Abstract
A valuable resource available in the search for new natural products is the diverse microbial life that spans the planet. A large subset of these microorganisms synthesize complex specialized metabolites exhibiting biomedically important activities. A limiting step to the characterization of these compounds is an elucidation of the genetic regulatory mechanisms that oversee their production. Although proteins that control transcription initiation of specialized metabolite gene clusters have been identified, those affecting transcription elongation have not been broadly investigated. In this study, we analysed the phylogenetic distribution of the large, widespread NusG family of transcription elongation proteins and found that it includes a cohesive outgroup of paralogues (herein coined LoaP), which are often positioned adjacent or within gene clusters for specialized metabolites. We established Bacillus amyloliquefaciens LoaP as a paradigm for this protein subgroup and showed that it regulated the transcriptional readthrough of termination sites located within two different antibiotic biosynthesis operons. Both of these antibiotics have been implicated in plant-protective activities, demonstrating that LoaP controls an important regulon of specialized metabolite genes for this microorganism. These data therefore reveal transcription elongation as a point of regulatory control for specialized metabolite pathways and introduce a subgroup of NusG proteins for this purpose.
After nearly a century of searching for bioactive natural products, bacteria still constitute a major target of modern drug discovery1,2. This is in part because most bacteria can synthesize at least a few complex specialized metabolites, among which a subset exhibit biomedically relevant activities. One general approach is to screen for bioactive molecules from culture supernatants of random environmental bacterial isolates. However, the characterization of the biochemical pathways of these molecules remains a bottleneck to their development. One of the key restrictions is a shortage of knowledge about the range of genetic mechanisms that can affect them. In particular, an incomplete understanding of the genetic regulatory mechanisms that affect specialized metabolite pathways stifles attempts at expressing them within heterologous hosts. Therefore, the discovery of new classes of genetic regulatory mechanisms is likely to impact the future development of natural products.
Characterization of the genetic mechanisms affecting specialized metabolite pathways has been generally restricted to transcription initiation factors3. However, transcription initiation is only the first stage of gene expression. The stages that follow include transcription elongation, transcription termination, translation and mRNA degradation. Notably, each can be subjected to genetic regulatory control, often involving different types of regulatory RNAs4. Yet, while many post-initiation regulatory mechanisms have been identified, few have been characterized for secondary metabolite pathways. Indeed, it is possible that the unusually long transcription units for many specialized metabolite gene clusters present challenges that can be resolved by post-initiation regulatory mechanisms. Specifically, we hypothesize that some specialized metabolite synthesis gene clusters may rely on regulatory mechanisms that improve the efficiency of transcription elongation and ensure transcript completion. One manner in which the efficiency of transcription elongation may be affected is through processive antitermination (PA) mechanisms5–7.
In PA mechanisms, antitermination factors associate with a bacterial RNA polymerase (RNAP) elongation complex, leading to the bypass of terminator sites. Termination signals normally induce rapid dissociation of the transcription elongation complex (TEC) and are most often located at the ends of operons. However, when placed within operons, they can serve as key points of regulatory control. In general, there are two classes of terminators: intrinsic and Rho-dependent terminators8. Intrinsic terminators consist of a GC-rich RNA hairpin followed by a poly-uridine tract. Alone9, or enhanced by a factor such as NusA (refs 10,11), these RNA elements promote pausing of the TEC, followed by release of the nascent transcript and dissociation of polymerase. In contrast, Rho-dependent termination calls upon the adenosine triphosphate (ATP)-dependent translocase Rho to traverse the nascent RNA and promote TEC dissociation. Both classes of termination sites can be specifically regulated within the context of signal-responsive riboswitches12,13. However, whereas riboswitches exert control over a single intrinsic terminator site, or a single entry point for Rho, PA systems differ in that they create modified TECs that have been rendered generally resistant to downstream terminator sites7. PA systems, therefore, are capable of causing readthrough of multiple termination sites over long distances. Several classes of bacterial and phage PA mechanisms have been discovered5–7, although none have been definitively shown to affect secondary metabolite pathways.
In this study we present the discovery of a NusG specialized paralogue in Bacillus amyloliquefaciens, which we name LoaP, for ‘long operon associated protein’. LoaP promotes readthrough of intrinsic terminators located within the polyketide synthase (PKS) gene cluster encoding for the antibiotic difficidin (dfn). LoaP can also promote readthrough of heterologous intrinsic terminators through a mechanism that requires a portion of the dfn 5′ leader region. LoaP also promotes PA of a second PKS gene cluster, encoding for a different antibiotic, macrolactin (mln). LoaP therefore affects a selective regulon of antibiotic gene clusters. A broad-scale phylogenetic analysis revealed that the LoaP protein is widespread in Firmicutes, Actinobacteria and Spirochaetes, and is often associated with specialized metabolite gene clusters or polysaccharide biosynthesis operons. These data demonstrate that transcription elongation is subject to regulatory control by PA for some specialized metabolite gene clusters, and implicate a cohesive subgroup of NusG paralogues for this purpose.
Results
mRNA abundance of the dfn operon is dependent on RBAM_022090
B. amyloliquefaciens FZB42 contains multiple specialized metabolite gene clusters, which encode for the production of polyketides, non-ribosomally produced lipopeptides and bacteriocins14. These gene clusters range from 5 to 10 kilobases (kb) for bacteriocins, 12 to 40 kb for non-ribosomal lipopeptides and up to 75 kb for polyketide-producing gene clusters14,15. The 70 kb dfn operon encodes for the production of the polyketide antibiotic difficidin15. Immediately upstream is the gene RBAM_022090, hereafter referred to as loaP, encoding for a protein distantly related to the elongation factor NusG (Fig. 1a). This paralogue is distinct from core nusG, which is also present in B. amyloliquefaciens but located elsewhere in the genome. Due to its proximity, we hypothesized that loaP may regulate the dfn operon.
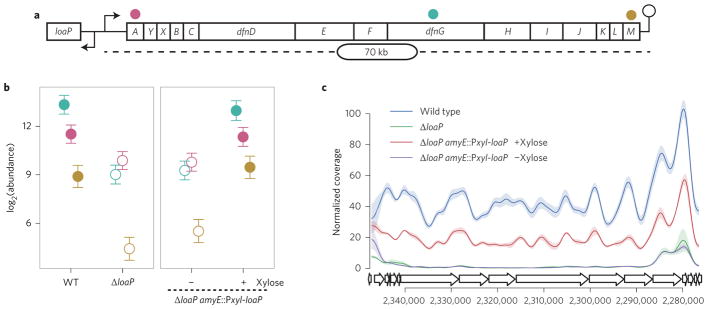
Figure 1. LoaP is required for expression of the B. amyloliquefaciens difficidin gene cluster.
a, Schematic depiction of the dfn gene cluster, including the general location of dfnA, dfnG and dfnM amplicons used for quantification by qRT–PCR. b, Normalized transcript abundance at the beginning, middle and end of the dfn operon (dfnA, dfnG, dfnM) as measured by qRT–PCR. Filled symbols represent samples with loaP expression and open symbols represent samples with no or minimal loaP expression. Colours correspond to amplicon locations in a, with dfnA in pink, dfnG in teal and dfnM in gold. Error bars represent Bayesian 95% highest posterior density estimates of mean expression. Data resulted from biological triplicate cultures with qPCR technical duplicates. WT, wild type. c, RNA-seq coverage across the dfn gene cluster normalized with DESeq2 normalization factors. Traces represent coverage data smoothed with Gaussian smoothing with a bandwidth of 500 nt. Shading represents standard deviation from libraries from three independent cultures for each condition.
To test this hypothesis, loaP was replaced with an antibiotic resistance gene and transcript levels were monitored by quantitative reverse transcription PCR (qRT–PCR) at the beginning (dfnA), middle (dfnG) and end (dfnM) of the operon. Deletion of loaP resulted in a threefold decrease at dfnA and a 20-fold reduction at dfnG and dfnM (Fig. 1b). A xylose-inducible copy of loaP (Pxyl-loaP) was then ectopically integrated into the genome at a non-essential locus (amyE) for both wild-type and ΔloaP strains. Corresponding measurements of transcript abundance revealed that levels of dfnA, dfnG and dfnM are unchanged in the uninduced complementation strain, but are restored to wild-type levels when loaP is induced (Fig. 1b).
Induction of loaP elevates mRNA abundance across the full dfn operon
Whole-transcriptome RNA-seq analyses were conducted on wild-type, ΔloaP and loaP complementation strains. Analysis of differential expression between wild-type and ΔloaP showed a 14-fold decrease in dfnA, which increased to 30-fold towards the middle and end of the operon (Fig. 1c and Supplementary Table 1). The majority of the observable decrease occurs in the first 8–10 kb of the dfn transcript. Specifically, there is a rapid drop to ~10% of wild-type for the first few kilobases, with a more gradual reduction over the next few kilobases to 3–4% of wild-type. Interestingly, the amount of dfn in the deletion strain increases slightly beginning at dfnJ and continuing to dfnM, implying the possible presence of an internal promoter near the end of the gene cluster.
Induction of loaP significantly increased transcription across the entire length of dfn, restoring transcript levels to 35–50% of wild-type levels. The gradual drop within the first 10 kb of the transcript was also eliminated upon complementation of loaP (Fig. 1c). These data demonstrate that loaP has a dramatic effect on dfn mRNA abundance and suggest it may use a post-initiation regulatory mechanism.
LoaP promotes processive antitermination within the dfn mRNA leader region
Transcription start sites (TSSs) have previously been mapped across the B. amyloliquefaciens FZB42 genome16. Our inspection of these data identified only a single TSS hundreds of nucleotides upstream of dfnA (Fig. 2a). Any 5′ leader regions longer than 100 nucleotides in length are unusual; most mRNA leader regions in Bacillus species are only long enough to permit translation initiation (~35 nucleotide (nt))16,17. Correspondingly, unusually long leader regions are typically involved in post-initiation regulation of their downstream genes. At 417 nt, the dfnA leader region is exceptionally long and we therefore hypothesized that it is involved in regulation.
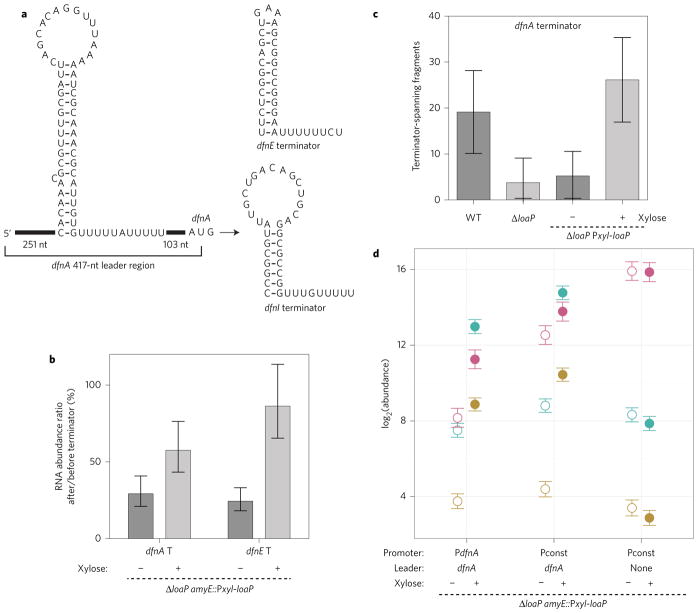
Figure 2. LoaP promotes readthrough of intrinsic terminator sites.
The dfnA leader region contains determinants for LoaP-mediated processive antitermination. a, Schematics of intrinsic terminator candidates identified within the dfnA leader region, or within coding sequences of dfnE or dfnI. b, Estimated transcription termination efficiencies for putative intrinsic terminators (T) in the dfnA leader region and dfnE coding sequence. Efficiencies are calculated as the ratio of transcript abundance immediately before and after the terminator sequences measured by qRT–PCR. Error bars represent 95% highest posterior density of ratios calculated directly from posterior estimates of normalized transcript abundance. Experiments were performed with four independent cultures for each condition and qPCR technical duplicates. c, Normalized count of RNA-seq read pairs spanning the termination site of the dfnA leader intrinsic terminator. Error bars represent standard deviation (n = 3). d, Normalized transcript abundance at the beginning, middle and end of the dfn operon in strains where the dfnA promoter has been replaced by the constitutive promoter Pconst, with or without the dfnA leader region. Colours correspond to amplicon locations in Fig. 1a, with dfnA in pink, dfnG in teal and dfnM in gold. All strains contain a marker-replacement of loaP and an ectopic xylose-inducible loaP. Error bars represent Bayesian 95% highest posterior density estimates of mean expression. Data represents six biological replicates with technical duplicates. Open symbols represent uninduced cultures and filled symbols represent cultures with loaP induction.
RNA-seq coverage data revealed that abundance across the dfnA leader decreased in the 5′ → 3′ direction (Supplementary Fig. 1). A particularly acute drop occurred at the midway point of the leader, where its sequence exhibited an ability to form an intrinsic terminator candidate (Fig. 2a). Therefore, although interpretation of coverage profiles at high resolution can be complicated by regions containing inverted repeats18, we hypothesized that the decrease resulted from premature termination. Indeed, this terminator corresponded to a permanent drop in coverage for loaP-deficient samples. In contrast, the moderate drop in coverage at this site for the wild-type sample was temporary and recovered shortly thereafter. To further test the transcript coverage patterns in this region, we performed qRT–PCR using amplicons located before and after the putative intrinsic terminator. Induction of loaP led to an increase after the terminator, with a minimal change in upstream abundance (Fig. 2b). Moreover, examination of the Illumina RNA-seq data revealed that the number of paired cDNA reads spanning the putative terminator site was significantly increased with loaP induction (Fig. 2c). Our search for intrinsic terminator sites also identified several downstream candidates, which share no substantial sequence similarity to the terminator within the dfn leader. Analysis of a putative intrinsic terminator within dfnE (Fig. 2a,b and Supplementary Fig. 2) revealed that readthrough is also dependent on expression of LoaP, suggesting that LoaP exhibits a general ability to bypass intrinsic terminators.
To examine whether LoaP antitermination determinants involve the promoter region, we replaced the native dfnA promoter with a constitutively active promoter (Pconst) for strains that either contained or lacked the dfnA leader. We quantified transcript levels at dfnA, dfnG and dfnM for these strains, in the presence and absence of loaP (Fig. 2d). With the leader region under control of Pconst, transcript level changes appeared to closely resemble wild-type, albeit with slightly higher basal transcript levels. In contrast, removal of the leader region resulted in complete loss of loaP-induced transcript levels.
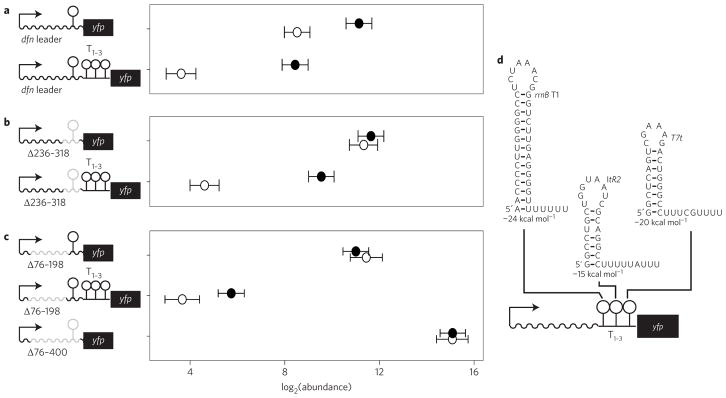
The dfnA leader region (including its intrinsic termination site) was then placed upstream of a yfp gene to create a reporter construct for antitermination. Induction of loaP resulted in an approximately eightfold increase in yfp transcript abundance (Fig. 3a). In a separate construct, we added an array of three validated but completely unrelated intrinsic termination sites19, located downstream of the dfnA leader region but upstream of yfp (Fig. 3d). Again, induction of loaP significantly increased the yfp reporter gene, despite the heterologous terminator sites. As a negative control, a separate reporter lacked the dfnA intrinsic termination site (Fig. 3b). Transcript levels for this construct remained high in the presence and absence of loaP. Addition of the three heterologous termination sites to this terminator-less construct restored dependency on loaP for transcription of yfp. We also tested a few large truncations within the dfnA leader and found they eliminated terminator readthrough, suggesting that LoaP determinants may be within the 5′ portion of the dfnA leader (Fig. 3c). Finally, we integrated the dfnA-yfp reporter into a heterologous host (Bacillus subtilis) that also contained an inducible copy of B. amyloliquefaciens loaP. Quantification of yfp expression by fluorescence microscopy showed a comparable increase upon induction of LoaP, suggesting that LoaP antitermination does not require any additional factors specific to B. amyloliquefaciens (Supplementary Fig. 2).
Figure 3. LoaP mediates transcription antitermination in reporter constructs.
Normalized transcript abundance of yfp mRNA measured by qRT–PCR. All strains contain a marker-replacement of loaP and an ectopic xylose-inducible loaP integrated into amyE. In addition, all strains contain a single copy of the PdfnA promoter transcriptionally fused to yfp with different modified dfnA leader regions. a, In these constructs, a wild-type dfnA leader region was included upstream of yfp. A variant of this construct contained a dfnA leader region followed by an array of three tandem intrinsic terminators. b, In these constructs, the region of the dfnA leader containing an intrinsic terminator was deleted, but they were otherwise identical to constructs in a. c, Deletions were introduced into the dfnA leader region of the yfp reporter fusions for constructs with and without the terminator array. In all rows, open symbols represent conditions without xylose induction and filled symbols represent conditions with 1% xylose induction of loaP. All conditions were measured with duplicate independent cultures and qPCR technical duplicates. Error bars in a–c represent Bayesian 95% highest posterior density estimates of mean expression. Data resulted from biological duplicate cultures with qPCR technical duplicates d, Sequences of the tandem intrinsic terminators that were incorporated into some yfp reporter constructs, as indicated by schematics.
LoaP regulates a second polyketide synthase gene cluster
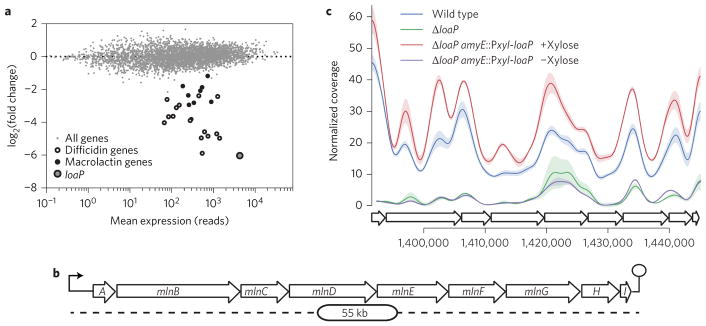
Deletion of loaP resulted in significant (adjusted P-value <0.01) differential expression of only 30 genes (Fig. 4a and Supplementary Tables 1 and 2). With very few exceptions, the differentially expressed genes (P < 0.01) belonged to either the difficidin operon or a second polyketide biosynthesis operon (mln) that encodes for the antibiotic macrolactin (Fig. 4b,c)20. Across its length, the mln operon was reduced between 5-fold and 13-fold for the loaP-deficient strain (Supplementary Fig. 3).
Figure 4. LoaP expression affects transcription of both difficidin and macrolactin operons.
a, MA-plot showing mean expression and log-fold-changes for all genes between wild-type and ΔloaP strains from RNA-seq analysis. Large open points represent dfn synthesis genes and large filled points represent mln synthesis genes. Data represents the average expression of three (wild-type) and two (ΔloaP) replicates. b, Schematic of the mln macrolactin synthesis operon. c, RNA-seq coverage across the mln gene cluster normalized with DESeq2 normalization factors. Traces represent coverage data smoothed with Gaussian smoothing with a bandwidth of 500 nt. Shading represents standard deviation from libraries from three independent cultures for each condition.
Statistical analysis of the loaP complementation strain RNA-seq showed many more differentially expressed genes (373), although many were annotated either specifically as xylose metabolism genes or with other carbohydrate metabolism functions (Supplementary Table 2). Therefore, we conclude that most differentially expressed genes were altered from the use of xylose as an inducer molecule for controllable expression of loaP. However, the remaining differentially expressed genes agreed well with analysis of the loaP deletion strain. Of the 33 genes differentially expressed in the loaP deletion strain, 28 were also differentially expressed for the complementation strain upon induction of loaP, including the mln pathway. From these transcriptomic data, we conclude that loaP specifically affects transcript abundance of a regulon of B. amyloliquefaciens antibiotic biosynthesis genes.
Production of difficidin and macrolactin depends on LoaP
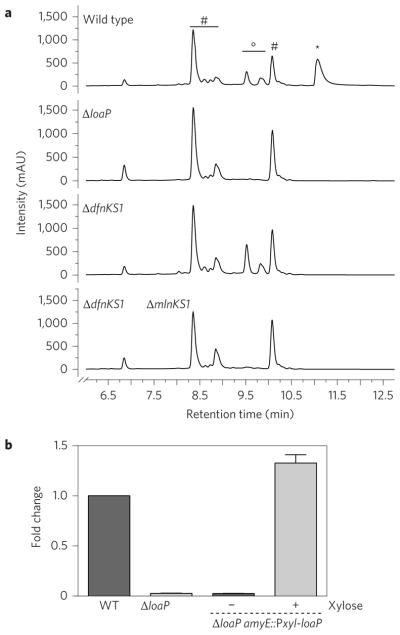
The production of polyketides (difficidin, macrolactin and bacillaene) was measured by high-performance liquid chromatography (HPLC) analysis for extracts of B. amyloliquefaciens culture supernatants. The ΔloaP strain showed near-complete elimination of both difficidin and macrolactin, with no effect on bacillaene production (Fig. 5a). Induction of loaP restored metabolite production, confirming that the loaP-dependent changes in gene expression indeed correlated with antibiotic production (Fig. 5b and Supplementary Table 3).
Figure 5. LoaP-dependent production of difficidin and macrolactin.
a, A comparison of ΔloaP to wild-type B. amyloliquefaciens FZB42 production of difficidin, macrolactin and bacillaene by HPLC. Deletion of loaP specifically disrupts production of difficidin and macrolactin, while bacillaene production is maintained. HPLC peaks corresponding to difficidin (*), macrolactin (°) and bacillaene (#) are labelled for reference on the chromatographs. The Δpks3KS1 strain is deficient in difficidin production and the Δpks2KS1, Δpks3KS1 double mutant strain is deficient in both difficidin and macrolactin53. The mutant strains serve as reference controls for specificity of HPLC peaks. Metabolites were detected at λ = 280 nm. Representative traces for each genotype are shown. mAU, milli-absorbance units. b, Quantitative comparison of difficidin production by B. amyloliquefaciens FZB42 strains, wild-type (WT), ΔloaP and ΔloaP, amyE::Pxyl-loaP (+ and −1% xylose). Relative production of difficidin was compared between the wild-type and mutant strains. Peak values were compared to bacillaene as a reference. Data and error bars represent the average and standard deviation of two biological replicates.
LoaP is a member of the NusG-homologue subfamily and is broadly conserved
To investigate whether LoaP is present and performing similar functions in other organisms, we initially searched for close homologues using phmmer from the HMMER3 software suite, manually checking genomes of the resulting hits for the presence of a core nusG gene in addition to a loaP homologue21. The most highly homologous protein sequence hits were generally distributed among other species of Bacillus, Brevibacillus and Paenibacillius. Other closely related homologues were also found among Clostridia, including Clostridium and Thermoanaerobacter species.
To gain insight into the relationship between LoaP proteins and the larger NusG family, we combined a few LoaP sequences with selected examples of other NusG family member proteins—Spt5, NusG, RfaH, ActX, UpxY and TaA—and constructed a structure-assisted multiple sequence alignment and maximum-likelihood phylogenetic tree22 (Supplementary Fig. 4a). The LoaP sequences formed a monophyletic clade separate from NusG and from the other specialized paralogues. We began to differentiate the top scoring hits between LoaP and other NusG subfamilies by sequentially adding the sequences to the reference alignment and reconstructing the phylogenetic tree with the additional sequence to determine where the protein fit in the reference tree, to preliminarily classify the protein sequences.
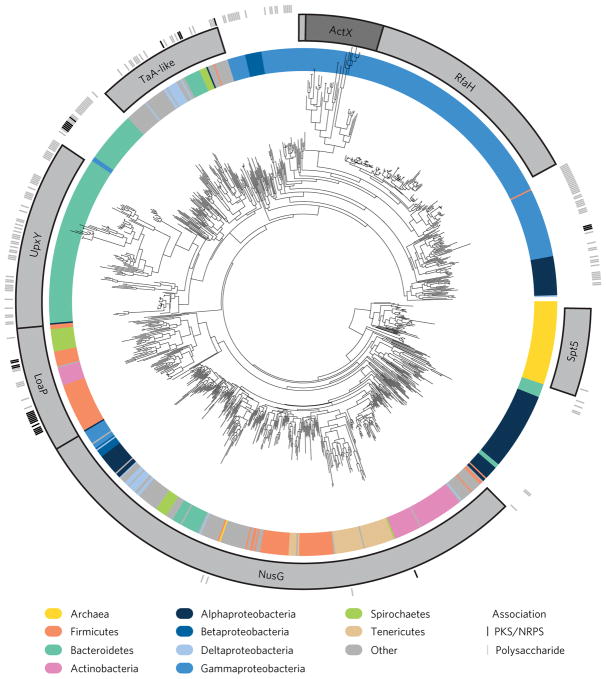
For a broad picture of the NusG family we extracted all NusG protein sequences (defined by the presence of the characteristic N-terminal domain) from the Uniprot complete database (14,435 sequences in total). To reduce the number of nearly identical protein sequences while maintaining the majority of sequence diversity, we used sequence-similarity clustering. This limited putative core NusG sequences to a set of at most 60% identical sequences and NusG paralogues to at most 95% sequence identical (1,205 total). We constructed a large-scale multiple alignment using the accurate multidomain progressive alignment algorithm of the MAFFT software and constructed a maximum-likelihood phylogenetic tree of the trimmed alignment using RAxML. The underlying topology of the large tree (Fig. 6) matched very closely the topology of the small reference tree (Supplementary Figs 4 and 5). Protein subfamilies were labelled by assigning a subtype to the monophyletic group formed by the most recent ancestor of all of the curated protein examples found in the small tree analysis. In general, additional protein sequences corresponding to putative LoaP homologues were identified in Bacilli and Clostridia classes, as well as the Coriobacteriia class of Actinobacteria and in a variety of Spirochaetes. Not all proteins were assigned a type. Some subtrees were found adjacent to known paralogue groups and exhibit distinct characteristics and may represent additional subtypes.
Figure 6. Large-scale phylogenetic analysis of NusG family proteins reveals several subclasses of specialized paralogues.
A large-scale phylogenetic tree composed on 1,205 representatives of NusG homologues. The bacterial phylum (or class for Proteobacteria) of the organism containing each protein sequence is represented by colour on the inner ring. Subtrees formed from the most recent ancestor of curated subgroups are labelled with grey boxes in the middle ring. Tick marks representing the association of particular sequences with PKS or NRPS gene clusters (black) or polysaccharide gene clusters (grey) are found in the outer ring. The ActX label is darker to clarify the distinction between the ActX subtree and adjacent RfaH sequences.
LoaP is associated with polysaccharide and specialized metabolite biosynthesis gene clusters
We manually surveyed a selection of close loaP homologues to look for large gene clusters in the genomic region. Almost all loaP homologues were located adjacent to large gene clusters containing either polyketide synthase genes or sugar-related enzymes consistent with polysaccharide synthesis operons. We then took a more rigorous approach and collected all 1,205 NusG family members from the large phylogenetic tree and, using the antiSMASH pipeline, searched their surrounding genomic sequence for putative specialized metabolite gene clusters beginning or ending within 5 kb of the NusG family gene sequence (Fig. 6 and Supplementary Fig. 6). Most loaP sequences were found immediately adjacent to the large gene clusters encoding synthesis of specialized metabolites or polysaccharides. Interestingly, the different NusG paralogue groups showed distinct patterns of gene cluster association. UpxY, as previously shown23 is very commonly associated with polysaccharide gene clusters in Bacteroidetes. Yet there also appeared to be a separate UpxY/TaA-like cluster found in Bacteroidetes associated with gene clusters for either polysaccharides or specialized metabolites. RfaH and ActX appear to be rarely associated with antiSMASH-identifiable gene clusters, although there are some RfaH-like clusters found in both Alpha- and Gammaproteobacteria associated with long gene clusters, as well as a subgroup found largely in Alphaproteobacteria independent of putative gene clusters.
Discussion
Bacteria appear to have evolved distinct PA mechanisms for different circumstances. The best-characterized PA examples are mediated by phage lambda proteins N and Q, which primarily prevent Rho termination for early and late transcripts, respectively5–7. For the related phage HK022, a ~65-nucleotide, cis-acting RNA sequence (‘PUT’) can protect against Rho termination in lieu of N or Q (ref. 24). Another cis-acting RNA capable of promoting PA is the ~125-nucleotide ‘EAR’ element, which promotes readthrough of termination sites in operons encoding exopolysaccharide biosynthesis19. In certain Gram-negative bacteria, the NusG paralogue RfaH prevents Rho termination sites within horizontally acquired operons22,25,26. RfaH is recruited to the RNAP elongation complex by a characteristic nontemplate DNA sequence and does not require additional factors, making it a dedicated antitermination factor. Another NusG paralogue, UpxY, is widespread in Bacteroidetes and is also presumed to trigger PA (ref. 23). Interestingly, these bacteria encode several UpxY proteins, each associated with a distinct polysaccharide pathway, although the molecular mechanism of UpxY regulation has yet to be revealed. Similarly, mutational disruption of a Myxococcus xanthus NusG paralogue, TaA, resulted in decreased production of a polyketide, myxovirescin27. Although antitermination activity has yet to be observed for TaA, this discovery suggested proof in principle that transcription elongation factors may affect specialized metabolite pathways. In a separate study, general overexpression of a NusG homologue led to discovery of a new Clostridium cellulolyticum natural product28. Inspired by these findings, we hypothesized that more antitermination systems await discovery and that they will prove to be essential for the transcription of important pathways, including those encoding specialized metabolites.
An abbreviated search for NusG paralogues proximal to specialized metabolite gene clusters led us to the discovery of B. amyloliquefaciens loaP. Deletion of loaP dramatically affected transcript abundance of its neighbouring gene cluster (dfn), which could be re-established with loaP complementation. With very few exceptions, the genes differentially expressed in the presence or absence of LoaP belonged to PKS biosynthesis operons for difficidin or macrolactin. In contrast, a third PKS cluster, encoding bacillaene production, was unaffected.
Replacement of the dfnA promoter with a constitutive promoter resulted in no loss of dependency for LoaP, demonstrating that LoaP determinants are downstream of the promoter and transcription start site, consistent with a transcription elongation mechanism. Interestingly, both dfn and mln have a feature in common—an unusually long 5′ leader region. Within each dfn and mln leader region is a moderately strong intrinsic termination site that is bypassed upon LoaP induction. Indeed, LoaP promotes general readthrough of intrinsic terminators, including other intrinsic terminators in the dfn and mln operons, as well as unrelated terminators introduced into reporter fusions. B. amyloliquefaciens LoaP was also able to promote antitermination within the dfnA leader region in the heterologous host B. subtilis (Supplementary Fig. 2), suggesting that additional B. amyloliquefaciens factors are not required. However, addition of purified LoaP to transcription reactions with purified B. subtilis RNA polymerase resulted in no change in transcription termination within the dfnA leader region (Supplementary Fig. 6), suggesting that LoaP was purified in an inactive form or that additional elongation factors are still required.
NusG family proteins are composed of two domains separated by an unstructured linker26,29. The N-terminal portion is responsible for binding to RNAP and competes with the sigma subunit for access26,29. RfaH (ref. 25), core NusG (ref. 30) and Spt5 (ref. 31) have all been shown to interact with the nontemplate DNA strand when associated with RNAP. This DNA recognition is particularly significant for RfaH, which is recruited to RNAP by a characteristic nontemplate sequence called ops. Therefore, the simplest prediction is that nontemplate DNA determinants are required for LoaP antitermination, located somewhere within the dfn and mln 5′ leader regions. However, a preliminary search for sequence features common to dfn and mln revealed something potentially different: each leader contains a similar inverted repeat sequence (Supplementary Fig. 2) that corresponds well to a UNCG-type RNA tetraloop32.
UNCG tetraloops are widespread RNA secondary structure elements33. We changed the penultimate nucleotide of the putative dfn tetraloop from G to C, which is predicted to destabilize the UNCG tetraloop. This mutation resulted in a dramatic decrease in antitermination activity (Supplementary Fig. 2). Interestingly, lambda N protein associates with the related but structurally distinct GNRA tetraloop hairpin (boxB)34, or, for some phages, an alternative sequence that mimics the GNRA fold35. The boxB hairpin, in combination with an adjacent unstructured sequence (boxA), acts as loading region for N-protein and bacterial elongation factors. The proteins bound to boxB/boxA RNA elements remain in a complex, associated with one another and with the elongation complex, while the emerging transcript loops from the exit channel5. By extension of this model it is possible that the putative dfn and mln UNCG tetraloop may serve as a loading site for elongation factors other than LoaP. Alternatively, LoaP itself may interact specifically with the UNCG hairpin as a crucial part of its recruitment to dfn and mln.
Although its N-terminal domain associates with RNAP, the core NusG C-terminal Kyprides–Onzonis–Woese domain (KOW) interacts with other factors. One of these partners is ribosomal protein S10 (NusE). This interaction may improve coupling of transcription and translation machinery26,29. A second KOW-binding factor is Rho, whose termination activity is either enhanced or reduced upon interaction with NusG (refs 26,29). To inhibit Rho termination, RfaH outcompetes and thereby excludes the NusG:Rho complex from RNAP. However, Rho is dispensable in B. subtilis26,29. Moreover, LoaP is the first NusG/Spt5 family member shown to promote readthrough of intrinsic terminators. Therefore, it is unclear if LoaP will share a similar relationship with Rho or if the LoaP KOW domain plays a different role.
Protein sequences clustering with LoaP formed a monophyletic group distinct from core NusG. Moreover, the LoaP subfamily of NusG paralogues is related but distinct from RfaH in Proteobacteria and UpxY in Bacteroidetes. Highly conserved LoaP sequences were detected for Paenibacillus and Brevibacillus species, as well as other Firmicutes, including but not limited to Clostridium, Thermoanaerobacter and Pelosinus. Although RfaH and UpxY are each restricted to a single phylum, LoaP proteins may also be represented in at least one order of both Spirochaetes and Actinobacteria. The relatively limited distribution of loaP genes among members of each phylum may indicate horizontal gene transfer has played a role in its apportionment among bacterial genomes. For example, many non-pathogenic Clostridia encode an abundance of cryptic PKS gene clusters that exhibit evidence of horizontal gene transfer36, a subset of which are associated with a loaP homologue. In an earlier study, a novel C. cellulolyticum poly-thioamide (closthioamide) was produced upon overexpression of a NusG homologue28, which our phylogenetic analysis revealed to be within the LoaP subclass, providing further evidence for a regulatory relationship between loaP and PKS genes.
There are additional subfamilies of NusG paralogues yet to be significantly explored. For example, M. xanthus TaA is a member of another subgroup that is often adjacent to specialized metabolite gene clusters. Furthermore, an uncharacterized RfaH-associated subgroup is widely associated with polysaccharide pathways. There also appear to be two distinct groups of Alphaproteobacteria NusG paralogues that form a distinct clade and are frequently found to be associated with gene clusters for polysaccharides and specialized metabolites. Therefore, there is a broad role in microbial biology for the regulation of transcription elongation via NusG-like proteins, although their mechanistic diversity remains to be explored. In general, elucidation of these regulatory mechanisms will assist analyses on the regulation of specialized metabolites, while also broadening the genetic tools that can be used for improving their production from within heterologous hosts.
Methods
Construction of B. amyloliquefaciens FZB42 plasmids and strains
To construct a marker-replacement of loaP, a backbone plasmid derived from pUC19 was digested with BamHI/EcoRI into which a PCR product for the B. amyloliquefaciens loaP region was subcloned via Gibson assembly37. A PCR product containing an erythromycin resistance cassette was then inserted into the above plasmid using restriction-free cloning to construct plasmid pJG030. This plasmid was transformed into B. amyloliquefaciens subsp. plantarum FZB42 (obtained from the Bacillus Genetic Stock Center) using a B. subtilis transformation protocol, which resulted in the construction of strain JG091 (ref. 38). Plasmid sequences were verified by Sanger sequencing and the genome sequence of JG091 was verified by analysis of Illumina RNA-seq data.
To construct a plasmid for inducible expression of loaP, the B. subtilis integration vector pDG1662 (obtained from the Bacillus Genetic Stock Center) was modified for integration into B. amyloliquefaciens. Specifically, the B. subtilis amyE homology arms were replaced with the corresponding amyE homology arms from B. amyloliquefaciens by Gibson assembly of amyE homology PCR products with pDG1662 backbone PCRs, resulting in plasmid pJG031. Upon publication of this article, pJG031 will be submitted to the Bacillus Genetic Stock Center for distribution upon request. A B. amyloliquefaciens region encompassing loaP was then PCR-amplified and subcloned into a NheI/BamHI digestion of plasmid pSWEET-III (ref. 39) via Gibson assembly. The resulting plasmid was digested with BamHI/HindIII, thereby releasing a restriction fragment containing the xylose-inducible promoter region followed by loaP, which was subcloned into pJG031, resulting in construction of pJG032 (ref. 39). This plasmid was integrated into wild-type B. amyloliquefaciens and JG091 for construction of overexpression and complementation strains.
Promoter replacement strains were constructed using variants of plasmid pJG030, which was designed for marker replacement of loaP (loaP::erm). Specifically, in plasmid pJG105, the promoter region for dfnA was replaced by a semisynthetic constitutive promoter, Pconst. Similarly, in plasmid pJG102, the dfnA leader region was included downstream of the Pconst promoter. These plasmids were integrated into strain JG098 (containing amy::Pxyl-loaP). All plasmid sequences are available as described in Supplementary Table 4.
For construction of yfp reporter plasmids, we modified a base B. subtilis plasmid, pJG019, which contains a semisynthetic constitutive promoter expressing yfp. To construct yfp reporter strains the dfnA promoter and leader region was subcloned in front of the yfp sequence to give an intermediate plasmid. The dfnA-yfp reporter construct was then varied by mutagenesis. For example, some of the modifications included removal or addition of terminator sequences, which was accomplished by Q5 Site-Directed Mutagenesis (NEB). All yfp reporter cassettes were subcloned into plasmid pJG031 to create combination yfp reporter/inducible loaP plasmids. These plasmids were transformed into strain JG091 to generate reporter strains with inducible loaP.
The plasmids used to construct each B. amyloliquefaciens strain are detailed in Supplementary Table 4. All plasmids were verified by Sanger sequencing of the inserted region.
Construction and analysis of yfp reporter sequences in B. subtilis
The B. amyloliquefaciens dfnA leader-yfp reporter sequence was PCR-amplified and subcloned behind a strong, constitutive promoter (Pconst) into pDG1662 (Bacillus Genetic Stock Center), resulting in pJG019, which could be used for integration of the reporter construct into B. subtilis amyE. A xylose-inducible copy of B. amyloliquefaciens loaP was subcloned into pDG1664 (Bacillus Genetic Stock Center) for integration into B. subtilis thrC. Separately, the Pconst-dfnA leader-yfp reporter sequence was further modified using Q5 Site-Directed Mutagenesis (NEB) to alter the putative UUCG tetraloop within the dfnA leader sequence to UUCA, as described in the text. The two reporter strains, together with wild-type B. subtilis 168 lacking a yfp reporter sequence, were cultured in rich medium overnight. Three independent cultures for each strain were inoculated 1:100 into fresh medium and cultured with shaking at 37 °C until reaching an optical density at 600 nm (OD600) of 0.8. Xylose was added to a final concentration of 1%, or, for a negative control, an equal volume of water was added and the cultures were incubated for an additional 3 h. Aliquots of these cultures were diluted 1:1 in phosphate-buffered saline (PBS) and placed on 1.5% low-melting agarose PBS pads, which were allowed to equilibrate for 15 min. The agarose pads were then inverted onto a glass bottom Petri dish and imaged (phase contrast and YFP fluorescence) using a Zeiss Axio Observer Z1 inverted fluorescence microscope with a Rolera EM-C2 electron-multiplying charge-coupled device camera. Three representative fields of view were imaged for each replicate culture. Fluorescence intensity was quantified using Oufti analysis software, and background autofluorescence was measured from a negative control strain lacking a yfp reporter sequence and subtracted from quantified values.
Extraction of total RNA for qRT–PCR and Illumina sequencing
Total RNA was extracted from B. amyloliquefaciens cultures by bead-beating using 300 μm acid-washed glass beads, followed by extraction with TRI Reagent RT (MRC) according to the manufacturer’s protocol. Total RNA was treated with RQ1 RNase-Free DNase I (Promega) or PerfeCTa DNase I (Quanta Bio), re-purified using Zymo Research Direct-Zol columns, and the overall integrity of the extracted RNA was assessed by agarose gel electrophoresis and concentration by Nanodrop (Thermo Scientific).
Quantification of transcript abundance by qRT–PCR
DNA-depleted total RNA was converted to cDNA with Quanta Bio qScript cDNA Supermix. The cDNA was diluted in tris-EDTA buffer and added to qPCR reactions made with Quanta Bio PerfeCTa SYBR Green FastMix. All qPCR reactions were prepared for 18 μl volumes in 96- or 384-well plates and analysed using a Roche Lightcycler 480 with the recommended three-step fast cycle. All oligonucleotides used for each amplicon are listed in Supplementary Table 5. All experiments used negative RNA-only controls for DNA contamination using reference gene amplicons. Quantitation cycle (Cq) values and amplification efficiencies were determined using LinRegPCR (ref. 40). Relative quantification analysis and statistics were performed using the MCMC.qpcr R library using efficiency and Cq values from LinRegPCR (ref. 41). Relative quantification was performed relative to three reference genes (rpoB, gyrB and dnaG) using the soft-normalization approach in MCMC.qpcr, and significance testing was performed using two-sided MCMC posterior sampling in MCMC.qpcr (Bayesian P values). Mean expression values are reported with per-condition 95% posterior credible intervals representing all sources of between-sample variability. All experiments used, at a minimum, three biological replicates to achieve a power sufficient to control the false-negative rate to 5% at an effect size of less than twofold between samples, given an estimate of the average between-sample variance, except for the yfp reporter samples, which targeted an effect size of fourfold. Diagnostic plots from MCMC.qpcr were used to assess the model assumptions of normality, homoscedasticity and linearity.
Construction, sequencing and analysis of Illumina libraries
We constructed Illumina transcriptome libraries from RNA extracted from three independent cultures of each strain using Illumina ScriptSeq. We subjected these to paired-end 75 bp sequencing on a NextSeq 500 and obtained high-quality sequence for all libraries, with the exception that one library of the JG091 marker-replacement strain did not provide sufficient reads for analysis and was excluded from further analysis. FASTQ reads were quality-filtered using fastq-mcf and aligned to the FZB42 reference genome using BWA’s bwa-mem algorithm42,43. Normalization and differential expression analysis and statistics were performed using the DEseq2 R library44. Significance testing was performed using multiple-testing adjusted Wald test P values in DEseq 2. Coverage plots with standard deviations were constructed using custom Python scripts using the normalization factors calculated by DEseq2.
Bioinformatic analysis of NusG family protein sequences
A representative phylogenetic tree was constructed by obtaining, via manual curation of the literature, three to eight sequences representing each NusG family subgroup. These sequences were aligned using T-COFFEE in Expresso mode using three-dimensional structure information for alignment45. A maximum-likelihood phylogenetic tree was constructed using RAxML with automatic evolution model selection and 1,000 rapid bootstraps46.
A comprehensive list of prokaryotic NusG protein sequences was obtained by searching the full UniprotKB protein sequence database with the NusG N-terminal domain PFAM HMM using HMMER 3.0 with the default PFAM score cutoff and filtering for proteins from prokaryotic organisms47. Sequences were assigned a preliminary subgroup classification by addition to the small representative alignment and rapid tree construction using FastTree and assigned to the nearest subgroup48,49. Near-duplicate sequences were removed by UCLUST clustering preliminary core-NusG sequences to 60% sequence identity and non-core-NusG sequences to 95% sequence identity, resulting in 1,205 sequences50. These were aligned using MAFFT in E-INS-I local domain alignment mode and a phylogenetic tree was constructed using RAxML with automatic evolution model selection and 1,000 rapid bootstraps49.
The representative genomic sequences for each protein were obtained from the Uniprot database and the surrounding region for each sequence was searched for biosynthetic gene clusters using antiSMASH 3.0.5 with additional ClusterFinder search51. Each protein coding sequence was checked for proximity to detected gene clusters and this information was mapped onto the large-scale phylogenetic tree. Labelling of NusG paralogue subtypes was performed by identification of the sub-tree corresponding to the most recent common ancestor of curated representative sequences.
Extraction and detection of difficidin, macrolactin and bacillaene
To investigate the effects of loaP on metabolites, overnight cultures of B. amyloliquefaciens FZB42 strains were diluted to an OD600 of 0.08, growing in 25 ml Landy medium52 for 8 h as described in ref. 53. The supernatant from each culture was collected. A 25 ml volume of each culture was centrifuged for 30 min at 11,000 r.p.m. and loaded onto an SPE column (3M, Empore, C18-SD). After loading, the columns were washed once with dH2O and once with 20% methanol. Samples were eluted using 2 ml 100% methanol followed by 1 ml 100% ethanol. The eluates were dried in a rotary evaporator and samples were re-dissolved in 100 μl 90% methanol.
Metabolite analysis was performed using HPLC with an Agilent 1200 device. A 10 μl volume of each sample was injected onto a ZORBAX Eclipse Plus C18 column (4.6 × 100 mm, 5 μm; Agilent). The run was performed with a flow rate of 1.0 ml min−1 at 30 °C. Samples were eluted with a gradient of 20% CH3CN and 80% of 0.1% vol/vol HCOOH, which reached 95% CH3CN and 5% of 0.1% vol/vol HCOOH after 12 min. The 95% CH3CN–5% HCOOH step was maintained for a further 5 min. The column was equilibrated with 20% CH3CN–80% HCOOH for 2 min. Difficidin and macrolactin peaks were detected at 1,280 nm as previous reported53. We confirmed the specificity of difficidin, macrolactin and bacillaene peaks in the HPLC chromatographs by comparison to samples from B. amyloliquefacians Δpks3KS1 (abolished difficidin biosynthesis) and Δpks2KS1, Δpks3KS1 (abolished difficidin and macrolactin biosynthesis) strain and by liquid chromatography–tandem mass spectrometry.
Quantification analysis of difficidin and macrolactin
For preparation of the samples used for quantification analysis, B. amyloliquefaciens FZB42 strains were grown in lysogeny broth (LB) (1% tryptone (Bacto), 0.5% yeast extract (BBL), 0.5% sodium chloride (Sigma)). Overnight cultures of B. amyloliquefaciens strains were diluted to an OD600 of 0.08, cultured to an OD600 of ~0.2, and rediluted to an OD600 of 0.08. This step was repeated three times to generate a uniform population of cells. Experimental cultures were inoculated in 25 ml of LB ± 0.5% xylose with an OD600 of 0.08, and additional 0.5% xylose was added at an OD600 of 3. The supernatant from each culture was collected at an OD600 of 4.0±0.1.
The extraction method, HPLC column and detection wavelength were as described in the previous section. For quantitative analysis, samples were eluted with a gradient of 30% CH3CN and 70% of 0.1% vol/vol HCOOH, which reached 70% CH3CN and 30% of 0.1% vol/vol HCOOH after 6 min. The 70% CH3CN–30% HCOOH step was maintained for a further 3 min. The column was equilibrated with 30% CH3CN–70% HCOOH for 2 min. Quantitation for each sample was determined by integrating the area under the relevant peaks on the elution chromatography. Relative metabolite values were determined first by calculating the ratio of difficidin or macrolactin to bacillaene as a reference, then by normalizing to the wild-type sample value. Values are reported as the average of two biological replicates and standard deviation.
Purification of LoaP and addition to dfn transcription reactions in vitro
The B. amyloliquefaciens loaP open reading frame was subcloned in-frame with an N-terminal hexahistidine tag in pHis-parallel2. Six 500 ml cultures in 2 l flasks were inoculated 1:100 from an overnight culture into LB containing carbenicillin and cultured, shaking at 37 °C, until reaching an OD600 of 0.8. The cultures were then transferred to room temperature, at which point 100 μM isopropyl-β-D-thiogalactoside (IPTG) was added and cultures were incubated, with shaking, at room temperature overnight. Cells were collected by centrifugation, resuspended in 120 ml of lysis buffer (50 mM sodium phosphate pH 7.5, 300 mM sodium chloride, 1× ProBlock Gold protease inhibitor, 500 μg ml−1 lysozyme), incubated at room temperature for 15 min, and lysed on ice by sonication. The lysate was clarified by centrifugation at 4 °C for 40 min at 14,000g. Clarified lysate was incubated with gentle shaking with 1 ml Ni-NTA resin (Qiagen) for 1 h at 4 °C. Three batch washes were performed using lysis buffer without lysozyme and 40 mM imidazole. The resin was transferred to a column for gravity flow and washed with 30 volumes of lysis buffer containing 80 mM imidazole. Protein was eluted using 250 mM imidazole, and quantified by ultraviolet spectroscopy and Bradford assay and visualized using SDS–PAGE.
For in vitro transcription reactions the dfnA leader region, including the endogenous promoter, was PCR amplified using a forward oligo upstream of the either a 314 nt transcript (in the event of premature transcription termination) or a 420-nt transcript (in the event of run-off transcription). Both transcripts could be observed in 5 μl reactions containing 2 pmol PCR-generated DNA template (Supplementary Fig. 6). Also included in these reactions were 20 mM Tris-HCl pH 8.0, 15 mM NaCl, 4 mM MgCl2, 0.1 mM EDTA, 5 mM dithiothreitol (DTT), 0.01% Triton X100, 1 mM NTPs, 1.5 pmol (5 μCi) α-32P-UTP and 160 nM σA-saturated B. subtilis RNAP (gift from P. Babitzke). As shown in Supplementary Fig. 6, LoaP was added to final concentrations of 300, 150, 75 and 3.7 nM. Purified hexahistidine-tagged B. subtilis S10 was added (to 150 nM), and purified hexahistidine-tagged B. subtilis NusA was added (to 50 nM), as indicated. The reactions were incubated at 37 °C for 1 h, and products were resolved alongside dfnA size markers by 6% urea-denaturing PAGE.
Data availability
R and Python code to analyse qPCR, RNA-seq and the large-scale phylogenetic tree are available online (https://github.com/jgoodson/LoaP-2016). Full sequences of all plasmids are available from GenBank and the accession numbers for plasmids are detailed in Supplementary Table 4. Sequencing data are available from NCBI SRA (BioProject PRJNA327241). Other data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
Research on this project was supported by the National Science Foundation (MCB1051440 to W.C.W. and NSF-CAREER MCB1253215 to P.S.).
Footnotes
Author contributions
The experimental plan was devised by J.R.G., P.S. and W.C.W. All authors assisted in the collection and interpretation of data. J.R.G., P.S. and W.C.W. wrote the manuscript.
Supplementary information is available for this paper.
Competing interests
The authors declare no competing financial interests.
References
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2016;79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 2.Sidebottom AM, Carlson EE. A reinvigorated era of bacterial secondary metabolite discovery. Curr Opin Chem Biol. 2015;24:104–111. doi: 10.1016/j.cbpa.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 3.Walsh C. Antibiotics: Actions, Origins, Resistance. ASM; 2013. [Google Scholar]
- 4.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weisberg RA, Gottesman ME. Processive antitermination. J Bacteriol. 1999;181:359–367. doi: 10.1128/jb.181.2.359-367.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts JW, Shankar S, Filter JJ. RNA polymerase elongation factors. Annu Rev Microbiol. 2008;62:211–233. doi: 10.1146/annurev.micro.61.080706.093422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santangelo TJ, Artsimovitch I. Termination and antitermination: RNA polymerase runs a stop sign. Nat Rev Microbiol. 2011;9:319–329. doi: 10.1038/nrmicro2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ray-Soni A, Bellecourt MJ, Landick R. Mechanisms of bacterial transcription termination: all good things must end. Annu Rev Biochem. 2016;85:319–347. doi: 10.1146/annurev-biochem-060815-014844. [DOI] [PubMed] [Google Scholar]
- 9.Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- 10.Greenblatt J, McLimont M, Hanly S. Termination of transcription by nusA gene protein of Escherichia coli. Nature. 1981;292:215–220. doi: 10.1038/292215a0. [DOI] [PubMed] [Google Scholar]
- 11.Mondal S, Yakhnin AV, Sebastian A, Albert I, Babitzke P. NusA-dependent transcription termination prevents misregulation of global gene expression. Nat Microbiol. 2016;1:15007. doi: 10.1038/nmicrobiol.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Proshkin S, Mironov A, Nudler E. Riboswitches in regulation of Rho-dependent transcription termination. Biochim Biophys Acta. 2014;1839:974–977. doi: 10.1016/j.bbagrm.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Chowdhury SP, Hartmann A, Gao X, Borriss R. Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42—a review. Front Microbiol. 2015;6:780. doi: 10.3389/fmicb.2015.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen XH, et al. Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB 42. J Bacteriol. 2006;188:4024–4036. doi: 10.1128/JB.00052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan B, et al. dRNA-Seq reveals genomewide TSSs and noncoding RNAs of plant beneficial rhizobacterium Bacillus amyloliquefaciens FZB42. PLoS ONE. 2015;10:e0142002. doi: 10.1371/journal.pone.0142002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irnov I, Sharma CM, Vogel J, Winkler WC. Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res. 2010;38:6637–6651. doi: 10.1093/nar/gkq454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura K, et al. Sequence-specific error profile of Illumina sequencers. Nucleic Acids Res. 2011;39:e90. doi: 10.1093/nar/gkr344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irnov I, Winkler WC. A regulatory RNA required for antitermination of biofilm and capsular polysaccharide operons in Bacillales. Mol Microbiol. 2010;76:559–575. doi: 10.1111/j.1365-2958.2010.07131.x. [DOI] [PubMed] [Google Scholar]
- 20.Chen XH, et al. Genome analysis of Bacillus amyloliquefaciens FZB42 reveals its potential for biocontrol of plant pathogens. J Biotechnol. 2009;140:27–37. doi: 10.1016/j.jbiotec.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 21.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomar SK, Artsimovitch I. NusG-Spt5 proteins—universal tools for transcription modification and communication. Chem Rev. 2013;113:8604–8619. doi: 10.1021/cr400064k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatzidaki-Livanis M, Weinacht KG, Comstock LE. Trans locus inhibitors limit concomitant polysaccharide synthesis in the human gut symbiont Bacteroides fragilis. Proc Natl Acad Sci USA. 2010;107:11976–11980. doi: 10.1073/pnas.1005039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King RA, Banik-Maiti S, Jin DJ, Weisberg RA. Transcripts that increase the processivity and elongation rate of RNA polymerase. Cell. 1996;87:893–903. doi: 10.1016/s0092-8674(00)81996-0. [DOI] [PubMed] [Google Scholar]
- 25.Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- 26.NandyMazumdar M, Artsimovitch I. Ubiquitous transcription factors display structural plasticity and diverse functions: NusG proteins—shifting shapes and paradigms. BioEssays. 2015;37:324–334. doi: 10.1002/bies.201400177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paitan Y, Orr E, Ron EZ, Rosenberg E. A NusG-like transcription antiterminator is involved in the biosynthesis of the polyketide antibiotic TA of Myxococcus xanthus. FEMS Microbiol Lett. 1999;170:221–227. doi: 10.1111/j.1574-6968.1999.tb13377.x. [DOI] [PubMed] [Google Scholar]
- 28.Behnken S, Lincke T, Kloss F, Ishida K, Hertweck C. Antiterminator-mediated unveiling of cryptic polythioamides in an anaerobic bacterium. Angew Chem Int Ed. 2012;51:2425–2428. doi: 10.1002/anie.201108214. [DOI] [PubMed] [Google Scholar]
- 29.Yakhnin AV, Babitzke P. NusG/Spt5: are there common functions of this ubiquitous transcription elongation factor? Curr Opin Microbiol. 2014;18:68–71. doi: 10.1016/j.mib.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yakhnin AV, Murakami KS, Babitzke P. NusG is a sequence-specific RNA polymerase pause factor that binds to the non-template DNA within the paused transcription bubble. J Biol Chem. 2016;291:5299–5308. doi: 10.1074/jbc.M115.704189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crickard JB, Fu J, Reese JC. Biochemical analysis of yeast suppressor of Ty 4/5 (Spt4/5) reveals the importance of nucleic acid interactions in the prevention of RNA polymerase II arrest. J Biol Chem. 2016;291:9853–9870. doi: 10.1074/jbc.M116.716001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thapar R, Denmon AP, Nikonowicz EP. Recognition modes of RNA tetraloops and tetraloop-like motifs by RNA-binding proteins. Wiley Interdiscip Rev RNA. 2014;5:49–67. doi: 10.1002/wrna.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang HC, Nagaswamy U, Fox GE. The application of cluster analysis in the intercomparison of loop structures in RNA. RNA. 2005;11:412–423. doi: 10.1261/rna.7104605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fiore JL, Nesbitt DJ. An RNA folding motif: GNRA tetraloop–receptor interactions. Q Rev Biophys. 2013;46:223–264. doi: 10.1017/S0033583513000048. [DOI] [PubMed] [Google Scholar]
- 35.Cilley CD, Williamson JR. Structural mimicry in the phage φ 21 N peptide–boxB RNA complex. RNA. 2003;9:663–676. doi: 10.1261/rna.2189203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Behnken S, Hertweck C. Cryptic polyketide synthase genes in non-pathogenic Clostridium spp. PLoS ONE. 2012;7:e29609. doi: 10.1371/journal.pone.0029609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 38.Jarmer H, Berka R, Knudsen S, Saxild HH. Transcriptome analysis documents induced competence of Bacillus subtilis during nitrogen limiting conditions. FEMS Microbiol Lett. 2002;206:197–200. doi: 10.1111/j.1574-6968.2002.tb11009.x. [DOI] [PubMed] [Google Scholar]
- 39.Bhavsar AP, Zhao X, Brown ED. Development and characterization of a xylose-dependent system for expression of cloned genes in Bacillus subtilis: conditional complementation of a teichoic acid mutant. Appl Environ Microbiol. 2001;67:403–410. doi: 10.1128/AEM.67.1.403-410.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 41.Matz MV, Wright RM, Scott JG. No control genes required: Bayesian analysis of qRT–PCR data. PLoS ONE. 2013;8:e71448. doi: 10.1371/journal.pone.0071448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aronesty E. Comparison of sequencing utility programs. Open Bioinform J. 2013;7:1–8. [Google Scholar]
- 43.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinforma Oxf Engl. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Armougom F, et al. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 2006;34:W604–W608. doi: 10.1093/nar/gkl092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 47.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Price MN, Dehal PS, Arkin AP. Fasttree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinform Oxf Engl. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 51.Weber T, et al. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Landy M, Warren GH. Bacillomycin; an antibiotic from Bacillus subtilis active against pathogenic fungi. Proc Soc Exp Biol Med. 1948;67:539–541. doi: 10.3181/00379727-67-16367. [DOI] [PubMed] [Google Scholar]
- 53.Chen XH, et al. Difficidin and bacilysin produced by plant-associated Bacillus amyloliquefaciens are efficient in controlling fire blight disease. J Biotechnol. 2009;140:38–44. doi: 10.1016/j.jbiotec.2008.10.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
R and Python code to analyse qPCR, RNA-seq and the large-scale phylogenetic tree are available online (https://github.com/jgoodson/LoaP-2016). Full sequences of all plasmids are available from GenBank and the accession numbers for plasmids are detailed in Supplementary Table 4. Sequencing data are available from NCBI SRA (BioProject PRJNA327241). Other data that support the findings of this study are available from the corresponding author upon request.