Abstract
To expand our capacity to discover venom sequences from the genomes of venomous organisms, we applied targeted sequencing techniques to selectively recover venom gene superfamilies and nontoxin loci from the genomes of 32 cone snail species (family, Conidae), a diverse group of marine gastropods that capture their prey using a cocktail of neurotoxic peptides (conotoxins). We were able to successfully recover conotoxin gene superfamilies across all species with high confidence (> 100× coverage) and used these data to provide new insights into conotoxin evolution. First, we found that conotoxin gene superfamilies are composed of one to six exons and are typically short in length (mean = ∼85 bp). Second, we expanded our understanding of the following genetic features of conotoxin evolution: 1) positive selection, where exons coding the mature toxin region were often three times more divergent than their adjacent noncoding regions, 2) expression regulation, with comparisons to transcriptome data showing that cone snails only express a fraction of the genes available in their genome (24–63%), and 3) extensive gene turnover, where Conidae species varied from 120 to 859 conotoxin gene copies. Finally, using comparative phylogenetic methods, we found that while diet specificity did not predict patterns of conotoxin evolution, dietary breadth was positively correlated with total conotoxin gene diversity. Overall, the targeted sequencing technique demonstrated here has the potential to radically increase the pace at which venom gene families are sequenced and studied, reshaping our ability to understand the impact of genetic changes on ecologically relevant phenotypes and subsequent diversification.
Keywords: Conidae, comparative phylogenetics, diversification
Introduction
Understanding the molecular basis for adaptation and speciation is a central goal in evolutionary biology. Past studies have described several genetic characteristics that seem to be associated with rapidly radiating clades or the evolution of novel phenotypes, including evidence for diversifying selection, gene gains and losses, and accelerated rates of sequence evolution (Floudas et al. 2012; Brawand et al. 2014; Guillén et al. 2014; Cornetti et al. 2015; Malmstrøm et al. 2016; Pease et al. 2016). Although large-scale comparative genomic studies have vastly increased our knowledge of the genetic changes associated with diversification, the link between genotype and ecologically relevant phenotypes frequently remains unclear. Often, the functional consequences of genetic patterns such as an excess of gene duplicates or regions under positive selection are unknown (Brawand et al. 2014; Cornetti et al. 2015; Pease et al. 2016), limiting our ability to understand how genetic changes shape the evolutionary trajectory of species.
Animal venoms provide an excellent opportunity to study the interplay between genetics and adaptation because of the relatively simple relationship between genotype, phenotype, and ecology. Venoms have evolved multiple times throughout the tree of life (e.g., spiders, snakes, and snails) and play a direct role in prey capture and survival (Barlow et al. 2009; Casewell et al. 2013). Venoms are composed of mixtures of toxic proteins and peptides that are usually encoded directly by a handful of known gene families (Kordis and Gubensek 2000; Fry et al. 2009; Casewell et al. 2013). Exceptionally high estimated rates of gene duplication and diversifying selection across these venom genes families are thought to contribute to the evolution of novel proteins and thus changes in venom composition (Duda and Palumbi 1999; Gibbs and Rossiter 2008; Chang and Duda 2012), allowing venomous taxa to specialize and adapt onto different prey species (Kohn 1959a; Daltry et al. 1996; Li et al. 2005; Barlow et al. 2009; Chang and Duda 2016; Phuong et al. 2016). Therefore, the study of venomous taxa can facilitate understanding of the genetic contributions to ecologically relevant traits and subsequent diversification.
A fundamental challenge associated with the study of venom evolution is the inability to rapidly obtain sequences from venomous multigene families. Traditionally, venom genes were sequenced through cDNA cloning techniques, which can be labor intensive and time-consuming (Gibbs and Rossiter 2008; Chang et al. 2015). Although transcriptome sequencing has greatly increased the pace of venom gene sequencing and the discovery of previously undescribed gene families (Casewell et al. 2009; Phuong et al. 2016), transcriptome sequencing still requires fresh RNA extracts from venom organs, which may be difficult to obtain for rare and/or dangerous species. Targeted sequencing approaches have vastly improved the capacity to obtain thousands of markers across populations and species for ecological and evolutionary studies (Bi et al. 2012; Faircloth et al. 2012). Until now, these approaches have not been applied to selectively sequence venomous genomic regions. This may be in part, due to the extraordinary levels of sequence divergence exhibited by venom loci (Gibbs and Rossiter 2008; Chang and Duda 2012), potentially rendering probes designed from a single sequence from one gene family unable to recover any other sequences in the same family (fig. 1). However, past studies have shown that noncoding regions (i.e., introns, untranslated regions [UTRs]) adjacent to hypervariable mature toxin exons are conserved between species (Nakashima et al. 1993, 1995; Gibbs and Rossiter 2008; Wu et al. 2013), suggesting that these conserved regions can be used for probe design to potentially recover all venom genes across clades of venomous taxa.
Fig. 1.
A superfamily conotoxins from Conus lividus described in the transcriptome from Phuong et al. (2016). Protein alignment generated using Geneious. Amino acids are highlighted based on disagreement to a consensus sequence generated from the alignment, not shown. Cysteines are highlighted and bolded. Signal region and mature toxin coding region are annotated based on the presence of these functional regions at a particular position in the alignment in any of the sequences.
Here, we used a targeted sequencing approach to recover venom genes and study the evolution of venom gene families across 32 species of cone snails from the family, Conidae. Cone snails are a hyper diverse group of carnivorous marine gastropods (> 700 spp.) that capture their prey using a cocktail of venomous neurotoxins (Puillandre et al. 2014). Cone snail venom precursor peptides (conotoxins) are typically composed of three regions: the signal region that directs the protein into the secretory pathway, the prepro region that is cleaved during protein maturation, and the mature region that ultimately becomes the mature peptide (Robinson and Norton 2014). In some instances, there exists a “post” region of the peptide following the mature region that is also cleaved during protein processing (Robinson and Norton 2014). Conotoxins are classified into > 40 gene superfamilies (e.g., A superfamily and O1 superfamily) based on signal sequence identity, though some gene superfamilies were identified based on domain similarities to proteins from other venomous taxa (Robinson and Norton 2014). To examine the evolution of conotoxin gene superfamilies from genomic DNA, we designed probes targeting over 800 nonconotoxin genes for phylogenetic analyses and conotoxins from 12 previously sequenced Conidae transcriptomes (Phuong et al. 2016). With the recovered conotoxin loci, we describe several features of conotoxin genes, including its genetic architecture, molecular evolution, expression patterns, and changes in gene superfamily size. Finally, we use comparative phylogenetic methods to test whether diet specificity or dietary breadth can explain patterns of gene superfamily size evolution.
Results
Exon Capture Results
We used custom designed 120-bp baits (custom MYbaits-1 kit, 20,000 bait sequences; Arbor Biosciences, Ann Arbor, MI) to selectively target phylogenetic markers and conotoxin genes from 32 Conidae species (table 1). We sequenced all samples on a single Illumina HiSeq2000 lane, producing an average of 12.8 million reads per sample (table 1). After redefining exon boundaries for the phylogenetic markers, we generated a reference that consisted of 5,883 loci. We recovered an average of 5,335 loci (90.7%) across all samples representing ∼0.66 Mb (Megabases) on an average (table 1). For the conotoxin loci, given that conotoxin introns can be several kilobases in length (Wu et al. 2013) and the average insert size of the libraries was ∼350 bp, we were only able to assemble conotoxin exon fragments (conotoxin exons with any adjacent noncoding regions). The number of sequences we assembled containing a conotoxin exon ranged from 281 fragments in Conus papilliferus to 2,278 fragments in C. atristophanes (table 1). Approximately 48.8% of the reads mapped to both the phylogenetic markers and venom genes with 52.3% of these reads being marked as duplicates (table 1). Average coverage across the phylogenetic markers was 95.9×, whereas the average coverage for the conotoxin exons was 149.6× (table 1).
Table 1.
Sample Information and Exon Capture Statistics.
| Species | No. of Reads | % Reads on Target | % Duplicates | No. of Bases Recovered | No. of Exons Recovered | Exon Coverage | No. of Conotoxin Fragments Recovered | Conotoxin Coverage | Collection Source | Year Collected |
|---|---|---|---|---|---|---|---|---|---|---|
| magus | 10,348,142 | 48.17 | 56.18 | 652,113 | 5,258 | 79.0 | 332 | 136.9 | Australian Museum (Catalog No. C487620) | 2009 |
| textile | 8,130,024 | 47.86 | 51.79 | 649,193 | 5,236 | 64.8 | 764 | 109.9 | Australian Museum (Catalog No. C487632) | 2009 |
| lischkeanus | 8,942,888 | 45.30 | 52.64 | 627,806 | 5,024 | 64.2 | 834 | 127.1 | Australian Museum (Catalog No. C480270) | 2011 |
| aristophanes | 15,276,200 | 52.96 | 53.68 | 655,444 | 5,310 | 92.9 | 2,278 | 159.5 | Australian Museum (Catalog No. C480269) | 2011 |
| papilliferus | 9,733,028 | 46.97 | 46.97 | 659,001 | 5,318 | 82.5 | 281 | 114.1 | Australian Museum (Catalog No. C469190) | 2011 |
| californicus | 11,257,992 | 40.29 | 46.23 | 581,181 | 4,659 | 89.2 | 649 | 167.1 | Monterey Bay, California (provided by WF Gilly) | 2014 |
| virgo | 8,319,348 | 51.90 | 44.74 | 659,419 | 5,312 | 84.9 | 406 | 151.9 | Bali, Indonesia (Field collection) | 2013 |
| quercinus | 12,260,196 | 47.95 | 53.40 | 667,897 | 5,409 | 92.6 | 500 | 154.6 | Bali, Indonesia (Field collection) | 2013 |
| ebraeus | 16,311,454 | 49.57 | 55.37 | 669,852 | 5,427 | 120.7 | 849 | 203.9 | Bali, Indonesia (Field collection) | 2013 |
| flavidus | 12,175,022 | 46.25 | 41.37 | 671,106 | 5,446 | 110.0 | 614 | 174.8 | Bali, Indonesia (Field collection) | 2013 |
| lividus | 13,561,122 | 51.65 | 54.67 | 663,079 | 5,365 | 93.5 | 987 | 159.4 | Bali, Indonesia (Field collection) | 2013 |
| miles | 18,577,352 | 50.13 | 49.06 | 680,191 | 5,541 | 175.9 | 594 | 200.5 | Bali, Indonesia (Field collection) | 2013 |
| rattus | 16,948,836 | 52.08 | 55.04 | 677,626 | 5,497 | 139.7 | 727 | 183.5 | Bali, Indonesia (Field collection) | 2013 |
| distans | 12,706,016 | 50.85 | 57.47 | 658,791 | 5,411 | 100.0 | 384 | 124.1 | Bali, Indonesia (Field collection) | 2013 |
| sponsalis | 11,168,242 | 46.70 | 44.51 | 657,141 | 5,313 | 77.8 | 2,033 | 125.4 | Bali, Indonesia (Field collection) | 2013 |
| imperialis | 8,838,484 | 43.70 | 50.19 | 652,250 | 5,242 | 69.0 | 397 | 96.8 | Bali, Indonesia (Field collection) | 2013 |
| marmoreus | 11,494,484 | 52.20 | 50.27 | 669,298 | 5,430 | 103.8 | 524 | 195.7 | Bali, Indonesia (Field collection) | 2013 |
| varius | 15,035,404 | 50.78 | 52.57 | 666,670 | 5,387 | 113.6 | 743 | 209.0 | Bali, Indonesia (Field collection) | 2013 |
| musicus | 12,761,042 | 46.59 | 56.15 | 649,666 | 5,245 | 71.9 | 1,970 | 108.7 | Bali, Indonesia (Field collection) | 2013 |
| planorbis | 13,623,080 | 44.80 | 58.94 | 658,461 | 5,330 | 76.0 | 899 | 116.7 | Bali, Indonesia (Field collection) | 2013 |
| miliaris | 13,039,246 | 52.88 | 55.59 | 640,651 | 5,152 | 67.2 | 2,106 | 142.5 | Bali, Indonesia (Field collection) | 2013 |
| vexillum | 12,197,882 | 49.41 | 52.89 | 668,942 | 5,417 | 97.6 | 603 | 132.6 | Bali, Indonesia (Field collection) | 2013 |
| arenatus | 14,607,148 | 47.23 | 48.61 | 665,490 | 5,397 | 98.3 | 1,466 | 147.4 | Bali, Indonesia (Field collection) | 2013 |
| muriculatus | 17,328,206 | 47.25 | 59.85 | 662,812 | 5,359 | 102.3 | 1,072 | 137.6 | Bali, Indonesia (Field collection) | 2013 |
| chaldaeus | 11,778,640 | 50.84 | 55.12 | 661,691 | 5,371 | 91.3 | 704 | 131.8 | Bali, Indonesia (Field collection) | 2013 |
| coronatus | 14,873,018 | 49.92 | 45.09 | 670,729 | 5,442 | 100.8 | 2,158 | 179.3 | Bali, Indonesia (Field collection) | 2013 |
| moreleti | 10,843,692 | 49.41 | 57.47 | 653,436 | 5,282 | 78.8 | 485 | 142.8 | Bali, Indonesia (Field collection) | 2013 |
| anemone | 10,662,156 | 42.78 | 41.61 | 670,558 | 5,440 | 94.2 | 321 | 130.6 | Museum Victoria (Registration No. F237402) | 2014 |
| mustelinus | 15,317,812 | 50.86 | 58.11 | 668,295 | 5,416 | 115.5 | 679 | 153.8 | Australian Museum (Catalog No. C487551) | 2014 |
| litteratus | 13,002,630 | 51.11 | 56.64 | 673,310 | 5,451 | 99.8 | 538 | 148.4 | Australian Museum (Catalog No. C487581) | 2014 |
| capitaneus | 16,366,304 | 51.30 | 57.35 | 672,762 | 5,461 | 120.4 | 909 | 151.9 | Australian Museum (Catalog No. C487552) | 2014 |
| emaciatus | 13,391,964 | 51.38 | 54.88 | 665,699 | 5,387 | 101.7 | 554 | 168.3 | Australian Museum (Catalog No. C487593) | 2014 |
We recovered representative exons from all 49 conotoxin gene superfamilies targeted, plus exons from the Q gene superfamily which we did not explicitly target (supplementary fig. S1, Supplementary Material online). Of the 49 targeted gene superfamilies, “capture success” (defined in Materials and Methods) was 80% or above for 34 gene superfamilies, even though we did not explicitly target every single transcript (supplementary table S1, Supplementary Material online). For example, we only targeted one sequence of the A gene superfamily from C. varius, but we recovered sequences that showed high identity to every single transcript from the A gene superfamily discovered in the C. varius transcriptome (supplementary table S1, Supplementary Material online). We assessed the ability of targeted sequencing to recover conotoxins from species that were not explicitly targeted in the bait sequences by calculating the number of previously sequenced conotoxins (obtained via Genbank and Conoserver; Kaas et al. 2010) recovered in our data set (supplementary table S2, Supplementary Material online). We recovered a higher percentage of previously sequenced conotoxins if the species was included in the bait design (52.35%, supplementary table S2, Supplementary Material online) compared with species not included in the bait design (39.5%, supplementary table S2, Supplementary Material online).
Conotoxin Genetic Architecture
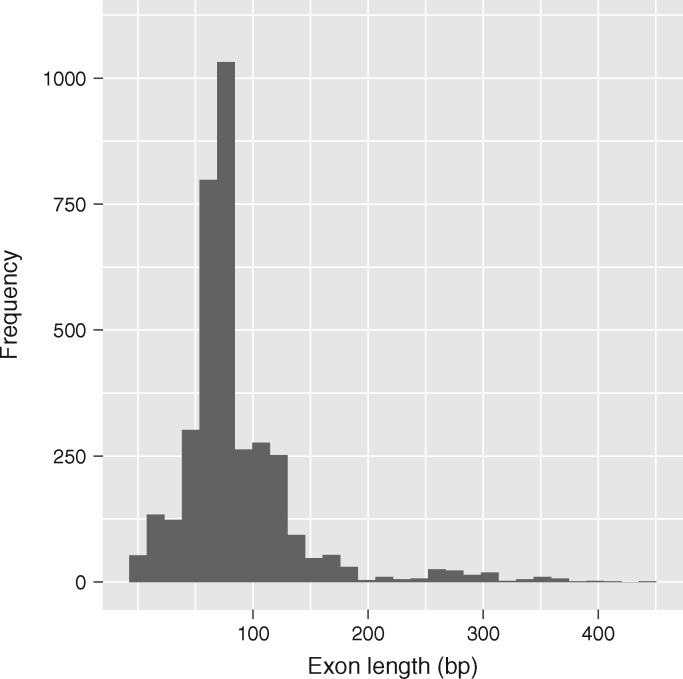
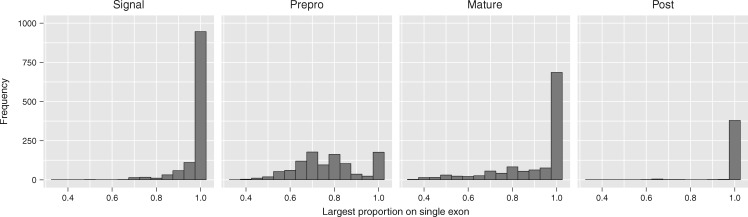
Through analyses of conotoxin genetic structure across species, we found that the number of exons that comprise a conotoxin transcript ranged from one to six exons and exon size ranged from 5 to 444 bp, with an average length of 85.2 bp (fig. 2 and supplementary table S3, Supplementary Material online). Whether or not UTRs were adjacent to terminal exons was dependent on the gene superfamily, with some gene superfamilies always having both 5′- and 3′-UTRs adjacent to terminal exons and some where the 5′- or 3′-UTRs cannot be found directly adjacent to the terminal exons (supplementary table S3, Supplementary Material online). Regions in conotoxin transcripts identified as the signal region, the mature region, or the postregion were most often confined to a single exon (fig. 3). In contrast, the prepro region was more frequently distributed across more than one exon (fig. 3).
Fig. 2.
Exon length distribution. Exon length distribution across all conotoxin gene superfamilies sequenced in this study. Analysis only includes sequences from species that had transcriptome data available.
Fig. 3.
Histograms showing the frequency of the largest proportion of each conotoxin precursor peptide region found on a single exon in Conidae genomes. Analysis only includes sequences from species that had transcriptome data available.
Conotoxin Molecular Evolution
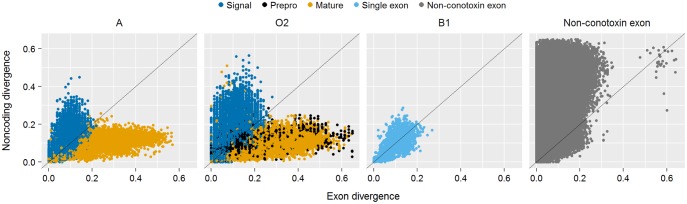
To determine if there are differences in divergence depending on what conotoxin precursor peptide region each exon contains, we calculated uncorrected pairwise differences to quantify the level of sequence divergence between exons and immediately adjacent noncoding regions. Exons containing the signal region were more conserved than their adjacent noncoding regions (average relative ratio < 1, supplementary table S4, Supplementary Material online, fig. 4, and supplementary fig. S2, Supplementary Material online). In contrast, all other exon classifications generally showed the opposite pattern, where the exons were typically more divergent relative to their adjacent noncoding regions (average relative ratio > 1, supplementary table S4, Supplementary Material online, fig. 4, and supplementary fig. S2, Supplementary Material online). The largest contrast in divergence between exons and adjacent noncoding regions came from exons containing the mature region, where the coding region was on an average 2.9 times more divergent than regions surrounding the exon (supplementary table S4, Supplementary Material online, fig. 4, and supplementary fig. S2, Supplementary Material online). For comparison, exons from nonconotoxin genes were more conserved than their adjacent noncoding regions (average relative ratio < 1, fig. 4).
Fig. 4.
Scatterplot of uncorrected pairwise distances for select gene superfamilies and nonconotoxin loci between exons and adjacent noncoding regions. Each point on the graph represents a unique pairwise comparison and points are highlighted by conotoxin functional region. x = y line is shown.
Conotoxin Expression
To examine expression regulation across gene superfamilies and species, we compared transcriptomes we previously sequenced (Phuong et al. 2016) to the targeted sequencing data. The proportion of conotoxin genes expressed per gene superfamily was highly variable (supplementary table S5 and fig. S3, Supplementary Material online) and the exact proportion depended on the gene superfamily and the species. In several cases, all gene copies of a gene superfamily were not expressed in the transcriptome (e.g., Conus ebraeus, A gene superfamily, 0/9 copies expressed, supplementary table S5 and fig. S3, Supplementary Material online), and in other cases, all copies were expressed in the transcriptome (e.g., C. californicus, O3 gene superfamily, 3/3 copies expressed, supplementary table S5 and fig. S3, Supplementary Material online). The average proportion of gene copies expressed per gene superfamily per species was 45% (range: 24–63%, supplementary table S5, Supplementary Material online).
Conotoxin Gene Superfamily Size Evolution
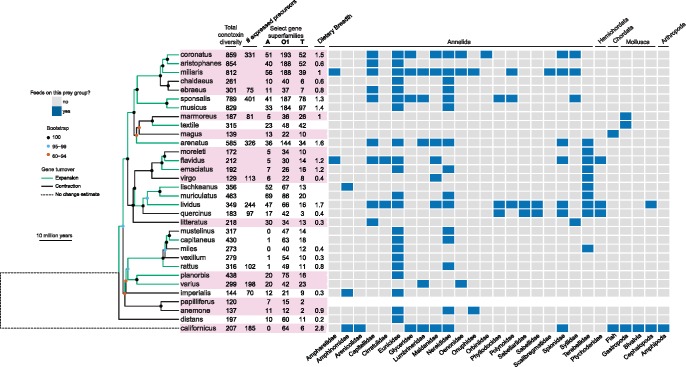
With a concatenated alignment of 4,441 exons representing 573,854 bp, we recovered a highly supported phylogeny with all but four nodes having ≤ 95% bootstrap support (fig. 5). Total conotoxin gene diversity ranged from as low as 120 in C. papilliferus to as high as 859 in C. coronatus (fig. 5). 25 gene superfamilies showed evidence of phylogenetic signal in gene superfamily size, such that closely related species tended to have similar gene superfamily sizes (supplementary table S6, Supplementary Material online). For example, a clade consisting of C. coronatus, C. aristophanes, and C. miliaris contains nearly 5 times more gene copies of the O1 superfamily than their immediate sister clade (fig. 5). CAFE v3.1 (Han et al. 2013) estimates of net gene gains and losses showed that species-specific net conotoxin expansions and contractions are scattered throughout the phylogeny (fig. 5 and supplementary fig. S4, Supplementary Material online).
Fig. 5.
Diet and conotoxin evolution in a phylogenetic context. Time-calibrated maximum likelihood phylogeny of 32 Conidae species generated from concatenated alignment of 4,441 exons. Phylogeny is rooted with Californiconus californicus. Branches are colored based on net gains or losses in total conotoxin diversity based on CAFE v3.1 analyses. Recognized subgenera are alternately colored pink. Total conotoxin diversity, the number of expressed precursors for species with transcriptomes, size estimates for commonly studied gene superfamilies, and dietary breadth displayed next to tip names. Recorded observations of each species preying on each of the 27 represented prey families shown in the matrix adjacent to the phylogeny, with cells colored based on whether or not a species has been observed to feed on that prey family (gray = no, blue = yes). Phylum level classifications are shown at the top of the diet matrix and family level classifications are shown at the bottom of the diet matrix.
Diet and Conotoxin Gene Superfamily Evolution
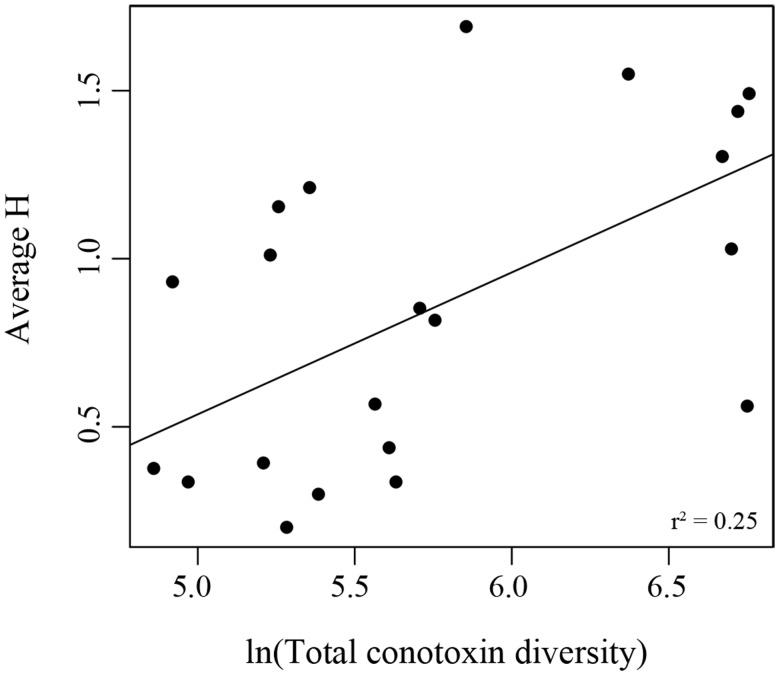
We used comparative phylogenetic methods and extensive prey information from the literature to examine the impact of diet specificity (i.e., what prey a cone snail feeds upon) and dietary breadth (i.e., how many prey species a cone snail feeds upon) on two measures of conotoxin composition: 1) gene superfamily size and 2) total conotoxin diversity. Neither diet specificity nor dietary breadth was correlated in changes with gene superfamily size (distance-based phylogenetic generalized least squares [D-PGLS], P > 0.05). Although diet specificity did not predict changes in total conotoxin diversity (PGLS, P > 0.05), we found a significant positive relationship between dietary breadth and total conotoxin diversity in both the full conotoxin data set (PGLS, P < 0.05, fig. 6) and the conotoxin data set containing gene superfamilies that had > 80% capture success (PGLS, P < 0.001).
Fig. 6.
Scatterplot of total conotoxin gene diversity and dietary breadth. Each point represents a unique species. Graphs are labeled with a regression line and Pearson’s correlation coefficient generated from a phylogenetic generalized least squares (PGLS) analysis. Asterisk denotes significant correlation from the PGLS analysis.
Discussion
Targeted Sequencing and Conotoxin Discovery
Through targeted sequencing of conotoxins in cone snails, we demonstrate the potential to rapidly obtain venom sequences at high coverage (> 100×, table 1) from species for which no venom information is available and without the need of RNA from the venom duct. This is remarkable, given that alignments in amino acid sequences between mature regions of a single gene superfamily within a single individual can be incomprehensible (fig. 1) due to the rapid evolution of the mature region (Duda and Palumbi 1999). Effective capture of conotoxin gene superfamilies was possible in part because conotoxin exons were often directly adjacent to conserved UTRs, which were targeted in the design (supplementary table S3, Supplementary Material online). Although it has been recognized for decades that cone snails collectively harbor tens of thousands of biologically relevant proteins for fields such as molecular biology and pharmacology in their venoms (Olivera and Teichert 2007; Lewis 2009), traditional techniques for conotoxin sequencing (e.g., cDNA cloning) have barely begun to uncover and characterize the full breadth of conotoxin diversity. The targeted sequencing technique presented in this study adds an additional tool to increase the speed at which conotoxins are discovered. Specifically, this technique will allow the rapid toxin sequencing of genetic samples housed in museum collections, which often contain a large proportion of the species diversity for venomous taxonomic groups that have been amassed through several decades of intensive field expeditions. In addition, targeted sequencing approaches are not limited by the venom transcripts that are expressed at any one particular time point. As demonstrated here, on an average, over half of the conotoxin genes available in the genome are not expressed in adult individuals (supplementary table S5 and fig. S3, Supplementary Material online). Therefore, sequencing conotoxins from genomic samples effectively doubles the number of conotoxin sequences that can be recovered.
Overall, the proportion of reads that mapped to our targeted sequences (mean = 48.8%, table 1) is on par with studies that employed similar techniques in Anuran frogs (mean = 60.2%; Portik et al. 2016) and Salamanders (mean = 18.21%; McCartney-Melstad et al. 2016). The proportion of reads marked as duplicates were higher than previous studies (mean = 24.5%; McCartney-Melstad et al. 2016, mean = 17.5%; Portik et al. 2016). The high duplication levels in our data set may have been a function of our small target size (∼0.8 Mb) or a sign that we overamplified our posthybridization product. To reduce the duplication levels in the future, we may reduce the number of posthybridization PCR cycles. Given the high coverage on both the phylogenetic markers and conotoxin sequences (average cov > 95×, table 1), future sequencing experiments should be able to multiplex more than 32 samples on a single lane.
Although most of the gene superfamilies had high capture success, some gene superfamilies performed poorly (supplementary table S1, Supplementary Material online). Variation in overall capture success can be attributed to several factors: first, a lack of diversity in bait sequences for a particular gene superfamily may have impeded effective capture. For example, we only had bait sequences designed from two species for the divMKFPLLFISL gene superfamily and we were unable to recover full sequences from any of the other species (supplementary table S1, Supplementary Material online). Second, the genetic organization of gene superfamilies my hinder capture success. For example, the mature toxin exon for the T gene superfamily is not readily recoverable because it is not adjacent to a conserved UTR that is discoverable through transcriptome sequencing (supplementary tables S1 and S3 and fig. S1, Supplementary Material online). Finally, conotoxin sequence properties may hinder capture, as it has been documented that high or low GC content values can depress capture efficiency statistics (Gnirke et al. 2009). To increase capture success of gene superfamilies in the future, we recommend including a large diversity of sequences from several species for gene superfamilies that had low capture success. In addition, bait sequences should be redesigned for gene superfamilies in which the prepro region or the mature region were not immediately adjacent to the conserved UTRs. We recovered intron sequences in this study that can be used in future bait designs to effectively recover the entire coding region because adjacent noncoding regions are often evolving at a much slower rate than the coding region containing the prepro or mature region (supplementary table S4, Supplementary Material online, fig. 4, and supplementary fig. S2, Supplementary Material online).
Although targeted sequencing can increase the speed at which conotoxins are sequenced, we note several limitations with this approach. First, venom sequencing is limited to the sequences used in the bait design—only known venom gene families can be recovered. Therefore, approaches such as RNAseq are still necessary to identify undiscovered venom gene families. However, broad-scale discovery of venom gene superfamilies through transcriptome sequencing can be performed prior to applying target capture techniques (as done in this study) to obtain target sequences for most of the major venom components. Second, for some gene families, annotation of the exact mature toxin coding region may be difficult if the mature toxin is separated across multiple exons and if there is no closely related reference to accurately define the mature protein. Thus, expressed data are still necessary in some cases to study the mature toxin. Finally, depending on the level of sample multiplexing, targeted sequencing approaches can be more expensive than using RNAseq for venom discovery ($230.65 per RNAseq sample vs. $285.62 per targeted sequencing sample, supplementary table S7, Supplementary Material online). Therefore, the choice between two sequencing strategies will depend on the overall goal of the research project, as no one next-generation sequencing method is suited for all research applications (Jones and Good 2016).
When compared with conotoxin sequences available on Genbank and ConoServer, we found that we were able to recover a larger proportion of previously sequenced conotoxins if species were explicitly targeted with the baits (supplementary table S2, Supplementary Material online). Although this comparison is biased by unequal conotoxin discovery effort across species, nearly half of previously sequenced conotoxins were not recovered in this study. We performed a coarse investigation of database conotoxins and determined potential reasons for why we were not able to recover certain previously sequenced conotoxins. These reasons include: 1) the species in the database was misidentified, which was extensively documented in (Phuong et al. 2016), 2) the database conotoxin had no reliable reference in the literature, 3) the conotoxin was present in our species, but we could not recover it with the current bait design or the conotoxin was filtered during the bioinformatics processing of the data, and 4) the conotoxin was recoverable (i.e., high sequence similarity to bait sequences designed in this study), but the gene was not present in the genome. Future work integrating both population level RNAseq and targeted sequencing data may account for the large proportion of unrecovered conotoxins.
Conotoxin Genetic Architecture
Conotoxin exon length (range = 5–444 bp, supplementary table S3, Supplementary Material online) and the number of exons per gene (range = 1–6 exons, supplementary table S3, Supplementary Material online) are not unusual and fall within the range of variation seen in the genomes of other organisms (Deutsch and Long 1999; Sakharkar et al. 2004). In addition, the number of exons per gene within a gene superfamily align with results previous studies based on a relatively smaller number of sequences (Wu et al. 2013; Barghi et al. 2015a). For example, Barghi et al. (2015a) found that the J superfamily consisted of a single exon and (Wu et al. 2013) found several gene superfamilies (i.e., I1, I2, and M) consisting of three exons, which are identical to the results presented here.
A previous study suggested that rate variation among conotoxin functional regions (i.e., signal, prepro, mature) may be partially enabled by separation onto distinct exons in the genome (Olivera et al. 1999). Our results partially support this earlier hypothesis, given that the signal region and mature region were often confined to single exons (fig. 3). However, we found that the prepro region was distributed across multiple exons, conflicting with earlier hypotheses. Although not explicitly quantified, these results are also seen in earlier work examining the genomic architecture from conotoxin genes (Wu et al. 2013; Barghi et al. 2015a).
Conotoxin Molecular Evolution
A previous analysis of patterns of conotoxin divergence suggested that introns within contoxoin gene superfamilies were similar across species within a gene superfamily (Wu et al. 2013). Our results partially corroborate this suggestion, as the ratio of exon to noncoding divergence depended on what conotoxin region was encoded by the exon. Specifically, the exon containing the signal region was conserved and evolved much more slowly than adjacent noncoding regions (supplementary table S4, Supplementary Material online, fig. 4, and supplementary fig. S2, Supplementary Material online). This is similar to the pattern found in nonconotoxin exons (fig. 4), indicative of purifying selection removing deleterious mutations from coding regions of critically important proteins (Hughes and Yeager 1997). In contrast, the exon diverges faster than the noncoding regions in all other exons, with the clearest difference between exon and noncoding region divergence seen in the exon containing most or all of the mature toxin region. This pattern is indicative of positive selection and is the same pattern is also seen in other genes under positive selection, such as PLA2 genes in snakes (Nakashima et al. 1993, 1995; Gibbs and Rossiter 2008) and fertilization genes in abalone (Metz et al. 1998). Overall, the patterns reported in this study aligns with previous work characterizing rate variation in snake venom proteins (Nakashima et al. 1993, 1995; Gibbs and Rossiter 2008). Although we did not use traditional methods to test for positive selection (e.g., MK tests), positive selection is well documented in cone snails (Duda 2008; Duda and Remigio 2008; Puillandre et al. 2010) and is therefore inferred to shape patterns of increased divergence in coding regions relative to noncoding regions. In addition, this genomic divergence pattern is consistent with a recent analysis suggesting that the rapid evolution of conotoxin mature regions is due to positive selection (Roy 2016).
We found that on an average, cone snails only express a fraction of the conotoxin genes available in their genomes (supplementary table S5 and fig. S3, Supplementary Material online), concurring with similar reports from smaller sets of gene superfamilies (Chang and Duda 2012, 2014; Barghi et al. 2015a). Several reasons could lead to this pattern. First, it is known that expression changes throughout an individual’s lifetime in cone snails (Barghi et al. 2015b; Chang and Duda 2016), suggesting that the complement of genes expressed in the transcriptomes from Phuong et al. (2016) represent the adult conotoxins, and genes not discovered in the transcriptome but recovered from the genome are genes that are expressed in other life stages. Second, prey taxa available to cone snail species change with geography and so do the conspecifics it must compete against (Kohn 1959a, 1978; Kohn and Nybakken 1975; Duda and Lee 2009; Chang et al. 2015); therefore, different genes may be turned on or off in different geographic localities depending on the prey resources available and the composition of competitors in an individual’s environment. Third, some of the conotoxin genes in the genome may not be expressed because they are no longer functional and have become psuedogenized. Finally, conotoxin gene expression may be regulated by defensive strategies against predators, as cone snails have been documented to release different conotoxins based on differing external stimuli (presentation of a prey item vs. physical agitation through poking, Dutertre et al. 2014). However, this hypothesis remains to be tested as there exists no ecological information to suggest that cone snails use their venom for defense—observations in the literature show that often, cone snails will hide in their shell or become completely devoured when confronted with an aggressor (Kohn 1959b). Future work comparing patterns of expression relative to genomic availability will be able to disentangle the impact of conotoxin expression on changes to the venom phenotype.
We detected evidence for phylogenetic signal in the membership size of 25 gene superfamilies (supplementary table S6, Supplementary Material online and fig. 5), suggesting that history plays a role in shaping gene gains and losses in cone snails. We note that uncovering evidence for phylogenetic signal in gene superfamily size does not imply that natural selection has not played a role in the evolution of venom as implied in (Gibbs et al. 2013). As described in Revell et al. (2008), evolutionary processes should not be inferred from patterns of phylogenetic signal because several contrasting models of trait evolution can lead to similar amounts of phylogenetic signal. Through CAFE v3.1 analyses, we also showed that venom composition is shaped by both net gains and losses in the entire genomic content of conotoxins (fig. 5 and supplementary fig. S4, Supplementary Material online). This result is in line with past studies showing that gene turnover is a fundamental characteristic shaping species’ genic venom content (Duda and Palumbi 1999; Chang and Duda 2012; Dowell et al. 2016). We note a few limitations to the data used to examine gene turnover. First, total conotoxin gene diversity may be underestimated if large undiscovered gene superfamilies are present in specific clades of cone snails. Second, we only used one sample per species and technical variability in sequence capture and sample quality may have impacted our total conotoxin gene diversity estimates.
Diet and Venom Evolution
Why do cone snails vary in conotoxin gene superfamily size? Contrary to the popular assumption that particular gene superfamilies are associated with certain prey items (e.g., Kaas et al. 2010; Jin et al. 2013), prey families did not predict changes in gene superfamily size or total conotoxin diversity. This result aligns with a growing body of literature suggesting that the specific prey a species feeds upon may not accurately predict conotoxin gene superfamily composition (Puillandre et al. 2012; Chang et al. 2015; Phuong et al. 2016). Although this study, along with previous studies, did not find a correlation between prey families and measures of conotoxin composition, this does not imply a lack of a relationship between diet specificity and conotoxin evolution for the following reasons. Characterizing the functional aspects of conotoxins is critical to understanding the relationship between diet specificity and conotoxin evolution because prey specialization exists at the level of protein function. However, it is known that conotoxin gene superfamilies are poor predictors of protein function and conotoxins with similar functions can convergently evolve in different gene superfamilies (Kaas et al. 2010; Puillandre et al. 2012; Robinson and Norton 2014). Therefore, if the functional aspects of cone snail venom repertoires are examined, a correlation between diet specificity and conotoxin composition may appear, such as in the case with cone snail insulins (Safavi-Hemami et al. 2016). We also acknowledge that our sampling of dietary diversity is not comprehensive (mollusc-hunting and fish-hunting species are undersampled) and this may limit our ability to fully examine diet specificity and conotoxin composition.
Although dietary breadth also did not predict changes in gene superfamily size, we found a significant positive relationship with total conotoxin diversity (fig. 6), aligning with several studies showing a coupling between dietary breadth and venom gene diversity in cone snails at nearly all biological scales of organization (Duda and Lee 2009; Chang et al. 2015; Chang and Duda 2016; Phuong et al. 2016). The correlation coefficient in this study between dietary breadth and total conotoxin diversity was weaker (r2=0.25) than in our previous study (r2=0.75, Phuong et al. 2016), possibly due to examining all of the conotoxin genes in the genome rather than just expressed transcripts. Conotoxin expression is known to change throughout an individual’s lifetime (Barghi et al. 2015b; Chang and Duda 2016) and these changes in expression have been shown to track dietary breadth (Chang and Duda 2016). Therefore, the weaker relationship could possibly be explained by using dietary breadth values measured from adult populations and examining its relationship to the total conotoxin repertoire an individual may draw from throughout its lifetime. In addition, we note that we were not able to distinguish between pseudogenes and functional genes, and this may contribute to the weaker relationship between total conotoxin diversity and dietary breadth. Another limitation of these analyses is that the number of individuals sampled per H index calculation was uneven and H index values may be biased toward species that had populations that were more extensively sampled.
The importance of dietary breadth shaping venom evolution remains underappreciated and untested in other venomous systems despite signals across several studies in cone snails. Future work examining the role of dietary breadth in shaping the evolution of venom in other venomous taxa will greatly advance our understanding between the interplay between diet and venom. The lack of a relationship between dietary breadth and changes in conotoxin gene superfamily size suggests that venom should be characterized as an aggregate trait rather than decomposed into individual parts to fully assess the impact of dietary breadth on conotoxin evolution. Further, studies have documented synergistic and complementary effects of conotoxins on prey species, suggesting that selection may act on the entire cocktail rather than individual components (Olivera 1997).
Conclusions
Through targeted sequencing of conotoxin genes, we provided comprehensive analyses of the gene structure of conotoxin gene superfamilies. In addition, we improved understanding of conotoxin molecular evolution, including examining how positive selection impacts patterns of genomic divergence, how expression regulation of gene superfamilies varies across species, and how total conotoxin diversity changes through time. In addition, we found that variation in conotoxin diversity tracks changes in dietary breadth, suggesting that species with more generalist diets contain a greater number of conotoxin genes in their genome. Given that increased gene diversity is thought to confer an increased capacity for evolutionary change and species diversification (Kirschner and Gerhart 1998; Yang 2001; Malmstrøm et al. 2016), generalist species may speciate at faster rates than species with specialist diets. The targeted sequencing technique presented in this paper provides the necessary methodological advancement to rapidly sequence toxin genes across diverse clades of species, allowing tests of the relationship between ecology, toxin gene diversity, and higher order biodiversity patterns to be realized in future work.
Materials and Methods
Bait Design and Data Collection
To recover markers for phylogenetic analyses, we targeted 886 protein coding genes representing 728,860 bp. 482 of these genes were identified to be orthologous in Pulmonate gastropods (Teasdale et al. 2016) and we identified the remaining 404 genes using a reciprocal blast approach with 12 Conidae transcriptomes from (Phuong et al. 2016). For each gene, we chose the longest sequence from 1 of the 12 Conidae transcriptomes as the target sequence. For 421 of these genes, we used the entire length of the sequence as the target sequence, while for the remaining genes, we sliced the target sequences into smaller components based on exon/intron boundaries inferred with EXONERATE v2.2.0 (Slater and Birney 2005) using the Lottia gigantea genome as our reference. EXONERATE v2.2.0 was run under default parameters and under the est2genome model. We chose to use the L. gigantea genome as our reference because it is highly contiguous (scaffold N50 = 1.87 Mb) and well annotated (Simakov et al. 2013), as a Conidae genome of comparable quality was not available at the time of the bait design. Often, exon/intron boundaries are conserved across fairly divergent taxa and can be used to define exon/intron boundaries (Bi et al. 2012); therefore, the L. gigantea genome was an appropriate choice to define exon/intron boundaries here. If exons were <120 bp in length (i.e., our desired bait length), but longer than 50 bp, we generated chimeric target sequences by fusing immediately adjacent exons. We tiled bait sequences every 60 bp across each target sequence. We note that the split bait design was due to an internal communication error and was not for a prespecified purpose. For the conotoxin genes, we targeted 1,147 conotoxins discovered from an early analysis of the 12 transcriptomes described in (Phuong et al. 2016). These sequences represent regions targeting 49 gene superfamilies and we included the full protein coding region plus 100 bp of the 5′- and 3′-UTRs in our bait design when possible (supplementary table S1, Supplementary Material online). We tiled bait sequences every 40 bp across each conotoxin target sequence.
We obtained tissue samples preserved in 95% ethanol for 32 Conidae species through field collections in Australia and Indonesia and from the collections at the Australian Museum in Sydney, Australia (table 1). We verified species identities using shell morphology and by sequencing CO1 prior to any next-generation sequencing laboratory work. We extracted genomic DNA from foot tissue using an EZNA Mollusc DNA kit (Omega Bio-Tek, Doraville, GA) and used 1,500 ng of total DNA to prepare index-specific libraries following the Meyer and Kircher (2010) protocol. To increase the probability of obtaining sequence information beyond the targeted regions, we performed 1× bead purifications after all enzymatic steps to remove fragments <250 bp. Fragment lengths of DNA samples ranged from 300 to 1,000 bp, with an average length of 450 bp prior to hybridizations. We performed capture reactions following the MYbaits (v2) with the following specifications:
We pooled eight samples at a total concentration of 1.6 µg DNA per capture reaction and allowed the baits to hybridize with the DNA for ∼24 hours.
We substituted the universal blockers provided with the MYbaits kit with xGEN blockers (Integrated DNA Technologies).
We executed the “stringent wash” protocol during the recovery of the captured targets.
After hybridization, we sequenced all 32 samples on a single lane HiSeq2000 lane with 100 bp paired-end reads. Fragment lengths prior to hybridization were identical to the fragment length distributions posthybridization that were submitted for sequencing.
Data Assembly, Processing, and Filtration
We trimmed reads for quality and adapter contamination using Trimmomatic v0.33 (Bolger et al. 2014) under the following conditions: 1) we used the ILLUMINACLIP option to trim adapters with a seed mismatch threshold of 2, a palindrome clip threshold of 40, and a simple clip threshold of 15, 2) we performed quality trimming used the SLIDINGWINDOW option with a window size of 4 and a quality threshold of 20, 3) we removed reads <36 bp by setting the MINLEN option to 36, and 4) we removed leading and trailing bases under a quality threshold of 15. We merged reads using FLASH v1.2.8 (Magoč and Salzberg 2011) with a min overlap parameter of 5, a max overlap parameter of 100, and a mismatch ratio of 0.05. We generated assemblies for each sample using SPAdes v3.1.0 (Bankevich et al. 2012) under default parameters. We reduced redundancy in the assemblies with cap3 (Huang and Madan 1999) under default parameters and cd-hit v4.6 (Li and Godzik 2006) using a sequence identity threshold of 99%.
For the phylogenetic markers, we used BLAST+ v2.2.31 (Altschul et al. 1990) with an evalue threshold of 1×10−10 and a word size value of 11 to associate assembled contigs with the target sequences. We used EXONERATE v2.2.0 under default parameters and used the est2genome model to redefine exon/intron boundaries because either 1) exon/intron boundaries were never denoted or 2) previously defined exons were actually composed of smaller exons. For each sample, we used bowtie2 v2.2.6 (Langmead and Salzberg 2012) using the very sensitive local alignment option and not allowing for discordant pair mapping (unexpected paired read orientation during mapping) to map reads to a reference containing only the contigs associated with the original target sequences. We marked duplicates using picard-tools v2.0.1 (http://broadinstitute.github.io/picard) using default parameters. We masked all positions that were <5× coverage and removed the entire sequence if > 30% of the sequence was masked. To filter potential paralogous sequences in each species, we calculated heterozygosity (number of heterozygous sites/total number of sites) for each locus by identifying heterozygous positions using samtools v1.3 using default parameters and bcftools v1.3 (Li et al. 2009) using the call command and removed loci that were at least two SDs away from the mean heterozygosity.
For the conotoxin sequences, it is known that traditional assemblers perform poorly in reconstructing all potential conotoxin gene copies (Lavergne et al. 2015; Phuong et al. 2016). To ameliorate this issue, we reassembled conotoxin genes using the assembler PRICE v1.2 (Ruby et al. 2013), which employs iterative mapping and extension using paired read information to build out contigs from initial seed sequences. To identify potential seed sequences for contig extension, we first mapped reads to the entire assembly outputted by SPAdes using bowtie2 v2.2.6 with previously stated parameters for each program. Then, we identified all sequence regions that locally aligned to any part of the original conotoxin target sequences via blastn v2.2.31 using previously stated parameters; these regions represented our preliminary seed sequences. We kept all preliminary seed sequences that were at least 100 bp (read length of samples in this study) and extended these seeds to 100 bp if the alignable region was below that threshold. When extending these initial regions, we used Tandem Repeats Finder v4.09 (Benson 1999) to identify simple repeats and minimize the presence of these genomic elements in the preliminary seed sequences. We executed Tandem Repeats Finder v4.09 with default parameters except for the Minscore parameter, which we set at 12, and the Maxperiod parameter, which we set at 2. Often, only a subset of conotoxins are fully assembled with traditional assemblers (Phuong et al. 2016). However, when reads are mapped to these assemblies, unique conotoxin loci are similar enough to each other that relaxed mapping parameters will allow multiple copies to map to the contigs that were assembled. Therefore, multiple conotoxin copies will often map to each preliminary seed sequence. To generate seed sequences for all unique conotoxin loci, we used the python module pysam (https://github.com/pysam-developers/pysam) to pull all reads that mapped to regions of contigs representing the preliminary seed sequence and we reconstructed contigs from these reads using cd-hit v4.6 (percent identity = 98%) and cap3 (overlap percent identity cutoff =99%). From these reconstructed contigs, we used blastn v2.2.31 using previously described parameters to identify >100 bp regions that matched the original preliminary seed and used these hits as our final seeds. We merged all final seeds that were 100% identical using cd-hit v4.6, mapped reads to these seeds using bowtie2 v2.2.6 with previously described parameters, and used PRICE v1.2 to reassemble and extend each seed sequence under 5 minimum percentage identity (MPI) values (90%, 92%, 94%, 96%, 98%) with only the set of reads that mapped to that initial seed. Sequences were assembled using a minimum overlap length value of 40 and a threshold value of 20 for scaling overlap for contig-edge assemblies. A sequence was successfully reassembled if it shared ≥ 90% identity to the original seed sequence and if the final sequence was longer than the initial seed. For each seed sequence, we only retained the longest sequence out of the five MPI iterations for downstream filtering. We illustrated and described this workflow in supplementary figure S5, Supplementary Material online.
In order to generate a conotoxin reference database containing sequences that included both exons and adjacent noncoding regions, we used blastn v2.2.31 and EXONERATE v2.2.0 (using parameters described earlier) on species that were used in the bait design to 1) perform species-specific searches between our reassembled contigs and reference conotoxin sequences from (Phuong et al. 2016) and 2) define exon/intron boundaries on our reassembled contigs. We chose to constrain our blast searches to species-specific searches in order to improve accuracy and decrease the complexity of the data processing. In cases where a predicted terminal exon (i.e., the first or last exon of a conotoxin) was short (< 40 bp) and did not blast to any reassembled contig in our exon capture data set, we replaced the reference conotoxin from Phuong et al. (2016) with the identical conotoxin containing the adjacent UTR regions to aid in the sequence searches. We generated conotoxins with UTR regions using the PRICE v1.2 algorithm as described earlier because the reference conotoxins from the final data set in Phuong et al. (2016) did not include the UTR regions. We concatenated all annotated sequences into a single file to create the final conotoxin reference, which consisted of sequences with exons and introns defined from all species that were initially used in the bait design.
With the final conotoxin reference, we used blastn v2.2.31 to associate contigs with this reference in every species and used EXONERATE v2.2.0, blastx v2.2.31, and tblastn v2.2.31 to define exon/intron boundaries. For the BLAST+ v2.2.31 software, we used parameters previously described earlier and for EXONERATE v2.2.0, we used the protein2genome model and an alignment score threshold value of 50. When exon/intron boundaries could not be defined through these methods, we guessed the boundaries by aligning the assembled contig to the reference sequence using MAFFT v7.305 b (Katoh et al. 2005) and denoted the boundaries across the region of overlap with the exon in the reference sequence. For each sample, we mapped reads using bowtie2 v2.2.6, accounted for duplicates using picard-tools v2.0.1, and retained sequences that had at least 10× coverage across the exons defined within each contig. We increased mapping stringency for bowtie2; in addition to using the parameters previously described, we modified the alignment score threshold (–score-min L, 70,1). We masked regions <10× coverage and used cd-hit v4.6 to merge contigs that were ≥ 98% similar, generating our final conotoxin gene models. Finally, we used HapCUT v0.7 (Bansal and Bafna 2008) under default parameters to generate all unique haplotypes across coding regions. We note that our final conotoxin data set consists of exon fragments (conotoxin exons with any assembled adjacent noncoding regions), rather than full conotoxin genes. As described in (Wu et al. 2013), conotoxin introns can often be several kilobases in length, which is much longer than the average insert size of our sequencing experiment (∼350 bp).
To assess the overall effectiveness of our targeted sequencing experiment, we calculated 1) percent of reads aligned to intended targets, 2) percent duplicates, and 3) average coverage across targeted regions. To assess capture success of conotoxins, we divided the number of conotoxin transcripts successfully recovered in the exon capture data set by the number of conotoxin transcripts discovered in Phuong et al. (2016) for each gene superfamily. We defined a conotoxin transcript to be successfully sequenced if > 80% of the transcript was recovered in the exon capture experiment with > 95% identity. To assess the ability of targeted sequencing to recover gene superfamily sequences from species that were not explicitly targeted in the bait sequences, we calculated the number of previously sequenced conotoxins that match contigs recovered in our data set. We gathered conotoxin sequences from Genbank and ConoServer (Kaas et al. 2010) with species names that correspond to species in this study, merged sequences with 98% identity using cd-hit v4.6, and used blastn v2.2.31 under previously stated parameters to perform species-specific searches. We defined a conotoxin as successfully sequenced if the hypervariable mature toxin coding region aligned with ≥ 95% identity to a sequence in our data set.
Conotoxin Genetic Architecture
To characterize conotoxin genetic architecture, we quantified the following values: 1) the number of exons comprising a conotoxin transcript, 2) average length of each exon, and 3) the size range of exon length. We also determined the proportion of terminal exons adjacent to UTRs by conducting sequence searches (via blastn v2.2.31 under previously stated parameters) between contigs containing terminal exons against a database of conotoxins from Phuong et al. (2016) that were reassembled to contain the UTRs. To determine how traditional conotoxin precursor peptide regions are distributed among exons, we calculated the average proportion of each conotoxin region found on each exon in every gene superfamily. We defined regions of the Phuong et al. (2016) transcripts using ConoPrec (Kaas et al. 2012). We restricted these conotoxin genetic structure analyses to transcripts from Phuong et al. (2016) that were successfully recovered in the exon capture data set and that were retained after clustering with cd-hit v4.6 (similarity threshold = 98%). We performed clustering to avoid overinflating estimates because unique transcripts from Phuong et al. (2016) may have originated from the same gene.
Conotoxin Molecular Evolution
We first classified all exons into conotoxin precursor peptide regions. For species with transcriptome data, we first labeled exons as either the signal region or the mature region by identifying the exons containing the largest proportion of these separate regions. Then, exons between the signal and mature exon were labeled as the prepro exon(s) and exons after the mature region were labeled as the postexons. Gene superfamilies containing only a single exon were denoted as such. We then used blastn v2.2.31 under previously stated parameters to classify sequences without transcriptome data into these conotoxin precursor peptide regions. For each functional category within each gene superfamily, we calculated uncorrected pairwise distances between all possible pairwise comparisons. To avoid spurious alignments, we only considered comparisons within clusters that clustered with cd-hit v4.6 at an 80% threshold and we excluded comparisons if 1) the alignment length of the two exons was 20% greater than the longer exon, 2) the align-able nocoding region was <50 bp, or 3) the shorter exon’s length was <70% of the length of the longer exon. We calculated separate pairwise distance estimates for regions of the alignment that contained the exon and regions of the alignment that contained the noncoding DNA. We excluded region-labeled exons within superfamilies from this analysis that had <50 possible comparisons. For comparison, we also calculated pairwise distances between exons and noncoding regions across our phylogenetic markers which represent nonconotoxin exons, filtered with similar criteria described earlier.
Conotoxin Expression
To characterize variation in expression patterns among species per gene superfamily, we calculated the number of conotoxin genes expressed in species with transcriptome data divided by the number of genes available in the genome. We restricted these analyses to instances where 90% of the unique mature toxins were recovered for a gene superfamily within a species. To estimate gene superfamily size, we used the exon labeled as containing most or all of the mature region. We used the mature region because it is unique between sequences discovered from the transcriptome. We could not, for example, use a signal region sequence, as they may map to several sequences from the target capture data given that they are highly conserved within a gene superfamily. We defined a conotoxin gene as expressed if we retained a blast hit with 95% identity to a unique mature toxin sequence found in the transcriptome.
Gene Superfamily Size Change Estimation
To compare and contrast gene superfamily size changes between species, we used the total number of exons containing most or all of the signal region as our estimate of gene superfamily size because exons containing the signal regions are relatively conserved across species (Robinson and Norton 2014) and thus have the highest confidence of being recovered through exon capture techniques. To quantify and test the amount of phylogenetic signal in conotoxin gene diversity, we estimated Pagel’s lambda (Pagel 1997) in the R package phytools (Revell 2012). Lambda values range from 0 (phylogenetic independence) to 1 (phylogenetic signal) and P values < 0.05 represent significant departure from a model of random trait distribution across species with respect to phylogeny. To estimate conotoxin gene superfamily gains and losses along every branch, we used the program CAFEv3.1 (Han et al. 2013), which uses a stochastic gene birth–death process to model the evolution of gene family size. As input, we used a time-calibrated phylogeny and estimates of gene superfamily size for 37 superfamilies that were present in at least 2 taxa. To estimate a time-calibrated phylogeny, we aligned loci that had at least 26 species using MAFFT v7.305 b under default parameters and used a concatenated alignment to build a phylogeny in RAxML under a GTRGAMMA model of sequence evolution (Stamatakis 2006). We performed a maximum likelihood search of the phylogenetic tree and the rapid bootstrapping analyses with 100 replicates. We time-calibrated the phylogeny with the program r8s (Sanderson 2003) under default parameters and using two previous fossil calibrations described in cone snails (Duda et al. 2001). We excluded Californiconus californicus from the CAFE v3.1 analysis due to optimization failures.
Diet and Conotoxin Gene Superfamily Size Evolution
To examine the role of diet specificity and dietary breadth on conotoxin gene superfamily size evolution and total conotoxin diversity, we retrieved prey information from the literature (Kohn 1959a,b, 1966, 1968, 1978, 1981, 2001, 2003, 2015; Marsh 1971; Kohn and Nybakken 1975; Taylor 1978, 1984, 1986; Taylor and Reid 1984; Nybakken and Perron 1988; Kohn and Almasi 1993; Reichelt and Kohn 1995; Kohn et al. 2005; Nybakken 2009; Chang et al. 2015). For diet specificity, we classified prey items into 1 of 27 different prey categories (supplementary table S8, Supplementary Material online). For dietary breadth, we retrieved estimates of the Shannon’s diversity index (H′) or calculated it if there were at least five prey items classified to genus with a unique species identifier. When multiple H′ values were obtained for a species, we averaged them because species will consume different sets of prey taxa depending on geography. Raw data are available in supplementary table S8, Supplementary Material online. To examine the impact of prey group and dietary breadth on changes in gene superfamily size, we used D-PGLS, a phylogenetic regression method capable of assessing patterns in high-dimensional data sets (Adams 2014). To reduce redundancy among prey group variables, we removed variables that were 80% correlated with each other using the redun function in the R package Hmisc. We used the total number of exons containing the signal region as our estimate of gene superfamily size. To convert gene superfamily size counts into continuous variables, we transformed the data into χ2 distances between species in “conotoxin gene superfamily space” using the deostand function in the R package vegan (Oksanen et al. 2016). To examine the impact of diet specificity and dietary breadth on total conotoxin diversity, we used a PGLS analysis implemented in the caper package within R (Orme 2013). We ln-transformed total conotoxin diversity for the PGLS analysis. We performed all analyses with the full data set and a subset of the data that only included gene superfamilies with > 80% capture success. We did not perform any analyses with C. californicus because it is regarded as an outlier species among the cone snails due to its atypical diet and its deep relationship with the rest of Conidae (Kohn 1966; Puillandre et al. 2014).
Data Availability
Raw read data can be found at the National Center for Biotechnology Information associated with BioProject Accession #PRJNA437715. Scripts used to process the data can be found on Github (https://github.com/markphuong/venom.targetcapture.pilot and https://github.com/markphuong/phylogenetics.targetcapture.pilot). The bait sequences, final phylogenetic alignment, and all conotoxin sequences can be found on Dryad (doi:10.5061/dryad.vk245pd).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank DST Hariyanto, MBAP Putra, MKAA Putra, and the staff at the Indonesian Biodiversity Research Center in Denpasar, Bali for assistance in the field in Indonesia; F Criscione, F Köhler, A Moussalli, A Hogget, and L Vail for logistical assistance for fieldwork at the Lizard Island Research Station in Australia; M Reed, A Hallan, and J Waterhouse for access to specimens at the Australian Museum in Sydney, Australia; J Finn, M Mackenzie, and M Winterhoff for access to specimens at the Museum Victoria in Melbourne, Australia; WF Gilly for access to the C. californicus specimen, K Bi, L Smith, and A Moussalli for advice on bait design; A. Devault and MYcroarray for great service and technical support for bait synthesis; EM McCartney-Melstad, the B Shaffer lab, and the Evolutionary Genetics Lab at UC Berkeley for laboratory support; A. Kohn for providing cone snail diet data from several publications; MCW Lim and J Chang for thoughtful advice and discussions; N Puillandre, S Prost, S Robinson, and six anonymous reviewers for insightful comments on earlier versions of this manuscript. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number ACI-1053575. This work was supported by a Grants-in-Aid of research from Sigma Xi, a Grants-in-Aid of Research from the Society for Integrative and Comparative Biology, a research grant from the Society of Systematic Biologists, a Student Research Award from the American Society of Naturalists, a National Science Foundation Graduate Research Opportunities Worldwide to Australia, the Lerner Gray Fund for Marine Research from the American Museum of Natural History, research grants from the Department of Ecology and Evolutionary Biology at UCLA, a small award from the B Shaffer Lab, a National Science Foundation Graduate Research Fellowship, an Edwin W. Pauley fellowship, and a Chateaubriand fellowship awarded to M.A.P. This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 Instrumentation Grants S10RR029668 and S10RR027303. We thank the Indonesian Ministry of State for Research and Technology (RISTEK, permit number 277/SIP/FRP/SM/VIII/2013) for providing permission to conduct fieldwork in Bali. The C. californicus specimen was collected under a California Department of Fish and Wildlife collecting permit granted to WF Gilly (SC-6426).
References
- Adams DC. 2014. A method for assessing phylogenetic least squares models for shape and other high-dimensional multivariate data. Evolution 689:2675–2688. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic Local AlignmentSearch Tool. J Mol Biol. 2153:403–410. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD et al. , 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 195:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal V, Bafna V.. 2008. HapCUT: an efficient and accurate algorithm for the haplotype assembly problem. Bioinformatics 2416:i153–i159. [DOI] [PubMed] [Google Scholar]
- Barghi N, Concepcion GP, Olivera BM, Lluisma AO.. 2015a. Structural features of conopeptide genes inferred from partial sequences of the Conus tribblei genome. Mol Genet Genomics 291:411–422. [DOI] [PubMed] [Google Scholar]
- Barghi N, Concepcion GP, Olivera BM, Lluisma AO.. 2015b. Comparison of the venom peptides and their expression in closely related Conus species: insights into adaptive post-speciation evolution of Conus exogenomes. Genome Biol Evol. 7:1797–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow A, Pook CE, Harrison RA, Wüster W.. 2009. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc R Soc B 2761666:2443–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem Repeats Finder: a program to analyse DNA sequences. Nucleic Acids Res. 272:573–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi K, Vanderpool D, Singhal S, Linderoth T, Moritz C, Good JM.. 2012. Transcriptome-based exon capture enables highly cost-effective comparative genomic data collection at moderate evolutionary scales. BMC Genomics 13:403.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 3015:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, Wagner CE, Li YI.. 2014. The genomic substrate for adaptive radiation in African cichlid fish. Nature 51:375–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casewell NR, Harrison RA, Wüster W, Wagstaff SC.. 2009. Comparative venom gland transcriptome surveys of the saw-scaled vipers (Viperidae: echis) reveal substantial intra-family gene diversity and novel venom transcripts. BMC Genomics 10:564.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casewell NR, Wüster W, Vonk FJ, Harrison RA, Fry BG.. 2013. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol Evol. 284:219–229. [DOI] [PubMed] [Google Scholar]
- Chang D, Duda TF.. 2012. Extensive and continuous duplication facilitates rapid evolution and diversification of gene families. Mol Biol Evol. 298:2019–2029. [DOI] [PubMed] [Google Scholar]
- Chang D, Duda TF.. 2014. Application of community phylogenetic approaches to understand gene expression: differential exploration of venom gene space in predatory marine gastropods. BMC Evol Biol. 14:123.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D, Duda TF.. 2016. Age-related association of venom gene expression and diet of predatory gastropods. BMC Evol Biol. 16:27.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D, Olenzek AM, Duda TF Jr.. 2015. Effects of geographical heterogeneity in species interactions on the evolution of venom genes. Proc R Soc B 2821805:20141984.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornetti L, Valente LM, Dunning LT, Quan X, Black RA, Hébert O, Savolainen V.. 2015. The genome of the “great speciator” provides insights into bird diversification. Genome Biol Evol. 79:2680–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daltry JC, Wüster W, Thorpe RS.. 1996. Diet and snake venom evolution. Nature 3796565:537–540. [DOI] [PubMed] [Google Scholar]
- Deutsch M, Long M.. 1999. Intron – exon structures of eukaryotic model organisms. Nucleic Acids Res. 2715:3219–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell NL, Giorgianni MW, Kassner VA, Selegue JE, Sanchez EE, Carroll SB.. 2016. The deep origin and recent loss of venom toxin genes in rattlesnake. Curr Biol. 2618:2434–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda TF. 2008. Differentiation of venoms of predatory marine gastropods: divergence of orthologous toxin genes of closely related Conus species with different dietary specializations. J Mol Evol. 673:315–321. [DOI] [PubMed] [Google Scholar]
- Duda TF Jr, Kohn AJ, Palumbi SR.. 2001. Origins of diverse feeding ecologies within Conus, a genus of venomous marine gastropods. Biol J Linn Soc. 734:391–409. [Google Scholar]
- Duda TF, Lee T.. 2009. Ecological release and venom evolution of a predatory marine snail at Easter Island. PLoS One 45:e5558.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda TF, Palumbi SR.. 1999. Molecular genetics of ecological diversification: duplication and rapid evolution of toxin genes of the venomous gastropod Conus. Proc Natl Acad Sci U S A. 9612:6820–6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda TF, Remigio EA.. 2008. Variation and evolution of toxin gene expression patterns of six closely related venomous marine snails. Mol Ecol. 1712:3018–3032. [DOI] [PubMed] [Google Scholar]
- Dutertre S, Jin A-H, Vetter I, Hamilton B, Sunagar K, Lavergne V, Dutertre V, Fry BG, Antunes A, Venter DJ et al. , 2014. Evolution of separate predation- and defence-evoked venoms in carnivorous cone snails. Nat Commun. 5:3521.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faircloth BC, McCormack JE, Crawford NG, Harvey MG, Brumfield RT, Glenn TC.. 2012. Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Syst Biol. 615:717–726. [DOI] [PubMed] [Google Scholar]
- Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, Martínez AT, Otillar R, Spatafora JW, Yadav JS et al. , 2012. The Paleozoic origin of enzymatic from 31 fungal genomes. Science 3366089:1715–1719. [DOI] [PubMed] [Google Scholar]
- Fry BG, Roelants K, Champagne DE, Scheib H, Tyndall JDA, King GF, Nevalainen TJ, Norman JA, Lewis RJ, Norton RS et al. , 2009. The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu Rev Genomics Hum Genet. 10:483–511. [DOI] [PubMed] [Google Scholar]
- Gibbs HL, Rossiter W.. 2008. Rapid evolution by positive selection and gene gain and loss: pLA(2) venom genes in closely related Sistrurus rattlesnakes with divergent diets. J Mol Evol. 662:151–166. [DOI] [PubMed] [Google Scholar]
- Gibbs HL, Sanz L, Sovic MG, Calvete JJ.. 2013. Phylogeny-based comparative analysis of venom proteome variation in a clade of rattlesnakes (Sistrurus sp.). PLoS One 86:e67220.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C et al. , 2009. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 272:182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén Y, Rius N, Delprat A, Williford A, Muyas F, Puig M, Casillas S, Ràmia M, Egea R, Negre B et al. , 2014. Genomics of ecological adaptation in cactophilic Drosophila. Genome Biol Evol. 71:349–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MV, Thomas GWC, Lugo-Martinez J, Hahn MW.. 2013. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol Biol Evol. 308:1987–1997. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A.. 1999. CAP3: a DNA sequence assembly program. Genome Res. 99:868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Yeager M.. 1997. Comparative evolutionary rates of introns and exons in murine rodents. J Mol Evol. 452:125–130. [DOI] [PubMed] [Google Scholar]
- Jin A-h, Dutertre S, Kaas Q, Lavergne V, Kubala P, Lewis RJ, Alewood PF.. 2013. Transcriptomic messiness in the venom duct of Conus miles contributes to conotoxin diversity. Mol Cell Proteomics 1212:3824–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M, Good J.. 2016. Targeted capture in evolutionary and ecological genomics. Mol Ecol. 251:185–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas Q, Westermann J-C, Craik DJ.. 2010. Conopeptide characterization and classifications: an analysis using ConoServer. Toxicon 558:1491–1509. [DOI] [PubMed] [Google Scholar]
- Kaas Q, Yu R, Jin A-H, Dutertre S, Craik DJ.. 2012. ConoServer: updated content, knowledge, and discovery tools in the conopeptide database. Nucleic Acids Res. 40(Database issue):D325–D330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K-I, Toh H, Miyata T.. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 332:511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner M, Gerhart J.. 1998. Evolvability. Proc Natl Acad Sci U S A. 9515:8420–8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn A, Almasi K.. 1993. Comparative ecology of a biogeographically heterogeneous Conus assemblage. Proceedings of the Fifth International Marine Biological Workshop: The Marine Flora and Fauna of Rottnest Island. pp. 509–521.
- Kohn AJ. 1959a. Ecological notes on Conus (Mollusca: gastropoda) in the Trincomalee region of Ceylon. Ann Mag Nat Hist. 2:308–320. [Google Scholar]
- Kohn AJ. 1959b. The ecology of Conus in Hawaii. Ecol Monogr. 29:47–90. [Google Scholar]
- Kohn AJ. 1966. Food specialization in Conus in Hawaii and California. Ecology 476:1041–1043. [Google Scholar]
- Kohn AJ. 1968. Microhabitats, abundance and food of Conus on atoll reef’s in the Maldive and Chagos islands. Ecology 496:1046–1062. [Google Scholar]
- Kohn AJ. 1978. Ecological shift and release in an isolated population: Conus miliaris at Easter Island. Ecol Monogr. 483:323–336. [Google Scholar]
- Kohn AJ. 1981. Abundance, diversity, and resource use in an assemblage of Conus species in Enewetak Lagoon. Pac Sci. 34:359–369. [Google Scholar]
- Kohn AJ. 2001. Maximal species richness in Conus: diversity, diet and habitat on reefs of northeast Papua New Guinea. Coral Reefs 20:25–38. [Google Scholar]
- Kohn AJ. 2003. Biology of Conus on shores of the Dampier Archipelago, Northwestern Australia. In: Wells FE, Walker DI and Jones DS, editors. The Marine Flora and Fauna of Dampier, Western Australia. Perth: Western Australian Museum.
- Kohn AJ. 2015. Ecology of Conus on Seychelles reefs at mid-twentieth century: comparative habitat use and trophic roles of co-occurring congeners. Mar Biol. 16212:2391–2407. [Google Scholar]
- Kohn AJ, Curran KM, Mathis BJ.. 2005. Diets of the predatory gastropods Cominella and Conus at Esperance, Western Australia In: Wells FE, Walker DI and Kendrick GA, editors. 2005. The Marine Flora and Fauna of Esperance, Western Australia. Perth: Western Australian Museum. pp. 235–244. [Google Scholar]
- Kohn AJ, Nybakken JW.. 1975. Ecology of Conus on eastern Indian Ocean fringing reefs: diversity of species and resource utilization. Mar Biol. 293:211–234. [Google Scholar]
- Kordis D, Gubensek F.. 2000. Adaptive evolution of animal toxin multigene families. Gene 2611:43–52. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 94:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavergne V, Harliwong I, Jones A et al. , 2015. Optimized deep-targeted proteotranscriptomic profiling reveals unexplored Conus toxin diversity and novel cysteine frameworks. Proc Natl Acad Sci U S A. 112:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RJ. 2009. Conotoxins: molecular and therapeutic targets Springer pp. 45–65. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R.. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2516:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Fry BG, Kini RM.. 2005. Eggs-only diet: its implications for the toxin profile changes and ecology of the marbled sea snake (Aipysurus eydouxii). J Mol Evol. 601:81–89. [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A.. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2213:1658–1659. [DOI] [PubMed] [Google Scholar]
- Magoč T, Salzberg SL.. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2721:2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrøm M, Matschiner M, Tørresen OK, Star B, Snipen LG, Hansen TF, Baalsrud HT, Nederbragt AJ, Hanel R, Salzburger W et al. , 2016. Evolution of the immune system influences speciation rates in teleost fishes. Nat Genet. 4810:1204. [DOI] [PubMed] [Google Scholar]
- Marsh H. 1971. Observations on the food and feeding of some vermivorous Conus on the Great Barrier Reef. Veliger 14:45–55. [Google Scholar]
- McCartney-Melstad E, Mount GG, Shaffer HB.. 2016. Exon capture optimization in amphibians with large genomes. Mol Ecol Resour. 165:1084–1094. [DOI] [PubMed] [Google Scholar]
- Metz EC, Robles-Sikisaka R, Vacquier VD.. 1998. Nonsynonymous substitution in abalone sperm fertilization genes exceeds substitution in introns and mitochondrial DNA. Proc Natl Acad Sci U S A. 9518:10676–10681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Kircher M.. 2010. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 20106:pdb.prot5448.. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Nobuhisa I, Deshimaru M, Nakai M, Ogawa T, Shimohigashi Y, Fukumaki Y, Hattori M, Sakaki Y, Hattori S et al. , 1995. Accelerated evolution in the protein-coding regions is universal in crotalinae snake venom gland phospholipase A2 isozyme genes. Proc Natl Acad Sci U S A. 9212:5605–5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Ogawa T, Oda N, Hattori M, Sakaki Y, Kihara H, Ohno M.. 1993. Accelerated evolution of Trimeresurus flavoviridis venom gland phospholipase A2 isozymes. Proc Natl Acad Sci U S A. 9013:5964–5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybakken J. 2009. Ontogenetic change in the conus radla, its form, distribution among the radula tpes, and significance in systematics and ecology. Malacologia 32:35–54. [Google Scholar]
- Nybakken J, Perron F.. 1988. Ontogenetic change in the radula of Conus magus (Gastropoda). Mar Biol. 982:239–242. [Google Scholar]
- Oksanen J, Blanchet FG, Friendly M et al. , 2016. vegan: Community Ecology Package, R package.
- Olivera BM. 1997. Conus venom peptides, receptor and ion channel targets, and drug design: 50 million years of neuropharmacology. Mol Biol Cell 811:2101–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera BM, Teichert RW.. 2007. Diversity of the neurotoxic Conus peptides. Mol Interv. 75:251–260. [DOI] [PubMed] [Google Scholar]
- Olivera BM, Walker C, Cartier GE, Hooper D, Santos AD, Schoenfeld R, Shetty R, Watkins M, Bandyopadhyay P, Hillyard DR et al. , 1999. Speciation of cone snails and interspecific hyperdivergence of their venom peptides: potential evolutionary significance of introns. Ann N Y Acad Sci. 870:223–237. [DOI] [PubMed] [Google Scholar]
- Orme D. 2013. The caper package : comparative analysis of phylogenetics and evolution in R. pp. 1–36. [Google Scholar]
- Pagel M. 1997. Inferring evolutionary processes from phylogenies. Zool Scripta 264:331–348. [Google Scholar]
- Pease JB, Haak DC, Hahn MW, Moyle LC.. 2016. Phylogenomics reveals three sources of adaptive variation during a rapid radiation. PLoS Biol. 142:e1002379–e1002324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuong MA, Mahardika GN, Alfaro ME.. 2016. Dietary breadth is positively correlated with venom complexity in cone snails. BMC Genomics 17:401.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portik DM, Smith LL, Bi K.. 2016. An evaluation of transcriptome-based exon capture for frog phylogenomics across multiple scales of divergence (Class : amphibia, Order : Anura). Mol Ecol Resour. 165:1069–1083. [DOI] [PubMed] [Google Scholar]
- Puillandre N, Bouchet P, Duda TF, Kauferstein S, Kohn AJ, Olivera BM, Watkins M, Meyer C.. 2014. Molecular phylogeny and evolution of the cone snails (Gastropoda, Conoidea). Mol Phylogenet Evol. 78:290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puillandre N, Koua D, Favreau P, Olivera BM, Stöcklin R.. 2012. Molecular phylogeny, classification and evolution of conopeptides. J Mol Evol. 74(5–6):297–309. [DOI] [PubMed] [Google Scholar]
- Puillandre N, Watkins M, Olivera BM.. 2010. Evolution of Conus peptide genes: duplication and positive selection in the A-superfamily. J Mol Evol. 702:190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt R, Kohn A.. 1995. Feeding and distribution of predatory gastropods on some great barrier reef platforms. Proceedings of the Fifth International Coral Reef Congress. pp. 191–196. [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 32:217–223. [Google Scholar]
- Revell L, Harmon L, Collar D.. 2008. Phylogenetic signal, evolutionary process, and rate. Syst Biol. 574:591–601. [DOI] [PubMed] [Google Scholar]
- Robinson S, Norton R.. 2014. Conotoxin gene superfamilies. MarDrugs 1212:6058–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW. 2016. Is mutation random or targeted ?: No evidence for hypermutability in snail toxin genes. Mol Biol Evol. 3310:2642–2647. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Bellare P, Derisi JL.. 2013. PRICE: software for the targeted assembly of components of (Meta) genomic sequence data. G3 (Bethesda) 35:865–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safavi-Hemami H, Lu A, Li Q, Fedosov AE, Biggs J, Showers Corneli P, Seger J, Yandell M, Olivera BM.. 2016. Venom insulins of cone snails diversify rapidly and track prey taxa. Mol Biol Evol. 3311:2924–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar MK, Chow VTK, Kangueane P.. 2004. Distributions of exons and introns in the human genome. In Silico Biol. 44:387–393. [PubMed] [Google Scholar]
- Sanderson MJ. 2003. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 192:301–302. [DOI] [PubMed] [Google Scholar]
- Simakov O, Marletaz F, Cho S-J, Edsinger-Gonzales E, Havlak P, Hellsten U, Kuo D-H, Larsson T, Lv J, Arendt D et al. , 2013. Insights into bilaterian evolution from three spiralian genomes. Nature 4937433:526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater GSC, Birney E.. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6:31.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2221:2688–2690. [DOI] [PubMed] [Google Scholar]
- Taylor J. 1986. Diets of sand-living predatory gastropods at Piti Bay, Guam. Asian Mar Biol. 3:47–58. [Google Scholar]
- Taylor JD. 1978. Habitats and diet of predatory gastropods at addu atoll, maldives. J Exp Mar Biol Ecol. 311:83–103. [Google Scholar]
- Taylor JD. 1984. A partial food web involving predatory gastropods on a pacific fringing reef. J Exp Mar Biol Ecol. 743:273–290. [Google Scholar]
- Taylor JD, Reid DG.. 1984. The abundance and trophic classification of molluscs upon coral reefs in the Sudanese Red Sea. J Nat Hist. 182:175–209. [Google Scholar]
- Teasdale LC, Köhler F, Murray KD, O’Hara T, Moussalli A.. 2016. Identification and qualification of 500 nuclear, single-copy, orthologous genes for the Eupulmonata (Gastropoda) using transcriptome sequencing and exon capture. Mol Ecol Resour. 165:1107–1123. [DOI] [PubMed] [Google Scholar]
- Wu Y, Wang L, Zhou M, You Y, Zhu X, Qiang Y, Qin M, Luo S, Ren Z, Xu A.. 2013. Molecular evolution and diversity of Conus peptide toxins, as revealed by gene structure and intron sequence analyses. PLoS One 812:e82495.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AS. 2001. Modularity, evolvability, and adaptive radiations: a comparison of the hemi- and holometabolous insects. Evol Dev. 32:59–72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw read data can be found at the National Center for Biotechnology Information associated with BioProject Accession #PRJNA437715. Scripts used to process the data can be found on Github (https://github.com/markphuong/venom.targetcapture.pilot and https://github.com/markphuong/phylogenetics.targetcapture.pilot). The bait sequences, final phylogenetic alignment, and all conotoxin sequences can be found on Dryad (doi:10.5061/dryad.vk245pd).