Abstract
Although a distinct karyotype with defined chromosome number and structure characterizes each biological species, it is intrinsically labile. Polyploidy or whole-genome duplication has played a pervasive and ongoing role in the evolution of all eukaryotes, and is the most dramatic force known to cause rapid karyotypic reconfiguration, especially at the initial stage. However, issues concerning transgenerational propagation of karyotypic heterogeneity and its translation to phenotypic diversity in nascent allopolyploidy, at the population level, have yet to be studied in detail. Here, we report a large-scale examination of transgenerationally propagated karyotypic heterogeneity and its phenotypic manifestation in an artificially constructed allotetraploid with a genome composition of AADD, that is, involving two of the three progenitor genomes of polyploid wheat. Specifically, we show that 1) massive organismal karyotypic heterogeneity is precipitated after 12 consecutive generations of selfing from a single euploid founder individual, 2) there exist dramatic differences in aptitudes between subgenomes and among chromosomes for whole-chromosome gain and/or loss and structural variations, 3) majority of the numerical and structural chromosomal variations are concurrent due to mutual contingency and possible functional constraint, 4) purposed and continuous selection and propagation for euploidy over generations did not result in enhanced karyotype stabilization, and 5) extent of karyotypic variation correlates with variability of phenotypic manifestation. Together, our results document that allopolyploidization catalyzes rampant and transgenerationally heritable organismal karyotypic heterogeneity that drives population-level phenotypic diversification, which lends fresh empirical support to the still contentious notion that whole-genome duplication enhances organismal evolvability.
Keywords: allopolyploidy, meiotic chromosome instability, karyotypic heterogeneity, phenotypic diversity, progenitor genomes, wheat
Introduction
Every species has a defined karyotype with an evolved combination of chromosome number and structure, known as karyotype (Stebbins 1950). Thus, persistently ongoing alterations of chromosome number and structure, that is, numerical and structural chromosome instability (CIN) (Dion-Cote et al. 2015; Bakhoum and Landau 2017), has severe phenotypic and fitness consequences and may lead to species extinction or catalyze saltational evolution. Somatic CIN is increasingly recognized as a central player in tumorigenesis and cancer metastasis (Dewhurst et al. 2014). Thus, understanding causes and consequences of somatic CIN is of vital importance in cancer research.
In contrast to the immediate physiological and phenotypic manifestations of somatic CIN in cancer, only organismal level CIN is likely consequential to plant evolution. Consequently, for CIN to play a role in plant evolution, it needs to occur in gametophytes and is meiotically heritable.
Polyploidy, or whole-genome duplication (WGD), is a driving force in the evolution of all eukaryotes but especially pervasive in flowering plants (Otto and Whitton 2000; Jiao et al. 2011; Soltis et al. 2015; Wendel 2015; Van de Peer et al. 2017). Polyploidy encompasses two broad categories, autopolyploidy and allopolyploidy, with the former referring to doubling of a single species genome and the latter to duplication of merged genomes from two or more species. Thus, while autopolyploidy in theory can be a genetically pure line, allopolyploidy is an interspecific hybrid at an elevated ploidy level. Both types of polyploidy are known to instigate the occurrence of CIN and foster their perpetuation (Chen 2007; Otto 2007; Doyle et al. 2008; Feldman and Levy 2009, 2012; Parisod et al. 2009; Geiser et al. 2016). Conceivably, while numerical chromosome alterations (i.e., aneuploidy) are often, and perhaps inevitable, incidents associated with both categories of allopolyploidy, structural chromosome alterations occur more frequently in allopolyploidy due to more prone occurrence and less catastrophic consequences of rearrangements between homeologous chromosomes than between nonhomologous chromosomes. The primary cause for polyploidy-generated or -facilitated CIN in both number and structure is due to compromised functionality of the otherwise fine-tuned meiotic machinery under the acutely doubled genomic environment (Lloyd and Bomblies 2016). Nevertheless, the success of vast polyploid species suggests that meiotic machinery per se is evolvable and can be adapted to handle a suddenly doubled chromosome complement (Bomblies et al. 2015; Hollister 2015), as empirically documented recently (Yant et al. 2013; Wright et al. 2015).
Genome evolution due to WGD has become a burgeoning research field in recent years (Van de Peer et al. 2017). It becomes increasingly clear that WGD is more than doubling the nuclear genome dosage alone; rather, it generates extensive alterations de novo in genome and transcriptome, and their attendant downstream alterations in nearly all aspects of molecular, cellular, physiological and morphological phenotypes (Schoenfelder and Fox 2015). In particular, it has been shown in diverse plant taxa that allopolyploidization in particular may trigger rapid and extensive genetic and epigenetic changes at the molecular level primarily due to the effect of hybridization (Doyle et al. 2008), consistent with the “genome shock” hypothesis proposed by McClintock (1984) decades ago. Disproportionate to the rapid advances in genomic and molecular investigations, the issue concerning karyotype evolution immediately following allopolyploidy is less explored. For example, although it was found that rapid changes in chromosome number and/or structure may occur in both synthetic and naturally occurred nascent plant allopolyploids (Pires et al. 2004; Lim et al. 2008; Xiong et al. 2011; Chester et al. 2012, 2013; Zhang, Bian, Gou, Zhou et al. 2013), issues concerning origin, prevalence and transgenerational propagation of karyotypic alterations, as well as their effects on population-level phenotypic diversification have not been investigated in detail. However, understanding the immediate impacts of polyploidy on karyotypic evolution is important because it can be a driving force underpinning the widely observed molecular level genetic, epigenetic and gene expression changes (Chen 2007; Doyle et al. 2008; Van de Peer et al. 2017). An added significance to explore the transgenerational dynamics of karyotypic alteration (i.e., the evolutionary trajectories of karyotypic evolution) lies in the fact that evolved karyotypic stabilization is an essential property for nascent polyploidy to be persistent over evolutionary timescales and contribute to species diversification. Furthermore, knowledge gained regarding the extent and trend of karyotypic evolution, its transgenerational heritability and phenotypic consequences, as well as the mechanisms whereby karyotype stabilization can be achieved in newly formed polyploids may provide useful clues to more judicious utilization of the synthetic hybridization/allopolyploidization strategy for crop improvement (Mason and Batley 2015).
Common wheat (Triticum aestivum L., genome BBAADD) is a young allohexaploid species harboring three subgenomes originated from three diploid progenitor species of the Triticum–Aegilops complex (Feldman et al. 1995; Dubcovsky and Dvorak 2007). There are two sequential allopolyploidization events associated with the speciation of common wheat. The first event occurred ca. 0.5 Mya involving allotetraploidization between two diploid species, Triticum urartu (AA) and a yet unidentified or extinct goat–grass species closely related to the Sitopsis section (SS ≈ BB) of Aegilops (El Baidouri et al. 2017), which gave rise to the speciation of tetraploid emmer wheat, T. turgidum (Dvořák 1976). Then, the second event occurred only ca. 8,000 years ago involving hybridization of a primitive domesticated form of T. turgidum (BBAA) with the Tausch’s goatgrass, Ae. tauschii (DD), led to the speciation of common wheat (Kilian et al. 2007). We have reported recently that while T. aestivum as an evolved species contains a highly stabilized karyotype constituted by the three distinct subgenomes, only one of the three artificially constructed two progenitor–genome combinations at the tetraploid level is karyotypically stable (Zhang, Bian, Gou, Dong et al. 2013). Specifically, only synthetic allotetraploid wheats with the genome combination of SSAA (S ≈ B) is stable in karyotype while those with either SSDD or AADD genome combinations are highly unstable in both chromosome number and structure (Zhang, Bian, Gou, Dong et al. 2013). However, many important questions concerning numerical and structural chromosome instabilities in nascent allopolyploidy remain unclear. For example, how prevalent are the numerical and structural chromosome variations? Are the two types of chromosomal instabilities intrinsically correlated due to common origin and/or functional connectivity? Are the karyotypic variations heritable and transgenerationally accumulative? What is the phenotypic manifestation of the transgenerationally precipitated karyotypic heterogeneity? Is it possible for a karyotypic stabilization mechanism to evolve rapidly in the initially unstable genome combinations?
Here, we sought to address some of these questions using an artificially constructed allotetraploid wheat with a genome combination of two of the three polyploid wheat progenitor genomes, A and D. We present the results of in-depth characterization of transgenerationally precipitated numerical and structural chromosome variations in a large cohort containing 1,462 random individuals descended from a single S2 euploid founder plant of this synthetic allotetraploid wheat. Together with karyotyping of an independent single seed-descended multigenerational lineage constituted by intentional selecting and propagating of one euploid individual at each generation, our results document the remarkably exacerbated karyotypic evolvability of newly formed allopolyploidy and its highly penetrating phenotypic effects. The constructed transgenerational lineage for a synthetic allotetraploid involving two progenitor genomes of polyploid wheat (A and D) with exact pure-line diploid parents and a single euploid founder origin may serve as a uniquely tractable system to address fundamental issues germane to polyploid genome evolution and speciation.
Results
Massive Karyotypic Variations Are Precipitated after 12 Consecutive Generations of Selfing from a Single Euploid Founder Individual of the Synthetic Allotetraploid Wheat with a Genome Combination of AADD
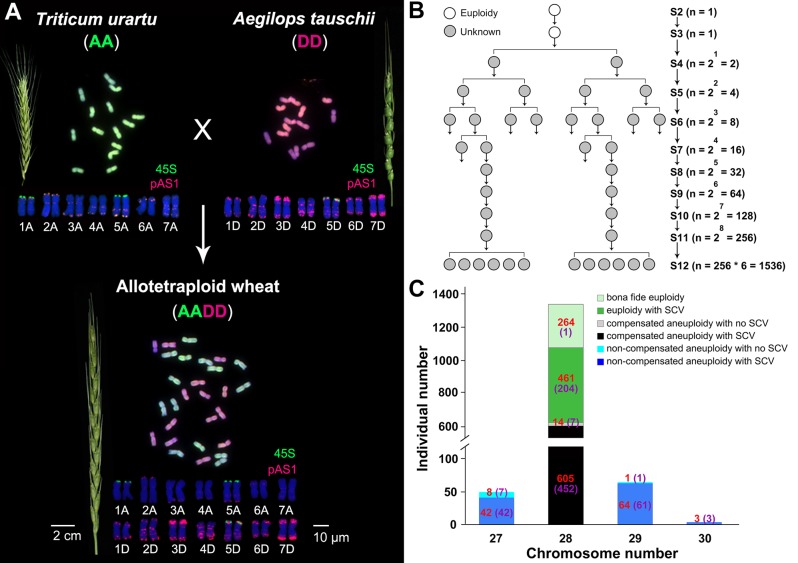
The study system concerns a synthetic allotetraploid wheat with genome AADD, produced by hybridization followed by WGD of two diploid progenitor species of polyploid wheat, Triticum urartu (genome AA) and Aegilops tauschii (genome DD) (fig. 1A). A cohort of 1,462 random individual plants at the S12 generation, which were descended from a single S2 euploid founder plant of the synthetic allotetraploid wheat (genome AADD) (fig. 1B) were karyotyped by using a sequential FISH and GISH protocol (Zhang, Bian, Gou, Zhou et al. 2013). Notably, in no case somatic CIN, that is, occurrence of CIN among the somatic cells within a given individual, was detected, indicating normal mitosis in all these tetraploid plants, and therefore any chromosomal variation detected should be rooted to abnormal meiosis. Of these plants, only 264 (18.1%) were bona fide euploids with a karyotype identical to the founder plant containing the expected parental chromosomal additivity (2n = 28) with no discernible structural changes at the molecular cytogenetic resolution (fig. 1A and C). The majority of the remaining individuals were either euploidy (2n = 28) but containing readily detectable structural alterations (461 individuals, 31.5%), or various types of numerical chromosome changes (aneuploidy) together with structural alterations (605 + 42 + 64 +3 = 714 individuals, 48.8%) (fig. 1C). Notably, a predominate proportion of the aneuploidies (14 + 605 = 619, 42.3% of the 1,462 plants) were “compensated polyploidy” (Chester et al. 2012) in the sense that they contained the euploid chromosome number (2n = 28) but with at least one chromosome being deviated from the modal two copies (fig. 1C). Importantly, the vast majority of these compensated aneuploidies also contained structural variations, and only 14 plants showed numerical changes only (i.e., containing no detectable structural changes). Relatively small proportions of aneuploid plants contained outlier chromosome numbers from the modal number of 28, with 50 (3.4%), 65 (4.4%), and 3 (0.2%) plants harboring chromosome numbers of 2n = 27, 29, and 30, respectively (fig. 1C).
Fig. 1.
The plant system, standard allotetraploid and parental karyotypes, propagation strategy, and variable karyotypic compositions of a cohort of 1,462 randomly sampled individuals at the 12th selfed generation (S12) propagated from a single S2 founder euploid individual of a synthetic allotetraploid wheat. (A) Spike morphology and FISH/GISH-based standard karyotypes of the S2 founder euploid individual of the artificially constructed allotetraploid wheat (genome AADD) and its diploid parental species, T. urartu (genome AA) and Ae. tauschii (genome DD). (B) Diagrammed strategy for transgenerational propagation of a random population containing a cohort of 1,536 plant individuals constituting the S12 population, which were descended from the single euploid founder plant (karyotype shown in A) of the S2 generation. (C) Karyotypic compositions of a cohort of 1,462 (successfully karyotyped among the 1,536 individuals, shown in B) individuals at the 12th selfed generation (S12) of the allotetraploid wheat, which could be categorized into six distinct karyotypic groups. Numerical values in parenthesis denote the numbers of distinct karyotypes identified in each karyotypic group.
Strikingly, the numerical and structural chromosomal alterations as well as their combinations have led to extensive karyotypic heterogeneity at the organismal level in these plants descended from a single euploid founder individual after just 12 generations of propagation via selfing without selection (fig. 1B). Specifically, out of the 1,462 analyzed individuals, there were 15 karyotypes with different chromosomal contents (aneuploidies) only, 204 karyotypes containing different structural chromosome alterations only, and 558 karyotypes containing different combinations of numerical and structural chromosome alterations (fig. 1C;supplementary table S1, Supplementary Material online). Altogether, 777 distinct karyotypes with regard to either or both numerical and cytologically discernible SCVs were generated among a cohort of 1,462 allotetraploid plants that were randomly propagated from a single euploid karyotype over the span of only 12 successive generations of selfing. These observations point to the remarkable karyotypic evolvability due to ongoing meiotic CIN when the A and D progenitor genomes of polyploid wheat were brought into a common nucleus by artificial allotetraploidization.
Different Aptitudes between Subgenomes and among Chromosomes for Whole-Chromosome Gain and/or Loss
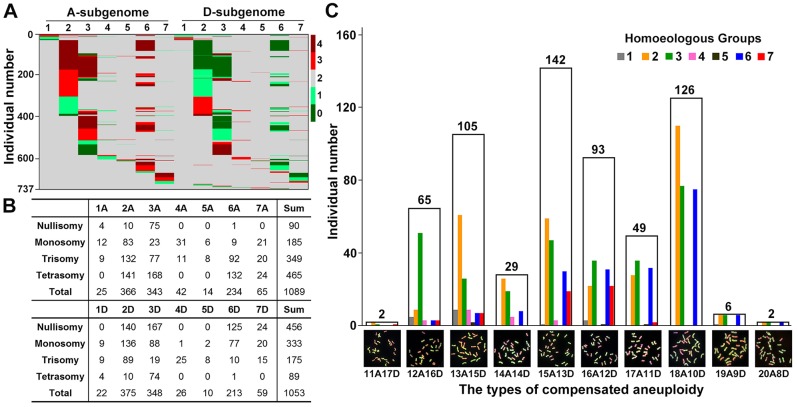
The foregoing results demonstrated that massive alterations of chromosomal copy number, that is, aneuploidy, occurred in half (50.4%) of the 1,462 karyotyped synthetic allotetraploid wheat individuals (fig. 1C), and all 14 chromosomes of the two subgenomes (A and D) were involved in numerical changes in the range of 0 to 4 copies (fig. 2A). A question to ask is: Are the two subgenomes each as a whole and their respective constituting chromosomes (seven for each subgenome) possess similar or different propensities towards whole-chromosome gain and/or loss, that is, numerical chromosome variation (NCV)? Results showed that there existed dramatic differences both between the two subgenomes each as a whole and among all the chromosomes of both subgenomes with respect to NCV. First, subgenome A showed a significantly higher aptitude for chromosome gain (trisomy + tetrasomy) than loss (monosomy + nullisomy) (Chi-squared test, P = 1.05E−65) while subgenome D showed the opposite trend, that is, more chromosome loss than gain (Chi-squared test, P = 1.05E−65) (fig. 2A and B). Second, chromosomes within a given subgenome were markedly variable towards gain and/or loss. Specifically, 1) among the seven subgenome A chromosomes, 2A (33.5%), 3A (30.1%), and 6A (27.5%) showed significantly higher frequencies for gain than the remaining four chromosomes, 1A (1.1%), 4A (1.4%), 5A (1%), and 7A (5.4%) (Chi-squared test, P = 3.86E−168), and chromosomes 2A (33.8%) and 3A (35.6%) showed significantly higher frequencies of loss than the rest five chromosomes (Chi-squared test, P = 5.70E−48) (fig. 2B); 2) among the seven subgenome D chromosomes, 2D (35.0%), 3D (32.3%), and 6D (25.6%) showed significantly higher frequencies for loss than the rest four chromosomes, 1D (1.1%), 4D (0.1%), 5D (0.3%), and 7D (5.6%) (Chi-squared test, P = 5.59E−179), and chromosomes 2D (37.5%) and 3D (35.2%) showed higher frequencies of gain than the rest five chromosomes (Chi-squared test, P = 1.23E−52) (fig. 2B); 3) across both subgenomes, two pair of homoeologous chromosomes, 4A/4D and 5A/5D, showed the lowest frequencies for both gain and loss, especially, no plant was found to be nullisomy or tetrasomy for any of these two pairs of homeologous chromosomes (fig. 2B), suggesting either these four chromosomes are fully stable during meiosis or they carry essential genes (which are dosage-sensitive) for normal gametophytic or zygotic development, with null or double-dosage of these genes in each of these four chromosomes leading to inviability of gamete or zygote.
Fig. 2.
NCVs in the 1,462 karyotyped cohorts, differential propensities for NCVs across the 14 chromosomes, and distribution of chromosome constitutions of 619 “compensated” aneuploidies (2n = 28) identified among the total karyotyped cohorts, at the 12th selfed generation (S12) of the allotetraploid wheat. (A) A heatmap depicting gain or loss of whole-chromosome copies among the 14 chromosomes of the A and D subgenomes. (B) Distribution of the numbers of plants showing variable copies (from 0 to 4) of each of the 14 chromosomes. (C) Number of plants with differential chromosome constitutions of the 619 “compensated” aneuploidies (fig. 1C) with concerted whole-chromosome gain and loss, thus maintaining a total chromosome number of 2n = 28. In both (A) and (C), the y-axes denote individual numbers. The numbers above the white columns in (C) refer to plant individuals belonging to each of the designated “compensated” aneuploid types. The colored vertical bars refer to the numbers of plant individuals falling to each of the seven homeologous chromosome groups within each of the “compensated” aneuploid types. GISH images (green and red colorations denote for chromosomes of subgenomes A and D, respectively) of one representative individual for each of the 10 identified types of “compensated” aneuploidies were shown.
Differences between the two subgenomes and among the chromosomes for NCV was also reflected by the chromosome compositions of the 619 (605 + 14) compensated aneuploidies with 2n = 28 euploid chromosomes (fig. 1C). Results showed that a significantly larger number of individuals contained concerted gain of subgenome A chromosomes and loss of the corresponding subgenome D chromosomes than the alternative possibility (individuals with ≥ 15A chromosomes vs. those with ≤ 13A chromosomes = 418 vs. 172, Chi-squared test, P = 4.16E−24) (fig. 2C). The sharply differential gain and/or loss frequencies among the individual chromosomes within these compensated aneuploidies showed the same trend (fig. 2C) as the whole cohort of individuals (fig. 2B), described above. These results further confirm the dramatically different aptitudes both between subgenomes and among chromosomes for NCV in the allotetraploid progenies with a genome composition of AADD.
Different Propensities between Subgenomes and among Chromosomes for SCVs
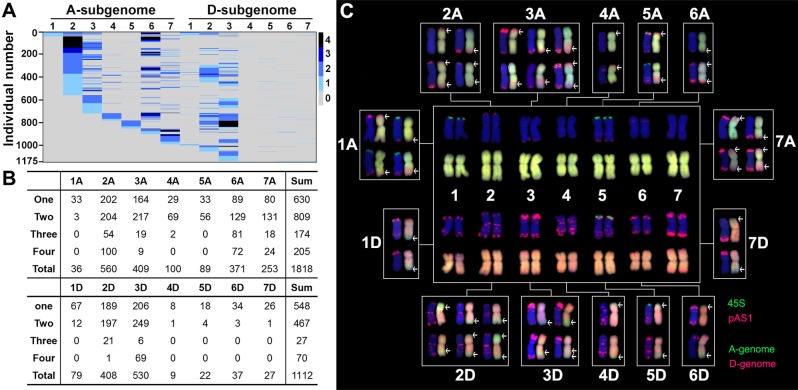
Similar to the situation of NCV, rampant SCV also occurred in a significant proportion (80.4%) of the 1,462 karyotyped allotetraploid individuals (fig. 1C), and all 14 chromosomes of the two subgenomes (A and D) underwent SCV in the range of 0–4 copies (fig. 3A). Also similar to the characteristics of NCV, described above, dramatic differences were found both between the two subgenomes each as a whole and among the 14 individual chromosomes for SCV in this set of allotetraploid plants. First, subgenome A showed an overly higher frequency of SCV than subgenome D (binomial test, P = 1.46E−39) (fig. 3A and B), as expected given the higher propensities for gain of additional A subgenome chromosomes and loss of D subgenome chromosomes by these plants (fig. 2). Second, chromosomes within a given subgenome manifested significant differences in the frequencies of SCV. Specifically, 1) among the seven subgenome A chromosomes, 2A (30.8%), 3A (22.5%), and 6A (20.4%) showed significantly higher frequencies of SCV than the rest four chromosomes, 1A (2%), 4A (5.5%), 5A (4.9%), and 7A (13.9%) (Chi-squared test, P = 1.08E−187) (fig. 3A and B); and 2) among the seven subgenome D chromosomes, 2D (36.7%) and 3D (47.7%) showed significantly higher frequencies of SCV than the rest five chromosomes, 1D (7.1%), 4D (0.8%), 5D (2.0%), 6D (3.3%), and 7D (2.4%) (Chi-squared test, P = 0) (fig. 3A and B). Clearly, the trend of differential probabilities in SCV both between subgenomes and among chromosomes largely accords with that for NCV, suggesting common origin and/or functional connectivity between the two types of chromosome variations, as being further detailed later. A notable feature of SCV was that plants bearing three or four copies of restructured chromosomes were markedly less than those containing one or two restructured chromosomes (fig. 3A and B). This later observation suggests that either the meiotic CIN is still ongoing, that is, SCVs being generated in a continuum from normal chromosomes, or higher copies of restructured chromosomes engenders fitness cost and being selected against.
Fig. 3.
Differential propensities for SCVs among the 14 chromosomes of the A and D subgenomes detected in the 1,462 randomly karyotyped cohorts at the 12th selfed generation (S12) of the synthetic allotetraploid wheat, which was depicted by a heatmap (A), all the individuals with SCVs (B), and representative SCVs (rearrangements) (C), identified among the karyotyped cohorts. For visual comparison, the standard FISH/GISH-based karyotype for a bona fide allotetraploid wheat individual (parental additivity) is arranged in the center of (C). The y-axis in (A) denotes the 1,175 individuals in total, with each row depicting one individual’s karyotype. The white arrows in (C) denote rearranged segments between the A and D subgenome chromosomes. For FISH, the 45S ribosomal gene (green) and pAS1 repeat (red) were used as probes. For GISH, the genomic DNA of T. urartu (genome AA) and Ae. tauschii (genome DD) were used as probes, and which generate green and red signals, respectively.
We further scrutinized characteristics of the SCVs, which led to the following observations: 1) as already described above (fig. 3A and B), all 14 chromosomes underwent structural alterations; 2) all discernible SCVs were inter-subgenome rearrangements or translocations (presumbly between homeologous chromosomes); 3) the rearranged segments spanned variable sizes, and which appeared to occur at any foci along a given chromosome; and 4) both terminal and intercalary translocations were detected, which mostly occurred independently but occasionally juxtaposed to each other. Together with the rampant NCVs, these characteristics of SCV could largely explain the massive organismal karyotypic heterogeneity in this propagated allotetraploid population (fig. 1C).
Concurrence of Numerical and Structural Chromosomal Variations
The above results indicated that only small proportions of the karyotypically altered allotetraploid plants contained in isolation NCV (23 out of 1,462 = 1.6%) or SCV (461 out of 1,462 = 31.5%), while the majority (48.8%) harbored both NCV and SCV together (fig. 1C). One apparent cause for the concurrence of both kinds of chromosomal alterations lies in their common origin, that is, both resulted from abnormal meiosis, such as multivalent formation and homeologous recombination, followed by segregation error (to be detailed in later section). However, given that any SCV event initially should have occurred in only one chromosome copy then being assorted randomly during meiotic anaphase, such a high level of concurrency of both NCV and SCV might suggest their functional connectivity. To test this possibility, we conducted the following two aspects of analysis.
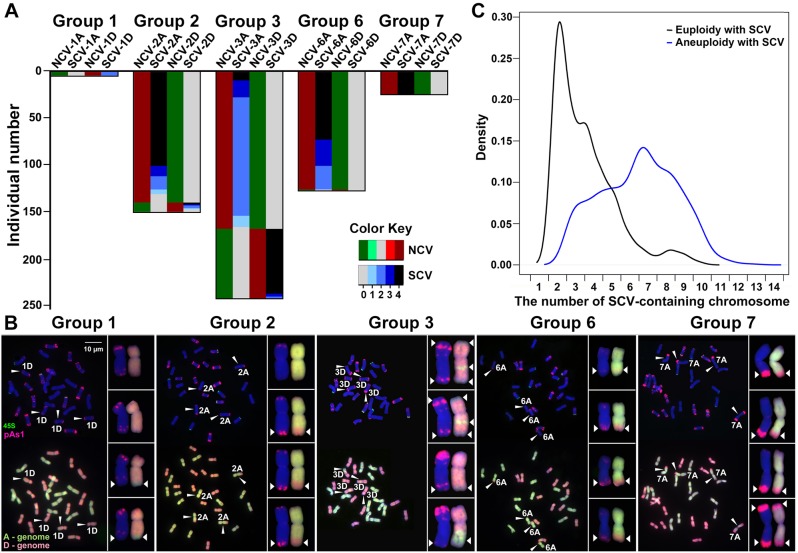
First, we analyzed occurrence of SCV in a subset (n = 544, supplementary table S1, Supplementary Material online) of the 1,462 karyotyped allotetraploid plants, which had karyotypic configurations containing nullisomy-tetrasomy for a given homeologous chromosome pair, that is, with concomitant loss of a pair of homologous chromosomes and gain of an extra pair of its corresponding homeologous chromosomes. The nullisomy–tetrasomy configurations concerned five of the seven homeologous chromosome groups (i.e., groups 1, 2, 3, 6, and 7), because no plant was found to be nullisomic or tetrasomic for homeologous chromosome groups 4 and 5 (fig. 2B). We found that in 92–100% of these plants, from one to all four copies of the tetrasomic chromosome(s) contained one or more translocated segments (of variable sizes) from the alternate subgenome most likely from their homeologous nullisomic chromosomes (fig. 4A and B;supplementary table S2, Supplementary Material online). Given the above consideration (occurrence of SCV and meiotic chromosomal segregation), this observation suggests that the extra homeologous chromosomes (tetrasomy) alone are probably insufficient to functionally compensate for nullisomy of their alternate subgenome counterparts, thus only gametes containing translocated segments (presumably carrying the otherwise missing essential genes or regulatory modules) of the nullisomic chromosomes are viable or functionally normal to generate the next filial generation plants. Notably, at the microscopic resolution, we cannot determine whether the translocated intercalary segments represent unidirectional insertions (i.e., no loss of genetic material from the recipient chromosome) or reciprocal homeologous recombinations, however, the terminal ones are most likely reciprocal (fig. 4B).
Fig. 4.
Concurrence of numerical and structural chromosome variations (NCVs and SCVs). (A) Numbers of plant individuals among a total of 544 (y-axis) showing nullisomic–tetrasomic chromosome configurations for each of the five homeologous chromosome groups, that is, 1, 2, 3, 6, and 7 (no plant was found to show a nullisomic–tetrasomic configuration for homeologous chromosome groups 4 and 5) and the number of plants in each homeologous chromosome group with concurrent NCVs and SCVs (the relative proportions were detailed in supplementary table S2, Supplementary Material online). The color keys were denoted the chromosomal copy number. (B) Examples for each of the five homeologous chromosome groups showing concurrent NCVs and SCVs. One to all four copies of the tetrasomic chromosomes carrying translocated chromosomal segments (most likely from their homeologous counterparts) are denoted by arrowheads and highlighted in the insets. (C) A density plot showing the distribution of restructured chromosomes harbored by all the euploid individuals (euploidy with SCVs, n = 461) versus all the aneuploid individuals (aneuploidy with SCVs, n = 714) identified from the 1,462 karyotyped cohorts, which are significantly different (F-test, P = 3.31E−11). The x-axis refers to the number of SCV-containing chromosomes by each of the individuals belonging to either of the two groups.
Next, we compared the distributions of restructured chromosomes harbored by all the euploid individuals (n = 461) versus all the aneuploid individuals (n = 714) of the 1,462 karyotyped plants, and we detected significant difference (F-test, P = 3.31E−11) between the two distributions (fig. 4C). Specifically, 1) >50% of the euploid individuals harbored <4 restructured chromosomes, with the majority containing only 1–3 and none containing >11 (maximum 10) restructured chromosomes; 2) in contrast, >50% of the aneuploid individuals harbored >5 restructured chromosomes, with 0.8% plants containing >11 (maximum 14) restructured chromosomes (fig. 4C).
Taken both aspects of analysis together, it appears clear that concurrence and concerted perpetuation of the two kinds of chromosome variation, NCV and SCV, in the S12 allotetraploid population, probably cannot be attributed to their common origin alone; instead, the phenomenon is at least in part constrained by their functional connectivity, that is, either unidirectional or mutual functional compensation is required for gamete viability or fitness (i.e., fertility) of the filial generation plants.
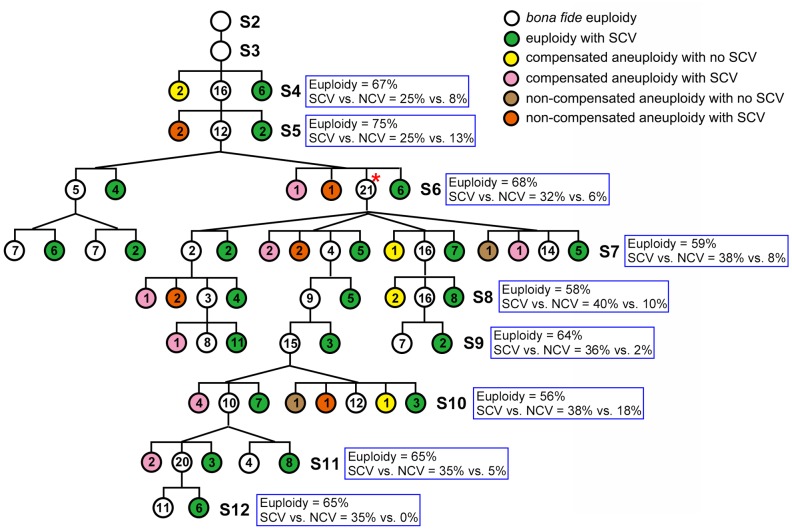
Transgenerational Assay of Numerical and Structural Chromosomal Variations in Euploidy-Derived Progenies
All preceding results concern a cohort of 1,462 random individuals propagated by selfing for 12 consecutive generations without any selection (fig. 1B). A pertinent question to ask is what would be the consequence if bona fide euploidy was purposely selected for at each generation? To address this question, we conducted a transgenerational karyotyping analysis of progenies descended from a single euploid mother individual at each selfed generation for nine consecutive generations (from S4 to S12), starting with an independent euploid founding individual of the same cross to generate the synthetic allotetraploid wheat (fig. 1B). Results showed that although the immediate progenies showed higher proportions of bona fide euploidy than the randomly propagated cohorts (fig. 1C), as expected, the persistent selection for euploidy over nine consecutive generations did not result in a significant increment of euploidy proportion with the progression of generations (variable from 58% to 75% across the generations, fig. 5). Moreover, all types of NCV and SCV that were detected in the randomly propagated plants (fig. 1C) were also seen in these euploidy-derived progenies (fig. 5). Notably, however, the relative proportions of the two types of chromosomal variations, NCVs and SCVs, generated immediately from euploidy in each generation are significantly different, with SCVs being several folds higher than NCVs, 25–40% versus 0–18% across S4–S12, and collectively 33.8% versus 7.8%, (paired Wilcoxon signed-rank test, P < 0.004) (fig. 5). Regardless, results of this pedigree analysis for euploidy-derivatives suggested that karyotypic stabilization towards euploidy cannot be accomplished swiftly based on transgenerational selection for euploidy alone under the standard growing conditions.
Fig. 5.
Karyotypic composition of progenies of bona fide euploidy-descendants (being derived from one or more euploid parental individuals, as indicated by the number of branches) at each of the nine successive organismal generations (from S4 to S12). The six different colored circles represent six distinct karyotypic groups, numbers in the circles denote individual numbers within a given karyotypic group. Frequencies (%) of euploidy and proportions of SCV versus NCV were calculated and indicated at each generation (blue boxes). The bona fide euploid individuals at the S6 generation of this pedigree (marked by an asterisk in red) were used for meiosis study.
Meiotic Chromosomal Abnormality
Both NCVs and SCVs are organismal by nature, that is, somatic NCV and SCV heterogeneity (like those in cancer cells) does not exist in any of the karyotyped tetraploid individuals (fig. 1), including those at different generations of the cohorts for pedigree analysis (fig. 5). It is thus clear that all NCVs and SCVs are due to meiotic abnormality while mitosis is fully normal. To explore the nature of meiotic abnormality, we analyzed the meiotic chromosome behavior of this synthetic allotetraploid wheat by using only bona fide euploid individuals to avoid confounding factors due to preexisting NCVs and SCVs that may affect the meiotic process. We took advantage of the fact that euploid individuals produced the same types and similar extent of NCVs and SCVs in their progenies (fig. 5) as those of the randomly propagated individuals (fig. 1C). We conducted meiotic analysis using pollen mother cells (PMCs) taken from a set of ten bona fide euploid individuals at S6 of the pedigree, because these euploid plants produced all types of karyotypes in the following S7 generation (fig. 5). Results showed that of the 207 well-resolved metaphase I PMCs analyzed, only 122 (58.9%) showed normal chromosome configurations, that is, exclusive homologous pairing and bivalent formation; the rest cells contained one or more types of univalents (30.9% of the cells), hetero-bivalents (5.8% of the cells), trivalent (2.9% of the cells), and quadrivalents (8.2% of the cells) (fig. 6; supplementary table S3, Supplementary Material online). Furthermore, we examined 752 anaphase I PMCs, and found 166 (22.1%) cells containing lagging chromosomes (supplementary table S3, Supplementary Material online). These meiotic abnormalities would inevitably produce gametes containing chromosomal variations in either or both number and structure, and hence, organismal NCVs and/or SCVs, in the filial generation plants.
Fig. 6.
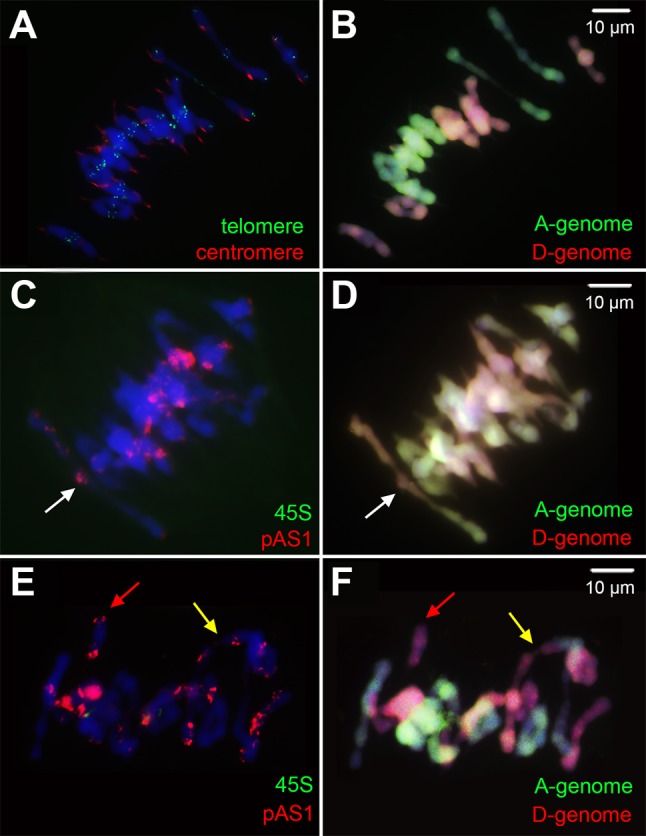
Meiotic chromosome behavior in bona fide euploidy of the allotetraploid wheat at metaphase I. (A) and (B) are examples showing normal chromosome pairing, that is, 14 bivalents were formed via exclusive homologous chromosome pairing. (C–F) are examples showing abnormal chromosome pairing. (A) is a FISH image with telomeric (green) and centromeric (red) probes. (C) and (E) are FISH images probed by the 45S ribosomal gene (green) and pAS1 repeat (red). (B, D, and F) are GISH images showing the A subgenome (green) and D subgenome (red) chromosomes. White arrows indicate hetero-bivalent formation (most likely between homeologous chromosomes). Yellow arrows denote multivalents involving chromosomes of both subgenomes. Red arrows indicate univalents.
Because identity for each of the 14 chromosomes can be unequivocally diagnosed by the combined FISH/GISH analysis when they were as univalents, we further quantified univalent frequency for each chromosome. Results indicated that the 14 chromosomes are dramatically different in univalent frequencies (supplementary fig. S1, Supplementary Material online). Apparently, the meiotic univalent frequencies are discordant with frequencies of organismal NCVs precipitated by the filial generation plants (fig. 2). As the most extreme example, chromosome 4A showed the highest univalent frequency (supplementary fig. S1, Supplementary Material online), however, plants harboring 4A aneuploidies are among the lowest proportions, with no plant containing nullisomy or tetrasomy of this chromosome (fig. 2A and B). Thus, the meiotic results confirmed the above speculation that the precipitated organismal NCVs (and probably also SCVs) are more affected by their variable fitness effects than differential frequencies of occurrence.
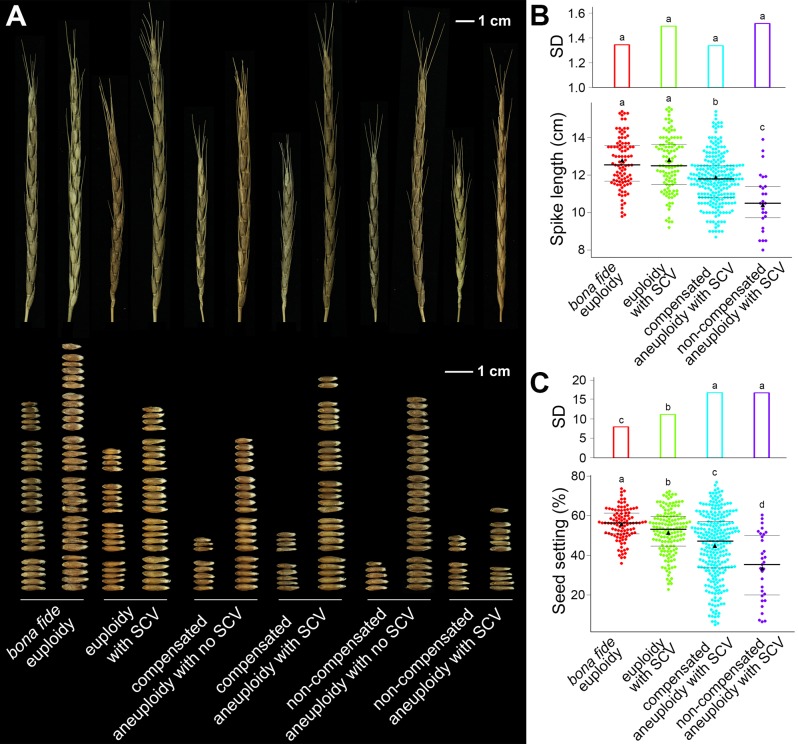
Population-Level Phenotypic Manifestation of the Karyotypic Heterogeneity
In total, ten phenotypic traits were measured for most of the karyotyped plants at the S12 generation, descended from a single S2 euploid founder plant of the synthetic allotetraploid wheat (genome AADD) (fig. 1). The phenotyped traits, including plant height, tiller number, spike length, spikelet number/spike, grain number/spike, spikelet density, grain length, grain width, seed setting, and 100-kernel weight, reflect both growth/development and reproductive fitness. To assess possible associations between the variations in karyotype and phenotype, we categorized the plants into four karyotypic groups such that each group contained sufficient number of individuals to allow statistical analysis. The four groups are: 1) bona fide euploidy (2n = 28), 2) euploidy with SCVs (2n = 28), 3) “compensated aneuploidy” with SCVs (2n = 28), and 4) noncompensated aneuploidy with SCVs (2n ≠ 28) (fig. 1C). We quantified both the mean phenotype (the median) and variability of phenotype (range of standard deviation, SD) of each group for each of the ten quantified traits. As anticipated, for most traits the three groups with karyotypic heterogeneity manifested significantly reduced mean phenotypes (medians) but increased variability (range of SD) of phenotype (fig. 7; supplementary fig. S2, Supplementary Material online; pairwise Wilcoxon signed-rank test, P < 0.05). Differences between bona fide euploidy and euploidy with SCVs suggest that SCVs alone generate extensive phenotypic variations, which could be either exacerbated or mitigated by the concurrent NCVs in the group of “compensated aneuploidy” with SCVs. Actually, the most unexpected result lies in the group of “compensated aneuploidy” with SCVs, as which produced the greatest phenotype variability in most of the quantified traits among the four karyotypic groups (fig. 7; supplementary fig. S2, Supplementary Material online). Collectively, the phenotypic analysis demonstrates that the transgenerationally precipitated organismal karyotypic heterogeneity has profound phenotypic consequences. An additional interesting observation is that when plants of the “compensated aneuploidy” with SCVs group were stratified according to the relative numbers of A-chromosomes vs. D-chromosomes they contain, two traits related reproductive fitness, that is, seed-setting and spikelet density, scaled with concomitant increase of the A-subgenome chromosomes and decrease of D-subgenome chromosomes (supplementary fig. S3, Supplementary Material online). This observation further testifies that the gain of even balanced numbers of additional homeologous chromosomes is insufficient to functionally compensate for loss of homologous ones in this allotetraploid wheat.
Fig. 7.
Phenotypic diversity manifested by different karyotypic groups of the 1,462 individual plants at the 12th selfed generation (S12) of the synthetic allotetraploid wheat. (A) Examples of variation in spike morphology and seed-setting of the different karyotypic groups. In each karyotypic group two of the main spikes of two random individuals were chosen, and all seeds produced by each spike were harvested and shown. (B) Quantification of spike-length among the four karyotypic groups that have sufficient individual numbers to enable the statistical analysis of medians and variability (SD) of this trait. (C) Quantification of seed-setting among the four karyotypic groups that have sufficient individual numbers to enable the statistical analysis of medians and variability (SD) of this trait. Different small letters in (B and C) indicate significant statistical differences (F-test and pairwise Wilcoxon signed-rank test, respectively) between the karyotypic groups in the respective trait. The black triangles indicate the mean of phenotypic data. The black lines indicate the median and the upper- and lower-quartile, respectively.
Discussion
In contrast to the cryptic or low-penetrating phenotypic effects of many, if not most, single-nucleotide variants when they occur individually, karyotypic alteration due to either or both numerical and structural chromosomal changes, often produce larger and immediate phenotypic effects and fitness consequences due to simultaneous changes in the expression of hundreds to thousands of genes resulting from altered gene dosage and/or rejuxtaposition of genomic loci (Otto 2007; Weischenfeldt et al. 2013; Bakhoum and Landau 2017). Complex karyotypic heterogeneity in human somatic cells, usually including intertwined numerical and structural chromosome abnormalities, is a defining feature of many types of cancers, and is increasingly recognized as a principal driver of both tumorigenesis and metastasis (Burrell et al. 2013). Notably, evolved cancer cells usually converge to a near-triploid chromosome constitution with numerous structural changes, and which is often preceded by a WGD event (Dewhurst et al. 2014; Bakhoum and Landau 2017). Not surprisingly, efforts to elucidate the mechanistic basis of karyotypic instability as well as its biological impacts on cancer genesis and progression has become a central theme in biomedical research (Burrell et al. 2013; Bakhoum and Landau 2017).
It has long been established that organismal numerical and structural chromosome changes (karyotype alterations) are large-effect genetic mutations that play significant roles in adaptation, speciation and may lead to saltational evolutionary leaps (Otto 2007). Over evolutionary timescales, many factors are known to underlie chromosome instabilities, such as catastrophic environmental conditions and mutation in critical genes responsible for maintaining genome stability. Among the factors, polyploidy or WGD stands out as the most pervasive en route to large-scale karyotype alterations and hence catalyzes rapid genome evolution. In plants, investigations in diverse taxa have documented that newly formed polyploid genomes are intrinsically unstable with nascent WGDs being often accompanied by extensive karyotypic repatterning (Pires et al. 2004; Han et al. 2005; Otto 2007; Lim et al. 2008; Xiong et al. 2011; Chester et al. 2012; Zhang, Bian, Gou, Zhou et al. 2013; Chester et al. 2015). However, many issues regarding nascent WGD-invoked karyotypic instability remain to be fully explored. For example, the meiotic origin of chromosomal instability, extent of instabilities and their transgenerational inheritance, relationships between structural and numerical chromosome alterations, as well as the phenotypic and fitness consequences of transgenerationally precipitated chromosomal instabilities are all prominent issues that are yet to be investigated in detail. Here, we systematically addressed some of these important issues using an artificially constructed allotetraploid wheat with a genome composition of AADD, and obtained several lines of new insights.
First, we document that massive karyotypic heterogeneity can be precipitated without selection following successive organismal generations from a single euploid founder individual of the synthetic allotetraploid wheat. The karyotypic heterogeneity has cumulated to 777 distinct karyotypes out of the 1,462 random individuals interrogated with regard to either or both numerical and structural chromosome alterations at the microscopic level. Thus, given that only two seeds were randomly selected at each generation to be propagated for the filial generation, the potential for karyotypic heterogeneity in this tetraploid genome combination is virtually unlimited. Nevertheless, it can be imaged that irrespective of population size the transgenerationally precipitated karyotypic diversities are only a fraction of those generated, which happened not to engender severe fitness cost at both the gametophytic and organismal levels (Otto 2007).
Second, we show that the occurrence of both NCVs and SCVs exhibits significant subgenome- and chromosome-bias. For NCVs, subgenome A showed an overly higher propensity towards chromosome gain than loss while subgenome D showed the opposite trend. Since no difference was detected between the A and D subgenome chromosomes in terms of their frequencies to form univalents during meiosis (detailed in later section), these opposing trends between the A and D subgenomes each as a whole in chromosome gain vs. loss is apparently due to their differential effects on fitness of gametes and/or plants. Similarly, the dramatic differences in NCVs among the individual chromosomes are most likely due to the same reason, that is, differential impact of gain or loss of a chromosome copy on fitness of gametes and/or plants. In this respect, it is interesting to note that there is a unique, though mechanistically as yet unelucidated, pattern of genome evolution subsequent to allopolyploidy in the Triticum/Aegilops complex, which concerns the Pivotal-Differential Genome Evolution hypothesis (Kihara 1954; Zohary and Feldman 1962; Kimber and Yen 1988; Feldman and Levy 2012; Mirzaghaderi and Mason 2017). According to this hypothesis, in many allopolyploid species of the Triticum/Aegilops complex, one genome is pivotal and remains unaltered, while the other is modified or differentiated. Three pivotal genomes were recognized, that is, the A, D and U genomes (Kihara 1954; Zohary and Feldman 1962). Thus, an interesting question to ask is what will be the genomic consequence if two pivotal genomes are brought together by allopolyploidization? Here, we show that combination of the A and D genomes into the AADD allotetraploid wheat catalyzes rampant meiotic chromosomal instability and fuels rapid karyotypic evolution, with the two subgenomes manifesting contrasted trends with respect to retention vs. loss of individual chromosomes. It would be interesting to further explore whether these trends are related to relative genome expression dominance (Woodhouse et al. 2014; Wendel et al. 2016) at critical developmental stages and/or tissues by the two subgenomes. For SCVs, subgenome A showed a generally higher frequency than subgenome D, which is understandable because there are more A chromosomes than D chromosomes in the aneuploid individuals, and therefore on a per cell (plant) basis, there are more SCVs events associated with the A chromosomes than the D chromosomes.
Third, we document that the NCVs and SCVs events rarely exist in isolation; instead, they show strong concurrence. A straightforward cause for this phenomenon is due to their common genesis, that is, unstable meiosis. For example, both multivalent and hetero-bivalent formation may cause homeologous recombination (hence SCVs) and mis-segregation (NCVs). This suggests that the elevated occurrence of NCV events that have been precipitated in the nonselected S12 generation cohorts should have been facilitated by SCVs in each of the preceding generations. Specifically, homeologous chromosomes already harboring reciprocal or unidirectional translocated segments are apparently more similar than intact ones, and hence, more prone to form multivalents or hetero-bivalents, and which would result in more segregation errors, and hence NCVs. Retrospectively, NCVs (aneuploidy) are also known to induce or facilitate genetic variations (Sheltzer et al. 2011), and conceivably hypomorphic or loss-of-function mutations affecting genes essential for maintaining meiosis fidelity would inevitably increase the SCVs rate. Thus, this snowballing-like effect of NCVs and SCVs drives the ever-stronger interrelatedness between the two types of chromosome variation with generations. However, it is also clear that common genesis is probably not the only cause for the high concurrence NCVs and SCVs. Specifically, we found that in the subset of the 544 plants that harbor a karyotypic configuration of nullisomy-tetrasomy for a given homeologous chromosome pair, great majority (92–100%) are with one to all four copies of the chromosomes in the tetrasomic state containing translocated segments from the alternate subgenome. Although we cannot ascertain at the cytogenetic resolution if the translocated segments are of the homeologous chromosome origin, this is intuitively the case. Given that any recombination event should have initially occurred in only one chromosome copy, this high incidence of translocated chromosomes being in the tetrasomic state cannot be due to random assortment alone. Rather, it is more plausible that gaining of the extra pair of homeologous chromosomes (tetrasomy) alone are insufficient to functionally compensate for nullisomy of a pair of chromosomes of gamete survival and/or organismal fitness, but this could be amended if one or more segments carrying the essential genes from the otherwise nullisomic chromosomes be retained. Naturally, this would result in the intermingling of NCVs and SCVs seen in these plants. Additional support for the functional connectivity between the NCVs and SCVs events lies in the observation that there exist sharp differences in the occurring frequencies of SCVs between the euploid plants and the aneuploid plants, with the former being strikingly lower than the later, suggesting that apart from the mutual facilitating of NCVs and SCVs, NCVs (i.e., aneuploidies) are more permissive to capacitate SCVs, as already been documented in cancer cells (Dewhurst et al. 2014).
Finally, we show that the phenotypic and fitness landscapes are dramatically altered at the population level by the vast karyotypic heterogeneity precipitated. Consequently, both the mean and the variability of phenotypes were profoundly altered by the karyotypic heterogeneity. A striking observation is that the group of “compensated aneuploidy” with SCVs manifested the largest phenotype variability in majority of the quantified traits. This suggests that the extra homeologous chromosomes clearly cannot fully compensate for the loss of homologous chromosomes at the level phenotypic manifestation. Alternatively but inclusively, the compensated aneuploidy genomic configurations might be more permissive to heterogenic SCVs, or some SCVs have larger effects under some of the “compensated aneuploidy” genomic environments, or the combinations of multiple or all of the above factors. Regardless, the remarkable ongoing as well transgenerationally heritable karyotypic heterogeneity due to allopolyploidy may provide a rich repertoire for adaption to sample under natural settings, thus circumvents the genetic bottleneck that is intrinsically associated with polyploidy, and hence, constraining the founding of new lineages (Han et al. 2015). Our results therefore lend new empirical support to the notion that evolvability can be profoundly enhanced in a polyploid than in its cognate diploid progenitor(s).
Materials and Methods
Plant Materials
The original seeds of the synthetic allotetraploid wheat (genome AADD) at the 2nd selfed generation (S2) together with those of the parental accessions were protracted from Dr. Moshe Feldman (Weizmann Institute of Science, Israel). This allotetraploid wheat (AADD) was produced by intergeneric hybridization between T. urartu (genome AA) and Ae. tauschii (genome DD) followed by colchicine-mediated genome doubling (Ozkan et al. 2001). We used a single S2 euploid seed (determined by FISH/GISH karyotyping, fig. 1A) as the founder to generate the S12 population for study. Specifically, a single euploid plant (karyotyped) was chosen from the S3 generation parented by the S2 euploid plant. Then, from S4 to S11, two plants were randomly chosen at each generation without karyotyping, and this strategy produced 256 plants in total. Finally, six seeds were randomly chosen from each of the 256 S11 plants to generate the S12 population which contained a total of 256×6 = 1,536 plants (fig. 1B). All 1,536 plants were subjected to fluorescence in situ hybridization (FISH) and genomic in situ hybridization (GISH)-based karyotyping. Consequently, 1,462 individuals were successfully karyotyped. Detailed information regarding karyotypes of all the 1,462 individual plants was provided in supplementary table S1, Supplementary Material online.
An independent euploid individual at S2 of this synthetic allotetraploid wheat (AADD) was used to generate another pedigree for which one bona fide euploid individual was arbitrarily chosen at each selfed generation to generate the filial generation (fig. 5). Karyotypic composition of progenies descended from true euploidy was transgenerationally assessed from S4 to S12. Ten bona fide euploid individuals at S6 of this pedigree (denoted by an asterisk in fig. 5) were chosen for meiotic analysis.
Mitotic and Meiotic Karyotyping
Mitotic karyotypes of all the 1,462 individual plants of the S12 population were generated on well-spread metaphase cells of root-tips by the sequential FISH and GISH karyotyping protocol originally reported by Han et al. (2004) and Kato et al. (2004) with minor modifications (Zhang, Bian, Gou, Dong et al. 2013). Specifically, for FISH, 45 S ribosomal DNA (rDNA) was labeled by nick translation with Alexa Fluor 488-5-dUTP (green), Ae. tauschii clone pAS1 (Rayburn and Gill 1986) was labeled with Texas red-5-dCTP (red). For GISH, genomic DNA from T. urartu (AA) and Ae. tauschii (DD) were labeled by nick translation with Alexa Fluor 488-5-dUTP and Texas red-5-dCTP, respectively. Slide denaturation, hybridization, and washing conditions were carried out following the manufacturer’s recommendations (Invitrogen; no. C11397). Slides were examined with an Olympus BX61 fluorescence microscope and digitally photographed. The images were captured using the Olympus IPP software package, and visualized and processed as entirety in Photoshop CS 6.0 version.
For meiotic karyotyping, the same sequential FISH and GISH protocol (Zhang, Bian, Gou, Dong et al. 2013) was used on pollen mother cells of anthers (Armstrong 2013). In total, ten bona fide euploid plants at S6 of the pedigree (fig. 5) were analyzed. Young inflorescences were fixed in Carnoy’s solution, and anthers were dissected to spread meiocytes at meiotic metaphase I and anaphase I. Meiotic chromosome behavior and configurations were tabulated. Representative images were collected using an Olympus BX61 fluorescence microscope equipped with a Retiga-SRV camera and controlled Olympus IPP software. Images were processed as entirety using Adobe Photoshop CS 6.0 version.
Phenotyping
In this study, ten phenotypic traits reflecting both growth/development and reproductive fitness were measured at the fully ripened stage for the karyotyped plant individuals at the S12 population, which included plant height, tiller number, spike length, spikelet number per spike, grain number per spike, spikelet density, grain-length, grain-width, seed setting and 100-kernel weight. Specifically, 1) plant height refers to the length from the base of a given plant to its apical meristem excluding awns; 2) tiller number refers to all tillers grown from each individual plant; 3) spike length refers to length of the main spike of (excluding awn) of the plants; 4) spikelet number per spike refers to the total number of spikelets on the main spike of each plant; 5) grain number per spike refers to the total number of grains produced by the main spike of each plant; 6) spikelet density refers to the value of spikelet number of a given main-spike divided by its length (spike length); 7) grain-length refers to ten randomly chosen seeds from each plant being lined up in an end-to-end manner and the total length was measured on a measuring paper (with scales) and then divided by 10; 8) grain-width refers to ten randomly chosen seeds from each plant being arranged on a measuring paper (with scales) to measure the total width and then divided by 10; 9) seed setting refers to percentage of filled seeds divided by the total number of kernels of five spikes of a given plant; and 10) one-hundred kernel weight refers to the weight of 100 filled seeds after being dried to constant values.
Statistics
The statistical significance of each comparison and graphical analyses were executed in R (version 3.2.2). To estimate the statistical significance of biases between the A and D subgenomes, and among the chromosomes with regard to both NCVs and SCVs, Chi-squared test and/or binomial test were applied with a P value 0.05 as cutoff. To compare the differences in the distributions of all the euploid individuals with SCVs (n = 461) versus all the aneuploid individuals with SCVs (n = 714), F-test was applied. To analyze the frequencies of NCV and SCV from euploidy of the independently constructed pedigree, the paired Wilcoxon signed-rank test was used at the threshold of P value = 0.05. To compare the phenotypic difference between four karyotype groups, the pairwise Wilcoxon signed-rank test was used to analyze the median phenotypes, and F-test was used to test for differences in the ranges of standard deviation, the pairwise Student’s t-test was used among the phenotypic comparison between different A- and D-subgenome combination of compensated aneuploidy with SCV, P < 0.05 as the criterion.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
B.L. conceived and supervised the study; X.W.G., Y.B., H.K.Z., B.W., R.L.L. and B.Z. carried out the experiments; X.W.G., A.Z., J.Z.L., and L.G. analyzed the data; X.W.G. and B.L. interpreted the results and wrote the manuscript, with contributions and approval from all authors.
Supplementary Material
Acknowledgments
We thank Dr Moshe Feldman (The Weizmann Institute of Science, Israel) for providing the initial seeds. This study was supported by the National Key Research and Development Program of China (2016YFD0102003), the National Natural Science Foundation of China (31290210), and the Program for Introducing Talents to Universities (B07017).
References
- Armstrong S. 2013. Spreading and fluorescence in situ hybridization of male and female meiocyte chromosomes from Arabidopsis thaliana for cytogenetical analysis. Methods Mol Biol. 990:3–11. [DOI] [PubMed] [Google Scholar]
- Bakhoum SF, Landau DA.. 2017. Chromosomal instability as a driver of tumor heterogeneity and evolution. Cold Spring Harb Perspect Med. 76:a029611.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Higgins JD, Yant L.. 2015. Meiosis evolves: adaptation to external and internal environments. New Phytol. 2082:306–323. [DOI] [PubMed] [Google Scholar]
- Burrell RA, McGranahan N, Bartek J, Swanton C.. 2013. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 5017467:338–345.http://dx.doi.org/10.1038/nature12625 [DOI] [PubMed] [Google Scholar]
- Chen ZJ. 2007. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol. 58:377–406.http://dx.doi.org/10.1146/annurev.arplant.58.032806.103835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester M, Gallagher JP, Symonds VV, Cruz da Silva AV, Mavrodiev EV, Leitch AR, Soltis PS, Soltis DE.. 2012. Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). Proc Natl Acad Sci U S A. 1094:1176–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester M, Lipman MJ, Gallagher JP, Soltis PS, Soltis DE.. 2013. An assessment of karyotype restructuring in the neoallotetraploid Tragopogon miscellus (Asteraceae). Chromosome Res. 211:75–85. [DOI] [PubMed] [Google Scholar]
- Chester M, Riley RK, Soltis PS, Soltis DE.. 2015. Patterns of chromosomal variation in natural populations of the neoallotetraploid Tragopogon mirus (Asteraceae). Heredity (Edinb) 1143:309–317.http://dx.doi.org/10.1038/hdy.2014.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhurst SM, McGranahan N, Burrell RA, Rowan AJ, Gronroos E, Endesfelder D, Joshi T, Mouradov D, Gibbs P, Ward RL et al. , 2014. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov. 42:175–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion-Cote AM, Symonova R, Rab P, Bernatchez L.. 2015. Reproductive isolation in a nascent species pair is associated with aneuploidy in hybrid offspring. Proc Biol Sci. 2821802:20142862.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis DE, Soltis PS, Wendel JF.. 2008. Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet. 42:443–461. [DOI] [PubMed] [Google Scholar]
- Dubcovsky J, Dvorak J.. 2007. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 3165833:1862–1866.http://dx.doi.org/10.1126/science.1143986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvořák J. 1976. The Relationship between the genome of Triticum urartu and the a and B genomes of Triticum aestivum. Can J Genet Cytol. 182:371–377. [Google Scholar]
- El Baidouri M, Murat F, Veyssiere M, Molinier M, Flores R, Burlot L, Alaux M, Quesneville H, Pont C, Salse J.. 2017. Reconciling the evolutionary origin of bread wheat (Triticum aestivum). New Phytol. 2133:1477–1486. [DOI] [PubMed] [Google Scholar]
- Feldman M, Levy AA.. 2012. Genome evolution due to allopolyploidization in wheat. Genetics 1923:763–774.http://dx.doi.org/10.1534/genetics.112.146316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M, Levy AA.. 2009. Genome evolution in allopolyploid wheat—a revolutionary reprogramming followed by gradual changes. J Genet Genomics 369:511–518. [DOI] [PubMed] [Google Scholar]
- Feldman M, Lupton FGH, Miller TE.. 1995. Wheats In: Smartt J, Simmonds NW, editors. Evolution of crop plants. 2nd ed.London: Longman Scientific. [Google Scholar]
- Geiser C, Mandakova T, Arrigo N, Lysak MA, Parisod C.. 2016. Repeated whole-genome duplication, karyotype reshuffling, and biased retention of stress-responding genes in Buckler Mustard. Plant Cell 281:17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han FP, Fedak G, Guo WL, Liu B.. 2005. Rapid and repeatable elimination of a parental genome-specific DNA repeat (pGc1R-1a) in newly synthesized wheat allopolyploids. Genetics 1703:1239–1245.http://dx.doi.org/10.1534/genetics.104.039263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han FP, Liu B, Fedak G, Liu ZH.. 2004. Genomic constitution and variation in five partial amphiploids of wheat–Thinopyrum intermedium as revealed by GISH, multicolor GISH and seed storage protein analysis. Theor Appl Genet. 1095:1070–1076. [DOI] [PubMed] [Google Scholar]
- Han TS, Wu Q, Hou XH, Li ZW, Zou YP, Ge S, Guo YL.. 2015. Frequent introgressions from diploid species contribute to the adaptation of the tetraploid Shepherd's purse (Capsella bursa-pastoris). Mol Plant. 83:427–438.http://dx.doi.org/10.1016/j.molp.2014.11.016 [DOI] [PubMed] [Google Scholar]
- Hollister JD. 2015. Polyploidy: adaptation to the genomic environment. New Phytol. 2053:1034–1039. [DOI] [PubMed] [Google Scholar]
- Jiao YN, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang HY, Soltis PS et al. , 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 4737345:97–100. [DOI] [PubMed] [Google Scholar]
- Kato A, Lamb JC, Birchler JA.. 2004. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc Natl Acad Sci U S A. 10137:13554–13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara H. 1954. Considerations on the evolution and distribution of Aegilops species based on the analyser-method. Cytologia 194:336–357.http://dx.doi.org/10.1508/cytologia.19.336 [Google Scholar]
- Kilian B, Ozkan H, Deusch O, Effgen S, Brandolini A, Kohl J, Martin W, Salamini F.. 2007. Independent wheat B and G genome origins in outcrossing Aegilops progenitor haplotypes. Mol Biol Evol. 241:217–227. [DOI] [PubMed] [Google Scholar]
- Kimber G, Yen Y.. 1988. Analysis of pivotal-differential evolutionary patterns. Proc Natl Acad Sci U S A. 8523:9106–9108.http://dx.doi.org/10.1073/pnas.85.23.9106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KY, Soltis DE, Soltis PS, Tate J, Matyasek R, Srubarova H, Kovarik A, Pires JC, Xiong Z, Leitch AR.. 2008. Rapid chromosome evolution in recently formed polyploids in Tragopogon (Asteraceae). PLoS ONE 310:e3353.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A, Bomblies K.. 2016. Meiosis in autopolyploid and allopolyploid Arabidopsis. Curr Opin Plant Biol. 30:116–122.http://dx.doi.org/10.1016/j.pbi.2016.02.004 [DOI] [PubMed] [Google Scholar]
- Mason AS, Batley J.. 2015. Creating new interspecific hybrid and polyploid crops. Trends Biotechnol. 338:436–441.http://dx.doi.org/10.1016/j.tibtech.2015.06.004 [DOI] [PubMed] [Google Scholar]
- McClintock B. 1984. The significance of responses of the genome to challenge. Science 2264676:792–801.http://dx.doi.org/10.1126/science.15739260 [DOI] [PubMed] [Google Scholar]
- Mirzaghaderi G, Mason AS.. 2017. Revisiting pivotal-differential genome evolution in wheat. Trend Plant Sci. 228:674–684.http://dx.doi.org/10.1016/j.tplants.2017.06.003 [DOI] [PubMed] [Google Scholar]
- Otto SP. 2007. The evolutionary consequences of polyploidy. Cell 1313:452–462.http://dx.doi.org/10.1016/j.cell.2007.10.022 [DOI] [PubMed] [Google Scholar]
- Otto SP, Whitton J.. 2000. Polyploid incidence and evolution. Annu Rev Genet. 34:401–437.http://dx.doi.org/10.1146/annurev.genet.34.1.401 [DOI] [PubMed] [Google Scholar]
- Ozkan H, Levy AA, Feldman M.. 2001. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell. 138:1735–1747.http://dx.doi.org/10.1105/tpc.13.8.1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisod C, Salmon A, Zerjal T, Tenaillon M, Grandbastien MA, Ainouche M.. 2009. Rapid structural and epigenetic reorganization near transposable elements in hybrid and allopolyploid genomes in Spartina. New Phytol. 1844:1003–1015. [DOI] [PubMed] [Google Scholar]
- Pires JC, Zhao J, Schranz ME, Leon EJ, Quijada PA, Lukens LN, Osborn TC.. 2004. Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae). Biol J Linn Soc. 824:675–688. [Google Scholar]
- Rayburn AL, Gill BS.. 1986. Molecular identification of the D-genome chromosomes of wheat. J Hered. 774:253–255.http://dx.doi.org/10.1093/oxfordjournals.jhered.a110231 [Google Scholar]
- Schoenfelder KP, Fox DT.. 2015. The expanding implications of polyploidy. J Cell Biol. 2094:485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, Humpton TJ, Brito IL, Hiraoka Y, Niwa O, Amon A.. 2011. Aneuploidy drives genomic instability in yeast. Science 3336045:1026–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis PS, Marchant DB, Van de Peer Y, Soltis DE.. 2015. Polyploidy and genome evolution in plants. Curr Opin Genet Dev. 35:119–125.http://dx.doi.org/10.1016/j.gde.2015.11.003 [DOI] [PubMed] [Google Scholar]
- Stebbins GL. 1950. Chapter XII: the karyotype In: Dunn LC, editor. Variation and evolution in plants. New York: Columbia University Press; p. 442–472. [Google Scholar]
- Van de Peer Y, Mizrachi E, Marchal K.. 2017. The evolutionary significance of polyploidy. Nat Rev Genet. 187:411–424.http://dx.doi.org/10.1038/nrg.2017.26 [DOI] [PubMed] [Google Scholar]
- Weischenfeldt J, Symmons O, Spitz F, Korbel JO.. 2013. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat Rev Genet. 142:125–138. [DOI] [PubMed] [Google Scholar]
- Wendel JF. 2015. The wondrous cycles of polyploidy in plants. Am J Bot. 10211:1753–1756. [DOI] [PubMed] [Google Scholar]
- Wendel JF, Jackson SA, Meyers BC, Wing RA.. 2016. Evolution of plant genome architecture. Genome Biol. 17:37.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse MR, Cheng F, Pires JC, Lisch D, Freeling M, Wang X.. 2014. Origin, inheritance, and gene regulatory consequences of genome dominance in polyploids. Proc Natl Acad Sci U S A. 11114:5283–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KM, Arnold B, Xue K, Surinova M, O’Connell J, Bomblies K.. 2015. Selection on meiosis genes in diploid and tetraploid Arabidopsis arenosa. Mol Biol Evol. 324:944–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong ZY, Gaeta RT, Pires JC.. 2011. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc Natl Acad Sci U S A. 10819:7908–7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L, Hollister JD, Wright KM, Arnold BJ, Higgins JD, Franklin FCH, Bomblies K.. 2013. Meiotic adaptation to genome duplication in Arabidopsis arenosa. Curr Biol. 2321:2151–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HK, Bian Y, Gou XW, Dong YZ, Rustgi S, Zhang BJ, Xu CM, Li N, Qi B, Han FP et al. , 2013. Intrinsic karyotype stability and gene copy number variations may have laid the foundation for tetraploid wheat formation. Proc Natl Acad Sci U S A. 110:19466–19471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HK, Bian Y, Gou XW, Zhu B, Xu CM, Qi B, Li N, Rustgi S, Zhou H, Han FP et al. , 2013. Persistent whole-chromosome aneuploidy is generally associated with nascent allohexaploid wheat. Proc Natl Acad Sci U S A. 110:3447–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohary D, Feldman M. 1962. Hybridization between amphidiploids and the evolution of polyploids in the wheat (Aegilops-Triticum) group. Evolution 16:44–61. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.