembled by Platanus 1.2.1.1 (Kajitani et al. 2014) after the removal of adaptor sequences and an error correction by SOAPec v2.01 using a k-mer size of 27, a low-frequency k-mer cut-off of 19 and manual correction methods. The sequences were ultimately assembled into 53,253 scaffolds that covered approximately 489 Mb of the California poppy genome (approximately 97% of the 502 Mb genome) (Cui et al. 2006) (Table 1). The N50 of the scaffolds consisted of approximately 753 kb, and the longest was 4 Mb. To validate the completeness of the sequences, the EST and RNA sequencing data of E. californica from NCBI (http://www.ncbi.nlm.nih.gov/) and PhytoMetaSyn (http://www.phytometasyn.ca/) databases were mapped onto the draft genome sequence. The mapping results indicated that >90% of the RNA sequences (approximately 80,000 in total) were mapped onto the draft genome and covered >50% of the known RNA sequences (Supplementary Table S2). Gene prediction was also performed with the RNA sequence data obtained from the NCBI Sequence Read Archive (SRA) database (accession: SRX096037) and by Augustus v3.0.3, as described in the Materials and Methods. The genes predicted by Augustus were further annotated by similarity searches against the NCBI non-redundant (NR) database. The gene set that included the genes predicted by Augustus was named CSE_r1.0_cds_braker1. Alternatively, the gene set excluding the genes without similarity matches against the NR database but with similarity matches against transposable elements and pseudogenes was named CSE_r1.0_cds. The numbers of the genes in the CSE_r1.0_cds_braker1 and CSE_r1.0_cds groups are shown in Table 1. The draft genome sequences were deposited in the DDBJ/GenBank/EMBL databases (accession Nos. BEHA01000001–BEHA01053253; 53,253 entries), and the annotated gene information is available at the Eschscholzia Genome DataBase (http://eschscholzia.kazusa.or.jp).
Table 1.
Eschscholzia californica draft genome assembly statistics
| Draft genome sequence (ECA r1.0) | Predicted genes | |||
|---|---|---|---|---|
| (ECA_r1.0_cds_braker1) | (ECA_r1.0_cds) | |||
| Total | No. of sequences | 53,253 | 93,339 | 41,612 |
| Total length (bases) | 489,064,912 | 86,538,158 | 42,739,276 | |
| Average (bases) | 9,184 | 927 | 1,027 | |
| Maximum (bases) | 4,249,866 | 16,281 | 16,281 | |
| Minimum (bases) | 300 | 30 | 150 | |
| N50 (bases) | 752,971 | 1,263 | 1,329 | |
| G + C% | 36.5 | 41.2 | 40.9 | |
| ≥500 bases | No. of sequences | 37,795 | 59,645 | 30,683 |
| Total length (bases) | 482,984,812 | 74,697,139 | 38,761,101 | |
| Average (bases) | 12,779 | 1,252 | 1,263 | |
| ≥1 kb | No. of sequences | 15,580 | 29,617 | 16,617 |
| Total length (bases) | 467,061,688 | 53,254,023 | 28,552,063 | |
| Average (bases) | 29,978 | 1,798 | 1,718 | |
| ≥5 kb | No. of sequences | 2,223 | 199 | 141 |
| Total length (bases) | 444,303,297 | 1,266,048 | 935,853 | |
| Average (bases) | 199,867 | 6,362 | 6,637 | |
We further searched the genome for previously known genes that are involved in BIA biosynthesis, including reticuline and sanguinarine biosynthesis. Queries with tblastn (Altschul et al. 1990) revealed that all the BIA biosynthetic enzyme genes except CYP719A3 were in the draft genome (Supplementary Table S3). However, a CYP719A3 gene with an unfixed N-sequence was also found in the draft genome, and the presence of this gene was confirmed by genomic PCR and sequencing. This finding indicates that the draft genome is a sufficient platform for further sequence analyses. In fact, when the coclaurine N-methyltransferase (CNMT) and dihydrobenzophenanthridine alkaloid oxidase (DBOX) genes, which have not been characterized in California poppy, were queried against sequences from Coptis japonica and Papaver somniferum (Choi et al. 2002, Hagel et al. 2012), putative orthologs of CjCNMT and PsDBOX were found in the draft genome. Thus, the sequences were useful for obtaining the full genomic information for EcCYP719A3 and other P450 genes, as mentioned below; however, the relatively abundant number of N-sequences produced due to the heterozygosity of California poppy (Supplementary Fig. S1) hindered the sequence analyses.
We searched for orthologs of the biosynthetic enzyme genes involved in the synthesis of morphinan alkaloids (e.g. CYP719B1; Gesell et al. 2009) and gene clusters associated with noscapine alkaloid biosynthesis (Winzer et al. 2012); such gene clusters are specific to the opium poppy. Our search did not identify any of these genes, indicating that morphinan and noscapine biosyntheses are specific to opium poppy at the genetic level. Additional searches for clusters of the biosynthetic enzyme genes involved in BIA biosynthesis also yielded no matches in the scaffolds (Supplementary Table S3), except for the identified CYP82 gene clusters, which are discussed later (Supplementary Table S5; Supplementary Fig. S2).
Analysis of P450 families
In BIA biosynthesis and many other specialized metabolite biosynthesis pathways, P450s are important for determining chemical diversity in metabolism (Mizutani and Sato 2011, Nelson and Werck-Reichhart 2011). Plants are highly enriched with P450s; thus far, 135 families (http://drnelson.uthsc.edu/biblioD.html) have been reported. Therefore, using tblastn searches, we estimated the amount of P450 family genes in the E. californica draft genome based on sequence similarity. We also compared the number of P450 family genes with that of A. thaliana, as only three P450 families, i.e. CYP80, CYP82 and CYP719, have been identified as components of BIA biosynthesis (CYP80B1, Pauli and Kutchan 1998; CYP719A1, Ikezawa et al. 2003; CYP719A5, Ikezawa et al. 2007; CYP80G2, Ikezawa et al. 2008; CYP719B1, Gesell et al. 2009; CYP719A2/A3, Ikezawa et al. 2009; CYP82N4 (MSH), Beaudoin and Facchini 2013; CYP82N2v2 (P6H), Takemura et al. 2013; CYP82Y1, Dang and Facchini 2014). As shown in Supplementary Table S4, the numbers of some of the P450 families differed between the plant species. Specifically, CYP80, CYP82 and CYP719, which are involved in BIA biosynthesis, were more enriched in E. californica than in A. thaliana; these families were named by the P450 nomenclature committee (Dr. D.R. Nelson, University of Tennessee, Memphis, TN, USA; Supplementary Table S5). Among these P450s, the CYP82 family was the most diversified.
The CYP80 family contained CYP80B1 (N-methylnorcoclaurine hydroxylase, Pauli and Kutchan 1998) and an uncharacterized CYP80 gene (named CYP80B5) that had a 70% amino acid sequence identity to CYP80B1. The CYP719 family consisted of seven genes, one of which exhibited 95% homology with CYP719A5. On the other hand, the CYP82 family consisted of 28 genes, of which only three (CYP82N2v2, CYP82N5 and CYP82Y1) have been biochemically characterized; another four genes could be potential homologs of previously identified genes. These results indicate that the CYP82 family is rich in uncharacterized biosynthetic enzyme genes that are involved in BIA biosynthesis. Many uncharacterized CYP82 genes also formed gene clusters (Supplementary Fig. S2). Although CYP82P3 contained an unfixed N-sequence in the draft genome, its full sequence was determined by genomic PCR/reverse transcription–PCR and nucleotide sequencing. PseudoCYP82 was determined to be a pseudogene due to the presence of a stop codon in the coding frame.
When the genome structure, including the presence of introns, was analyzed, all genes in the CYP719 family and some CYP82 genes, such as protopine 6-hydroxylase (CYP82N2v2 and its homologs), lacked introns (Supplementary Table S6). On the other hand, CYP80 and many CYP82 family genes contained an intron in the region of their fourth substrate recognition site.
The phylogenetic analysis of the CYP82 genes showed that they were quite diverse (Supplementary Fig. S3), and some CYP82 subfamily genes, e.g. CYP82P (P2, P4–P6), CYP82B (B2–B4) and CYP82D (D170, D171), clustered in scaffolds, including Eca_sc000081.1, Eca_sc007583.1 and Eca_sc066992.1 (Supplementary Fig. S2). To characterize the functions of these uncharacterized CYP82 genes, we analyzed their expression in both plant tissues and cell lines that exhibited different BIA production abilities.
CYP82 family genes show diverse expression and function
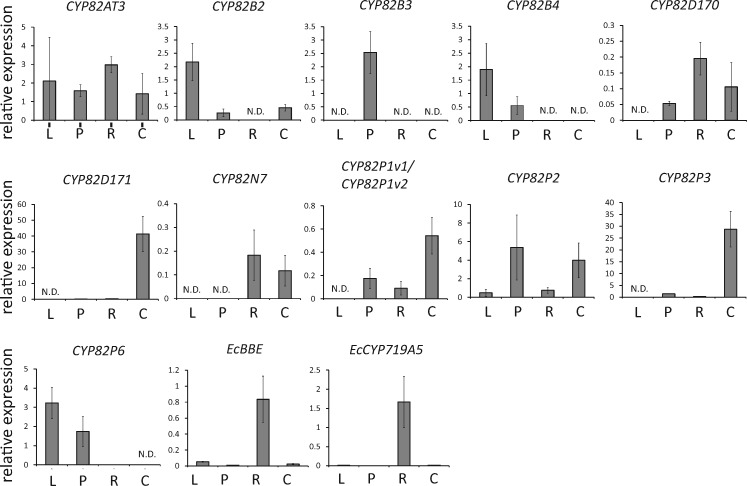
Using specific gene primers, we analyzed the expression of uncharacterized CYP80 and CYP82 genes, except for CYP80B1, CYP82N2v2 and CYP82N5 and their homologs (i.e. CYP80B5, CYP82N1, CYP82N2v3 and CYP82N2v4) (Supplementary Table S7). Due to their high sequence similarity, CYP82P1v1 and CYP82P1v2 were analyzed as a set of genes. First, we examined their expression in E. californica plant tissues that exhibited different alkaloid accumulation profiles (Supplementary Fig. S4). Our previous analysis indicated that roots, which accumulate benzophenanthridine alkaloids, showed high expression levels of sanguinarine biosynthetic genes such as CYP80B1, N-methylcoclaurine 4′-O-methyltransferase (4′OMT), berberine bridge enzyme (BBE), CYP719A5, CYP719A2 and CYP719A3 (Ikezawa et al. 2007, Ikezawa et al. 2009). Similarly, genes involved in macarpine biosynthesis, such as DB10H and DB12H, were expected to be expressed in the roots in which macarpine accumulated. Aerial parts, such as leaves and petioles, are known to accumulate pavine-type alkaloids such as escholtzine (Fabre et al. 2000), so pavine-type alkaloid biosynthetic enzyme-encoding genes were expected to be expressed in those tissues.
As shown in Fig. 2, each P450 gene showed a distinct expression profile. CYP82N7 was specifically expressed in the roots and cultured cells. CYP82D170, CYP82P1v1/CYP82P1v2 and CYP82P2 were expressed in the petioles, roots and cultured cells. In contrast, CYP82B2, CYP82B3, CYP82B4 and CYP82P6 were not expressed in the roots but were highly expressed in the aerial parts. CYP82D171 and CYP82P3 were preferentially expressed in the wild-type cultured cells. There was no accumulation of the CYP82AT2, CYP82AU1, CYP82AV1, CYP82D172, CYP82N6, CYP82P4 and pseudoCYP82 transcripts. The data for CYP80B5, CYP82AT1 and CYP82P5 transcripts were not obtained due to large replication errors.
Fig. 2.
Expression analysis of novel P450 genes in E. californica plant tissues. Transcript expression was measured by quantitative real-time PCR using the cDNAs of the plant tissues and wild-type cultured cells of E. californica. The relative expression levels were calculated against the values of the β-actin gene. Error bars indicate the SD calculated from three technical replicates.
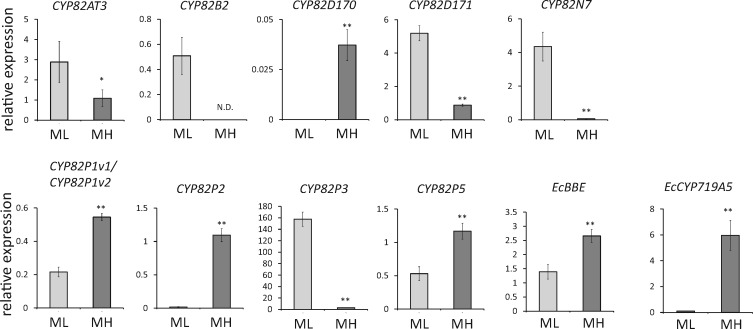
Next, we analyzed the expression of CYP82 genes in two cultured E. californica cell lines with differences in macarpine productivity: a low-macarpine-producing (ML) cell line and a high-macarpine-producing (MH) cell line (Supplementary Fig. S5). The MH cells showed exclusive or higher expression of CYP82D170, CYP82P1v1/CYP82P1v2, CYP82P2 and CYP82P5 than the ML cells, whereas the ML cells showed higher expression of CYP82AT3, CYP82B2, CYP82D171, CYP82N7, CYP82P3 and CYP82N7 than the MH cells (Fig. 3). No expression of CYP82AT1, CYP82AT2, CYP82AU1, CYP82AV1, CYP82B3, CYP82B4, CYP82P6 or pseudoCYP82 was observed in either cell line. The data for CYP82D172 and CYP82N6 were not obtained due to large replication errors.
Fig. 3.
Expression analysis of novel P450 genes in cultured E. californica cells. Transcript expression was measured by quantitative real-time PCR using the cDNAs of macarpine ML/MH cultured cells. The relative expression levels were calculated by against the values of the β-actin gene. Error bars indicate the SD calculated from three technical replicates. *P < 0.05, **P < 0.01 (statistical analysis followed by the t-test).
Enzymatic activity of novel P450s
Using the expression profile data, we characterized the enzymatic activity of the macarpine biosynthesis candidate genes, i.e. CYP82D170, CYP82N7, CYP82P1v1, CYP82P2 and CYP82P5, which were clearly expressed in the MH line and roots. Full-length cDNAs were prepared from the cDNA library by reverse transcription–PCR using gene-specific primer pairs (Supplementary Table S8) designed from the draft genome sequence; expression vectors of these cDNAs for Saccharomyces cerevisiae were constructed to prepare recombinant microsomal fractions to measure enzymatic activities.
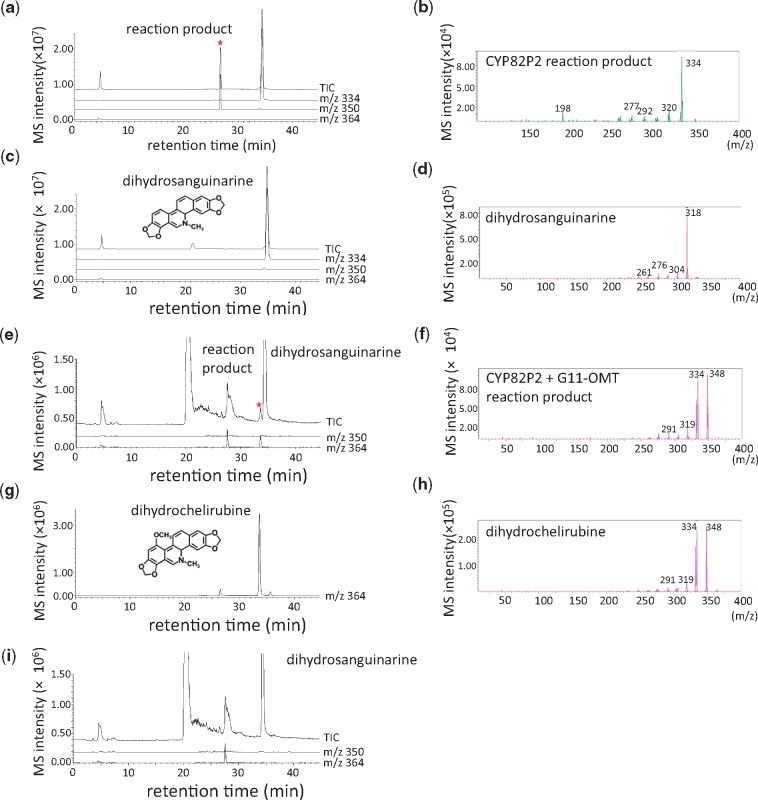
When the recombinant microsomal fractions reacted with 6-benzophenanthridine alkaloid substrates (dihydrochelerythrine, dihydrochelirubine, dihydrosanguinarine, chelerythrine, chelirubine and sanguinarine; Supplementary Fig. S6), only the microsomal fraction expressing CYP82P2 produced a new hydrophilic peak [mass-to-charge ratio (m/z) increase of 16] from dihydrosanguinarine (m/z 334), suggesting that a hydroxylation reaction occurred (Supplementary Fig. S7). Transgenic Pichia pastoris cells expressing CYP82P2 confirmed this reaction (Fig. 4a–d). The co-culture of transgenic Pichia cells expressing the CYP82P2 and G11 O-methyltransferase isolated from MH cells (Purwanto et al. 2017) produced another new peak that had an identical retention time and mass spectrometry (MS) fragment pattern to the dihydrochelirubine (Fig. 4e–h), and the dihydrosanguinarine 10-hydroxylase function of CYP82P2 was confirmed. However, a recombinant CYP82P2 neither hydroxylated dihydrochelerythrine nor converted dihydrochelirubine, indicating that other P450s, such as 12-hydroxylase and/or dihydrochelerythrine 10-hydroxylase, exist.
Fig. 4.
Enzymatic activity of CYP82P2 expressed in Pichia cells. LC-MS chromatogram data (a, c, e, g, i) and MS fragment patterns (b, d, f, h) are presented as follows: (a, b) reaction product of CYP82P2 incubated with dihydrosanguinarine; (c, d) vector control with dihydrosanguinarine; (e, f) reaction product of the co-reaction of CYP82P2 and G11-OMT with dihydrosanguinarine; (g, h) dihydrochelirubine prepared from the transgenic cultured E. californica cells; (i) reaction product of the vector control with dihydrosanguinarine. The molecular masses of dihydrosanguinarine and dihydrochelirubine are m/z 334, [M+H]+ and m/z 364 [M+H]+, respectively.
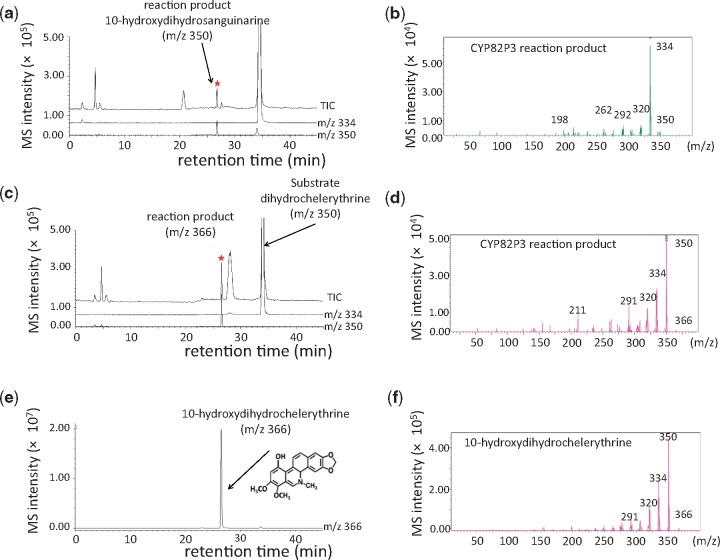
Based on the finding that CYP82P2 is a dihydrosanguinarine 10-hydroxylase, we searched for additional hydroxylases involved in macarpine biosynthesis and for dihydrochelerythrine 10-hydroxylases. Because none of the five candidates (CYP82D170, CYP82N7, CYP82P1v1, CYP82P2 and CYP82P5) showed hydroxylation activity for dihydrochelirubine, we searched for dihydrochelerythrine 10-hydroxylase, which produces 10-hydroxychelerythrine, in the ML cells. Although CYP82P4 was a good candidate for these reactions because it was located in the same scaffold as CYP82P2 and exhibits high sequence homology, the full-length CYP82P4 cDNA could not be isolated from the cDNA library. Nevertheless, the highly homologous CYP82P3 gene was amplified from 10-hydrochelerythrine-producing ML cells, and its enzymatic activity was examined.
When transgenic S. cerevisiae cells expressing CYP82P3 were prepared and reacted with the substrate alkaloids, the cells not only formed 10-hydroxydihydrosanguinarine but also formed 10-hydroxydihydrochelerythrine (m/z 366) from the corresponding substrates (Fig. 5). The products were identified by their retention times and MS fragment patterns in comparison with those of authentically prepared 10-hydroxydihydrochelerythrine. CYP82P3 did not show any enzymatic activity against other benzophenanthridine alkaloids, i.e. sanguinarine, chelerythrine, chelirubine or dihydrochelirubine.
Fig. 5.
Enzymatic activity of CYP82P3 expressed in yeast cells. LC-MS chromatogram (a, c, e, g) and MS fragment patterns (b, d, f, h) were presented. (a, b) Reaction product of CYP82P3 incubated with dihydrosanguinarine, (c, d) vector control with dihydrosanguinarine, (e, f) reaction product of CYP82P3 with dihydrochelerythrine, (g, h) 10-hydroxydihydrochelerythrine prepared from MH cell extracts. The molecular masses of dihydrosanguinarine, dihydrochelerythrine and 10-hydroxydihydrochelerythrine are m/z 334, [M+H]+, m/z 350, [M+H] and m/z 364, [M+H]+, respectively.
Sequence characteristics of uncharacterized P450s
Two isolated P450s, CYP82P2 and CYP82P3, shared high sequence similarity (Supplementary Figs. S3, S8) but showed different expression profiles and substrate specificities (Figs. 2–5). Furthermore, some P450 genes showed no expression in the examined plant tissues and cell cultures. Although heterologous expression in microbial cells is useful for examining the biological function of enzymes, recombinant protein production can be difficult. In fact, CYP82D171 was not transformed in the S. cerevisiae cells. Therefore, we examined the sequence characteristics of the uncharacterized P450s.
First, we examined the conservation of P450 signatures in several candidate CYP82 P450s that were found in this study, e.g. CYP82D170, CYP82D171, CYP82N3, CYP82N7, CYP82P1v1, CYP82P2, CYP82P3, CYP82P5 and CYP82P6. These P450s, except for CYP82N3, CYP82P1v1 and CYP82P6, contained the conserved domains of eukaryotic P450s, i.e. the K-helix region, aromatic region and heme-binding domain, and there was a conserved threonine residue in the I-helix region of the monooxygenase-type P450s (Supplementary Fig. S8). In CYP82N3, a conserved threonine was replaced with serine, and, in CYP82Pv1 and CYP82P6, a serine replacement occurred at a conserved glycine/alanine residue found in non-canonical methylenedioxy-forming enzymes (CYP719A1, CYP719A2, CYP719A3 and CYP719A5) and the C–C phenol-coupling enzyme (CYP80G2). These modifications suggest that CYP82N3, CYP82Pv1 and CYP82P6 may have unique functions. However, other P450s such as CYP82D170, CYP82D171, CYP82N7 and CYP82P5 had conserved threonine and glycine/alanine residues, and these enzymes, as well as CYP82P2 and CYP82P3, were expected to have general functions as hydroxylases. More biochemical and physiological characterizations are needed to identify the remaining hydroxylases involved in macarpine and pavine biosynthesis and their physiological roles in BIA biosynthesis.
Discussion
Due to the rapid progress of NGS technology, the whole-genome sequencing of non-model organisms is more achievable, although many obstacles still exist. In this study, we used a commercial variety of California poppy (E. californica cv. Hitoezaki) to determine the California poppy draft genome sequence. Despite the heterozygosity of the genome of this plant species, the genome sequence obtained by this study still contained many unfixed N-sequences and >53,000 scaffolds, and the draft sequence covered >95% of the genome (approximately 489 Mb of 502 Mb) and transcriptome data. Heterozygosity and unfixed N-sequences hindered the identification of genes, but the current data still sufficiently covered the biosynthetic enzyme genes involved in the BIA pathway (Table 1), and the N-sequences found in CYP719A3 and CYP82P3 could be fixed by the amplification of the genomic DNA after nucleotide sequencing. These results indicated that NGS-based draft genome sequencing can provide a sufficient platform for the characterization of specialized metabolism; specialized metabolism is diversified, and its expression is limited to specific cells or tissues and affected by environmental conditions.
The absence of both a gene cluster involved in noscapine biosynthesis (e.g. CYP82X2 and CYP82Y1, Winzer et al. 2013) and the morphinan alkaloid gene (e.g. CYP719B1, Gesell et al. 2008) in the draft genome of California poppy also indicated that noscapine and morphinan biosynthetic enzyme-encoding genes are specific to the opium poppy, despite the identification of some homologs in the California poppy genome. Our draft genome sequence is a good starting platform for dissecting the molecular evolution of BIA biosynthesis in Papaveraceae and comparing results with the recently published draft genome sequence of Macleaya cordata, which is also a member of the Papaveraceae family that produces BIAs (Liu et al. 2017). The M. cordata draft genome sequence also showed an absence of noscapine- and morphine-specific genes. The genome sequence of Aquilegia coerulea, an IQA-producing member of the Ranunculaceae family (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Acoerulea), was recently reported. These genome sequences enable future analyses of the evolution of BIA biosynthesis in the Papaveraceae and Ranunculaceae families.
As the present study indicates, draft genome sequencing is especially useful for analyzing individual genes, such as the P450s involved in BIA biosynthesis. We estimated all the possible P450 genes in the draft genome and clarified the diversification of P450 family genes, i.e. the CYP80, CYP82 and CYP719 genes involved in BIA biosynthesis in E. californica. In particular, the CYP82 genes formed gene clusters within scaffolds (Supplementary Fig. S2), and the evolution of these genes via gene duplication was estimated by their high sequence similarity and conservation of exon–intron junction sites (Supplementary Table S6). However, the results of the analysis of individual gene expression showed diverse expression profiles among the plant tissues and cultured cells (Figs. 2, 3). Furthermore, the functional analysis of P450 homologs showed functional diversification, similar to the case of CYP82P2 and CYP82P3, which function as dihydrosanguinarine 10-hydroxylase and dihydrosanguinarine/dihydrochelerythrine 10-hydroxylase, respectively, in BIA biosynthesis (Figs. 4, 5). Interestingly, the expression of the CYP82P2 and CYP82P3 genes was correlated with the alkaloid profiles in each cell; the high expression of CYP82P2 induced macarpine production in the MH cells, and the CYP82P3 expression induced 10-hydroxychelerythrine production in the ML cells. The relative expression levels of CYP82P2 and CYP82P3 in the roots were lower than those in the cultured cells, and the accumulation of macarpine was low in the roots. These data suggest that CYP82P2/P3 (DB10H) proteins are important key regulators in macarpine biosynthesis.
In this study, we characterized P450 genes based on a draft genome sequence and indicated the diversification of both their expression patterns and functions in plant specialized metabolism. Importantly, many genes were not expressed under the current analytical conditions. Functional analyses of these non-expressed and uncharacterized genes using our alkaloid production platform and microbial systems (Minami et al. 2008, Hori et al. 2016, Nakagawa et al. 2016, Ehrenwoth and Peralta-Yahya 2017) would be useful to understand the future molecular evolution of secondary metabolism and the development of more diversified metabolite production, as shown in microbial genomes by the activation of potential genes (Ozaki et al. 2013).
Materials and Methods
Plant materials
Eschscholzia californica (cultivar Hitoezaki, Takii Seed Co., Ltd.) plants were grown in soil at 23°C under white light (100–200 μmol photons m−2 s−1) and a 10 h light/14 h dark photoperiod, and were used for the genomic DNA extraction. The total RNA for the expression analysis was prepared from the leaf blades, petioles and roots of plants grown for 5 months. Suspension-cultured wild-type, S-38 line [ML cell line expressing the scoulerine 9-O-methyltransferase (SMT) gene of Coptis japonica (Takemura et al., 2010b)] and A5-1 line [MH cell line expressing the EcCYP719A5 gene (Takemura et al., 2010a) [E. californica cells were grown in Linsmaier–Skoog medium (pH 5.7) (Linsmaier and Skoog 1965) containing 10 μM naphthaleneacetic acid, 1 μM benzyladenine and 3% sucrose at 23°C in the dark at 3 week intervals. ML and MH cells grown for 2 d and wild-type cells grown for 2 weeks were used for the total RNA preparation.
Chemicals
Sanguinarine was purchased from Sigma-Aldrich Co., LLC. N-Methyltetrahydroberberine, protopine, allocryptopine and chelerythrine were retrieved from the collection of Dr. K. Iwasa. 10-Hydroxychelerythrine and chelirubine were isolated from the ML cells and the cultured E. californica cells with the ectopic expression of the C. japonica WRKY1 gene, respectively, using a CombiFlash® Rf+ (Teledyne Isco) purification system, and the identities of these compounds were determined based on their retention times and m/z values by liquid chromatography–MS (LC-MS), as previously described (Takemura et al. 2010b). 10-Hydroxydihydrochelerythrine, dihydrochelirubine, dihydrosanguinarine and dihydrochelerythrine were prepared by a NaBH4 reduction from 10-hydroxychelerythrine, chelirubine, sanguinarine and chelerythrine, respectively.
Metabolite analysis
Alkaloids were extracted in methanol with 0.01 N HCl and analyzed using an LC-MS 2020 or an LC-MS 8030 system (Shimadzu Corp.) under the following conditions: electrospray ionization (ESI)-MS, 1.5 kV, positive ion mode; column, TSkgel 80-TM (4.6X250 mm; TOSOH); temperature, 40°C; mobile phase, acetonitrile (A) and water containing 1% (v/v) acetic acid. The elution profile was as follows: 0–15 min, isocratic elution with 40% (v/v) A; 15–18 min, linear gradient from 40% to 80% (v/v) A; 18–40 min, isocratic elution with 80% (v/v) A; 40–43 min, linear gradient from 80% to 40% (v/v) A; and 43–45 min, re-equilibration with 40% (v/v) A. The MS fragment spectra of the alkaloids were detected using the LC-MS 8030 system under the following conditions: ESI-MS, product ion scan mode, m/z of 10.00–400.00 and collision energy at –35.0 V. The other conditions were the same as those described above.
Eschscholzia californica genome sequencing and assembly
The E. californica genomic DNA (50 μg at 100 ng μl−1) was prepared using a NucleoSpin® Plant Midi Kit (TAKARA BIO INC.). The genomic DNA was digested, and 150 bp of several PE and MP libraries (PE300, PE500, MP3k, MP5k, MP8k, MP10, MP15k and MP20k) containing average insert sizes of 342, 545, 2,820, 4,545, 7,926, 8,945p, 13,158 and 15,125 bp, respectively, were prepared using a TruSeq DNA Sample Prep Kit and a Nextera Mate Pair Sample Prep Kit (Illumina) according to the manufacturer’s instructions; these libraries were sequenced using an Illumina HiSeq 2500 sequencer 610 million, 510 million, 220 million, 250 million, 120 million, 360 million, 130 million and 170 million times, respectively, to produce a 790 times genome coverage (Supplementary Table S1). The raw sequence reads were filtered by both the trimming of adaptor sequences in the reads and error correction using SOAPec v2.01 with a k-mer size of 27 and a low-frequency k-mer cut-off value of 19. The sequence reads were assembled by PLATANUS v1.2.11 (Kajitani et al. 2014) and edited manually. Scaffolds longer than 300 bp were extracted. Scaffolds that matched the genome sequences of bacteria, fungi or humans in NCBI, vector sequences (UniVec in NCBI), PhiX sequences (Illumina) or the chloroplast (NC_000932.1) and mitochondrial (NC_001284.2) genomes of A. thaliana with an E-value ≤1E-10 and a length coverage ≥10% were excluded and considered to be probable contamination.
Gene prediction
The RNA sequencing (RNA-Seq) data sampled from the California poppy roots for gene prediction were obtained from the NCBI SRA database (accession: SRX096037). Three nucleotides from the 3′ terminus of all of the reads were trimmed because the average quality value (QV) for this region was >20. Fewer than 10 nucleotides at the 3′ terminus were further trimmed, and the reads with unknown nucleotides (Ns) were excluded by using PRINSEQ v0.20.4 (Schmieder and Edwards 2011). In addition, the reads that included >10 bases of nucleotides from the 5′ terminus of the adaptor sequence (AGATCGGAAGAGC) at the 3′ terminus were excluded by using the FASTX-toolkit v0.0.14 (http://hannonlab.cshl.edu/fastx_toolkit/). After being trimmed and filtered, the RNA-Seq reads were mapped onto the scaffolds using TopHat v2.0.14 (Trapnell et al. 2009), and the resulting BAM file was processed with BRAKER1 v1.3 (Hoff et al. 2016) to construct a training set for gene prediction. Gene prediction was performed by Augustus v3.0.3 (Stanke et al. 20) with the training set.
Annotation
The genes predicted by Augustus were subjected to similarity searches against the NCBI NR database (ftp://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/nr.gz) by BLAST, with an E-value cut-off of 1E-10; domain searches against the InterPro database (Finn et al. 2017) by InterProScan (Jones et al. 2014) using an E-value cut-off of 1.0 were also performed. According to their gene structure results, the genes were tagged as ‘partial’ (partial gene; gene with either a start or a stop codon or without both codons), ‘pseudo’ (pseudogene; gene including an in-frame stop codon) or ‘short’ (short gene; gene product shorter than 50 amino acid residues), and those without tags contained both start and stop codons. The genes with hits against transposable elements were tagged as ‘TE’. According to the similarity searches, the genes were tagged ‘f’ (hit against the NR database, with an E-value ≤1E-20 and a length coverage ≥70%), ‘p’ (hit against the NR database, with an E-value ≤1E-20 and length coverage <70%), ‘d’ (hit against the InterPro database, with an E-value ≤1.0) and ‘n’ (either no hits or hits against the NR database with an E-value >11E-20 and no hits against the InterPro database).
Estimation of P450 genes
Arabidopsis thaliana P450 sequences were obtained from The Arabidopsis Information Resource (http://www.arabidopsis.org/index.jsp). Eschscholzia californica P450s were estimated by tblastn searches using previously known P450 amino acid sequences as the queries, with the exception of sequences from the CYP95, 713, 737, 738, 740, 773–797 and 800–805 families, which were not available to the public. Partial sequences and N-containing sequences were removed from the results. The introns, start codons and stop codons of CYP80, CYP82 and CYP719 family genes were estimated based on comparisons with cDNA sequences. The CYP80 and CYP82 genes were estimated from the draft genome and were named by the P450 nomenclature committee (Dr. D.R. Nelson, University of Tennessee) (Supplementary Table S5). The nucleotide sequences for CYP80B5, CYP82B2–5, CYP82D170–172, CYP82N1, CYP82N2v3, CYP82N2v4, CYP82N5–7, CYP82P1v1, CYP82P1v2, CYP82P2–6, CYP82AT1–3, CYP82AU1, CYP82AV1 and CYP719C1 were submitted to DDBJ/GenBank/EMBL under the accession numbers LC316736–LC316762.
Genomic PCR
To determine the identity of the N-sequences, the E. californica genomic DNAs for CYP719A3, CYP82P2 and CYP82P4 were analyzed by genomic PCR using the primer sequences shown in Supplementary Table S9. CYP82P3 and CYP82P4 were amplified by nested PCR using gene-specific primer pairs.
Alignment analysis of the CYP82 family
Representative plant P450 amino acid sequences were obtained from GenBank for the tree construction, whereas EcCYP82N5 (N-methylstylopine hydroxylase) was newly identified in this study. The accession numbers for the amino acid sequences were as follows: AAC39454.1, E. californica EcCYP82B1 (wrongly annotated N-methylcoclaurine 3′-hydroxylase); BAK20464.1, EcCYP82N2v2 (P6H); AGC92397.1, P. somniferum PsCYP82N3 (P6H); AGC92398.1, PsCYP82N4 (MSH); AFB74614.1, PsCYP82X1 (P450 involved in noscapine biosynthesis); and uncharacterized P450s [AFB74616.1, PsCYP82X2; AFB74617.1, PsCYP82Y1 (N-methylstylopine 1-hydroxylase); BAF98472.1, C. japonica CjCYP82R1; NP_194925.1, A. thaliana AtCYP82C2; NP_194923.1, AtCYP82C3; NP_194922.1, AtCYP82C4; NP_180088.1, AtCYP82F1; NP_189154.1 and AtCYP82G1]. The CYP82 sequences were aligned by ClustalW (Thompson et al. 1994), and a phylogenetic tree was generated by MEGA6 (using the Neighbor–Joining statistical method, a bootstrap test involving 1,000 replicates and the Jones–Taylor–Thompson model) (Tamura et al. 2013).
Quantitative real-time PCR
The total RNA was extracted from 2-day-old cultured E. californica cells (ML and MH cells), 2-week-old cultured E. californica cells (wild types) and the tissues (leaf blade, petiole and roots) of the plants that had been growing for 5 months. RNA extraction was performed using an RNeasy plant Mini Kit (Qiagen). Single-stranded cDNAs were synthesized from 1 mg of total RNA using a PrimeScript™ RT reagent Kit (TAKARA BIO INC.). Quantitative real-time PCR was performed with gene-specific primer pairs (Supplementary Table S7) using the iQ™ SYBR Green Supermix and a CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories Inc.) under the following PCR conditions: 95°C for 3 min, followed by 40 cycles of 95°C for 10 s, 55°C for 20 s and 72°C for 20 s. The relative expression levels between the samples were calculated by using the standard curve method with the expression level of β-actin as an internal standard.
Isolation of the full-length cDNAs of novel P450s
The full-length cDNAs of novel P450s were amplified by PCR from the total cDNA that was used for the expression analysis using the primer pairs shown in Supplementary Table S7. CYP82D170, CYP82D171, CYP82P1v1, CYP82P2 and CYP82P3 were amplified with gene-specific primers designed from the coding sequences of the draft genome, whereas CYP82P5 was amplified using primers corresponding to its putative 5′-untranslated region (UTR)/3′-UTR sequences. The PCR for the CYP82N7 isolation was performed with a forward primer designed for its 5′-UTR and a reverse primer corresponding to an anchor sequence (5′-GCTGTCAACGATACGCTACGTAACGGCATGACAGTG-3′) added to the 3′ end of the template cDNA. All PCR products were re-amplified with primers to introduce an AvrII, NheI or XbaI site for the construction of yeast expression vectors (Supplementary Table S8). The sequences of the PCR products were confirmed after they were subcloned into pGEM T-easy or pT7 blue vectors but before their transformation.
Heterologous expression of novel P450s in S. cerevisiae or P. pastoris
The budding yeast expression vectors were constructed in pGYR-SpeI that contained the S. cerevisiae NADPH-P450 reductase (Ikezawa et al. 2008). The full-length cDNAs of CYP82D170, CYP82D171, CYP82P1v1, CYP81P2, CYP82P3, CYP82P5 and CYP82N7 were prepared from their subclones by the use of appropriate restriction enzymes (AvrII, NheI or XbaI) and were ligated into the SpeI site of the pGYR-SpeI vectors as previously described (Ikezawa et al. 2007). The expression vectors were introduced into the S. cerevisiae strain AH22, and microsomal fractions prepared from the cultured S. cerevisiae cells were used for the enzyme assay.
A P. pastoris expression vector was constructed by the insertion of the full-length cDNA of CYP82P2 prepared by AvrII into a pPIC3.5K vector (Invitrogen) and introduced into the P. pastoris strain GS115 as described previously (Hori et al. 2016).
Enzyme assays
Six benzophenanthridine alkaloids, i.e. sanguinarine, chelerythrine, chelirubine, dihydrosanguinarine, dihydrochelerythrine and dihydrochelirubine, were used as substrates (Supplementary Fig. S4), and the product formation was analyzed by LC-MS and LC-MS/MS as described in the metabolic analysis section.
In vitro assays were performed in a reaction mixture consisting of 100 mM HEPES/NaOH (pH 7.5), 500 μM NADPH, 100 μM substrate and 10 μg of the microsomal fractions of yeast cells expressing CYP82D170, CYP82P1v1, CYP81P2, CYP82P3, CYP82P5 or CYP82N7. After an incubation at 30°C for 2 h, the reaction was terminated by the addition of trichloroacetic acid (final concentration of 2%) and methanol (final concentration of 40%). After the proteins precipitated, the reaction products were analyzed using LC-MS and LC-MS/MS systems.
Regarding the in vivo assays, transgenic S. cerevisiae cells that contained the EcCYP82P3 gene and that had been resuspended in 1 ml of HEPES/NaOH buffer (pH 7.5) after a pre-culture period in concentrated SD medium (8% glucose, 1.36% yeast nitrogen base without amino acids and ammonium sulfate, 4% ammonium sulfate and 160 μg ml−1l-histidine) until OD600 = 10–15 were incubated together with the substrate (final concentration of 100 μM) at 30°C for 20 h. In vivo reactions using transgenic Pichia cells expressing CYP82P2 and co-cultures with Pichia cells expressing G11-OMT were performed as previously described, with minor modifications (Hori et al. 2016). The supernatant and methanol cell extracts (containing 0.01 N hydrochloride) of the yeast cell reactions were analyzed by LC-MS and LC-MS/MS as described above.
Database
The sequences of the draft genome (ECA_r1.0), the genes (ECA_r1.0_cds) and the proteins (ECA_r1.0_pep) are available from the Eschscholzia Genome DataBase (http://eschscholzia.kazusa.or.jp). The genome sequence is also available from DDBJ/GenBank/EMBL under the accession numbers BEHA01000001–BEHA01053253 (53,253 entries). The raw data of the PE and MP reads are available from the NCBI SRA database under the BioProject accession number PRJDB6318.
Author contributions
K.H. and F.S. conceived this work and designed the experiments. K.H. performed all the experiments. Y.M. and A.T. performed the draft genome sequencing, and Y.M., A.T., K.H., S.T., H.H., Y.Y. and F.S. analyzed the genomic data. R.P. provided G11OMT. K.H. and F.S. wrote the manuscript. All authors reviewed and approved the final version.
Supplementary Data
Supplementary data are available online.
Funding
This study was supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) [Grant-in-Aid for Scientific Research (S); 26221201 to F.S.]. The draft genome sequencing of E. californica was also supported by MEXT KAKENHI [No. 221S0002].
Supplementary Material
Acknowledgments
Computations were partially performed on the NIG supercomputer at the ROIS National Institute of Genetics.
Disclosures
The authors have no conflicts of interests to declare.
Glossary
Abbreviations
- BBE
berberine bridge enzyme
- BIA
benzylisoquinoline alkaloid
- Cj
Coptis japonica
- CNMT
coclaurine N-methyltransferase
- CYP80B1
N-methylnorcoclaurine hydroxylase
- CYP82N2v2
protopine 6-hydroxylase
- CYP82N4
N-methylstylopine 14-hydroxylase
- CYP719A2/A3
stylopine synthase/canadine synthase
- CYP719A5
cheilanthiforine synthase
- DB10H
dihydrobenzophenanthridine alkaloid 10-hydroxylase
- DB12H
dihydrobenzophenanthridine alkaloid 12-hydroxylase
- DBOX
dihydrobenzophenanthridine alkaloid oxidase
- Ec
Eschscholzia californica
- ESI
electrospay ionization
- EST
expressed sequence tag
- IQA
isoquinoline alkaloid
- LC
liquid chromatography
- MH
high-macarpine-producing
- ML
low-macarpine-producing
- MP
mate-pair
- MS
mass spectrometry
- MSH
N-methylstylopine 14-hydroxylase
- m/z
mass-to charge ratio
- NGS
next-generation sequencing
- NR
non-redundant
- OMT
O-methyltransferase
- 4′OMT
N-methylcoclaurine 4′-O-methyltransferase
- 7OMT
reticuline 7-O-methyltransferase
- 10OMT
10-hydroxydihydrobenzophenanthridine alkaloid 10-O-methyltransferase
- 12OMT
12-hydroxydihydrobenzophenanthridine alkaloid 12-O-methyltransferase
- P450
cytochrome P450
- PE
paired-end
- P6H
protopine 6-hydroxylase
- Ps
Papaver somniferum
- RNA-Seq
RNA sequencing
- SOMT
scoulerine 9-O-methyltransferase
- SRA
Sequence Read Archive
- UTR
untraslated region
Glossary
Footnotes
- The draft genome sequences were deposited in the DDBJ/GenBank/EMBL databases (accession numbers: BEHA01000001-BEHA01053253, 53,253 entries), and the annotated gene information is available at the Eschscholzia Genome DataBase (http://eschscholzia.kazusa.or.jp). The raw PE and MP read data are available from the NCBI SRA database under the BioProject accession number PRJDB6318. The nucleotide sequences of Eschscholzia californica for CYP80B5, CYP82B2-5, CYP82D170–172, CYP82N1, CYP82N2v3, CYP82N2v4, CYP82N5–7, CYP82P1v1, CYP82P1v2, CYP82P2–6, CYP82AT1–3, CYP82AU1, CYP82AV1 and CYP719C1 were submitted to DDBJ/GenBank/EMBL under the accession numbers LC316736–LC316762
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Beaudoin G.A., Facchini P.J. (2013) Isolation and characterization of a cDNA encoding (S)-cis-N-methylstylopine 14-hydroxylase from opium poppy, a key enzyme in sanguinarine biosynthesis. Biochem. Biophys. Res. Commun. 431: 597–603. [DOI] [PubMed] [Google Scholar]
- Choi K.B., Morishige T., Shitan N., Yazaki K., Sato F. (2002) Molecular cloning and characterization of coclaurine N-methyltransferase from cultured cells of Coptis japonica. J. Biol. Chem. 277: 830–835. [DOI] [PubMed] [Google Scholar]
- Cui L., Wall P.K., Leebens-Mack J.H., Lindsay B.G., Soltis D.E., Doyle J.H., et al. (2006) Widespread genome duplications throughout the history of flowering plants. Genome Res. 16: 738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De-Eknamkul W., Tanahashi T., Zenk M.H. (1992) Enzymic 10-hydroxylation and 10-O-methylation of dihydrosanguinarine in dihydrochelirubine formation by Eschscholzia. Phytochemistry 31: 2713–2717. [Google Scholar]
- Dittrich H., Kutchan T.M. (1991) Molecular cloning, expression, and induction of berberine bridge enzyme, an enzyme essential to the formation of benzophenanthridine alkaloids in the response of plants to pathogenic attack. Proc. Natl. Acad. Sci. USA 88: 9969–9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenworth A.M., Peralta-Yahya P. (2017) Accelerating the semisynthesis of alkaloid-based drugs through metabolic engineering. Nat. Chem. Biol. 13: 249–258. [DOI] [PubMed] [Google Scholar]
- Fabre N., Claparols C., Richelme S., Angelin M.L., Fouraste I., Moulis C. (2000) Direct characterization of isoquinoline alkaloids in a crude plant extract by ion-pair liquid chromatography–electrospray ionization tandem mass spectrometry: example of Eschscholtzia californica. J. Chromatogr. A 904: 35–46. [DOI] [PubMed] [Google Scholar]
- Facchini P.J. (2001) Alkaloid biosynthesis in plants: biochemistry, cell biology, molecular regulation, and metabolic engineering applications. Annu. Rev. Plant Biol. 52: 29–66. [DOI] [PubMed] [Google Scholar]
- Facchini P.J., Bird D.A., St-Pierre B. (2004) Can Arabidopsis make complex alkaloids? Trends Plant Sci. 9: 116–122. [DOI] [PubMed] [Google Scholar]
- Finn R.D., Attwood T.K., Babbitt P.C., Bateman A., Bork P., Bridge A.J., et al. (2017) InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res. 45: D190–D199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossati E., Ekins A., Narcross L., Zhu Y., Falgueyret J.-P., Beaudoin G.A.W., et al. (2014) Reconstitution of a 10-gene pathway for synthesis of the plant alkaloid dihydrosanguinarine in Saccharomyces cerevisiae. Nat. Commun. 5: 3283. [DOI] [PubMed] [Google Scholar]
- Gesell A., Rolf M., Ziegler J., Diaz C.M.L., Huang F.C., Kutchan T.M. (2009) CYP719B1 is salutaridine synthase, the C–C phenol-coupling enzyme of morphine biosynthesis in opium poppy. J. Biol. Chem. 284: 24432–24442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagel J.M., Beaudoin G.A.W., Fossati E., Ekins A., Martin V.J.J., Facchini P.J. (2012) Characterization of a flavoprotein oxidase from opium poppy catalyzing the final steps in sanguinarine and papaverine biosynthesis. J. Biol. Chem. 287: 42972–42983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagel J.M., Facchini P.J. (2013) Benzylisoquinoline alkaloid metabolism: a century of discovery and a brave new world. Plant Cell Physiol. 54: 647–672. [DOI] [PubMed] [Google Scholar]
- Hoff K.J., Lange S., Lomsadze A., Borodovsky M., Stanke M. (2016) BRAKER1: unsupervised RNA-seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32: 767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K., Okano S., Sato F. (2016) Efficient microbial production of stylopine using a Pichia pastoris expression system. Sci. Rep. 6: 22201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikezawa N., Iwasa K., Sato F. (2007) Molecular cloning and characterization of methylenedioxy bridge-forming enzymes involved in stylopine biosynthesis in Eschscholzia californica. FEBS J. 274: 1019–1035. [DOI] [PubMed] [Google Scholar]
- Ikezawa N., Iwasa K., Sato F. (2008) Molecular cloning and characterization of CYP80G2, a cytochrome P450 that catalyzes an intramolecular C–C phenol coupling of (S)-reticuline in magnoflorine biosynthesis, from cultured Coptis japonica cells. J. Biol. Chem. 283: 8810–8821. [DOI] [PubMed] [Google Scholar]
- Ikezawa N., Iwasa K., Sato F. (2009) CYP719A subfamily of cytochrome P450 oxygenases and isoquinoline alkaloid biosynthesis in Eschscholzia californica. Plant Cell Rep. 28: 123–133. [DOI] [PubMed] [Google Scholar]
- Itkin M., Heinig U., Tzfadia O., Bhide A.J., Shinde B., Cardenas P.D., et al. (2013) Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341: 175–179. [DOI] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H.-Y., Fraser M., Li W., McAnulla C., et al. (2014) InterProScan5: genome-scale protein function classification. Bioinformatics 30: 1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajitani R., Toshimoto K., Noguchi H., Toyoda A., Ogura Y., Okuno M., et al. (2014) Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24: 1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer L., De-Eknamkul W., Zenk M.H. (1994) Enzymic 12-hydroxylation and 12-O-methylation of dihydrochelirubine in dihydromacarpine formation by Thalictrum bulgaricum. Phytochemistry 36: 1409–1416. [Google Scholar]
- Kraus P.F., Kutchan T.M. (1995) Molecular cloning and heterologous expression of a cDNA encoding berbamunine synthase, a C–O phenol-coupling cytochrome P450 from the higher plant Berberis stolonifera. Proc. Natl. Acad. Sci. USA 92: 2071–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsmaier E.M., Skoog F. (1965) Organic growth factor requirements of tobacco tissue cultures. Physiol. Plant. 18: 100–127. [Google Scholar]
- Liscombe D.K., Facchini P.J. (2007) Molecular cloning and characterization of tetrahydroprotoberberine cis-N-methyltransferase, an enzyme involved in alkaloid biosynthesis in opium poppy. J. Biol. Chem. 282: 14741–14751. [DOI] [PubMed] [Google Scholar]
- Liscombe D.K., Ziegler J., Schmidt J., Ammer C., Facchini P.J. (2009) Targeted metabolite and transcript profiling for elucidating enzyme function: isolation of novel N-methyltransferases from three benzylisoquinoline alkaloid-producing species. Plant J. 60: 729–743. [DOI] [PubMed] [Google Scholar]
- Liu X., Liu Y., Huang P., Ma Y., Qing Z., Tang Q., et al. (2017) The genome of the medicinal plant Macleaya cordata provides new insights into benzylisoquinoline alkaloids metabolism. Mol. Plant. 10: 975–989. [DOI] [PubMed] [Google Scholar]
- Minami H., Kim J.-S., Ikezawa N., Takemura T., Katayama T., Kumagai H., et al. (2008) Microbial production of plant benzylisoquinoline alkaloids. Proc. Natl. Acad. Sci. USA 105: 7393–7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M., Sato F. (2011) Unusual P450 reactions in plant secondary metabolism. Arch. Biochem. Biophys. 507: 194–203. [DOI] [PubMed] [Google Scholar]
- Morishige T., Dubouzet E., Choi K.B., Yazaki K., Sato F. (2002) Molecular cloning of columbamine O-methyltransferase from cultured Coptis japonica cells. Eur. J. Biochem. 269: 5659–5667. [DOI] [PubMed] [Google Scholar]
- Morishige T., Tsujita T., Yamada Y., Sato F. (2000) Molecular characterization of the S-adenosyl-l-methionine:3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase involved in isoquinoline alkaloid biosynthesis in Coptis japonica. J. Biol. Chem. 275: 23398–23405. [DOI] [PubMed] [Google Scholar]
- Nakagawa A., Matsumura E., Koyanagi T., Katayama T., Kawano N., Yoshimatsu K., et al. (2016) Total biosynthesis of opiates by stepwise fermentation using engineered Escherichia coli. Nat. Commun. 7: 10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D., Werck-Reichhart (2011) A P450-centric view of plant evolution. Plant J. 66: 194–211. [DOI] [PubMed] [Google Scholar]
- Ozaki T., Nishiyama M., Kuzuyama T. (2013) Novel tryptophan metabolism by a potential gene cluster that is widely distributed among actinomycetes. J. Biol. Chem. 288: 9946–9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli H.H., Kutchan T.M. (1998) Molecular cloning and functional heterologous expression of two alleles encoding (S)-N-methylcoclaurine 3′-hydroxylase (CYP80B1), a new methyl jasmonate-inducible cytochrome P-450-dependent mono-oxygenase of benzylisoquinoline alkaloid biosynthesis. Plant J. 13: 793–801. [DOI] [PubMed] [Google Scholar]
- Purwanto R., Hori K., Yamada Y., Sato F. (2017) Unraveling additional O-Methylation steps in benzylisoquinoline alkaloid biosynthesis in California poppy (Eschscholzia californica). Plant Cell Physiol. 58: 1528–1540. [DOI] [PubMed] [Google Scholar]
- Rutledge P.J., Challis G.L. (2015) Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 13: 509–523. [DOI] [PubMed] [Google Scholar]
- Sato F. (2013) Characterization of plant functions using cultured plant cells, and biotechnological applications. Biosci. Biotechnol. Biochem. 77: 1–9. [DOI] [PubMed] [Google Scholar]
- Sato F. (2014) Plant secondary metabolism. eLS.
- Schmieder R., Edwards R. (2011) Quality control and preprocessing of metagenomic datasets. Bioinformatics 27: 863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaninová I., Slanina J., Táborská E. (2007). Quaternary benzo[c]phenanthridine alkaloids—novel cell permeant and red fluorescing DNA probes. Cytometry A 71: 700–708. [DOI] [PubMed] [Google Scholar]
- Stadler R., Zenk M.H. (1993) The purification and characterization of a unique cytochrome P-450 enzyme from Berberis stolonifera plant cell cultures. J. Biol. Chem. 268: 823–831. [PubMed] [Google Scholar]
- Stanke M., Diekhans M., Baerisch R., Haussler D. (2008) Using native and syntenically mapped cDNA alighnments to improve de novo gene finding. Bioinformatics 24: 637–644. [DOI] [PubMed] [Google Scholar]
- Takemura T., Chow Y.L., Todokoro T., Okamoto T., Sato F. (2010a) Over-expression of rate-limiting enzymes to improve alkaloid productivity. Methods Mol. Biol. 643: 95–109. [DOI] [PubMed] [Google Scholar]
- Takemura T., Ikezawa N., Iwasa K., Sato F. (2010b) Metabolic diversification of benzylisoquinoline alkaloid biosynthesis through the introduction of a branch pathway in Eschscholzia californica. Plant Cell Physiol. 51: 949–959. [DOI] [PubMed] [Google Scholar]
- Takemura T., Ikezawa N., Iwasa K., Sato F. (2013) Molecular cloning and characterization of a cytochrome P450 in sanguinarine biosynthesis from Eschscholzia californica cells. Phytochemistry 91: 100–108. [DOI] [PubMed] [Google Scholar]
- Takeshita N., Fujiwara H., Mimura H., Fitchen J.H., Yamada Y., Sato F. (1995) Molecular cloning and characterization of S-adenosyl-l-methionine:scoulerine-9-O-methyltransferase from cultured cells of Coptis japonica. Plant Cell Physiol. 36: 29–36. [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumer S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanahashi T., Zenk M.H. (1990) New hydroxylated benzo[c]phenanthridine alkaloids from Eschscholtzia californica cell suspension cultures. J. Nat. Prod. 53: 579–586. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009) TopHat: discovering spice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer T., Gazda V., He Z., Kaminski F., Kern M., Larson T.R.. et al. (2012) A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science 336: 1704–1708. [DOI] [PubMed] [Google Scholar]
- Wurtzel E.T., Kutchan T.M. (2016) Plant metabolism, the diverse chemistry set of the future. Science 353: 1232–1236. [DOI] [PubMed] [Google Scholar]
- Ziegler J., Facchini P.J. (2008) Alkaloid biosynthesis: metabolism and trafficking. Annu. Rev. Plant Biol. 59: 735–769. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.