Abstract
MicroRNA (miRNA) sponges are vital components of posttranscriptional gene regulation. Yet, only a limited number of miRNA sponges have been identified. Here, we show that the recently evolved noncoding tumor suppressor transcript, antisense RNA to TP73 gene (TP73-AS1), functions as a natural sponge of human-specific miRNA miR-941. We find unusually nine high-affinity miR-941 binding sites clustering within 1 kb region on TP73-AS1, which forms miR-941 sponge region. This sponge region displays increased sequence constraint only in humans, and its formation can be traced to the tandem expansion of a 71-nt-long sequence containing a single miR-941 binding site in old world monkeys. We further confirm TP73-AS1 functions as an efficient miR-941 sponge based on massive transcriptome data analyses, wound-healing assay, and Argonaute protein immunoprecipitation experiments conducted in cell lines. The expression of miR-941 and its sponge correlate inversely across multiple healthy and cancerous tissues, with miR-941 being highly expressed in tumors and preferentially repressing tumor suppressors. Thus, the TP73-AS1 and miR-941 duo represents an unusual case of the extremely rapid evolution of noncoding regulators controlling cell migration, proliferation, and tumorigenesis.
Keywords: microRNA sponge, young noncoding RNA, co-evolution, tumorigenesis
Introduction
miRNAs are well-recognized posttranscriptional regulators that fine-tune the expression of the majority of mammalian protein-coding genes (PCG) and long noncoding RNA (lncRNA). The main function of miRNAs is to guide RNA-Induced Silencing Complex (RISC) to transcript-containing complementary binding sites (Bartel 2004; Friedman et al. 2009; Lewis et al. 2003; Yoon et al. 2014). Some of the miRNA target transcripts have been recently recognized to function as miRNA sponges—transcripts that modulate the repression of other targets by sequestering RISC bound miRNAs (Ebert and Sharp 2010; Salmena et al. 2011; Thomson and Dinger 2016).
Whereas miRNA sponges containing single miRNA binding sites have been described (Wang et al. 2013), the majority of efficient miRNA sponge transcripts contain multiple high-affinity miRNA binding sites and are expressed at high levels (Ebert and Sharp 2010; Hansen et al. 2013; Memczak et al. 2013; Rani et al. 2016; Thomson and Dinger 2016). At present, 64 and 117 miRNA sponge transcripts with experimental evidence are documented in humans and mice, respectively (Wang et al. 2015). Examples include recently evolved pseudogene PTENP1, lncRNAs linc-RoR, and lncND specific to primates, as well as transcripts well conserved among vertebrate species: circular RNAs (circRNAs) CDR1as and sry RNA (Hansen et al. 2013; Memczak et al. 2013; Poliseno et al. 2010; Rani et al. 2016; Wang et al. 2013).
Functionally, miRNA sponges were shown to play diverse roles. The PTENP1 transcript regulates tumor suppressor gene (TSG) PTEN’s expression by sequestering miRNAs from broadly conserved miR-17 and miR-19 families in several cancers (Poliseno et al. 2010). linc-RoR plays roles in the control of self-renewal and pluripotency maintenance of human embryonic stem cells by regulating pivotal pluripotent cell-specific factors SOX2, OCT4, and NANOG by sequestering conserved mammalian miR-145 (Wang et al. 2013). lncND, which contains 16 binding sites for conserved mammalian miR-143-3p acquired on the evolutionary branch leading to old world monkeys (OWMs), apes, and humans, mediates the Notch signaling pathway during neuronal development (Rani et al. 2016). CDR1as, the antisense transcript to cerebellar degeneration-related protein 1, which contains 72 miR-7 binding sites, was shown to regulate midbrain size in zebrafish and sensorimotor gating and synaptic transmission in mice (Memczak et al. 2013; Piwecka et al. 2017). Notably, both miR-7 and 72 miR-7 binding sites within CDR1as are conserved among most vertebrate species (Memczak et al. 2013). sry RNA transcribed from sex-determining region Y was shown to sequester miR-138 with exact functions remaining to be elucidated. While miR-138 is conserved among vertebrate species, sry RNA contains 16 miR-138 binding sites in mice (Hansen et al. 2013) and one site in its human homolog, suggesting different roles in humans and rodents (Guo et al. 2014).
The abovementioned examples, as well as other known sponge transcripts target broadly conserved miRNA. Yet, miRNA have rapid evolutionary dynamics characterized by high birth and death rates (Lu et al. 2008; Meunier et al. 2013) resulting in a number of prominent species-specific members, including human-specific miRNA miR-941 (Hu et al. 2012). Here, we present analytical and experimental evidence that a recently evolved lncRNA transcript annotated as antisense RNA to TP73 gene (TP73-AS1) functions as a sponge of this human-specific miRNA.
Results
Computational Search for miR-941-Sponge Genes
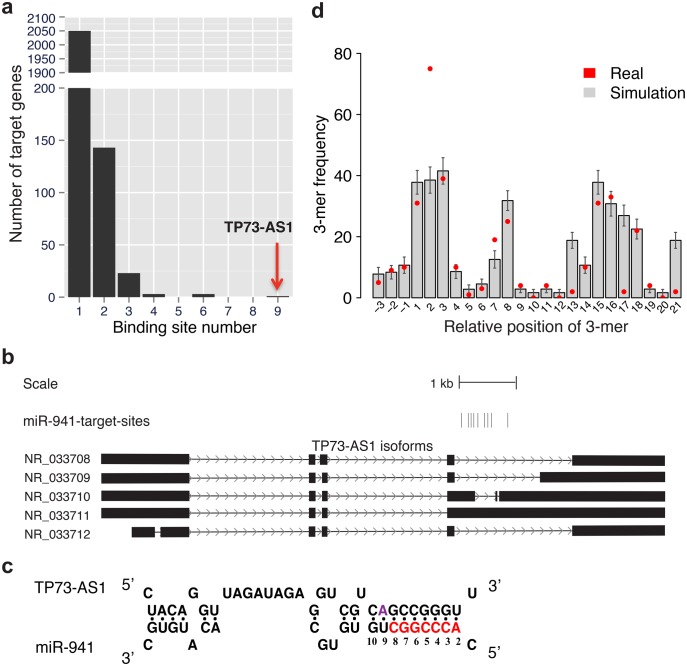
To identify putative miR-941-sponge genes, we screened human noncoding transcripts (NCTs) including 36,687 linear transcripts and 36,019 circRNAs. For this, we compiled known linear NCTs and identified putative circRNA using Ribo-minus sequencing data from four human prefrontal cortex (PFC) samples generated for this study and the re-analysis of 54 published Ribo-minus sequencing data sets (supplementary table S1, supplementary Data S1, Supplementary Material online). We identified miR-941 binding sites residing within NCTs using TargetScan (Grimson et al. 2007) with no site conservation requirement. Out of 72,706 NCTs examined in our study, 70,472 (96.9%) contained no miR-941 binding site and 2,050 (2.8%) contain one. Remarkably, a linear transcript annotated as antisense RNA to TP73 gene (TP73-AS1, also known KIAA0495 or PDAM) contained nine miR-941 binding sites (fig. 1a, supplementary table S2, Supplementary Material online). Finding nine binding sites within a 6.4-kb-long transcript is highly unexpected, based on the binding sites identification within shuffled NCTs sequences maintaining the original dinucleotide frequency (maximum N = 6, simulations, P < 0.001) and suggests that TP73-AS1 might function as a miR-941-sponge.
Fig. 1.
Sequence features of miR-941 sponge gene TP73-AS1. (a) Numbers of miR-941 binding sites within noncoding putative target transcripts. (b) Location of nine miR-941 binding sites within the TP73-AS1 gene shown by vertical bars. The five TP73-AS1 isoforms are based on RefSeq annotations: exons—boxes, introns—lines. (c) The schematic representation of base-pairing interaction between miR-941 and TP73-AS1. The interaction features the 9-nt-long extended seed region match, as well as the presence of adenosine at position 9 of the target site (purple). The canonical seed region is shown in red. (d) The enrichment of the miR-941 subseed motif in the vicinity of miR-941 binding sites. The x-axis shows the start position of each 3-mer motif relative to the miR-941 mature sequence. The background frequency of each 3-mer motif was estimated by 1,000 permutations of target noncoding transcript sequences retaining the same dinucleotide composition.
To validate our miRNA sponge identification method, we applied it to search for sponge transcripts for other human miRNAs. Using 505 high confidence human miRNA families constructed by intersecting annotation from miRbase, mirDB, and mirGeneDB, we identified 152 putative sponge transcripts for 121 miRNA families (see Materials and Methods, supplementary Data S2, Supplementary Material online). Notably, these putative miRNA sponges included both known human miRNA sponge transcripts containing high numbers of miRNA binding sites, Cdr1as and ZNF91 (Guo et al. 2014; Memczak et al. 2013). Similarly, our method classified well known mouse sponge transcript, Sry RNA, as a miRNA sponge (Hansen et al. 2013).
Within TP73-AS1, nine miR-941 binding sites are not distributed uniformly, but cluster within an ∼900-nt-long region, which forms miR-941 sponge region (fig. 1b). Each of the nine binding sites contains a 9-nt-long region of perfect complementarity to positions 2–10 at the 5′ end of miR-941 (fig. 1c, supplementary fig. S1, Supplementary Material online). The 9-nt-long target sites are more efficient in miRNA target repression experiments than the canonical 7- or 8-nt-long target sites (Hafner et al. 2010; Stark et al. 2003). Moreover, two additional sequence features further enhance the predicted binding efficiency of TP73-AS1: adenosine at position 9 of the target site associated with the most efficient 9-mer target site type (Lewis et al. 2005), and >2-fold overrepresentation of miR-941 subseed motif (a 3-mer sequence, GGT, matching to miR-941 positions 2–4) in the vicinity of miR-941 binding sites associated with an AGO2 initial target site search (Chandradoss et al. 2015; fig. 1c and d, supplementary fig. S2, Supplementary Material online). The enrichment of each 3-mer sequence was estimated by comparing its frequency with the chance frequency estimated by shuffling TP73-AS1 sponge region sequence, while maintaining the same dinucleotide frequency, 1,000 times. Notably, the miR-941 subseed motif is the only 3-mer sequence overrepresented in the vicinity miR-941 binding sites (Binomial test, Bonferroni corrected P < 0.0001, fig. 1d). Taken together, these sequence features result in the significantly higher binding affinity of TP73-AS1 compared with the other miR-941 target genes (Wilcoxon rank sum test, P < 0.01, supplementary fig. S3, Supplementary Material online), further indicating that the TP73-AS1 transcript represents a naturally occurring miR-941 sponge.
Evolution of TP73-AS1 Sponge Sequence
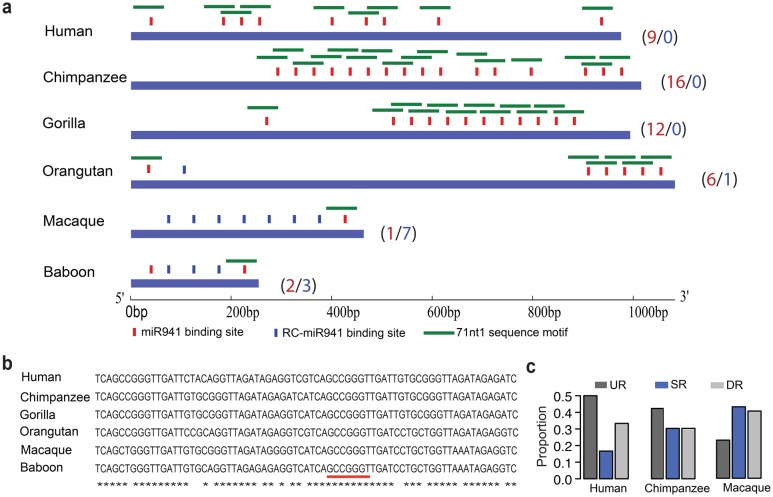
The emergence of miR-941 binding sites in TP73-AS1 transcripts precedes the evolution of miR-941. Tandem repeats containing a miR-941 binding site in either direct or reverse configuration (the reverse complementary of miR-941 binding site) are present within a specific region of TP73-AS1 in humans, apes and OWMs (fig. 2a). OWMs, such a macaques and baboons, have one and two miR-941 binding sites respectively, orangutans have six, gorillas have 12, chimpanzees have 16, and humans have 9. The increase of the miR-941 binding site number on the human and ape lineages can be traced to the tandem expansion of a 71-nt-long sequence containing a single miR-941 binding site in OWMs (fig. 2a and b).
Fig. 2.
The evolution of TP73-AS1 sponge region. (a) Location of predicted miR-941 binding sites within human TP73-AS1 sponge region and within the orthologous regions in apes and OWMs. The vertical bars and the numbers represent miR-941 binding sites (red) and the reverse complementary sequences of miR-941 binding site (blue). The horizontal green bars show the 71 nt-long repetitive sequence containing the miR-941 binding site. (b) The alignment of the 71 nt-long repetitive sequence unit among human, ape and OWM genomes. The red underscore indicates the region complementary to miR-941 canonical seed. Asterisks represent conserved nucleotides. (c) The SNP proportion within 976-nt-long sponge sequence region (SR), 976-nt-long upstream region (UR), and 976-nt-long downstream region (DR) in humans, chimpanzees, and macaques.
Notably, the 1-kb-long genomic region encompassing the “sponge” part of TP73-AS1 containing all miR-941 binding sites shows increased sequence conservation only in humans. Specifically, an analysis of common single nucleotide polymorphisms (SNPs) within the region and in the flanking 1-kb-long sequences showed a clear reduction of SNP occurrence in humans, but not in chimpanzees and macaques (fig. 2c).
Sequence and Expression of TP73-AS1
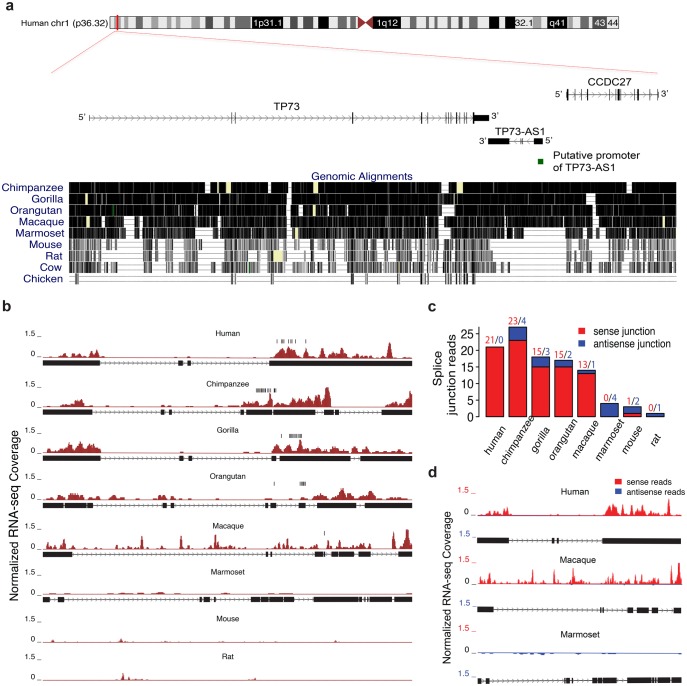
In the human genome, TP73-AS1 is annotated as a 6, 409-nt-lncRNA located between two PCGs, TP73 and CCDC27, in the p36.32 region of chr1 (fig. 3a). TP73-AS1 overlaps with TP73 in a tail-to-tail configuration with an overlap length of 216 nt. Our analysis of ribosome profiling data from GWIPS-viz (Michel et al. 2014) further confirmed that TP73-AS1 does not contain any active open reading frames (supplementary fig. S4, Supplementary Material online).
Fig. 3.
The sequence and RNA-seq read profile of TP73-AS1 transcript across species. (a) The sequence conservation of the genomic region containing TP73-AS1, CCDC27, and TP73 based on the genome alignment across 10 species. Each gene is represented by its longest isoform. The green box shows the location of the putative promoter region of TP73-AS1. (b) TP73-AS1 RNA-seq read profile shown as normalized RNA-seq coverage (dark red) and orthologous transcript structure. The orthologous regions in mice and rats are drawn based on genomic synteny. The vertical black bars indicate the location of the predicted miR-941 binding sites. (c) TP73-AS1 transcription in the eight species calculated based on reads that cross the splice junction within TP73-AS1 transcript in either sense (red) or antisense (blue) orientation. (d) TP73-AS1 RNA-seq read profile in humans, macaques, and marmosets, estimated using normalized RNA-seq coverage of sense and antisense reads based on strand-specific RNA-seq data.
While both TP73 and CCDC27 are conserved among mammals, sequence orthologs of human TP73-AS1 can be identified in apes, OWMs, and new world monkeys (NWMs), but not in mice, rats, cows and chickens (fig. 3a, supplementary fig. S5, Supplementary Material online). Furthermore, an examination of the putative promoter region of TP73-AS1 inferred using CpG-islands and chromatin states information(Rosenbloom et al. 2015) revealed the absence of the promoter region in NWMs and nonprimate species (fig. 3a, supplementary fig. S6, Supplementary Material online). This implies that transcription of TP73-AS1 is restricted to humans, apes and OWMs. Our assessment of transcriptional activity at the TP73-AS1 region based on sequencing reads from chimpanzees, gorillas, orangutans, macaques, marmosets, mice, and rats, as well as reads mapped to splice junctions and strand-specific reads, confirmed this notion (fig. 3b–d).
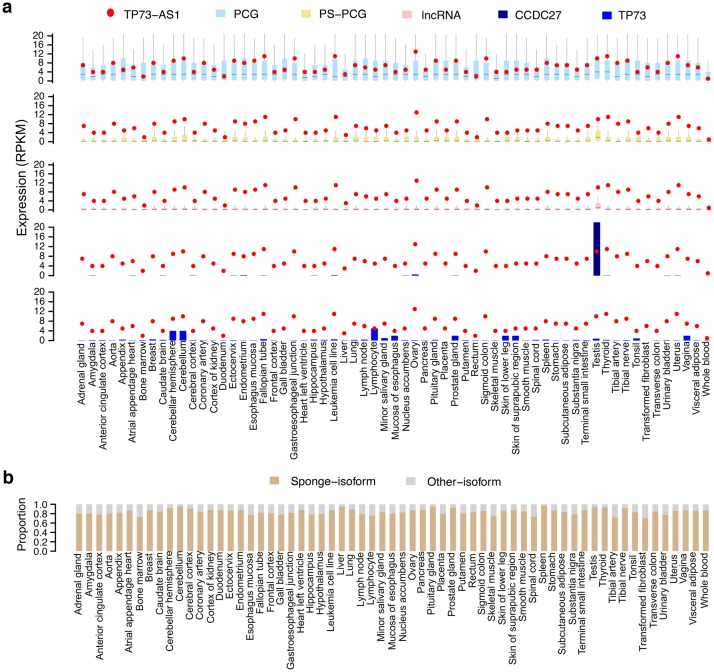
Although TP73-AS1 is a recently evolved lncRNA, it is ubiquitously and highly expressed across 63 human tissues (Consortium 2013; Petryszak et al. 2016; Uhlen et al. 2015; fig. 4a). In most tissues, TP73-AS1 expression is higher than the expression of almost all lncRNAs and primate-specific PCGs, as well as the median expression of PCGs detected in a tissue (fig. 4a). Furthermore, the TP73-AS1 expression is on average 50 times higher than the expression of its neighboring genes, TP73 and CCDC27 (fig. 4a). Notably, even though there are five annotated TP73-AS1 isoforms, an isoform containing all nine miR-941 binding sites constitutes nearly 80% of all TP73-AS1 expression in human tissues (fig. 4b).
Fig. 4.
TP73-AS1 expression in 63 human tissues. (a) The expression levels of TP73-AS1, protein-coding genes (PCG), primate-specific protein-coding genes (PS-PCG), long noncoding RNAs (lncRNA), and neighboring genes CCDC27 and TP73. (b) The proportion of reads mapping within TP73-AS1 isoforms containing miR-941 binding sites (sponge-isoform) and the other isoforms (other isoforms).
TP73-AS1 Is an Authentic Target of miR-941 in Human Tissues and Cells
Because of the inhibitory function of miRNA, its expression commonly inversely correlates with the expression of linear sponge transcripts (Thomson and Dinger 2016). To assess whether this is the case for miR-941 and TP73-AS1, we conducted cell line transfection and immunoprecipitation experiments, and we analyzed existing transcriptome data.
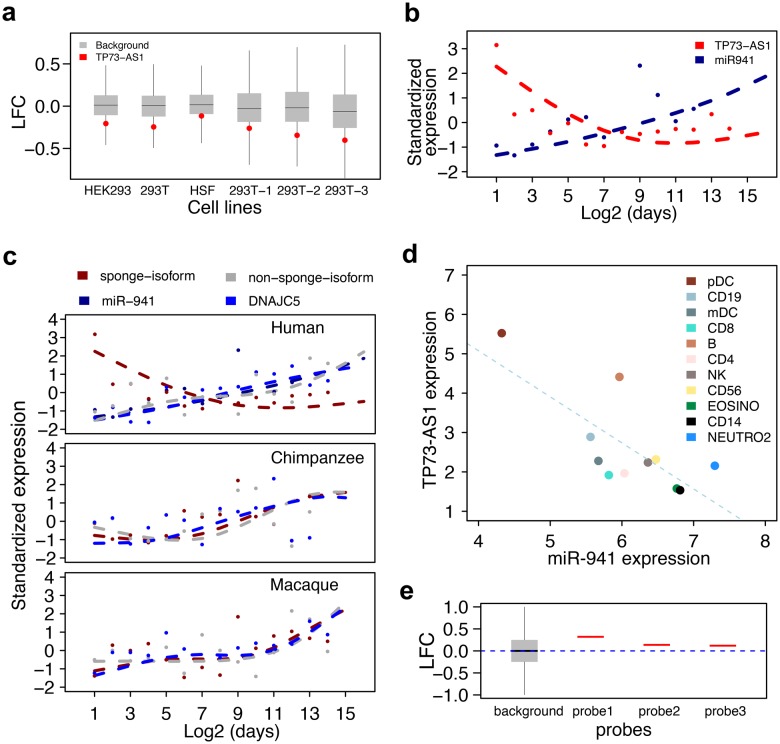
TP73-AS1 is among the predicted targets of miR-941, which was experimentally verified in transfection experiments in three human cell lines (HSF, HEK293, and 293T) measured using microarrays (Hu et al. 2012). To verify this, we transfected 293T cell line with miR-941 duplex or mock control in triplicates and assessed TP73-AS1 expression using RNA-seq. Compared with the mock, TP73-AS1 expression decreased significantly 24 h after miR-941 duplex transfection (fig. 5a, negative binomial test, P < 0.001, FDR < 0.1%). Thus, TP73-AS1 expression is consistently negatively regulated by miR-941 in various human cell lines.
Fig. 5.
TP73-AS1 is an authentic miR-941 target. (a) The log2-transformed gene expression fold-change (LFC) between the miR-941 duplex and mock duplex transfection in human cells lines. The red dots stand for the TP73-AS1 transcript and the gray boxes stand for nonmiR941 target transcripts. (b) The expression of TP73-AS1 and miR-941 in human PFC at different ages. (c) The expression of miR-941, DNAJC5, and TP73-AS1 isoforms containing and not containing miR-941 binding sites (sponge-isoform and nonsponge-isoform) in the PFC of humans, chimpanzees, and macaques at different ages. In (b) and (c), dots indicate individuals, lines represent cubic spline curves fitted with four degrees of freedom. (d) The expression of TP73-AS1 and miR-941 in 11 human immune cell types. (e) The log2-transformed gene expression fold-change (LFC) between AGO2-IP and IgG control after miR-941 duplex transfection. The red bars stand for three TP73-AS1 microarray probes and the gray box stands for probes complementary to nonmiR941 target transcripts.
We further examined miR-941 and TP73-AS1 expression relationship in vivo using data from the human PFC (Hu et al. 2014) and immune cells (Allantaz et al. 2012). Mature miR-941 is only expressed in humans (Hu et al. 2012), while expression of its host gene, DNAJC5, is conserved in chimpanzees and macaques. In human PFC, miR-941 expression rises and TP73-AS1 expression decreases over the lifespan (Pearson correlation, r = −0.61, P < 0.01, fig. 5b). Notably, the TP73-AS1 expression decrease is only found in humans and is restricted to isoform carrying miR-941 binding sites (fig. 5c).
Similarly, expression miR-941 and TP73-AS1 transcript inversely correlated across 11 types of human immune cells (Pearson correlation r = −0.74, P < 0.01, fig. 5d).
To assess whether TP73-AS1 physically interacts with miR-941 in RISC, we conducted AGO2 immunoprecipitation (AGO2-IP) experiment in 293T cell line transfected with miR-941 duplex. A microarray-based analysis of IP products showed consistent enrichment of TP73-AS1 in AGO2 complexes compared with IgG control for all three microarray probes specific to TP73-AS1 (fig. 5e), indicating direct interaction of TP73-AS1 and miR-941 in RISC. Altogether, the above-mentioned results strongly suggest that TP73-AS1 is an authentic target of miR-941.
TP73-AS1 Expression Blocks miR-941-Driven Migration Inhibition
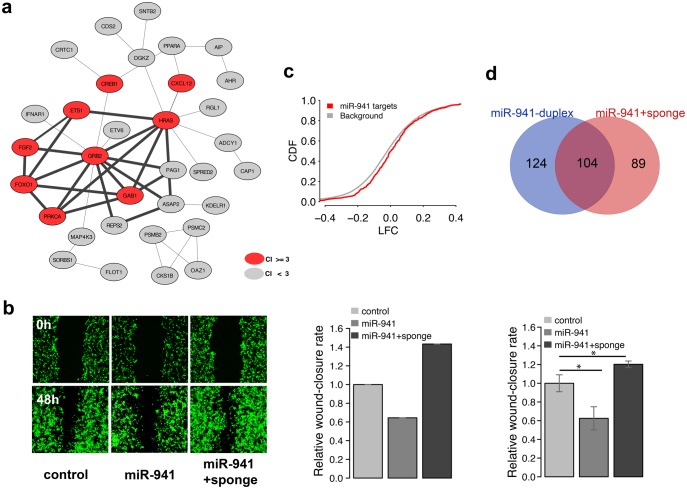
The overexpression of miR-941 was shown to cause significant inhibition of cell migration in hepatocellular, colorectal, and gastric cancer cells (Kim et al. 2014; Zhang et al. 2014). Our analysis of protein–protein interaction (PPI) networks based on experimentally verified miR-941 targets similarly singled out a network module significantly enriched in genes linked to the “cell migration” term using CoCiter (Qiao et al. 2013; fig. 6a, permutations, P < 0.005, supplementary table S4, Supplementary Material online). Furthermore, the “cell migration” term enrichment was 20-times greater among module genes compared with the other network genes (fig. 6a, Fisher’s exact test, P < 0.001, odds ratio = 20.9, supplementary table S3, Supplementary Material online).
Fig. 6.
TP73-AS1 expression blocks miR-941-driven migration inhibition. (a) The bold edges indicate protein–protein interaction network module constructed based on experimentally verified miR-941 targets. The red nodes indicate genes linked to cell migration terms by publications: citation index (CI) ≥ 3. (b) The microscope images of cell density in a wound healing assay (left) and quantified wound efficiency (middle and right) after transient transfection with empty SD1266 vector (control), vector carrying miR-941 cluster region containing seven miR-941 precursors (miR-941), and vector carrying both miR-941 cluster region and TP73-AS1 sponge region (miR-941 + sponge). The middle and right panels represent the results of two independent transfection experiments, the right one conducted in triplicates. The significance of wound closure difference estimated using a t-test is shown above the bars: * P < 0.05. (c) Cumulative distribution of log2-transformed gene expression fold-change (LFC) values for genes containing predicted miR-941 target sites (red) and all other expressed genes (grey) after transfection with control and miR941 + sponge vectors into 293T cells. The y-axis shows the cumulative distribution function (CDF) of LFC distribution. (d) The overlap between miR-941 target genes (purple) identified in miR-941 duplex transfection experiment (blue) and miR941 + sponge vector transfection experiment (red).
To assess whether TP73-AS1 can block miR-941 inhibition of cell migration, we conducted miR-941 and TP73-AS1 overexpression experiments followed by wound-healing assays in 293T cells. Consistent with previous studies, the transient transfection of 293T cells with vector carrying human miR-941 cluster region containing seven miR-941 precursors reproducibly inhibited cell migration. In contrast, parallel transfection with miR-941 vector and vector carrying human TP73-AS1 region containing nine miR-941 binding sites (TP73-AS1 sponge region) abolished the inhibition, and led to accelerated cell migration (fig. 6b). This result was further reproduced in an independent experiment conducted in triplicates (t-test, P < 0.05, fig. 6b and supplementary fig. S7, Supplementary Material online).
Gene expression measurements conducted using RNA-seq in transfected cells showed significantly higher expression of the predicted miR-941 targets after TP73-AS1 sponge region overexpression (Kolmogorov–Smirnov test, P < 0.001, fig. 6c). Furthermore, genes upregulated after TP73-AS1 sponge region overexpression overlapped significantly with the experimentally verified targets of miR-941 (hypergeometric test, P < 0.001, fig. 6d). These targets included TP73, an antisense gene of TP73-AS1 (negative binomial test, P < 0.002, FDR < 0.1%, supplementary fig. S8, Supplementary Material online). TP73 upregulation by TP73-AS1 was further confirmed using qPCR in U251 cells stably transfected with TP73-AS1 sponge region (t-test, P < 0.05, supplementary fig. S8, Supplementary Material online).
TP73-AS1 Acts as a Tumor Suppressor against Oncogenic miR-941
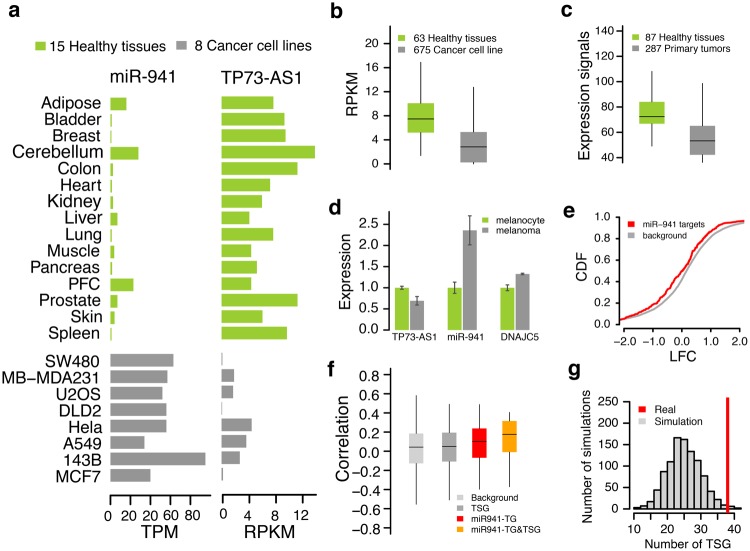
Previous studies demonstrated that TP73-AS1 functions as a tumor suppressor (Koga et al. 2009; Pang et al. 2010; Ronchetti et al. 2016; Wang et al. 2014; Wong et al. 2015), while miR-941 is preferentially expressed in proliferating and tumor cells (Hu et al. 2012). We showed that TP73-AS1 and miR-941 have strikingly opposite expression patterns in 15 healthy human tissues and 8 human cancer cell lines: miR-941 expression is ∼8-fold higher in tumors, TP73-AS1 expression—∼4-fold higher in healthy tissues (fig. 7a). Similarly, we find the elevated expression of TP73-AS1 in healthy tissues using an RNA-seq data set encompassing 63 healthy tissues and 675 cancer cell lines (Consortium 2013; Klijn et al. 2015; Petryszak et al. 2016; Uhlen et al. 2015; Wilcoxon rank sum test, P < 0.0001, fig. 7b), and a microarray data set surveying 87 healthy tissues and 287 primary tumors (Shaul et al. 2016; Wilcoxon rank sum test, P < 0.0001, fig. 7c).
Fig. 7.
TP73-AS1 and miR-941 properties in tumors and healthy tissues. (a) The expression level of TP73-AS1 and miR-941 in 15 healthy human tissues (green) and 8 human cancer cell lines (grey). (b) The distribution of TP73-AS1 expression levels in 63 healthy human tissues (green) and 675 human cancer cell lines (grey) based on RNA-seq data. (c) The distributions of TP73-AS1 expression levels in 87 healthy human tissues (green) and 287 human primary tumors (grey) based on microarray data. (d) The expression differences between melanocytes and melanoma of TP73-AS1, miR-941, and miR-941 host gene DNAJC5. (e) Cumulative distribution plots of log2-transformed gene expression fold-changes (LFC) between melanocytes and melanoma drawn for predicted miR-941 targets (red) and all other nonmiR-941 target genes (grey, background genes). The y-axis shows the cumulative distribution function (CDF) of LFC distribution. (f) The expression level correlations between TP73-AS1 transcript and four groups of genes computed using gene expression data from 82 primary and metastatic melanomas. The four groups are: nonmiR-941 target genes (background), annotated tumor suppressor genes (TSG), miR-941 target genes (miR-941-TG), miR-941-regulated tumor suppressor genes (miR-941-TG&TSG). (g) The enrichment of miR-941 target gene in annotated TSG. The background distribution of TSG numbers was computed by randomly sampling the same numbers of miRNA target genes 1,000 times.
The expression repression of TP73-AS1 was observed in many cancers, including melanoma, presumably due to DNA hypermethylation of the promoter region (Koga et al. 2009; Wang et al. 2014). Our results suggest that this might lead to miR-941 upregulation. To test this, we examined miR-941 and its target gene expression changes during the transformation of normal melanocytes to melanoma (Xiao et al. 2012). We found that while TP73-AS1 declined, miR-941 expression indeed increased more than 2-fold from melanocytes to melanoma (fig. 7d). Accordingly, expression of the miR-941 target genes was significantly inhibited in melanoma (Kolmogorov–Smirnov test, P < 0.005, fig. 7e). Furthermore, TP73-AS1 expression correlated positively with the expression of miR-941 target genes in 82 primary and metastatic melanoma cell lines (Kabbarah et al. 2010; Wilcoxon rank sum test, P < 1e-7, fig. 7f). Notably, among miR-941 targets, TSG showed even stronger correlations (Wilcoxon rank sum test, P < 0.001, fig. 7f, supplementary table S4, Supplementary Material online), including known TSG TP73, an antisense gene of TP73-AS1 (Pearson correlation, r = 0.40, P < 0.001, supplementary fig. S9, Supplementary Material online). In contrast, TSG not targeted by miR-941 did not show any significant correlations. Intriguingly, we found significantly more TSG among miR-941 targets (simulation, P < 0.02, fig. 7g).
Our analysis of a large cohort of MCTP cancer samples from nine different tissue types containing a total of 303 measurements yielded similar results (supplementary fig. S10 and supplementary table S4, Supplementary Material online). Thus, a strong link between the expression of miR-941 and TP73-AS1 that affects expression levels of multiple TSGs might indicate the importance of the role of transcripts in both tumorogenesis and tumor suppression.
Discussion
The pervasive transcription of mammalian genomes produces numerous noncoding RNAs (Awan et al. 2017; Rashid et al. 2016; Yu and Shan 2016). The functionality of these transcripts is far from being fully understood, and novel types of noncoding RNAs linked diverse functional mechanisms continue to emerge (Chen et al. 2015; Hu et al. 2016). Naturally occurring miRNA sponges represent a specific type of noncoding RNA that play important roles in the mediation of posttranscriptional regulation. While functionally annotated miRNA sponges are rare, the well known examples include human CDR1as, ZNF91 and mouse circular Sry transcripts (Guo et al. 2014; Hansen et al. 2013; Memczak et al. 2013). A characteristic feature of these transcripts is the large number of binding sites for a specific miRNA: 74, 24, and 16. Even though miRNA sponges containing only one miRNA binding site were revealed by functional studies (Wang et al. 2013, 2014), the presence of multiple miRNA binding sites, their high density and high binding affinity represent the major criteria for computational prediction of miRNA sponge transcripts. Our screen of 36,687 linear and 36,019 circRNAs singled out TP73-AS1 transcript as potential miR-941 sponge.
TP73-AS1 exhibits all known features of an efficient miRNA sponge. We showed that TP73-AS1 contains an unusually large number of high affinity miR-941 binding sties, as well as additional sequence features involved in AGO binding, such as the high density of subseed motifs: 8 per 100 bp. Furthermore, miRNA binding sites and subseed motifs cluster within a specific region of a transcript, with distances allowing them to act synergistically (Grimson et al. 2007; Sætrom et al. 2007). Another important feature increasing TP73-AS1 efficiency as miRNA sponge is its unusually high expression across human tissues. On the basis of data from 63 human tissues, TP73-AS1 ranks 23rd among all 1,085 expressed lncRNAs and ninth among 837 recently evolved ones (primate-specific lncRNA). TP73-AS1 expression is also ∼50 times higher than the expression its two neighboring PCGs, TP73, and CCDC27, indicating specific regulation rather than a general chromatin conformation change.
Our experiments conducted in cell lines, as well as analysis of in vivo expression patterns, demonstrate that TP73-AS1 is targeted by miR-941, interacts with miR-941 in RISC, and blocks miR-941-induced inhibition at the levels of transcriptome and physiological effects, such as cell migration rates. These findings confirm our computational assessment that TP73-AS1 functions as a miR-941 sponge. Recent reports show that TP73-AS1 can function as miR-200 sponge in human hepatocellular carcinoma cells and breast cancer cells (Li et al. 2017; Zou et al. 2018). TP73-AS1 contains a single miR-200 binding site located outside of miR-941-sponge region, but still within the same major isoform. This suggests that TP73-AS1, analogous to well-characterized sponge transcripts circHIPK3 and CDR1as (Guo et al. 2014; Zheng et al. 2016), can absorb multiple miRNAs. Intriguingly, the two miRNAs bound by TP73-AS1 appear to play opposite roles: while miR-941 is highly expressed in cancerous and proliferating cells, miR-200 functions as tumor suppressor (Hu et al. 2012; Li et al. 2017; Zou et al. 2018).
Both TP73-AS1 and miR-941 are recently evolved transcripts: miR-941 is specific to humans and TP73-AS1 is present in OWMs, apes, and humans. At the same time, both TP73-AS1 and miR-941 are highly expressed, ranking ninth and first among evolutionary novel lncRNAs and miRNAs respectively. We showed that the emergence of miR-941 in humans leads to increased constraint of the miR-941 binding sites’ sequence within TP73-AS1. These observations suggest the functional significance of TP73-AS1 and miR-941, as well as the potential importance of their interplay.
Previous studies demonstrated that TP73-AS1 has tumor-suppressive properties in gastric cancer, oligodendrogliomas, glioblastoma, and melanoma (Koga et al. 2009; Pang et al. 2010; Wang et al. 2014; Wong et al. 2015). Moreover, TP73-AS1 resides in 1p36.32 chromosome region that is frequently deleted in neuroblastoma and other tumors (Kaghad et al. 1997). Accordingly, we show that TP73-AS1 expression is inhibited in 675 cancer-derived cell lines and 287 primary tumors compared with healthy tissues. In contrast, miR-941 expression is on average 8-times higher in tumors and proliferating cells compared with healthy tissues. It is therefore appealing to speculate that in humans the TP73-AS1 tumor suppressor function is counterbalanced by miR-941. Indeed, we show that TSGs are overrepresented among miR-941 target genes, and the expression of these TSGs positively correlates with TP73-AS1 expression across multiple tumors. These observations also fit reports of higher prevalence of epidermal cancers, such as prostate, breast or lung carcinomas, in humans compared with nonhuman primates(Beniashvili 1989; McClure 1973; Puente et al. 2006; Waters et al. 1998).
Taken together, these observations allow us to propose a possible scenario of TP73-AS1 and miR-941 evolution. The rapid rise of TP73-AS1 expression after its emergence ∼40 million year ago might be due to its tumor suppressor function providing a fitness benefit to individuals who express it. In turn, the rapid rise of miR-941 expression after its emergence over the past 6 million years might have been facilitated by TP73-AS1 functioning as a miR-941 sponge and, therefore, reducing the potential deleterious effects of a new regulator. Yet, miR-941 appears to act as a TP73-AS1 antagonist and could represent a potent tumorigenesis factor. This apparently deleterious function of miR-941 must have been outweighed by the adaptive advantage it provided to its carriers, possibly linked to cell migration regulation during development. If these notions are correct, the regulatory balance of evolutionary novel TP73-AS1 and miR-941 transcripts might play an important role in tumorigenesis in human tissues. Moreover, this role would be missed in model systems based on species other than human.
Materials and Methods
Ethics Statement
Experiments were approved by the ethics committee of CAS-MPG Partner Institute for Computational Biology. Human brain tissues were obtained from the Wuhan brain bank in accordance with the brain bank protocol. Ethical agreements were obtained from the donors or their relatives by written informed consent.
RiboMinusSeq in Human PFC
The human PFC samples were taken from the frontal part of the superior frontal gyrus: a cortical region approximately corresponding to Brodmann Area 9, from two infant and two adult postmortem humans (supplementary table S1, Supplementary Material online). For all frozen PFC samples, similar proportions of grey and white matter were collected. Total RNA was isolated from the PFC samples using the Trizol (Invitrogen, USA) protocol with no modifications. Ribosomal RNA (rRNA) was depleted from the total RNA using the Ribominus kit (Invitrogen). The cDNA libraries were prepared from the rRNA-depleted RNA in accordance with the standard Illumina RNA-seq protocol. The generated cDNA library was sequenced using Illumina HiSeq2000 with 1 × 100 bp run (RiboMinusSeq). All original RiboMinusSeq data were deposited in the GEO database with accession number: GSE93708.
miR-941 Duplex Transfection and RNA-Seq Experiments
The miR-941 duplex (miR-941 mimics from Invitrogen) transfection experiments were conducted in a highly transfectable derivative of human embryonic kidney 293 cells (293T cells) in triplicates. Briefly, miR-941 duplex and Lipofectamine 2000 complexes (Invitrogen) were prepared before transfection freshly according to the manufacturer's protocol. The 293T cells were firstly plated 24 h before transfection in 0.5 ml culture medium DMEM with 10% fetal bovine serum (from HyClone) without antibiotics in six-well plates, and then transfected using miR-941 duplex and Lipofectamine 2000 with a final oligonucleotide concentration of 10 nmol/l. In parallel, negative control transfections with mock oligonucleotide (a scrambled oligo) were conducted according to the manufacturer's instructions. The transfection experiments with miR-941 duplex Lipofectamine 2000 and negative control oligonucleotides were carried out in three independent replicates. For each replicate, the transfected 293T cells were harvested after 24 h, total RNA were extracted with Trizol reagent (Invitrogen) and prepared into cDNA library according to the standard Illumina RNA-seq protocol. The generated cDNA library was sequenced using Illumina HiSeq2000 with 1 × 100 bp run. All original RNA-seq data are deposited in GEO database with accession number: GSE93717. To estimate the expression changes of predicted miR-941 targets after miR-941 duplex transfection, the obtained six RNA-seq data sets (three miR-941-transfection data sets and three mock-control-transfection data sets) were mapped to human genome (hg19) using STAR (Dobin et al. 2013) by requiring minimal mapped nucleotides no <90 nt. The targets of miR-941 were predicted using TargetScan without considering target site conservation. The log2-transformed gene expression fold-changes (LFC) and significantly differentially expressed miR-941 target genes between miR-941-transfection and mock-control-transfection were estimated using edgeR (Robinson et al. 2010) after TMM normalization (FDR < 0.1%).
AGO2 IP and Microarray Experiments
The AGO2-IP experiments after miR-941 duplex transfection were conducted in 293T cell line. Briefly, transfection experiments were performed in 293T cells with miR-941 duplex Lipofectamine 2000 and corresponding mock oligonucleotides (Invitrogen) at 30 nmol/l each (final concentration) per 1 × 106 cells per well in a six-well plate using DharmaFECT (GE Healthcare). After collecting 5 × 106 miR-941 duplex transfected cells, AGO2-IP was conducted using the RNA isolation kit human Ago2 (Wako Chemicals) in accordance with the manufacturer's protocol. For the negative control, immunoprecipitation was performed using non-immune IgG beads prepared with the antibody immobilization bead kit (Wako Chemicals). The IP pull down RNA was processed and hybridized to Affymetrix Human Genome U133 Plus 2.0 arrays following the manufacturer's instructions. The gene expression levels were determined using RMA in affy R package. All original microarray data are deposited in GEO database with accession number: GSE93707.
Cell Migration Assay
The cell migration status of transient-transfected 293T cells (miR-941 transient-transfected, miR-941 and TP73-AS1 transient-transfected, and SD1266 vector transient-transfected) was determined using a wound-healing assay. The detailed procedures of plasmid constructions for transient transfection were described in supplementary Methods, Supplementary Material online. In brief, for the wound-healing assay, cells were firstly seeded into 12-well tissue culture plates. When cells reached 70–80% confluence, a straight scratch wound was created by gently removing attached cells using a 1,000 µl standard pipette tip. The wounded monolayer cells were washed with fresh media once to remove floating cells, and then fresh gibco DMEM medium supplemented with 10% FBS was added. For transient-transfected 293T cells, the wound healing assay were photographed at 0 h and 48 h under a Nikon ECLIPSE Ti-s microscope. To measure miR-941 target gene expression changes after miR-941 and TP73-AS1 transient-transfection, the miR-941 and TP73-AS1 transfected 293T cells were harvested after 48 h. In parallel, SD1266 vector transient-transfected 293T cells were used as a negative control. Total RNA were extracted with Trizol reagent (Invitrogen) and prepared into cDNA library according to the standard Illumina RNA-seq protocol. The miR-941 and TP73-AS1 transient-transfection experiments were conducted in triplicates. The generated cDNA library was sequenced using Illumina HiSeq2000 with 1 × 100 bp run. All original RNA-seq data have been deposited in the GEO database with accession number: GSE93716.
Noncoding RNA Transcript (NCT) Data Set Construction
The NCT data set consists of transcripts of pseudogene, 3′UTR, lncRNA, and circRNA. The transcripts of pseudogene and 3′UTR were based on Ensembl annotation (Cunningham et al. 2015). The transcripts of lncRNAs were based on Refseq annotation (O'Leary et al. 2016) and supplemented by Ensembl annotation (Cunningham et al. 2015). The circRNA transcripts were identified based on 58 circRNA data including four human PFC samples generated in this study and 54 published circRNA sequencing data (supplementary table S1, Supplementary Material online). CircRNAs were identified using CIRI (Gao et al. 2015). Briefly, the raw circRNA sequencing data were mapped to the human genome (hg19) using BWA-MEM (Li and Durbin 2009) with default parameters except “-T 19” (filters out alignments with score < 19), as recommended by CIRI. To obtain more comprehensive circRNAs, we predicted circRNAs in each of the 58 circRNA sequencing data using CIRI with low stringency requirement with parameter (-low). The identified circRNAs were further merged to construct final circRNA data set (supplementary Data S1, Supplementary Material online). The sequences of pseudogene, 3′UTR and lncRNA were extracted from the human genome directly. The sequences of circRNA were inferred based on the circRNAs embraced exon sequences annotated by Refseq and human transcriptome assembly transcripts. The human transcriptome assembly transcripts were based on the merged data set of transcriptome assemblies from human PFC age-series data (Hu et al. 2014), human body map (Cabili et al. 2011) and Ulitsky’s lab (Hezroni et al. 2015).
miR-941 Sponge Gene Identification
To identify miR-941 sponge gene candidates from NCTs, TargetScan was used to predict miR-941 binding sites without considering site conservation. The miR-941 sponge gene candidates were classified as the NCTs that contain more miR-941 binding sites than any other shuffled NCTs. The shuffled NCT data set was generated by shuffling NCTs 1,000 times while keeping the same dinucleotide frequency. The same procedure was used to predict miRNA sponge transcripts for the other high-confidence miRNA family. We built high-confidence miRNA data set by intersecting annotated high-confidence miRNAs from three sources: miRBase (Kozomara and Griffiths-Jones 2014), mirDB (Wong and Wang 2015), and mirGeneDB (Fromm et al. 2015). The miRNAs supported by at least two sources were considered as high-confidence miRNAs in this study. The high-confidence miRNA was further merged into the high-confidence miRNA family based on the mature sequence seed region identity.
Enrichment Analysis for 3-mer Motif of miR-941
The frequency for the 3-mer motif of miR-941 mature sequence plus upstream two nucleotide sequence was counted in the vicinity miR-941 binding site region (chr1: 3,655,778-3,656,753, hg19, TP73-AS1 sponge region) on TP73-AS1. The background frequency for those 3-mer motifs were estimated based on the 1,000 shuffled sequences of TP73-AS1 sponge region by keeping the same dinucleotide frequency. For each 3-mer motif, expected frequency was estimated using the average frequency of 1,000 simulations; then, a binomial test after Bonferroni correction for multiple testing was used to estimate the enrichment significance (Bonferroni corrected, P < 0.05).
Evolution of TP73-AS1 Sponge Region
The orthologous sequences of the human TP73-AS1 sponge region were searched in ape (chimpanzees, gorillas, and orangutans) and OWM (macaques and baboons), NWM (marmosets), mammals (mice, rats, cows), and vertebrate (chickens) genomes using Liftover from UCSC (Rosenbloom et al. 2015). The miR-941 binding sites and the reverse complementary sequence of miR-941 binding sites (RC-miR-941 binding sites) were identified using TargetScan in the human TP73-AS1 sponge region and corresponding orthologous regions in apes and OWMs. To identify the sequence elements that drive miR-941 binding site expansion from OWMs to apes and human, we aligned the human TP73-AS1 sponge region and sponge orthologous regions from apes and OWMs using MUSCLE (Edgar 2004), and then searched for the longest continuously conserved sequence segment containing miR-941 binding site in human, apes and OWMs, which was defined as the putative sequence motif that may drive miR-941 sponge region formation.
The SNPs in the human TP73-AS1 sponge region and corresponding orthologous regions in chimpanzees and rhesus macaques were identified based on the human dbSNP138 database (Sherry et al. 2001), PanMap (http://panmap.uchicago.edu) and RhesusBase PopGateway (http://www.rhesusbase.org/popGateway), respectively. Since chimpanzee and rhesus macaque SNP data sets contain mainly common SNPs, only common SNPs were used for SNP analysis in human. The SNPs in the upstream and downstream flanking regions of sponge region (with the same size of sponge region) were used as background for estimating sponge region sequence constraints in humans, chimpanzees and rhesus macaques.
Sequence and Expression Analyses for TP73-AS1
The orthologous sequences of TP73-AS1, CCDC27, and TP73 were identified using both global alignment and local alignment across ape (chimpanzees, gorillas, orangutans), OWM (macaques), NWM (marmosets), mammal (mice, rats and cows), and vertebrate (chickens) species. The global alignment of TP73-AS1, CCDC27, and TP73 region was extracted from the genome alignment of 100 species downloaded from UCSC (Rosenbloom et al. 2015). The local alignment of TP73-AS1, CCDC27, and TP73 orthologous sequences was identified using BLAST with default parameters. The genome sequences of chimpanzees (panTro4), gorillas (gorGor3), orangutans (ponAbe2), macaques (rheMac3), marmosets (calJac3), mice (mm10), rats (rn5), cows (bosTau8), and chickens (galGal4) were downloaded from UCSC (Rosenbloom et al. 2015). The putative promoter of TP73-AS1 was inferred using the combination of CpG-islands and chromatin states annotations from UCSC (Rosenbloom et al. 2015).
The expression of TP73-AS1 across species was estimated by RNA-seq coverage and junction reads using standard RNA-seq data as well as sense and antisense RNA-seq coverage using strand-specific RNA-seq data in humans (SRR649361), chimpanzees (SRR306816), gorillas (SRR306801), orangutans (SRR306792), macaques (SRR630492), marmosets (SRR975174), mice (SRR649372), and rats (SRR1041767), downloaded from SRA (Leinonen et al. 2011). The total RNA-seq coverage was estimated using uniquely mapped reads and further normalized by total mapped reads as coverage per million mapped reads (normalized RNA-seq coverage). The junction reads were obtained in reads mapping procedure conducted using Tophat. The TP73-AS1 orthologous regions in mice and rats were obtained based on genomic synteny information.
The tissue expression pattern of TP73-AS1 in humans was estimated using the merged gene expression profiles of 63 human tissues from GTEx (Consortium 2013), HPA (Uhlen et al. 2015), and Expression Atlas (Petryszak et al. 2016). The information of the corresponding 63 tissues is listed in supplementary table S3, Supplementary Material online. The PCGannotation was based on the Refseq annotation. The primate-specific protein-coding genes (PS-PCG) were classified as the PCGs without detectable orthologous sequences in mice, rats, dogs, cows and chickens using BLASTP (E-value >10−5).
The Analyses of Regulatory Relationship between TP73-AS1 and miR-941
The regulatory relationship between TP73-AS1 and miR-941 in human tissue was analyzed using age-series RNA-seq data and small RNA sequencing data from PFC tissue in humans, chimpanzees and macaques, downloaded from SRP005169 and GSE18012. The expression level of gene and transcript isoform in human, chimpanzee and macaque PFC was quantified and normalized using RSEM in RPKM (Reads Per Kilobase of transcript per Million mapped reads). The miRNA expression level was quantified and normalized in TPM (Transcripts Per Million mapped reads), using small RNA sequencing data downloaded from GSE18069. The expression correlation between TP73-AS1 and miR-941 was calculated using the cubic smoothing spline curves, due to the difference of individuals used in 14 RNA-seq and 12 small RNA sequencing data. For curve fitting, the expression levels of TP73-AS1 and miR-941 were standardized to mean = 0 and SD = 1 (standardized expression) and further fitted as curves using cubic smooth spline with a degree of freedom 4. The same procedure was applied to the expression comparison between DNAJC5, miR-941, and the TP73-AS1 isoforms. The regulatory relationship between TP73-AS1 and miR-941 in human cells was analyzed using matched and processed mRNA and miRNA microarray data in 11 human immune cells, downloaded from GSE28492.
Cell Migration Associated Network Module Identification
The miR-941 target gene network was constructed using experimental verified miR-941 target genes by incorporating PPI information downloaded from geneMANIA (http://genemania.org/). Only PPIs with evidence of biological pathways and physical interactions were used for network construction. The network module was identified using CFinder (Adamcsek et al. 2006) requiring clique numbers of >2. The association between network module genes and cell migration function was estimated using Cociter with the term “cell migration” with P-value cutoff 0.01.
The Expression of miR-941 and TP73-AS1 in Healthy Human Tissues and Cancer Cell Lines
The miR-941 and TP73-AS1 expression levels in healthy human tissues and cancer cell lines (fig. 7a–c) were estimated using public RNA-seq and microarray data. Specifically, for miR-941, its expression abundance in normal and cancer cell lines was estimated using the RNA-seq data from GSE45159, GSE31616, GSE39162, GSE43550, ERP000773, GSE33858, SRA012516, GSE31037, GSE26545, and GSE16579. For TP73-AS1, its expression abundance in normal and cancer cell lines was estimated using RNA-seq data from GTEx, HPA, Expression Atlas, and E-MTAB-2706 and using microarray data queried from MERAV database (http://merav.wi.mit.edu/). The mRNA and miRNA expression profiles in normal melanocytes (HEMa-LP) and melanoma (A375), shown in figure 7d, were analyzed using microarray data that downloaded from GSE35389.
The mRNA gene expression profile of 82 primary and metastatic melanoma, shown in figure 7e–g, was analyzed using a microarray that downloaded from GSE46517. The gene expression profiles measured using strand-specific sequencing from 303 cancer tissue samples from the Michigan Center for Translational Pathology (MCTP) compendium representing cancer and benign conditions from nine different tissue types (breast adenocarcinoma, lung adenocarcinoma, lung squamous carcinoma, prostate cancer, ovarian cancer, pancreatic cancer, meningioma, rare cancers) were downloaded from GSE66729. We used the processed gene expression matrix based on sense strand reads for our analysis. The TSG list was downloaded from TSG database 2.0 (http://bioinfo.mc.vanderbilt.edu/TSGene/).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
H.H., P.K., H.Y. conceived and designed the experiments; H.H. conducted data analysis; J.L., Z.H., X.J., X.Y., J.X.L., and Y.Z. performed the experiments; P.K., H.H., and H.Y. wrote the paper.
Competing Financial Interests
The authors declare no competing financial interests.
Supplementary Material
Acknowledgments
We thank the Wuhan brain bank for providing human samples, Ingrid Burke for helpful comments on the manuscript, and all members of the Comparative Biology Group in Shanghai for helpful discussions and suggestions. This work was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (grant number XDB13010200), from National Natural Science Foundation of China (grant numbers 91331203 and 31420103920), from National One Thousand Foreign Experts Plan (grant number WQ20123100078), from Bureau of International Cooperation, Chinese Academy of Sciences (grant number GJHZ201313), and from Russian Science Foundation (grant number 16-14-00220) to P.K., from the National Natural Science Foundation of China (grant number 31601068) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) to H.H., and from the National Natural Science Foundation of China (grant number 31471187), from Science and Technology Key Project of Yunnan Province (grant number 2017FA009) and from fund of University Key Laboratory of Tumor Molecular Biology in Yunan to H.Y.
References
- Adamcsek B, Palla G, Farkas IJ, Derenyi I, Vicsek T.. 2006. CFinder: locating cliques and overlapping modules in biological networks. Bioinformatics 228:1021–1023. [DOI] [PubMed] [Google Scholar]
- Allantaz F, Cheng DT, Bergauer T, Ravindran P, Rossier MF, Ebeling M, Badi L, Reis B, Bitter H, D'Asaro M et al. , 2012. Expression profiling of human immune cell subsets identifies miRNA-mRNA regulatory relationships correlated with cell type specific expression. PLoS One 71:e29979.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awan HM, Shah A, Rashid F, Shan G.. 2017. Primate-specific long non-coding RNAs and microRNAs. Genomics Proteomics Bioinformatics. 153:187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 1162:281–297. [DOI] [PubMed] [Google Scholar]
- Beniashvili DS. 1989. An overview of the world literature on spontaneous tumors in nonhuman primates. J Med Primatol. 186:423–437. [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL.. 2011. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2518:1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandradoss SD, Schirle NT, Szczepaniak M, MacRae IJ, Joo C.. 2015. A dynamic search process underlies microRNA targeting. Cell 1621:96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Huang C, Wang X, Shan G.. 2015. Circular RNAs in eukaryotic cells. Curr Genomics. 165:312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium GT 2013. The genotype-tissue expression (GTEx) project. Nat Genet. 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S et al. , 2015. Ensembl 2015. Nucleic Acids Res. 43(Database issue):D662–D669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 291:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA.. 2010. MicroRNA sponges: progress and possibilities. RNA 1611:2043–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 325:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP.. 2008. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 191:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm B, Billipp T, Peck LE, Johansen M, Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E et al. , 2015. A uniform system for the annotation of vertebrate microRNA genes and the evolution of the human microRNAome. Annu Rev Genet. 49:213–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wang J, Zhao F.. 2015. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 16:4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP.. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 271:91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Agarwal V, Guo H, Bartel DP.. 2014. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 157:409.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M et al. , 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 1411:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J.. 2013. Natural RNA circles function as efficient microRNA sponges. Nature 4957441:384–388. [DOI] [PubMed] [Google Scholar]
- Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP, Ulitsky I.. 2015. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 117:1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HY, He L, Fominykh K, Yan Z, Guo S, Zhang X, Taylor MS, Tang L, Li J, Liu J et al. , 2012. Evolution of the human-specific microRNA miR-941. Nat Commun. 3:1145.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HY, He L, Khaitovich P.. 2014. Deep sequencing reveals a novel class of bidirectional promoters associated with neuronal genes. BMC Genomics. 15:457.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Wang X, Shan G.. 2016. Insertion of an Alu element in a lncRNA leads to primate-specific modulation of alternative splicing. Nat Struct Mol Biol. 2311:1011–1019. [DOI] [PubMed] [Google Scholar]
- Kabbarah O, Nogueira C, Feng B, Nazarian RM, Bosenberg M, Wu M, Scott KL, Kwong LN, Xiao Y, Cordon-Cardo C et al. , 2010. Integrative genome comparison of primary and metastatic melanomas. PLoS One 55:e10770.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X et al. , 1997. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 904:809–819. [DOI] [PubMed] [Google Scholar]
- Kim JG, Kim TO, Bae JH, Shim JW, Kang MJ, Yang K, Ting AH, Yi JM.. 2014. Epigenetically regulated MIR941 and MIR1247 target gastric cancer cell growth and migration. Epigenetics 97:1018–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klijn C, Durinck S, Stawiski EW, Haverty PM, Jiang Z, Liu H, Degenhardt J, Mayba O, Gnad F, Liu J et al. , 2015. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol. 333:306–312. [DOI] [PubMed] [Google Scholar]
- Koga Y, Pelizzola M, Cheng E, Krauthammer M, Sznol M, Ariyan S, Narayan D, Molinaro AM, Halaban R, Weissman SM.. 2009. Genome-wide screen of promoter methylation identifies novel markers in melanoma. Genome Res. 198:1462–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S.. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42(Database issue):D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen R, Sugawara H, Shumway M, International Nucleotide Sequence Database C 2011. The sequence read archive. Nucleic Acids Res. 39(Database):D19–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP.. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 1201:15–20. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB.. 2003. Prediction of mammalian microRNA targets. Cell 1157:787–798. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2514:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Huang Y, Huang Y, Fu Y, Tang D, Kang R, Zhou R, Fan XG.. 2017. The long non-coding RNA TP73-AS1 modulates HCC cell proliferation through miR-200a-dependent HMGB1/RAGE regulation. J Exp Clin Cancer Res. 361:51.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Shen Y, Wu Q, Kumar S, He B, Shi S, Carthew RW, Wang SM, Wu CI.. 2008. The birth and death of microRNA genes in Drosophila. Nat Genet. 403:351–355. [DOI] [PubMed] [Google Scholar]
- McClure HM. 1973. Tumors in nonhuman primates: observations during a six-year period in the Yerkes primate center colony. Am J Phys Anthropol. 382:425–429. [DOI] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M et al. , 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 4957441:333–338. [DOI] [PubMed] [Google Scholar]
- Meunier J, Lemoine F, Soumillon M, Liechti A, Weier M, Guschanski K, Hu H, Khaitovich P, Kaessmann H.. 2013. Birth and expression evolution of mammalian microRNA genes. Genome Res. 231:34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AM, Fox G, Kiran AM, De Bo C, O’Connor PBF, Heaphy SM, Mullan JPA, Donohue CA, Higgins DG, Baranov PV.. 2014. GWIPS-viz: development of a ribo-seq genome browser. Nucleic Acids Res. 42(D1):D859–D864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D et al. , 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang JC, Li KK, Lau KM, Ng YL, Wong J, Chung NY, Li HM, Chui YL, Lui VW, Chen ZP et al. , 2010. KIAA0495/PDAM is frequently downregulated in oligodendroglial tumors and its knockdown by siRNA induces cisplatin resistance in glioma cells. Brain Pathol. 206:1021–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryszak R, Keays M, Tang YA, Fonseca NA, Barrera E, Burdett T, Fullgrabe A, Fuentes AM, Jupp S, Koskinen S et al. , 2016. Expression Atlas update – an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 44(D1):D746–D752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Jara CAC, Fenske P et al. , 2017. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 3576357:eaam8526.. [DOI] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP.. 2010. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 4657301:1033–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente XS, Velasco G, Gutierrez-Fernandez A, Bertranpetit J, King MC, Lopez OC.. 2006. Comparative analysis of cancer genes in the human and chimpanzee genomes. BMC Genomics. 7:15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao N, Huang Y, Naveed H, Green CD, Han JDJ.. 2013. CoCiter: an efficient tool to infer gene function by assessing the significance of literature co-citation. PLoS One 89:e74074.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani N, Nowakowski TJ, Zhou HJ, Godshalk SE, Lisi V, Kriegstein AR, Kosik KS.. 2016. A primate lncRNA mediates notch signaling during neuronal development by sequestering miRNA. Neuron 906:1174–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid F, Shah A, Shan G.. 2016. Long non-coding RNAs in the cytoplasm. Genomics Proteomics Bioinformatics. 142:73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 261:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronchetti D, Agnelli L, Taiana E, Galletti S, Manzoni M, Todoerti K, Musto P, Strozzi F, Neri A.. 2016. Distinct lncRNA transcriptional fingerprints characterize progressive stages of multiple myeloma. Oncotarget 712:14814–14830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M et al. , 2015. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 43(Database issue):D670–D681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sætrom P, Heale BSE, Snøve O, Aagaard L, Alluin J, Rossi JJ.. 2007. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 357:2333–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP.. 2011. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell 1463:353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul YD, Yuan B, Thiru P, Nutter-Upham A, McCallum S, Lanzkron C, Bell GW, Sabatini DM.. 2016. MERAV: a tool for comparing gene expression across human tissues and cell types. Nucleic Acids Res. 44(D1):D560–D566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K.. 2001. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 291:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Russell RB, Cohen SM.. 2003. Identification of Drosophila microRNA targets. PLoS Biol. 13:E60.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson DW, Dinger ME.. 2016. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 175:272–283. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A et al. , 2015. Proteomics. Tissue-based map of the human proteome. Science 3476220:1260419.. [DOI] [PubMed] [Google Scholar]
- Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, Siu HC, Deng S, Chu KM, Law S et al. , 2014. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 466:573–582. [DOI] [PubMed] [Google Scholar]
- Wang P, Zhi H, Zhang YP, Liu Y, Zhang JZ, Gao Y, Guo MN, Ning SW, Li X.. 2015. miRSponge: a manually curated database for experimentally supported miRNA sponges and ceRNAs. Database 2015:bav098.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, Wu M, Xiong J, Guo X, Liu H.. 2013. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 251:69–80. [DOI] [PubMed] [Google Scholar]
- Waters DJ, Sakr WA, Hayden DW, Lang CM, McKinney L, Murphy GP, Radinsky R, Ramoner R, Richardson RC, Tindall DJ.. 1998. Workgroup 4: spontaneous prostate carcinoma in dogs and nonhuman primates. Prostate 361:64–67. [DOI] [PubMed] [Google Scholar]
- Wong KY, Li Z, Zhang X, Leung GK, Chan GC, Chim CS.. 2015. Epigenetic silencing of a long non-coding RNA KIAA0495 in multiple myeloma. Mol Cancer. 14:175.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong N, Wang X.. 2015. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 43(Database issue):D146–D152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao D, Ohlendorf J, Chen Y, Taylor DD, Rai SN, Waigel S, Zacharias W, Hao H, McMasters KM.. 2012. Identifying mRNA, microRNA and protein profiles of melanoma exosomes. PLoS One 710:e46874.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Gorospe M.. 2014. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 34:9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Shan G.. 2016. Functions of long noncoding RNAs in the nucleus. Nucleus 72:155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang PP, Wang XL, Zhao W, Qi B, Yang Q, Wan HY, Shuang ZY, Liu M, Li X, Li S et al. , 2014. DNA methylation-mediated repression of miR-941 enhances lysine (K)-specific demethylase 6B expression in hepatoma cells. J Biol Chem. 28935:24724–24735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G et al. , 2016. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 7:11215.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Q, Zhou E, Xu F, Zhang D, Yi W, Yao J.. 2018. A TP73-AS1/miR-200a/ZEB1 regulating loop promotes breast cancer cell invasion and migration. J Cell Biochem. 1192:2189–2199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.