Abstract
The DNA methyltransferase 2 (DNMT2) protein is the most conserved member of the DNA methyltransferase family. Nevertheless, its substrate specificity is still controversial and elusive. The genomic role and determinants of DNA methylation are poorly understood in invertebrates, and several mechanisms and associations are suggested. In Drosophila, the only known DNMT gene is Dnmt2. Here we present our findings from a wide search for Dnmt2 homologs in 68 species of Drosophilidae. We investigated its molecular evolution, and in our phylogenetic analyses the main clades of Drosophilidae species were recovered. We tested whether the Dnmt2 has evolved neutrally or under positive selection along the subgenera Drosophila and Sophophora and investigated positive selection in relation to several physicochemical properties. Despite of a major selective constraint on Dnmt2, we detected six sites under positive selection. Regarding the DNMT2 protein, 12 sites under positive-destabilizing selection were found, which suggests a selection that favors structural and functional shifts in the protein. The search for new potential protein partners with DNMT2 revealed 15 proteins with high evolutionary rate covariation (ERC), indicating a plurality of DNMT2 functions in different pathways. These events might represent signs of molecular adaptation, with molecular peculiarities arising from the diversity of evolutionary histories experienced by drosophilids.
Keywords: Drosophila, Dnmt2, positive selection, epigenetic, positive-destabilizing selection
Introduction
Methylation of cytosine to form 5-methylcytosine is one of the most important epigenetic marks acting in the control of gene expression without altering the DNA nucleotide sequence. Cytosine methylation plays a critical role in the regulation of gene expression in higher eukaryotes. It is established by DNA methyltransferases (DNMTs), classified into three subfamilies: DNMT1, DNMT2 e DNMT3. The smallest eukaryotic methyltransferase, DNMT2, is most widely distributed in animals, fungi, protists, and plants (Ponger and Li, 2005). Dnmt2 was first identified in mice and humans and appears to be well conserved among eukaryotes (Okano et al., 1998; Yoder and Bestor, 1998). This enzyme is the only DNMT found in dipterans, including Drosophila (organisms “Dnmt2-only”) (Kucharski et al., 2008). In agreement with this structural conservation, different methods in various organisms have shown DNMT2 to have DNA methyltransferase activity (Hermann et al., 2003; Tang et al., 2003; Fisher et al., 2004; Katoh et al., 2006). However, according to analyses of human and Entamoeba enzymes, its catalytic activity on DNA is very weak (Hermann et al., 2003; Fisher et al., 2004). In addition, cytosine methylation of non-coding RNA (ncRNA) plays an important role in the epigenetic landscape. The functions of some tRNA modifications remain obscure. However, Tuorto et al. (2012) have shown that cytosine-C5 methylation of tRNAs is associated with their structural stability and the rates of protein synthesis in mammals. Cytosine-C5 methylation is involved in protecting tRNA against degradation induced by cellular stress events (Saikia et al., 2012).
Furthermore, studies have associated DNMT2 with RNA interference in Dictyostelium (Kuhlmann et al., 2005) and covalent histone modification in Drosophila (Kunert et al., 2003), suggesting a role of DNMT2 in epigenetic regulation. Several studies have shown the occurrence of DNA methylation phenomena in Drosophila species (Lyko et al., 2000; Kunert et al., 2003; Marhold et al., 2004). However, recently, Takayama et al. (2014) showed that methylation in the genome of Drosophila melanogaster probably is independent of DNMT2 activity. These findings brought new questions about the epigenetic mechanisms involved in the methylation process in evolutionarily related species of drosophilids.
Garcia et al. (2007) compared the DNMT2 protein sequences of D. willistoni and D. melanogaster and found higher conservation at the domains putatively responsible for methyl transfer catalysis and variability in the region containing the specific target recognition domain (TRD). These findings may be indicative of variation in DNMT2 function among organisms, suggesting that the targets – or modularity - of methylation can also vary among species of the same genus. Furthermore, Garcia et al. (2007) described sex-specific methylation patterns in D. willistoni, not present in D. melanogaster. Using the Methylation-Sensitive Restriction Endonuclease (MSRE) technique and Southern blot analysis with specific probes, the results suggested that selection for different targets of methylation might occur between different, but closely related species (Garcia et al., 2007). Furthermore, D’Ávila et al. (2010) found phylogenetic correlations in the sex-specific methylation patterns in the species of the willistoni subgroup, where D. willistoni, D. tropicalis and D. insularis (closer related species) shared methylation patterns in ribosomal genes, whereas D. equinoxialis and D. paulistorum patterns apparently are not restricted to rDNA.
The presence of DNMT2 enzymes in Diptera was described by Marhold et al. (2004), revealing that DNMT2 protein sequences are highly conserved in D. virilis, D. hydei, D. simulans, D. melanogaster and D. pseudoobscura, primarily within the catalytic DNA methyltransferase motifs. The Drosophilidae family is among the most diverse of the Diptera, encompassing more than 4,200 species (Bächli, 2016). Species of this family, especially of the genus Drosophila, are widely used in many areas of contemporary biological research. However, only few have been investigated with respect to the occurrence of DNA methylation and the presence of the Dnmt2 gene. Thus, the present study objectives are: (i) improve the previous search for Dnmt2 (Marhold et al., 2004; Garcia et al., 2007), including a large number of Drosophila species and other Drosophilidae, to evaluate the conservation, or not, of Dnmt2 in the genus; (ii) test whether the gene and protein are evolving under relaxed selective constraint or positive selection; and (iii), given the current controversial and enigmatic scenario involving the role of DNMT2 among epigenetic mechanisms of drosophilids, an attempt to find potential protein interaction partners of DNMT2 by database searching for protein-protein interactions and via evolutionary rate covariation (ERC) analysis.
Our results indicate, as expected, substantial conservation of DNMT2 catalytic motifs. Nevertheless, the TRD and the connecting region of the two main domains (catalytic and TRD) show some variability among the species examined, including closely related species. We also detected that several sites are under positive selection. These are located in potential regions of protein-protein interaction. The multiplicity of proteins with high ERC values found in the present work supports the hypothesis of DNMT2 can be involved in several networks, through control of gene expression, genomic stability, and in response to stressor events in “Dnmt2-only” organisms, like drosophilids.
Material and Methods
Fly stocks
The conservation of DNMT2 in the family Drosophilidae was analyzed in 68 species of Drosophila, along with Zaprionus indianus, Z. tuberculatus, Scaptodrosophila latifasciaeformis and S. lebanonensis (Table S1 (542.9KB, pdf) ). Most strains were maintained in the laboratory by mass crosses and reared in corn flour culture medium (Marques et al., 1966) in a controlled environment chamber (17 ± 1 °C, 60% r.h.), except for those species for which sequences were obtained directly from GenBank.
PCR, cloning and sequencing
Genomic DNA was extracted from adult flies following Sassi et al. (2005). Primers initially used are described in Marhold et al. (2004) (5’ D. melanogaster Dnmt2-F: 5’ GTGGCATTGGCGGCATGCATTATGCCT 3’ and D. melanogaster Dnmt2-R: 5’ CGATACTTTTGTCGATTCGTTGTTTCTGGC 3’). This pair of primers was designed directed against conserved catalytic motifs of D. melanogaster and used to amplify Dnmt2 sequences from D. simulans, D. hydei and D. virilis (Marhold et al., 2004). In this work, specific primers were designed to D. willistoni Dnmt2 genes (wDnmt2A-F: 5’ TCACCCACAACCTTGACATT 3’ and wDnmt2C-R: 5’ ACCTTCTCGCAGACACCAA 3’). Both pairs of primers align in similar regions of the gene Dnmt2. PCRs were performed in 25 μL volumes containing 20 ng of genomic DNA, 1 U of Taq DNA polymerase (Invitrogen, Carlsberg, CA, USA), 1X reaction buffer, 200 μM of each nucleotide, 20 pmol of each primer and 1.5 mM MgCl2. The amplification sets consisted of a denaturation step of 95 °C for 5 min, followed by 30 cycles at 95 °C for 40 s, 55 °C to 60 °C for 40 s and 72 °C for 1 min, and then a final extension cycle at 72 °C for 5 min. Dnmt2 amplicons were directly purified by incubation at 37 ºC for 30 min with Exonuclease I and Shrimp Alkaline Phosphatase (SAP) (both from USB, Cleveland, OH, USA) followed by a 15 min inactivation step at 80 ºC. For the Dnmt2 amplicons of D. teissieri, D. ornatifrons, D. ornatipennis and D. tropicalis, fragments were excised of the agarose gel and purified using Illustra GFX PCR DNA kit (GE Healthcare, Pittsburgh, PA, USA). The purified fragments were cloned into pCR4-TOPO plasmids (Invitrogen). DNA sequencing was performed by Macrogen Inc. (South Korea) using the appropriate primers (forward and reverse). The sequences generated by PCR and sequencing were assembled using the GAP 4 software of the Staden Package (Staden, 1996) (Table S2 (245.4KB, pdf) ).
Dot blot analyses
For Dot blot hybridizations, samples of denatured DNA (1 μg) were transferred onto a nylon membrane (Hybond-N+; GE Healthcare Biosciences). The AlkPhos Direct Labelling and Detection System and the CDP-Star kit (GE Healthcare) were used to label and detect nucleic acids according to the manufacturer’s instructions. The Dnmt2 amplicon from D. melanogaster was used as a probe at the stringency temperature of 55 ºC.
Data collection from public databases
In silico searches were performed to identify the complete sequence homologs of Dnmt2 among 24 sequenced Drosophila genomes available in the FlyBase database, using the Dnmt2 of D. melanogaster (Accession number: AAF53163.2) as query (Table S2 (245.4KB, pdf) ). The D. buzzattii and D. suzukii Dnmt2 genes were obtained from the Drosophila buzzatii Genome Project server and Spotted Wing Fly Base, a dedicated online resource for D. suzukii genomics (Table S2 (245.4KB, pdf) ).
Evolutionary analysis
All sequences were aligned using the Muscle tool (Edgar, 2004). The evolutionary relationships among the Dnmt2 sequences were estimated using Bayesian analysis, which is implemented in MrBayes (Ronquist and Huelsenbeck, 2003), with the evaluation of at least 1,000,000 generations and a burn-in region of 2,500 trees. Each nucleotide sequence was individually translated into its corresponding proteins and aligned using the Muscle tool with default parameter values. For the evolutionary analysis of the amino acid sequences, the Jones-Taylor-Thornton (JTT+G) model was used, as suggested by ProtTest 2.2 (Abascal et al., 2005), in accordance with the Akaike information criterion (Akaike, 1974). A Bayesian analysis of the nucleotide sequences was performed with the general time reversible (GTR) model using the ratio of invariable sites (I) and the gamma distribution of the variable sites (G) model, as suggested in MrModel Test 2.3 (Nylander, 2004). The sequence of Spodoptera frugiperda was used as outgroup. For the nucleotide and amino acid divergence analyses, sequences were clustered within species groups to perform a p-distance analysis using the MEGA 7 package (Kumar et al., 2016).
Analysis of positive selection
To investigate probable selective pressures that shaped the evolution of drosophilid Dnmt2 genes, we performed a relaxed branch-site test and a strict branch-site test (Yang and Nielsen, 2002; Zhang et al., 2005) using the software CODEML (Yang, 2007) - PAML package. This software tests for positive selection by comparing a series of alternative hypothesis that differ in how variable dN/dS ratio can change in different branches and codons, in which dN/dS > 1 would indicate positive selection and dN/dS < 1 would indicate a purifying selection, due to a selective constraint at the codon level.
The complete sequences of Dnmt2 genes were used, obtained from in silico search, as previously described. The initial maximum likelihood (ML) phylogenetic trees were constructed using the 24 complete sequences (Table S1 (542.9KB, pdf) ) by the software PHYML 3.1 (Guindon and Gascuel, 2003). The analysis of the nucleotide sequences was performed with the general time reversible (GTR) model using the ratio of invariable sites (I) and the gamma distribution of the variable sites (G) model, as suggested by MrModel Test 2.3 software (Nylander, 2004).
With the relaxed branch-site test or strict branch-site test, phylogenetic trees are separated into foreground branches, at which positive selection is tested, and background branches, represented by the other lineages. Both tests use the alternative model (MA), in which the codons in foreground are allowed to have a dN/dS > 1, and the background codons, dN/dS ≤ 1. The relaxed branch-site test null model (M1a) assumes that evolutionary rates are the same for all sites and branches, with all sites varying dN/dS from 0 and 1. In the null model (restricted MA), the dN/dS > 1 category is fixed to 1, so all sites with dN/dS > 1 are forced to evolve neutrally (dN/dS = 1). We used a log likelihood ratio test (LRT) to infer the positive selection when these values result in a significant value. The significance of the LTR was verified by a χ2 2 null distribution, with critical values of 2.71 for 5% and 5.41 for 1% significance levels, respectively, originated from a null distribution composed of a 50:50 mixture of point mass 0 and χ1 2 (Zhang et al., 2005).
Additionally, we investigated positive selection with respect to several physicochemical properties of the datasets. The MM01 method of McClellan et al. (2005) evaluates whether nonsynonymous substitutions favored changes in protein, either structural or functional. The analyses were carried out by TreeSAAP 3.2 (McClellan and McCracken, 2001; Woolley et al., 2003; McClellan et al., 2005). First, global deviation from neutrality is verified by a goodness-of-fit test, in which a comparison of neutral expected distribution and observed distribution of the selected physicochemical properties is made. Positive selection is detected in TreeSAAP software when the number of inferred amino acid replacements significantly exceeds the number of expected replacements caused by chance alone, given positive z-scores. Stabilizing-selection can be visualized when the magnitude of chance is low (categories 1, 2 and 3), meaning it is a conservative process, while positive-destabilizing selection is represented as a high magnitude of change (categories 6, 7 and 8) (McClellan et al., 2005). Stabilizing selection is defined by McClellan et al. (2005) as a selection that tends to maintain the original biochemical attributes of the protein, and positive-destabilizing selection as a selection that favors structural and functional shifts in a region of a protein. In other words, positive-destabilizing selection represents signs of molecular adaptation. To verify which regions were under positive selection (stabilizing and destabilizing) we performed a sliding window analysis using the amino acid properties significant for this type of change (McClellan et al., 2005).
Potential protein-protein interaction partners
Using the database STRING (Szklarczyk et al., 2015) we conducted a search for predicted protein-protein interactions with DNMT2. To perform the search, the D. melanogaster DNMT2 sequence was used as query and data were collected from D. ananassae, D. grimshawi, D. pseudoobscura, D. virilis, D. willistoni and D. yakuba. We also attempted to evaluate the predicted protein-protein partners from the STRING search and find protein partners with DNMT2 by evolutionary rate covariation (ERC) using the ERC Analysis Webserver (Clark et al., 2012, 2013, Findlay et al., 2014).
Results
Detection of Dnmt2 sequences in Drosophilidae species
In a preliminary screen of the presence of Dnmt2 sequence homologs within the Drosophila genomes, we tested 56 species by dot blot analysis (Table S1 (542.9KB, pdf) and Figure S1 (369.2KB, pdf) ). Of these, 54 showed a positive signal for the Dnmt2 probe (amplicon from D. melanogaster), two of which had a weak signal (D. orena and D. polymorpha). In this assay we observed a strong hybridization signal, primarily in the melanogaster group. Nevertheless, hybridization was also detected in the other species, indicating that the Dnmt2 gene has related sequences in all species groups analyzed.
To verify this homology, we also performed a Dnmt2 homolog search by PCR amplification in a large number of Drosophilidae species from different Drosophila groups (Table S1 (542.9KB, pdf) ). Altogether, 61 species were tested by PCR for presence of a Dnmt2 gene. Thirty species tested positive for Dnmt2 by PCR. We achieved 20 amplicons for several species of the Drosophila genus, including the Drosophila subgenus (guarani, guaramunu, tripunctata, calloptera, immigrans, mesophragmatica, flavopilosa and repleta groups) and the Sophophora subgenus (melanogaster and willistoni groups). The representative species of the Dorsilopha subgenus did not show Dnmt2 amplification with the primers used. The annealing regions of the primers correspond to motif I (forward primer) and motif X (reverse primer) of Dnmt2, both belonging to the catalytic domain of the enzyme (Figure S2 (1MB, pdf) ). The length of the amplified fragments was around 800 bp.
DNMT2 conservation
The availability of several species maintained in culture chambers in our laboratory and genomic sequences in gene banks allowed us to search for open reading frames encoding DNMT2 in several genomes. For the analyses of gene conservation, all cloned sequences (obtained by direct PCR or cloned) (Table S3 (249.1KB, pdf) ) and those obtained by in silico search were used in the phylogenetic analysis to investigate the evolutionary pattern and conservation of Dnmt2 among Drosophilidae species. The sequences were aligned to build a phylogenetic matrix from the 44 analyzed species (Figure S2 (1MB, pdf) ).
To perform the analyses of Dnmt2 nucleotide and amino acid divergence, pairwise comparisons of the sequences were conducted (with species clustered by taxonomic group). The highest nucleotide divergence found was between the willistoni and tripunctata groups, with a p-distance value of 34.47%; whereas the lowest divergence (p-distance value of 10.70%) was detected between the guarani and calloptera groups (represented only by D. ornatipennis) (Table 1, standard error presented at Table S4 (285.2KB, pdf) ). The comparison of all nucleotide sequences between species showed that the sequences of D. willistoni and D. suzukii are more divergent than any other, with a p-distance value of 35.3% (data not shown). When we estimated the average divergence of amino acid sequence pairs within subgenus Drosophila and Sophophora, it revealed the almost the same internal divergence: 17,8% (S.E. 1.65) and 18.7% (S.E. 1.62), respectively. Pairwise alignment of the amino acid sequences between groups (Table 1) revealed that willistoni and mesophragmatica groups were more divergent (p-distance value of 31.84%).
Table 1. Estimates of evolutionary divergence between sequence pairs of different Drosophilidae species groups. The numbers of amino acid differences per site from the average over all sequence pairs between groups are given below the diagonal. The measures of nucleotide evolutionary divergence are provided above the diagonal. The p-distances are given in percentages.
| Groups | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | calloptera | 25.31 | 26.39 | 12.89 | 10.70 | 26.08 | 30.53 | 23.66 | 29.58 | 23.87 | 15.66 | 22.22 | 33.33 | |
| 2 | flavopilosa | 23.32 | 23.30 | 24.23 | 25.77 | 26.77 | 30.83 | 15.12 | 29.32 | 16.62 | 25.62 | 16.20 | 32.18 | |
| 3 | grimshawi | 24.22 | 18.83 | 25.31 | 26.13 | 26.23 | 28.62 | 22.79 | 27.11 | 23.15 | 26.39 | 20.37 | 33.95 | |
| 4 | guaramunu | 10.09 | 21.08 | 23.54 | 15.15 | 25.23 | 29.27 | 22.97 | 26.72 | 23.46 | 13.87 | 21.14 | 34.07 | |
| 5 | guarani | 7.92 | 24.81 | 23.92 | 11.29 | 25.36 | 30.54 | 23.56 | 29.15 | 24.74 | 17.16 | 22.89 | 32.95 | |
| 6 | immigrans | 17.94 | 20.63 | 21.08 | 17.60 | 18.76 | 29.89 | 25.69 | 28.50 | 27.93 | 25.31 | 25.08 | 30.83 | |
| 7 | melanogaster | 25.16 | 25.41 | 24.22 | 24.63 | 25.95 | 23.56 | 29.87 | 23.72 | 29.78 | 29.93 | 29.24 | 32.72 | |
| 8 | mesophragmatica | 20.78 | 15.70 | 19.13 | 18.68 | 21.72 | 19.58 | 25.29 | 28.46 | 15.35 | 24.34 | 15.64 | 33.64 | |
| 9 | obscura | 25.56 | 26.01 | 22.57 | 24.29 | 24.81 | 23.02 | 19.11 | 25.26 | 28.91 | 28.10 | 26.23 | 32.61 | |
| 10 | repleta | 21.82 | 14.95 | 18.09 | 19.88 | 22.77 | 20.40 | 25.15 | 11.61 | 25.66 | 25.31 | 18.57 | 33.02 | |
| 11 | tripunctata | 12.00 | 23.09 | 23.99 | 10.48 | 12.86 | 18.89 | 25.26 | 20.67 | 24.96 | 21.56 | 22.69 | 34.47 | |
| 12 | virilis | 18.39 | 11.66 | 15.25 | 15.92 | 18.98 | 15.70 | 22.22 | 10.31 | 21.52 | 12.41 | 17.94 | 30.56 | |
| 13 | willistoni | 31.17 | 31.17 | 28.48 | 30.72 | 31.76 | 27.91 | 29.55 | 31.84 | 27.20 | 30.19 | 31.22 | 28.92 |
Standard error values are shown in Table S5 (236.6KB, pdf) .
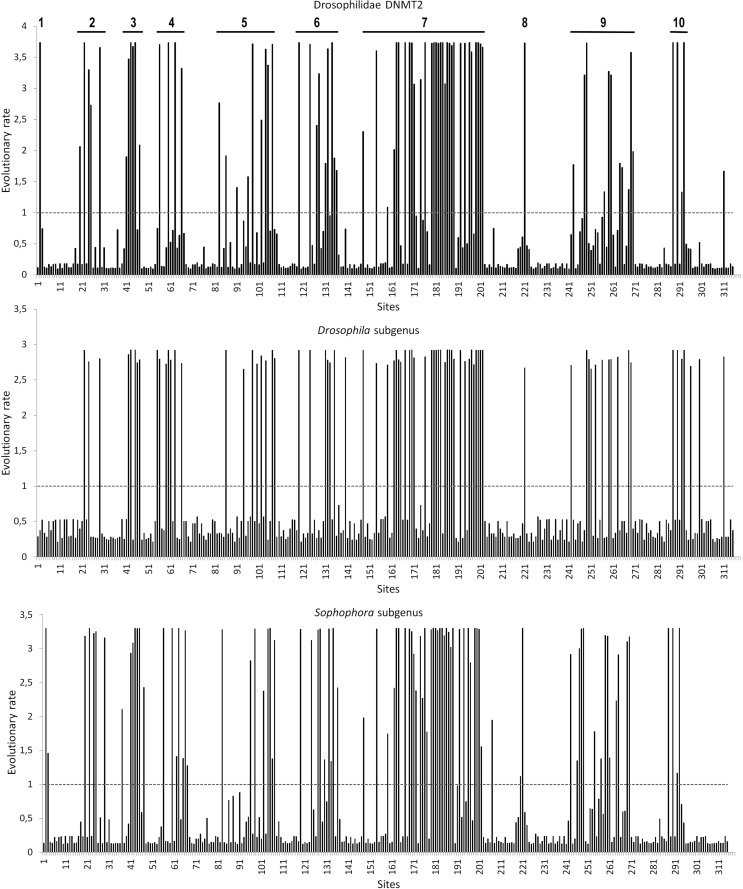
We used the MEGA7 package to compute the mean evolutionary rates at each DNMT2 site. Figure 1 shows the plots, which evolutionary rate are shown to each site for all Drosophilidae complete sequences and for each subgenus separately. Several DNMT2 sites are under evolutionary restrictions, and these sites are mainly part of the catalytic motifs of the enzyme. Nevertheless, 10 regions with sites having high evolutionary rates distributed throughout DNMT2 are evident (Figure 1). This means that sites showing a rate < 1 are evolving slower than average and those with a rate > 1 are evolving faster than average evolutionary rates. Evolutionary rates were estimated under the Jones-Taylor-Thornton (Jones et al., 1992) model (+G).
Figure 1. Evolutionary rate from all Drosophilidae DNMT2 complete sequences, Sophophora and Drosophila subgenus separately. The rates are scaled such that the average evolutionary rate across all sites is 1. All positions containing gaps and missing data are eliminated.
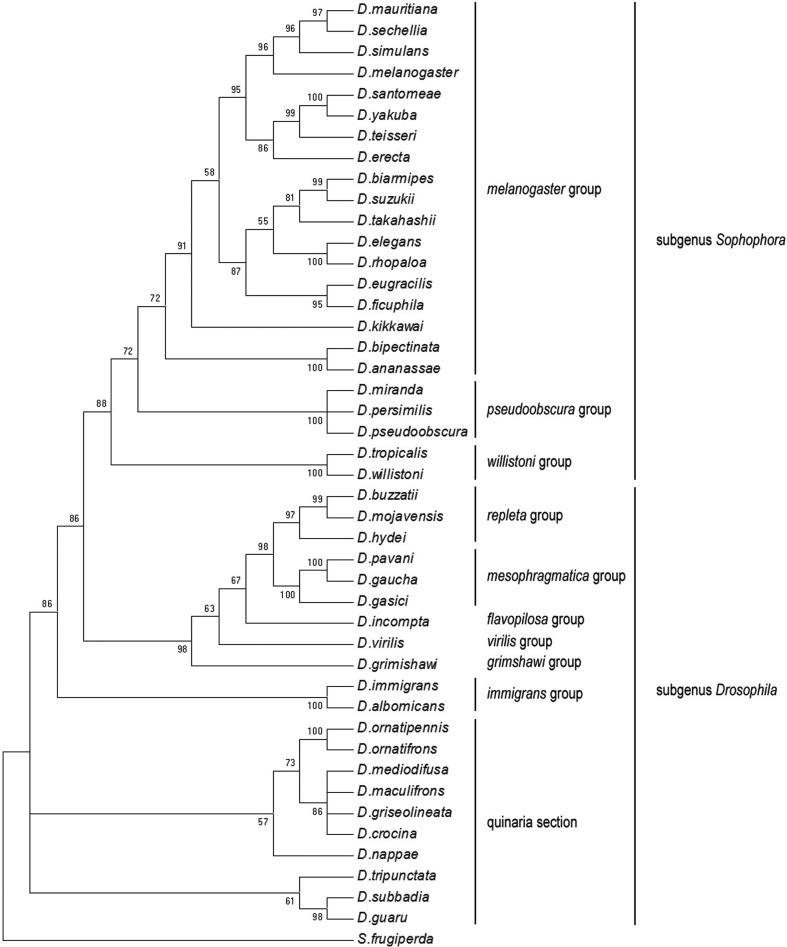
The phylogeny obtained by amino acid analysis recovered the evolutionary relationship between the subgenera Drosophila and Sophophora (Figure 2). The quinaria section (Drosophila subgenus) composed of guarani, guaramunu, calloptera and tripunctata species groups appears as basal radiation. A second cluster in Drosophila subgenus includes the virilis, repleta, flavopilosa and mesophragmatica species groups (virilis-repleta radiation). The Sophophora subgenus is composed of the melanogaster, willistoni and obscura groups, where the willistoni group appears as basal to the obscura-melanogaster radiation. D. ananassae and D. bipectinata appear more externally positioned within the melanogaster group, likely reflecting the ancestral condition of the ananassae subgroup (to which both species belong) within this group (Kopp, 2006; Clark et al., 2007). This placement is confirmed by the correct positioning of the remaining species of the melanogaster group, belonging to the melanogaster, takahashii, rhopaloa and elegans subgroups, consistent with previous studies (Lewis et al., 2005; Kopp, 2006).
Figure 2. Bayesian phylogenetic inference of the Dnmt2 gene in Drosophilidae species based on amino acid sequences alignment. Tree generated using the JTT model with a gamma distribution. Sequence of Spodoptera frugiperda was used as outgroup.
The nucleotide Bayesian inference tree of Dnmt2 (Figure S3 (336.1KB, pdf) ) showed congruence with the tree previously obtained for the Drosophilidae, which was based on nuclear genes and produced using similar methods (Gailey et al., 2000; Tarrio et al., 2001; Da Lage et al., 2007). However, D. immigrans and D. albomicans sequences were incorrectly positioned, grouping with the Sophophora subgenus, but with low statistical support. Also, the quinaria section presented polytomy. There is no strong evidence to sustain this finding, and the data from amino acids analysis confirm the correct positioning of this species, grouped with the rest of the Drosophila subgenus species, according to previous studies (Throckmorton, 1975; Russo et al., 1995; Kwiatowski and Ayala, 1999), however, again some clades showed low statistical support and the quinaria section species grouped with polytomy.
Long-term evolutionary analysis in drosophilids Dnmt2
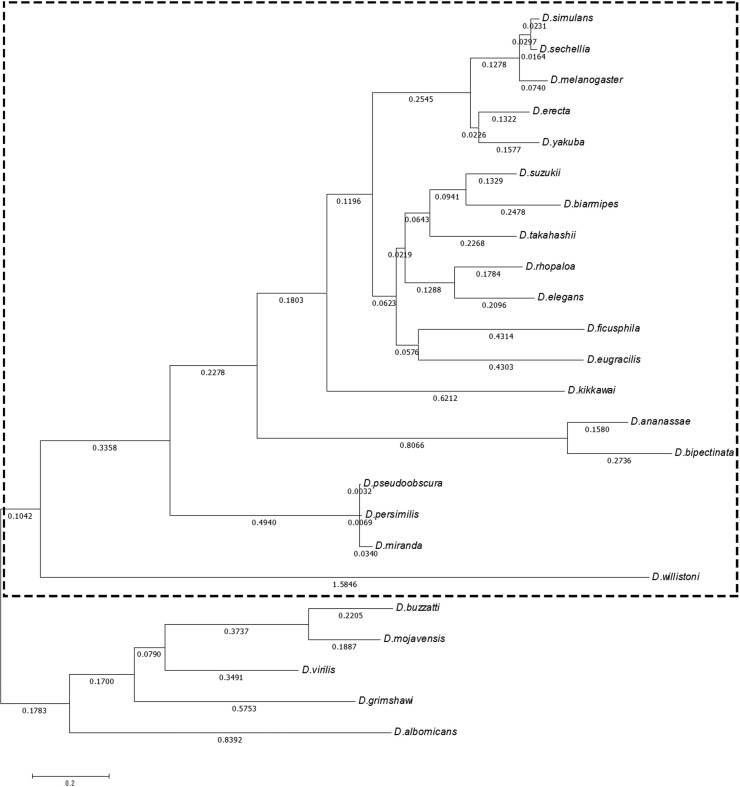
To evaluate positive selection in Dnmt2, we selected representative species of the main groups of drosophilids whose complete sequences could be obtained (Figure 3). Using the PAML package we obtained an initial tree that was later analyzed for positive selection in codeML. The initial ML tree recovers the evolutionary relationship between Drosophila and Sophophora subgenera (Figure 4), as well as the amino acid tree obtained from the Bayesian analysis (Figure 2). We used the results of the divergence analysis (Table 1) to establish which branch to use as foreground. When analyzing the results of divergence, we found that the divergence within the Sophophora subgenus is lower than the divergence between the Sophophora and Drosophila subgenera, as expected (Table S5 (236.6KB, pdf) ). Moreover, the Sophophora and Drosophila subgenera show different evolutionary rates, with the Sophophora subgenus having more sites with fast evolution (Figure 1). Therefore, we asked whether the divergence observed between the groups is random or driven by selection. Thus, we established the Sophophora subgenus as foreground and the Drosophila subgenus as background.
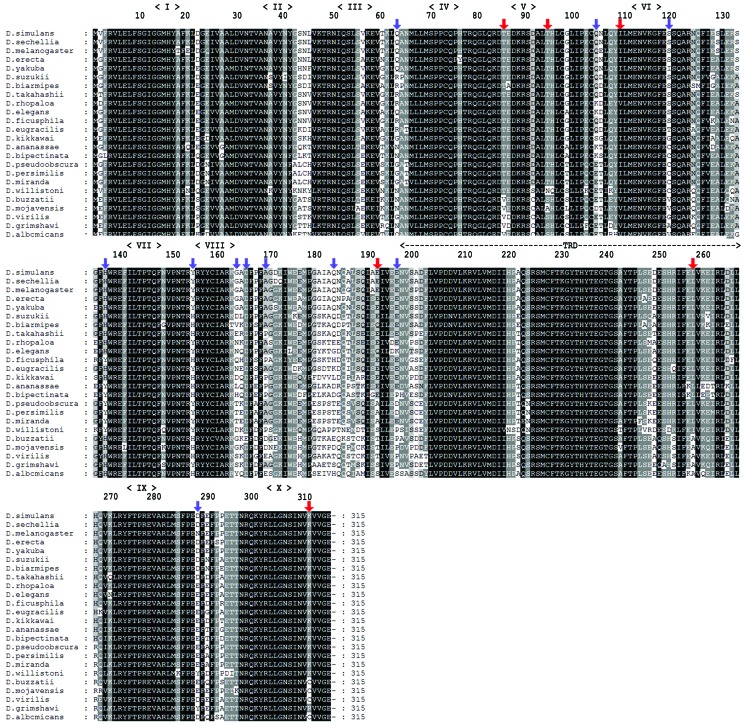
Figure 3. Multiple alignment of DNMT2 sequences used in the positive selection analysis with no gaps. Black boxes represent conserved 100% in all sequences, dark grey 80% and light grey 60%. Red arrows indicate sites under positive selection and purple arrows sites under positive-destabilizing selection. Roman letters indicate the conserved (cytosine-5) MT2 motifs. TRD indicates the target recognition domain.
Figure 4. ML phylogenetic tree of Dnmt2 used in analysis for detection of selection. The dotted frame highlights the foreground branch. The genetic distances are in nucleotide substitution per codon (number below the branches).
The selective pressures over the Dnmt2 sequence were investigated by the ratio of nonsynonymous to synonymous substitutions. Table 2 shows the parameters inferred for the null models (M1a and MA) and for the alternative MA model. The null model of selective constraint (0 < ω ≤ 1) was rejected by the relaxed branch-site test, indicating that the foreground branch (Figure 3) has diverged by relaxed selective constraint or by positive selection (Table 3). By the contrast of the restricted-MA versus MA models (strict branch-site), we could discriminate between the two hypotheses, where the null model of selective constraint was rejected (Table 3), in favor of the hypothesis that several sites of Sophophora DNMT2 (Figure 3 and Figure 4) differentiated by positive selection. We estimated that six sites (position 85, 94, 109, 192, 257 and 311) were evolving under positive selection (Table 3) (Figure 3).
Table 2. Parameters estimates and log likelihood values for branch-site M1a, MA and restricted MA model.
| Model | Parameters | lnL | ||
|---|---|---|---|---|
| M1a | ω0 = 0.04651 | p0 = 0.86743 | -11 046.650097 | |
| Relaxed branch-site test null model | ω1 = 1.00000 | p1 = 0.13257 | ||
| Restricted MA (ω2 = 1 fixed) | ω0 =0.04520 | ω2 = 1.000 | -11 039.638028 | |
| Strict branch-site test null model | p0 = 0.82646 | p1 = 0.12587 | (p2a + p2b) = 0.04767 | |
| MA | ω0 = 0.04596 | ω2 = 50.90099 | -11 032.575669 | |
| Alternative model | p0 = 0.84965 | p1 = 0.12542 | (p2a + p2b) = 0.02493 |
Lnl = log likelihood; ω0 = dN/dS values for sites with 0 < ω < 1; ω1 = dN/dS values fixed to 1; ω2 = dN/dS values for sites with ω > 1, which corresponds only to sites in the foreground branch.
Table 3. Comparison of null and alternative models by LRT and positively selected sites estimated by Bayes Empirical Bayes.
| Test | Contrast | LRT | D.F. | χ2- Probability | Positively selected sites |
|---|---|---|---|---|---|
| Relaxed branch-site | M1a MA | 25.20 | 2 | p < 7.7x10-7 | |
| Strict branch-site | Restricted MA MA | 12.41 | 1 | p < 0.001 | 85, 94, 109, 192, 257, 311 |
LRT - likelihood ratio test; D.F - Degrees of freedom.
To investigate selection on amino acid properties we used the software TreeSAAP, based on the global goodness-of-fit statistics calculated by the MM01 method. All physicochemical properties examined are significant (cutoff = 0.05) (Table 4). Seven properties demonstrated significantly positive z-scores under a trait of radical changes category between 6 and 8: α-helical tendencies (Pα), Equilibrium constant (ionization of COOH) (pK’), Polar requirement (Pr), Power to be at the C-terminal (αC), Power to be at the middle of alpha-helix (αm), Power to be at the N-terminal (αn) and Turn tendencies (Pt). Specific analysis with the sliding window in TreeSAAP showed that 12 amino acids were under positive-destabilizing selection (Figure 5a); the properties of most of these are related to the alpha-helix structures, mainly located in the catalytic domain.
Table 4. Amino acid physicochemical properties under positive destabilizing selection in DNMT2.
| Physicochemical property | Goodness-of-fit (neutral expectation) | radical change category (6, 7 and 8) | z-score |
|---|---|---|---|
| Alpha-helical tendencies (P a) | 33.339*** | 8 | 4.292*** |
| Average number of surrounding residues (N s) | 119.136*** | 6.442*** | |
| Beta-structure tendencies (P b) | 30.909*** | 2.804** | |
| Bulkiness (B l) | 35.052*** | 3.104*** | |
| Buriedness (B r) | 54.407*** | 2.580** | |
| Chromatographic index (R f) | 107.109*** | 3.190*** | |
| Coil tendencies (C t) | 18.839** | - | |
| Composition (C) | 43.179*** | 3.898*** | |
| Compressibility (K 0) | 22.548** | - | |
| Equilibrium constant (ionization of COOH) (pK’) | 79.39*** | 8 | 2.828** |
| Helical contact area (C a) | 81.079*** | 3.732*** | |
| Hydropathy (H) | 83.046*** | 3.885*** | |
| Isoelectric point (pH i) | 45.283*** | 4.261*** | |
| Long-range non-bonded energy (E l) | 78.459*** | 5.712*** | |
| Mean r.m.s. fluctuation displacement (F) | 117.708*** | 5.736*** | |
| Molecular volume (M v) | 77.479*** | 3.151*** | |
| Molecular weight (M w) | 69.846*** | 3.577*** | |
| Normalized consensus hydrophobicity (H nc) | 57.459*** | 2.062* | |
| Partial specific volume (V 0) | 61.97*** | 2.986** | |
| Polar requirement (P r) | 29.801*** | 7 | 2.341** |
| Polarity (P) | 59.713*** | 2.522** | |
| Power to be at the C-terminal (α C) | 118.516*** | 6 | 5.758*** |
| Power to be at the middle of alpha-helix (α m) | 49.271*** | 7 | 3.657*** |
| Power to be at the N-terminal (α n) | 32.449*** | 7 | 3.159*** |
| Refractive index (μ) | 47.548*** | 3.632*** | |
| Short and medium range non-bonded energy (Esm) | 67.785*** | 3.743*** | |
| Solvent accessible reduction ratio (Ra) | 83.159*** | 2.942** | |
| Surrounding hydrophobicity (H p) | 69.397*** | 2.345** | |
| Thermodynamic transfer hydrohphobicity (H t) | 50.917*** | 2.785** | |
| Total non-bonded energy (E t) | 109.548*** | 5.654*** | |
| Turn tendencies (P t) | 109.136*** | 6 | 6.198*** |
p < 0.05
p < 0.01
p < 0.001
Figure 5. DNMT2 structure predictions. (a) Sliding window plots of the z-scores of radically changed properties showing regions under positive-destabilizing selection in Dnmt2. Dashed horizontal line indicates the Bonferroni-corrected significant limit (z-score = 3.09, p < 0.001). Alpha-helical tendencies (P a) – red; Equilibrium constant (ionization of COOH) (pK’) – orange; Polar requirement (P r) – green; Power to be at the C-terminal (α C) – blue; Power to be at the middle of alpha-helix (αm) – pink; Power to be at the N-terminal (αn) – black; Turn tendencies (Pt) – purple. (b) Structure of sfDNMT2 (PDVB: 4H0N) (Li et al., 2012), ribbon (up) and surface (down) representations. The respective sites under positive selection in drosophilids are represented in Spodoptera frugiperda structure. Sites under positive selection colored red, positive-destabilizing selection purple, motif CFT from TRD and the catalytic cysteine yellow and cofactor s-adenosyl methionine (SAM) cyan.
The DNMT2 sites that are under positive selection and positive-destabilizing selection (as indicated by the CodeML and TreeSAAP analyses, respectively) are shown in Figure 5b. Most sites under selection are located in the catalytic domain and especially located on the molecular surface. The only crystallographic model of DNMT2 available for arthropods is from S. frugiperda and it is used just as a representative model only (Li et al., 2012).
Protein-protein interactions and co-evolutionary predictions
We accessed the predicted genes as interacting with D. melanogaster, D. ananassae, D. grimshawi, D. pseudoobscura, D. virilis, D. willistoni and D. yakuba Dnmt2 through the STRING database (Table 5). These were concatenated and the D. melanogaster gene annotations to ERC analysis were used, since the taxonomic group utilized in ERC Analysis Webserver is D. melanogaster. We also made an extensive search for genes that have significant ERC values with D. melanogaster Dnmt2 by ERC Analysis Webserver Top Genes tool, which retrieves the genes with the highest ERC values for a given query gene from the entire genome (Clark et al., 2012., 2013, Findlay et al., 2014).
Table 5. Gene annotation and sequence location from STRING and ERC webserver searching from D. melanogaster genome. Genes with high ERC value (> 0.400) and p-value < 0.05 are colored gray.
| Gene | Annotation | Description |
|---|---|---|
| Atac3 | CG32343 | Contributes to histone acetyltransferase activity. Involved chromatin remodeling. |
| Cap | CG18408 | Interacts selectively and non-covalently with vinculin, a protein found in muscle, fibroblasts, and epithelial cells that binds actin and appears to mediate attachment of actin filaments to integral proteins of the plasma membrane. |
| CG10262 | CG10262 | Proliferating Cell Nuclear Antigen (PCNA) domain. These polymerase processivity factors play a role in DNA replication and repair. |
| CG13035 | CG44836 | Uncharacterized protein involved in sensorial perception of pain. |
| CG14618 | CG14618 | Belongs to the class IV-like SAM-binding methyltransferase superfamily. tRNA (guanine(9)-N(1))-methyltransferase TRM10. Enzyme catalyzes the conversion of a guanosine residue to N1-methylguanine in position 37, next to the anticodon, in tRNA. |
| CG14906 | CG14906 | N-6 adenine-specific DNA methylases. Specifically methylate the amino group at the C-6 position of adenines in DNA. |
| CG16863 | CG16863 | The protein has zinc finger, BED-type, which is thought to be involved in chromatin insulation at BEAF (boundary element-associated factor) and DREF, a transcriptional regulator. |
| CG17124 | CG17124 | PKC-activated protein phosphatase-1 inhibitor. Stops, prevents or reduces the activity of a protein phosphatase, an enzyme that hydrolyzes phosphate groups from phosphorylated proteins. |
| CG6712 | CG6712 | Probably RNA binding inferred from sequence or structural similarity with Saccharomyces RPF1, involved in ribosome biogenesis. Belongs to Brix superfamily. |
| CG7470 | CG7470 | Predicted gamma-glutamyl phosphate reductase with delta1-pyrroline-5-carboxylate synthetase activity, glutamate 5-kinase activity. An inner mitochondrial membrane enzyme, is essential to the de novo synthesis of the amino acids proline and arginine. Involved in epithelium development and germarium-derived egg chamber formation. |
| CTCF | CG8591 | CTCF is a ubiquitous transcription factor that binds to insulators and domain boundaries. It mediates insulator function and blocks enhancers by binding to Cp190. It contributes to long-range chromatin interaction, organizes chromatin domain boundaries and coordinates nuclear architecture. |
| Dnmt2 | CG10692 | Methyltransferase 2 is a (cytosine-5) DNA/tRNA methyltransferase. It is involved in regulation of gene expression by cytosine-5 methylation. The major protein role is the modifications that protects tRNAs against endonucleolytic cleavage and contributes to stress resistance, protein translation and small RNA-mediated gene regulation. |
| Eggless | CG12196 | Belongs to the class V-like SAM-binding methyltransferase superfamily. Histone-lysine methyltransferase family. Involved in negative regulation of gene expression. |
| Eno | CG17654 | Responsible for the catalysis of the conversion of 2-phosphoglycerate (2-PG) to phosphoenolpyruvate (PEP), the ninth and penultimate step of glycolysis |
| Haywire | CG8019 | Helicase Ercc3, core RNA polymerase binding transcription factor activity. Involved in regulation of alternative mRNA splicing, via spliceosome, cell proliferation and growth. |
| Homer | CG11324 | Involved in the positive regulation of circadian sleep/wake cycle, sleep; response to ethanol, behavioral response to ethanol, regulation of locomotion and adult behavior. Activity in stress response. |
| hop | CG1594 | Signal transduction-non receptor tyrosine kinase. Members of the Janus kinase (JAK) family of cytoplasmic protein tyrosine kinases physically associate with ligand-bound receptors. The JAK-STAT pathway regulates the expression of pair rule gene even-skipped (a transcriptional repressor of a number of genes) early in embryogenesis. |
| Hsp22 | CG4460 | A key player in cell-protection mechanisms against oxidative injuries and aging in Drosophila. Activated in late third-instar larvae of Drosophila melanogaster in the absence of heat stress |
| MBD-like | CG8208 | Methyl Binding Protein 2/3, a co-repressor and an integral component of the nucleosome remodelling and deacetylase (NuRD) complex. Negative regulation of transcription, involved in organism development |
| mei-S332 | CG5303 | Acts to maintain sister-chromatid cohesion before anaphase II of meiosis in both males and females. |
| mms4 | CG12936 | Methyl methanesulfonate sensitivity 4 is the non-catalytic subunit of the mus81-mms4 structure-selective endonuclease that functions in DNA repair. |
| mus209 | CG9193 | Belongs to the PCNA family. Involved in eggshell chorion gene amplification, DNA replication, mismatch repair, neurogenesis and antimicrobial humoral response. |
| Nsun2 | CG6133 | tRNA (cytosine-5-)-methyltransferase activity. |
| Orc2 | CG3041 | Origin recognition complex subunit 2. It is involved in eggshell chorion gene amplification, chromatin silencing, cell proliferation, DNA replication initiation, chromosome condensation and neurogenesis. |
| Pnt | CG17077 | It is a sequence-specific DNA binding transcription factor activity, repressing transcription factor binding, involved in positive regulation of transcription. It is involved in organism developmental process, open tracheal system development, post-embryonic organ morphogenesis, regulation of developmental process, sensory organ development, cardiovascular system development, regulation of RNA metabolic process, positive regulation of cell proliferation, anterior/posterior axis specification, multi-organism reproductive process, muscle cell differentiation, compound eye photoreceptor development, regulation of neurogenesis. |
| Rel | CG11992 | Relish is a transcription factor and the downstream component of the Immune Deficiency pathway, which regulates the antibacterial response and other less characterized cellular processes. |
| RhoGEF4 | CG8606 | Rho family small GTPases act as molecular switches that regulate neuronal morphogenesis, including axon growth and guidance, dendritic spine formation, and synapse formation. These proteins are positively regulated by guanine nucleotide exchange factors (GEFs) of the Dbl family. Findings suggest that DRhoGEF4 may participate in cytoskeleton-related cellular events by specifically activating RhoA in neuronal morphogenesis. |
| Rpd3 | CG7471 | Histone deacetylase 1. Catalyzes the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4). Histone deacetylation may constitute a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. For instance, deacetylation of histone H3 may be a prerequisite for the subsequent recruitment of the histone methyltransferase Su(var)3-9 to histones. Involved in position-effect variegation (PEV). |
| Rpp20 | CG33931 | A subunit of the RNase P and RNase MRP holoenzymes, has interaction with the Drosophila SMN protein. Immunofluorescence results indicate that Rpp20 is diffusely distributed throughout the cytoplasm with higher concentration observed in the nucleus. However, in response to stress, SMN forms aggregates and redistributes Rpp20 into punctuated cytoplasmic SMN granules. |
| Sna | CG3956 | Snail is a transcription factor that contributes to embryonic mesoderm development, epithelial to mesenchymal transition and asymmetric cell division. |
| Su(var)2-5 | CG8409 | Suppressor of variegation 205 is a heterochromatin protein associated with the pericentric heterochromatin and telomeres in Drosophila. It is involved in the positive autoregulatory expression and can bind directly to nucleosomes. |
| Su(var)2-10 | CG8068 | It is responsible for establishing and maintaining chromosome organization in interphase nuclei, promoting chromosome structure and function. |
| Su(var)3-3 | CG17149 | Probable histone demethylase that specifically demethylates ‘Lys-4’ of histone H3, a specific tag for epigenetic transcriptional activation, thereby acting as a corepressor. Required for heterochromatic gene silencing |
| Su(var)3-9 | CG43664 | Histone methyltransferase that specifically trimethylates ‘Lys-9’ of histone H3 using monomethylated H3 ‘Lys-9’ as substrate. H3 ‘Lys-9’ trimethylation represents a specific tag for epigenetic transcriptional repression by recruiting Su(var)205/HP1 to methylated histones. Mainly functions in heterochromatin regions, thereby playing a central role in the establishment of constitutive heterochromatin at pericentric regions. Involved in heterochromatic gene silencing including the modification of position-effect-variegation. |
| Tet | CG9973 | Ten-Eleven Translocation (TET) family protein. Involved in positive regulation of DNA demethylation, inducing positive regulation of transcription from RNA polymerase II promoter. |
| Thor | CG8846 | Eukaryotic translation initiation factor 4E binding protein, controlled by tor. It contributes to translation regulation, response to environmental stress and cell growth regulation. |
Altogether, 551 genes were obtained with an ERC value > 0.400 (p-value < 0.05) as result of the search. We filtered the genes found according to the expression period from 00-12 hour of the embryonic stage, which corresponds to the expression of Dnmt2 in D. melanogaster according to Lyko et al. (2000). The biological role played by genes according to the biological process described for DNMT2 was also considered (i.e., epigenetic functions, transcriptional control, stress response, tRNA methylation, response to heat, positive regulation of innate immune response, telomere maintenance). At the end, we obtained a list of 35 genes and added the enolase enzyme, since this was the first DNMT2 interacting protein described (Tovy et al., 2010) (Table 5).
ERC values for the 36 genes were obtained by the Group ERC Analysis tool, which returns the ERC values between a group of genes and statistics for the strength of ERC between them (Table S6 (1MB, pdf) ). Interestingly, Dnmt2 presents high ERC values related with 10 genes (p < 0.05) (Table 6), in which four are related to transcriptional control and three have a chromatin-remodeling function. However, genes with protein interaction prediction coming from the STRING database showed low ERC values (Table 6). The 15 genes with an ERC value > 0.400 (p-value < 0.05) are distributed between the Muller’s elements A, B, C, D and E.
Table 6. ERC values between potential DNMT2 protein-protein partners. The matrix shows all pairwise ERC values between genes below the diagonal and respectively p-values above the diagonal. Cells are shaded red according to the intensity of their deviation from the null expectation.
| Protein | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | pnt | N/A | 0.0010 | 0.1445 | 0.0781 | 0.4435 | 0.1082 | 0.0950 | 0.1429 | 0.3571 | 0.1486 | 0.1924 | 0.3492 | 0.0145 | 0.4435 | 0.0207 | 0.0170 | 0.0241 | 0.0760 |
| 2 | Su(var)2-10 | 0.811 | N/A | 0.0578 | 0.0168 | 0.0143 | 0.0868 | 0.0891 | 0.0912 | 0.1791 | 0.1679 | 0.3720 | 0.5330 | 0.0185 | 0.0912 | 0.0493 | 0.0572 | 0.0383 | 0.2213 |
| 3 | CG14618 | 0.375 | 0.480 | N/A | 0.0093 | 0.0398 | 0.0541 | 0.0117 | 0.0249 | 0.0220 | 0.1015 | 0.1111 | 0.1780 | 0.0123 | 0.0732 | 0.0489 | 0.1203 | 0.0775 | 0.4843 |
| 4 | CG14906 | 0.473 | 0.597 | 0.731 | N/A | 0.0089 | 0.0125 | 0.0025 | 0.0008 | 0.0387 | 0.0459 | 0.0879 | 0.2154 | 0.0266 | 0.1378 | 0.1770 | 0.1251 | 0.0431 | 0.4492 |
| 5 | CG32343 | 0.000 | 0.608 | 0.593 | 0.782 | N/A | 0.0032 | 0.0070 | 0.0057 | 0.1500 | 0.0492 | 0.1193 | 0.3912 | 0.0180 | 0.0818 | 0.2100 | 0.1325 | 0.1437 | 0.3912 |
| 6 | CTCF | 0.426 | 0.429 | 0.555 | 0.758 | 0.801 | N/A | 0.0004 | 0.0032 | 0.0435 | 0.0238 | 0.1094 | 0.1260 | 0.0641 | 0.1074 | 0.1074 | 0.1087 | 0.0153 | 0.1688 |
| 7 | CG16863 | 0.447 | 0.424 | 0.713 | 0.844 | 0.763 | 0.932 | N/A | 0.0009 | 0.0304 | 0.0262 | 0.0448 | 0.0905 | 0.0267 | 0.0693 | 0.0804 | 0.0794 | 0.0165 | 0.2525 |
| 8 | sna | 0.377 | 0.421 | 0.650 | 0.874 | 0.773 | 0.833 | 0.912 | N/A | 0.0149 | 0.1107 | 0.0146 | 0.0721 | 0.0278 | 0.0655 | 0.0518 | 0.0411 | 0.0127 | 0.3520 |
| 9 | mei-S332 | 0.087 | 0.311 | 0.662 | 0.650 | 0.386 | 0.643 | 0.698 | 0.796 | N/A | 0.1072 | 0.0903 | 0.0312 | 0.0248 | 0.4685 | 0.0943 | 0.1754 | 0.0643 | 0.5163 |
| 10 | homer | 0.370 | 0.323 | 0.457 | 0.622 | 0.587 | 0.708 | 0.713 | 0.524 | 0.457 | N/A | 0.0026 | 0.0788 | 0.0196 | 0.1536 | 0.1112 | 0.0661 | 0.0397 | 0.0903 |
| 11 | Rpp20 | 0.307 | 0.109 | 0.441 | 0.513 | 0.440 | 0.506 | 0.651 | 0.797 | 0.489 | 0.777 | N/A | 0.0127 | 0.0186 | 0.1018 | 0.0495 | 0.0950 | 0.0703 | 0.1915 |
| 12 | RhoGEF4 | 0.097 | -0.025 | 0.337 | 0.313 | 0.000 | 0.480 | 0.541 | 0.602 | 0.638 | 0.498 | 0.716 | N/A | 0.0322 | 0.0241 | 0.0250 | 0.1788 | 0.1488 | 0.1462 |
| 13 | Mt2 | 0.644 | 0.590 | 0.711 | 0.693 | 0.699 | 0.589 | 0.712 | 0.741 | 0.663 | 0.650 | 0.680 | 0.577 | N/A | 0.0179 | 0.0049 | 0.0339 | 0.0392 | 0.1800 |
| 14 | Orc2 | 0.000 | 0.421 | 0.512 | 0.425 | 0.509 | 0.508 | 0.585 | 0.616 | 0.000 | 0.375 | 0.470 | 0.609 | 0.686 | N/A | 0.0044 | 0.0428 | 0.4939 | 0.1165 |
| 15 | hay | 0.609 | 0.497 | 0.572 | 0.369 | 0.297 | 0.508 | 0.563 | 0.653 | 0.481 | 0.440 | 0.582 | 0.605 | 0.771 | 0.741 | N/A | 0.0028 | 0.0082 | 0.0493 |
| 16 | mms4 | 0.631 | 0.481 | 0.426 | 0.445 | 0.418 | 0.507 | 0.565 | 0.686 | 0.344 | 0.521 | 0.483 | 0.313 | 0.622 | 0.542 | 0.831 | N/A | 0.0041 | 0.0273 |
| 17 | hop | 0.596 | 0.524 | 0.503 | 0.629 | 0.399 | 0.747 | 0.762 | 0.805 | 0.540 | 0.588 | 0.528 | 0.351 | 0.602 | 0.000 | 0.774 | 0.845 | N/A | 0.0940 |
| 18 | mus209 | 0.476 | 0.264 | 0.020 | 0.052 | 0.000 | 0.409 | 0.295 | 0.185 | -0.010 | 0.479 | 0.336 | 0.355 | 0.349 | 0.397 | 0.621 | 0.703 | 0.551 | N/A |
Discussion
Evolutionary scenario of drosophilids DNMT2
In our analyses of Dnmt2 homologs among species of Drosophilidae (Dot blot, Figure S1 (369.2KB, pdf) ), we detected a hybridization signal in the majority of the analyzed species. Not all species amplified using the D. melanogaster and D. willistoni primers, indicating that the degree of Dnmt2 similarity among Drosophilidae species varies (Table S1 (542.9KB, pdf) ). The representative species of the Dorsilopha subgenus did not present Dnmt2 amplification with the primers used, which could indicate that the 5’ and 3’ regions of the Dnmt2 gene must have certain variation in the species that did not show amplicons (Table S1 (542.9KB, pdf) ). However, the average divergence value did not exceed 22.5% for the amino acid sequences, or 26.57% for the nucleotide sequences (data not shown). Functionally important genes are often evolutionarily constrained because the amino acid sequence must be preserved for a protein’s catalytic or structural role to be maintained (Figures 3 and S2 (1MB, pdf) ).
Overall, the Neotropical willistoni species group presents the highest differences when compared to all other groups, both in the nucleotide and amino acid analysis (Table 1). Interestingly, the difference is reflected in the increased presence of basic amino acids in the DNMT2 of the willistoni subgroup. Such differences may result in changes in physicochemical properties of the enzyme, giving modulations and differential affinities between proteins of different groups. Shifts in codon preferences are described in D. willistoni (Singh et al., 2006; Vicario et al., 2007), and being a lineage-specific feature, it is suggested that the differential preference may influence the evolution of DNMT2 in the species of the willistoni group. Also, molecular evolution is atypical in this species of the willistoni group (and its sister group saltans), characterized by high rates of nucleotide substitution and low portions of G/C (DeSalle, 1992; Powell and DeSalle, 1995; Remsen and O’Grady, 2002).
Our phylogenetic analysis further showed that the DNMT2 relationships in Drosophilidae comprised three main clades: a virilis-repleta section, a quinaria-tripunctata section and the Sophophora subgenus (Figures 1 and S2 (1MB, pdf) ), consistent with previous studies regarding the phylogeny of the genus (Lewis et al., 2005; Robe et al., 2005, 2010; Wang et al., 2006; Hatadani et al., 2009), although for the quinaria-tripunctata section, the evolutionary relationships between the species did not have a strong support value and presented, in some cases, polytomy. Monophyly of the subgenus Sophophora has been confirmed as well by Tatarenkov et al. (1999) and Robe et al. (2005). These findings suggest that the DNMT2 sequences exhibit similar patterns to the species evolution.
Despite the conservation of drosophilid DNMT2s, at least 10 regions have a high evolutionary rate along the protein (Figure 1). Noteworthy, region 7 has a long sequence with high evolutionary rate. The structural location of this region corresponds to the connecting handle between the catalytic domain and TRD. When analyzed separately, the evolutionary rates of the Drosophila and Sophophora subgenera showed very similar patterns with regard to the distribution of sites that evolve faster. Nonetheless, evolutionary rate values in the Drosophila subgenus seem to be smaller than those of Sophophora (Figure 1). Thus, we question whether some of these sites with faster evolutionary rates may be under some lineage-specific adaptive selection event, or just correspond to a relaxed selection pressure.
Positively selected sites in DNMT2 suggest adaptation for protein-protein interactions
The importance of Dnmt2 for development in drosophilid species remains unclear. Marhold et al. (2004) compared DNMT2 sequences from different dipterans, showing high evolutionary conservation mainly in the catalytic domain. The occurrence of DNA methylation in various dipteran species was also demonstrated, suggesting that DNMT2-mediated DNA methylation has a deep evolutionary relationship of at least 250 million years. In any case, DNMT2-mediated DNA methylation remains an open issue.
By depletion of Dnmt2 with RNA interference, Kunert et al. (2003) demonstrated that DNMT2 is both necessary and sufficient for DNA methylation in D. melanogaster and suggest a different target sequence for DNA methylation: CpT/A. However, depletion of Dnmt2 had no detectable effect on embryonic development, despite complete loss of DNA methylation. On the other hand, DNMT2 has low catalytic activity on DNA (Hermann et al., 2003; Fisher et al., 2004), and it seems to have a preference for tRNA-Asp as a methylation target (Goll et al., 2006; Jurkowski et al., 2008; Krauss and Reuter 2011; Tuorto et al., 2012; Durdevic et al., 2013a; Shanmugam et al., 2014). More recently, Takayama et al. (2014) have brought another piece to the controversial and elusive puzzle of DNA methylation in the genome of D. melanogaster: lines deficient for DNMT2 retain genomic methylation, although with altered patterns. Adding up to this peculiar field, there is the phenomenon of sex-specific DNA methylation in species of the subgroup willistoni (Garcia et al., 2007; D’Ávila et al., 2010), which raises questions about the mechanisms that are directly involved in the sex-specific DNA methylation process in this drosophilid group.
These scenarios raise issues about the selective pressures acting on Dnmt2 and on how the DNMT2 protein may have evolved along the several Drosophilidae lineages. Considering that Dnmt2 was described as unnecessary for embryological development (Kunert et al., 2003) and no loss of fitness with depletion of DNMT2 was detected (Takayama et al., 2014) in D. melanogaster, one might expect a relaxed selective constraint. However, the present study demonstrated that Dnmt2 is evolutionarily conserved (Figures 2 and S2 (1MB, pdf) ), as previous studies have already shown (Marhold et al., 2004), and therefore, it could be suggested that the entire gene has evolved under purifying selection and that positive selection has played only a minor role in the evolution of Drosophilidae Dnmt2.
Since the catalytic domain (residues from the cofactor-binding pocket and catalytic residues in the motifs ENV and PPC) and the motif CFT in TRD are generally conserved (Figures 3 and 5b), the 10 sites with high evolutionary rates in some motifs in the catalytic domain and most of the TRD motif would be due to relaxed selective constraint or neutral evolution (Figure 1). On the other hand, this variation would be better explained by an adaptive selection. Nevertheless, when we employed analyses which considered variation in ω rates in branches and sites, we found several positively selected sites in the catalytic domain and in the TRD from Dnmt2 between foreground (subgenus Sophophora) and background (subgenus Drosophila) lineages (Table 3 and Figure 5b). Positive selection has been shown to act directly on amino acid residues exposed on the surface of proteins, while those in the core remain stable (Casewell et al., 2011), favoring the maintenance of catalytic function of the protein.
The majority of sites under positive-destabilizing selection were located at the protein surface, which suggests that these sites are potentially involved in interactions with the surrounding environment. The sites 63, 163 and 288 (Figure 4a,b) were noted as having radically changed properties, namely Power to be at the C-terminal (αC). This property relates to the ability of residues to interact with other molecules, especially protein-protein and subunit interactions. Also, sites 136 to 183 are located in the connecting region of the catalytic domain and TRD and could be a potential region of protein interaction.
DNMT2 has new potential partners that have not been considered yet
Tovy et al. (2010) established enolase as the first DNMT2 interacting protein and highlighted an unexpected role of a glycolytic enzyme in the modulation of DNMT2 activity. Other potential protein partners of DNMT2 have been described, such as EGGLESS (dSETDB1), which is a member of the family of SET/MBD proteins and methylates lysine 9 in histone H3, that mediates DNA methylation and is involved in silencing genes and retrotransposons (Gou et al., 2010). Another probable candidate is MBD-like, a methyl-DNA binding protein that keeps certain genes epigenetically silenced during genome activation (Marhold et al., 2004; Gutierrez and Sommer, 2004). Since DNMT2 has tRNA as a preferential target, NSUN2 (a tRNA methyltransferase which has the function of cytosine-C5 methylation) may also be a possible protein partner, cooperating with tRNA stability and protein synthesis (Tuorto et al., 2012), together with DNMT2.
ERC values are typically elevated between interacting proteins and can be used to establish molecular and functional interactions between a pair and/or a group of proteins. Therefore, here we tested 36 proteins retrieved from searches through the STRING database and the tool ERC Analysis Webserver Top Genes. Interestingly, we found that the protein-interactions predicted with DNMT2 (STRING database) have low ERC values, mostly with negative values (Table 6). We had expected that high ERC values would be found in the protein pairs that are known to belong to epigenetic mechanisms predicted to interact with DNMT2. However, only the CG6712, MDB-like, RPD3 and SU(VAR)2-5 proteins showed positive ERC values, but these were very low (ERC < 0.3). DNMT2 and mus209 showed a low ERC (0.369) but this was not significant (p < 0.18). Even enolase, which was described as the first DNMT2 interacting protein (Tovy et al., 2010), showed a low ERC value. Nonetheless, Clark et al. (2012) demonstrated that direct physical interaction is not required to establish a high evolutionary rate covariation, but that the two major components associated with ERC are cofunctionality (functional and physical interactions annotations) and the coevolution of expression levels. When we looked at the proteins with high ERC values, we observed a variety of proteins with different levels of gene expression regulation (Table 6). This was not surprising, given the previously known subtract duality and the diversity of biological processes involving DNMT2 (Kunert et al., 2003; Tang et al., 2003; Lin et al., 2005; Goll et al., 2006; Phalke et al., 2009; Schaefer et al., 2010; Durdevic et al., 2013b). This said, we can rank the proteins with high ERC values into six groups: (1) chromatin remodeling, (2) transcription factors, (3) expression regulation, (4) DNA replication, (5) stress response, and (6) RNA editing.
The first group shows that DNMT2 has a high ERC value with proteins related to chromatin remodeling, like ATAC3 that is an essential D. melanogaster histone acetyl transferase (HAT) complex, together with transcriptional cofactors GCN5 (KAT2), ADA3, ADA2A, ATAC1 and HCF. This complex does not work in nucleosome remodeling itself, but it stimulates nucleosome sliding by the ISWI, SWI-SNF and RSC complexes (Suganuma et al., 2008). The nucleosome remodeling process is required to expose sequences that may be a target for gene silencing.
In their review, Klose and Bird (2006) describe that in targeting de novo DNA methylation, transcription factors (TF) have the capacity to interact with DNMT enzymes and to promote cytosine-target methylation as a part of the molecular silencing repertoire. This specific DNMT-TF network may be responsible for promoting specific differentiation stages in different organs. The significant ERC values found between DNMT2 and TFs like PNT and SNAIL (Table 6) reinforce the DNMT-TF interaction findings.
The involvement of DNMT2 in gene expression regulation appears in the interaction with histone methyltransferases enzymes (HMT) with high ERC values between the heterochromatin protein (HP) SU(VAR)2-10. SU(VAR)2-10 encodes a member of the PIAS protein family that controls diverse functions and is involved in different aspects of chromosome structure and function by establishing/maintaining chromosome organization (Hari et al., 2001). Furthermore, DNMT-TF interaction and DNMT-HP interaction can produce a ternary complex composed of a DNMT, a MBD and an HP, promoting the recruitment of histone H3K9 methyltransferases (HMTs) (Rai et al., 2010). The CG16863 protein has a BED-type zinc finger domain and is not characterized yet. BED fingers are able to bind DNA and are present in some proteins like Drosophila boundary element-associated factor (BEAF), responsible by chromatin insulation and also required during early development (Gilbert et al., 2006). DREF (DNA replication-related element-binding factor) is another protein that contains BED fingers; it is a transcription regulatory factor and it interacts genetically and physically with regulatory factors related to chromatin structures. Matsukage et al. (2008) identified more than 150 genes carrying DRE sequences in their promoter regions, most of them related to DNA replication, transcriptional regulation, cell cycle regulation, growth signal transduction and protein metabolism. The high ERC value between DNMT2 and BED-finger domain (0.712) is very interesting (Table 6), because the DREF target sequence is 5’-TATCGATA-3’, carrying a CpG motif that can be a DNMT2 target for cytosine methylation.
Another protein group that shares a high ERC with DNMT2 is related with expression regulation: HOPSCOTCH and HAYWIRE. HOPSCOTCH is a protein pseudokinase and is involved in many biological processes, like cell proliferation, structure morphogenesis, and others. Hou et al. (1996) have described Hopscotch/JAK kinase as an invertebrate JAK/STAT system. HOPSCOTCH regulates the transcription of target genes, such as the pair-rule gene even-skipped. even-skipped is a transcriptional repressor of a number of genes during early embryogenesis. Thus, having its role in the epigenetic system may DNMT2 have coevolved with Hopscotch/JAK to rearrange the epigenetic marks during the early development of drosophilids.
We also found significant evolutionary rate covariation values with proteins involved in DNA replication like ORC2, MMS4 and even MUS209 (a Proliferating cell nuclear antigen – PCNA) (Table 6). This may be linked to DNMT2 maintenance methylation of DNA in the replication event, or to de novo methylation during embryonic development.
Usually present in the nucleus, under conditions of stress, DNMT2 reallocated to cytoplasmic stress granules and RNA processing bodies (P-bodies). Thiagarajan et al. (2011) describe DNMT2 as part of the RNA processing machinery during cellular stress. During heat shock conditions, DICER-2 degrades tRNA and tRNA fragments, so DNMT2 can limit the extent of tRNA fragmentation during a stress event, since long double-stranded RNAs (dsRNAs) inhibit DICER-2 activity. Hence, DNMT2 is essential for DICER-2 processing in Drosophila (Durdevic et al., 2013a).
Another role of DNMT2 against stress events is the silencing of retrotransposons and the control of RNA viruses in Drosophila (Phalke et al., 2009; Durdevic et al., 2013b). So, DNMT2 seems to be important as a control tool to various forms of stress that involve RNA, which can be triggered by an excess of endogenous (retrotransposons) or exogenous (viruses) RNAs. The presence of the proteins HOMER and Rpp20, with high ERC values, may contribute to the presence and function of DNMT2 involved in the response to stress events and opens new possible partner proteins. HOMER protein acts in response to ethanol (Urizar et al., 2007), controlling circadian cycles (Naidoo et al., 2012), and acts during the stress response. Rpp20 is a subunit of RNase P and RNase MRP that is involved in precursor rRNA processing (Li et al., 2002) and interacts with SMN protein in response to stress (Hua and Zhou 2004).
Finally, we denoted an evolutionary rate relationship between DNMT2 and two nucleotide modification enzymes: CG14906 e CG14618. The CG14906 gene product corresponds to a TM-A70-like factor, an mRNA (2-O-methyladenosine-N(6)-)-methyltransferase (Bujnicki et al., 2002), and CG14618 belongs to the class IV-like SAM-binding methyltransferase superfamily, tRNA methyltransferase TRM10 family. TRM10 catalyzes all known instances of m1G9 modification and, according to Swinehart et al. (2013), it is involved in different pathways beyond tRNA processing. The plurality and importance of RNA and tRNA modifications found in the last decade are well described (Chiari et al., 2010; Chan et al., 2012; Gu et al., 2014; Hori, 2014; Swinehart and Jackman., 2015; Tuorto et al., 2015). The pathway diversity and response plasticity to environmental modifications makes the tRNA/RNA processing enzymes a major link between the genome and environment.
Conclusions
Our study shows that, although Dnmt2 is highly conserved within the Drosophilidae family, it carries several nonsynonymous changes in some domains, which were shown to be maintained by positive and destabilizing selection. Purifying selection remains the major maintenance process of gene function(s), but positive selection appears to act on certain domains potentially involved with environment interactions. Thus, the findings suggest that the residues affected by positive selection may be involved in an interaction-driven co-evolution and the connection of regions of catalytic domains and TRD that probably would interact – direct or indirectly – with other proteins.
We suggest that the episodes of adaptive evolution in Dnmt2 could be related to the wide diversity of niches, behaviors, amplitude distribution of drosophilids, as well as with and other peculiarities, such as the presence of transposable elements, chromosomal inversions, chromosomal stability, sex-specific DNA methylation, responsive modulation of RNA methylation (coding and noncoding), and endosymbiotic interactions. The multiplicity of proteins having strong evolutionary rate covariation found in the present work supports the hypothesis of a plurality of DNMT2 functions in Dnmt2-only organisms, like drosophilids. Since epigenetic systems are dynamic and change with the environment and along the evolutionary history of the organisms, we think that a wide scenario was opened, and the next step will be to analyze probable Dnmt2 interactions with other genes along the evolution of different lineages in their ecological and evolutionary contexts.
Acknowledgments
We would like to thank two anonymous referees for helpful comments and suggestions. This study was supported by grants and fellowships from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Supplementary material
The following online material is available for this article:
Footnotes
Associate Editor: Loreta B. Freitas
References
- Abascal F, Zardoya R, Posada D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;19:716–723. [Google Scholar]
- Bujnicki JM, Feder M, Radlinska M, Blumenthal RM. Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA:m6A methyltransferase. J Mol Evol. 2002;55:431–444. doi: 10.1007/s00239-002-2339-8. [DOI] [PubMed] [Google Scholar]
- Casewell NR, Wagstaff SC, Harrison RA, Renjifo CL, Wüster W. Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Mol Biol Evol. 2011;28:2637–2649. doi: 10.1093/molbev/msr091. [DOI] [PubMed] [Google Scholar]
- Chan CTY, Pang YLJ, Deng W, Babu IR, Dyavaiah M, Begley TJ, Dedon PC. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun. 2012;3:937. doi: 10.1038/ncomms1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiari Y, Dion K, Colborn J, Parmakelis A, Powell JR. On the possible role of tRNA base modifications in the evolution of codon usage: queuosine and Drosophila. J Mol Evol. 2010;70:339–345. doi: 10.1007/s00239-010-9329-z. [DOI] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Clark NL, Alani E, Aquadro CF. Evolutionary rate covariation reveals shared functionality and co-expression of genes. Genome Res. 2012;22:714–720. doi: 10.1101/gr.132647.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NL, Alani E, Aquadro CF. Evolutionary rate covariation involving meiotic proteins results from fluctuating evolutionary pressure in yeasts and mammals. Genetics. 2013;193:529–538. doi: 10.1534/genetics.112.145979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ávila MF, Garcia RN, Panzera Y, Valente VLS. Sex-specific methylation in Drosophila: an investigation of the Sophophora subgenus. Genetica. 2010;138:907–913. doi: 10.1007/s10709-010-9473-9. [DOI] [PubMed] [Google Scholar]
- Da Lage JL, Kergoat GJ, Maczkowiak F, Silvain JF, Cariou ML, Lachaise D. A phylogeny of Drosophilidae using the Amyrel gene: Questioning the Drosophila melanogaster species group boundaries. J Zool Syst Evol Res. 2007;45:47–63. [Google Scholar]
- DeSalle R. The phylogenetic relationships of flies in the family Drosophilidae deduced from mtDNA sequences. Mol Phylogenet Evol. 1992;1:31–40. doi: 10.1016/1055-7903(92)90033-d. [DOI] [PubMed] [Google Scholar]
- Durdevic Z, Mobin MB, Hanna K, Lyko F, Schaefer M. The RNA methyltransferase Dnmt2 is required for efficient Dicer-2-dependent siRNA pathway activity in Drosophila . Cell Rep. 2013a;4:931–937. doi: 10.1016/j.celrep.2013.07.046. [DOI] [PubMed] [Google Scholar]
- Durdevic Z, Hanna K, Gold B, Pollex T, Cherry S, Lyko F, Schaefer M. Efficient RNA virus control in Drosophila requires the RNA methyltransferase Dnmt2. EMBO Rep. 2013b;14:269–275. doi: 10.1038/embor.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: Mmultiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay GD, Sitnik JL, Wang W, Aquadro CF, Clark NL, Wolfner MF. Evolutionary rate covariation identifies new members of a protein network required for Drosophila female post-mating responses. PLoS Genetics. 2014;10:e1004108. doi: 10.1371/journal.pgen.1004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher O, Siman-Tov R, Ankri S. Characterization of cytosine methylated regions and 5-cytosine DNA methyltransferase (Ehmeth) in the protozoan parasite Entamoeba histolytica . Nucleic Acids Res. 2004;32:287–297. doi: 10.1093/nar/gkh161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gailey DA, Ho SK, Ohshima S, Liu JH, Eyassu M, Washington MA, Yamamoto D, Davis T. A phylogeny of the Drosophilidae using the sex-behaviour gene fruitless. Hereditas. 2000;133:81–83. doi: 10.1111/j.1601-5223.2000.00081.x. [DOI] [PubMed] [Google Scholar]
- Garcia RN, D’Ávila MF, Loreto ELS, Panzera Y, Heredia FO, Valente VLS. First evidence of methylation in the genome of Drosophila willistoni . Genetica. 2007;131:91–105. doi: 10.1007/s10709-006-9116-3. [DOI] [PubMed] [Google Scholar]
- Gilbert MK, Tan YY, Hart CM. The Drosophila boundary element-associated factors BEAF-32A and BEAF-32B affect chromatin structure. Genetics. 2006;173:1365–1375. doi: 10.1534/genetics.106.056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh C-L, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- Gou D, Rubalcava M, Sauer S, Mora-Bermúdez F, Erdjument-Bromage H, Tempst P, Kremmer E, Sauer F. SETDB1 is involved in postembryonic DNA methylation and gene silencing in Drosophila. PLoS One. 2010;5:e10581. doi: 10.1371/journal.pone.0010581. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gu C, Begley TJ, Dedon PC. tRNA modifications regulate translation during cellular stress. FEBS Lett. 2014;28:4287–4296. doi: 10.1016/j.febslet.2014.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Gutierrez A, Sommer RJ. Evolution of dnmt-2 and mbd-2-like genes in the free-living nematodes Pristionchus pacificus, Caenorhabditis elegans and Caenorhabditis briggsae . Nucleic Acids Res. 2004;32:6388–6396. doi: 10.1093/nar/gkh982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari KL, Cook KR, Karpen GH. The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev. 2001;15:1334–1348. doi: 10.1101/gad.877901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatadani LM, McInerney JO, de Medeiros HF, Junqueira AC, de Azeredo-Espin AM, Klaczko LB. Molecular phylogeny of the Drosophila tripunctata and closely related species groups (Diptera: Drosophilidae) Mol Phylogenet Evol. 2009;51:595–600. doi: 10.1016/j.ympev.2009.02.022. [DOI] [PubMed] [Google Scholar]
- Hermann A, Schmitt S, Jeltsch A. The human Dnmt2 has residual DNA (cytosine-C5) methyltransferase activity. J Biol Chem. 2003;278:31717–31721. doi: 10.1074/jbc.M305448200. [DOI] [PubMed] [Google Scholar]
- Hori H. Methylated nucleosides in tRNA and tRNA methyltransferases. Front Genet. 2014;5:1–26. doi: 10.3389/fgene.2014.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou XS, Melnick MB, Perrimon N. Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell. 1996;84:411–419. doi: 10.1016/s0092-8674(00)81286-6. [DOI] [PubMed] [Google Scholar]
- Hua Y, Zhou J. Survival motor neuron protein facilitates assembly of stress granules. FEBS Lett. 2004;572:69–74. doi: 10.1016/j.febslet.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Jurkowski TP, Meusburger M, Phalke S, Helm M, Nellen W, Reuter G, Jeltsch A. Human DNMT2 methylates tRNA Asp molecules using a DNA methyltransferase-like catalytic mechanism. Rna. 2008;14:1663–1670. doi: 10.1261/rna.970408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M, Curk T, Xu Q, Zupan B, Kuspa A, Shaulsky G. Developmentally regulated DNA methylation in Dictyostelium discoideum . Eukaryot Cell. 2006;5:18–25. doi: 10.1128/EC.5.1.18-25.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. Genomic DNA methylation: Yhe mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Kopp A. Basal relationships in the Drosophila melanogaster species group. Mol Phylogenet Evol. 2006;39:787–798. doi: 10.1016/j.ympev.2006.01.029. [DOI] [PubMed] [Google Scholar]
- Krauss V, Reuter G. DNA methylation in Drosophila - a critical evaluation. Progr Mol Biol Transl Sci. 2011;110:177–191. doi: 10.1016/B978-0-12-387685-0.00003-2. [DOI] [PubMed] [Google Scholar]
- Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319:1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- Kuhlmann M, Borisova BE, Kaller M, Larsson P, Stach D, Na J, Eichinger L, Lyko F, Ambros V, Söderbom F, et al. Silencing of retrotransposons in Dictyostelium by DNA methylation and RNAi. Nucleic Acids Res. 2005;33:6405–6417. doi: 10.1093/nar/gki952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura, Stecher G. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0. Mol Biol Evol. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunert N, Marhold J, Stanke J, Stach D, Lyko F. A Dnmt2-like protein mediates DNA methylation in Drosophila . Development. 2003;130:5083–5090. doi: 10.1242/dev.00716. [DOI] [PubMed] [Google Scholar]
- Kwiatowski J, Ayala FJ. Phylogeny of Drosophila and related genera: Conflict between molecular and anatomical analyses. Mol Phylogenet Evol. 1999;13:319–328. doi: 10.1006/mpev.1999.0657. [DOI] [PubMed] [Google Scholar]
- Lewis RL, Beckenbach AT, Mooers A. The phylogeny of the subgroups within the melanogaster species group: Likelihood tests on COI and COII sequences and a Bayesian estimate of phylogeny. Mol Phylogenet Evol. 2005;37:15–24. doi: 10.1016/j.ympev.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Li S, Du J, Yang H, Yin J, Zhong J, Ding J. Functional and structural characterization of DNMT2 from Spodoptera frugiperda . J Mol Cell Biol. 2012;5:64–66. doi: 10.1093/jmcb/mjs057. [DOI] [PubMed] [Google Scholar]
- Li X, Frank DN, Pace N, Zengel JM, Lindahl L. Phylogenetic analysis of the structure of RNase MRP RNA in yeasts. RNA. 2002;8:740–751. doi: 10.1017/s1355838202022082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MJ, Tang LY, Reddy MN, Shen CKJ. DNA methyltransferase gene dDnmt2 and longevity of Drosophila . J Biol Chem. 2005;280:861–864. doi: 10.1074/jbc.C400477200. [DOI] [PubMed] [Google Scholar]
- Lyko F, Ramsahoye BH, Jaenisch R. DNA methylation in Drosophila melanogaster . Nature. 2000;408:538–540. doi: 10.1038/35046205. [DOI] [PubMed] [Google Scholar]
- Marhold J, Rothe N, Pauli A, Mund C, Kuehle K, Brueckner B, Lyko F. Conservation of methylation in dipteran insects. Insect Mol Biol. 2004;13:117–123. doi: 10.1111/j.0962-1075.2004.00466.x. [DOI] [PubMed] [Google Scholar]
- Marques EK, Napp M, Winge H. A corn meal, soybean flour, wheat germ medium for Drosophila. Drosoph Inf Serv. 1966;41:187. [Google Scholar]
- Matsukage A, Hirose F, Yoo MA, Yamaguchi M. The DRE/DREF transcriptional regulatory system: A master key for cell proliferation. Biochim Biophys Acta Gene Regul Mech. 2008;1779:81–89. doi: 10.1016/j.bbagrm.2007.11.011. [DOI] [PubMed] [Google Scholar]
- McClellan DA, McCracken KG. Estimating the influence of selection on the variable amino acid sites of the cytochrome b protein functional domains. Mol Biol Evol. 2001;18:917–925. doi: 10.1093/oxfordjournals.molbev.a003892. [DOI] [PubMed] [Google Scholar]
- McClellan DA, Palfreyman EJ, Smith MJ, Moss JL, Christensen RG, Sailsbery JK. Physicochemical evolution and molecular adaptation of the cetacean and artiodactyl cytochrome b proteins. Mol Biol Evol. 2005;22:437–455. doi: 10.1093/molbev/msi028. [DOI] [PubMed] [Google Scholar]
- Naidoo N, Ferber M, Galante RJ, McShane B, Hu JH, Zimmerman J, Maislin G, Cater J, Wyner A, Worley P, et al. Role of homer proteins in the maintenance of sleep-wake states. PLoS One. 2012;7:e35174. doi: 10.1371/journal.pone.0035174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander JAA. MrModeltest v2. Software distributed by the author. Evolutionary Biology Center; Uppsala University: 2004. [Google Scholar]
- Okano M, Xie S, Li E. Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res. 1998;26:2536–40. doi: 10.1093/nar/26.11.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phalke S, Nickel O, Walluscheck D, Hortig F, Onorati MC, Reuter G. Retrotransposon silencing and telomere integrity in somatic cells of Drosophila depends on the cytosine-5 methyltransferase DNMT2. Nat Genet. 2009;41:696–702. doi: 10.1038/ng.360. [DOI] [PubMed] [Google Scholar]
- Ponger L, Li W. Evolutionary diversification of DNA methyltransferases in eukaryotic genomes. Mol Biol Evol. 2005;22:1119–1128. doi: 10.1093/molbev/msi098. [DOI] [PubMed] [Google Scholar]
- Powell JR, DeSalle R. Drosophila molecular phylogenies and their uses. In: Hecht MK, MacIntyre RJ, Clegg MT, editors. Evolutionary Biology. Plenum Press; New York: 1995. pp. 87–138. [Google Scholar]
- Rai K, Jafri IF, Chidester S, James SR, Karpf AR, Cairns BR, Jones DA. Dnmt3 and G9a cooperate for tissue-specific development in zebrafish. J Biol Chem. 2010;285:4110–4121. doi: 10.1074/jbc.M109.073676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remsen J, O’Grady PM. Phylogeny of Drosophilidae (Diptera), with comments on combined analysis and character support. Mol Phylogenet Evol. 2002;24:248–263. doi: 10.1016/s1055-7903(02)00226-9. [DOI] [PubMed] [Google Scholar]
- Robe LJ, Cordeiro J, Loreto ELS, Valente VLS. Taxonomic boundaries, phylogenetic relationships and biogeography of the Drosophila willistoni subgroup (Diptera: Drosophilidae) Genetica. 2010;138:601–617. doi: 10.1007/s10709-009-9432-5. [DOI] [PubMed] [Google Scholar]
- Robe LJ, Valente VL, Budnik M, Loreto EL. Molecular phylogeny of the subgenus Drosophila (Diptera, Drosophilidae) with an emphasis on Neotropical species and groups: A nuclear versus mitochondrial gene approach. Mol Phylogenet Evol. 2005;36:623–640. doi: 10.1016/j.ympev.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes version 3.0: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Russo CA, Takezaki N, Nei M. Molecular phylogeny and divergence times of drosophilid species. Mol Biol Evol. 1995;12:391–404. doi: 10.1093/oxfordjournals.molbev.a040214. [DOI] [PubMed] [Google Scholar]
- Saikia M, Krokowski D, Guan B-J, Ivanov P, Parisien M, Hu G, Anderson P, Pan T, Hatzoglou M. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J Biol Chem. 2012;287:42708–42725. doi: 10.1074/jbc.M112.371799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassi AK, Herédia FO, Loreto ELS, Valente VLS, Rohde C. Transposable elements P and gypsy in natural populations of Drosophila willistoni . Genet Mol Biol. 2005;4:734–739. [Google Scholar]
- Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam R, Aklujkar M, Schäfer M, Reinhardt R, Nickel O, Reuter G, Lovley DR, Ehrenhofer-Murray A, Nellen W, Ankri S, et al. The Dnmt2 RNA methyltransferase homolog of Geobacter sulfurreducens specifically methylates tRNA-Glu. Nucleic Acids Res. 2014;42:6487–6496. doi: 10.1093/nar/gku256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh ND, Arndt PF, Petrov DA. Minor shift in background substitutional patterns in the Drosophila saltans and willistoni lineages is insufficient to explain GC content of coding sequences. BMC Biology. 2006;4:37. doi: 10.1186/1741-7007-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. The Staden sequence analysis package. Mol Biotechnol. 1996;5:233–241. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- Suganuma T, Gutiérrez JL, Li B, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat Struct Mol Biol. 2008;15:364–372. doi: 10.1038/nsmb.1397. [DOI] [PubMed] [Google Scholar]
- Swinehart W, Jackman J. Diversity in mechanism and function of tRNA methyltransferases. RNA Biol. 2015;6286:37–41. doi: 10.1080/15476286.2015.1008358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinehart WE, Henderson JC, Jackman JE. Unexpected expansion of tRNA substrate recognition by the yeast m1G9 methyltransferase Trm10. RNA. 2013;19:1137–46. doi: 10.1261/rna.039651.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S, Dhahbi J, Roberts A, Mao G, Heo SJ, Pachter L, Martin DIK, Boffelli D. Genome methylation in Drosophila melanogaster is found at specific short motifs and is independent of DNMT2 activity. Genome Res. 2014;24:821–30. doi: 10.1101/gr.162412.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang LY, Reddy MN, Rasheva V, Lee TL, Lin MJ, Hung MS, Shen CK. The eukaryotic DNMT2 genes encode a new class of cytosine-5 DNA methyltransferases. J Biol Chem. 2003;278:33613–33616. doi: 10.1074/jbc.C300255200. [DOI] [PubMed] [Google Scholar]
- Tarrio R, Rodriguez-Trelles F, Ayala FJ. Shared nucleotide composition biases among species and their impact on phylogenetic reconstructions of the Drosophilidae. Mol Biol Evol. 2001;18:1464–1473. doi: 10.1093/oxfordjournals.molbev.a003932. [DOI] [PubMed] [Google Scholar]
- Tatarenkov A, Kwiatowski J, Skarecky D, Barrio E, Ayala FJ. On the evolution of Dopa decarboxylase (Ddc) and Drosophila systematics. J Mol Evol. 1999;48:445–462. doi: 10.1007/pl00006489. [DOI] [PubMed] [Google Scholar]
- Thiagarajan D, Dev RR, Khosla S. The DNA methyltranferase Dnmt2 participates in RNA processing during cellular stress. Epigenetics. 2011;6:103–13. doi: 10.4161/epi.6.1.13418. [DOI] [PubMed] [Google Scholar]
- Throckmorton LH. The phylogeny, ecology and geography of Drosophila. In: King RC, editor. Handbook of Genetics. Vol. 3. Plenum Press; New York: 1975. pp. 421–469. [Google Scholar]
- Tovy A, Hofmann B, Helm M, Ankri S. In vitro tRNA methylation assay with the Entamoeba histolytica DNA and tRNA methyltransferase Dnmt2 (Ehmeth) enzyme. J Vis Exp. 2010;2:2–7. doi: 10.3791/2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol. 2012;19:900–905. doi: 10.1038/nsmb.2357. [DOI] [PubMed] [Google Scholar]
- Tuorto F, Herbst F, Alerasool N, Bender S, Popp O, Federico G, Reitter S, Liebers R, Stoecklin G, Gröne H-J, et al. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J. 2015;34:2350–2362. doi: 10.15252/embj.201591382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urizar NL, Yang Z, Edenberg HJ, Davis RL. Drosophila Homer is required in a small set of neurons including the ellipsoid body for normal ethanol sensitivity and tolerance. J Neurosci. 2007;27:4541–4551. doi: 10.1523/JNEUROSCI.0305-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicario S, Moriyama EN, Powell JR. Codon usage in twelve species of Drosophila . BMC Evol Biol. 2007;7:226. doi: 10.1186/1471-2148-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BC, Park J, Watabe HA, Gao JJ, Xiangyu JG, Aotsuka T, Chen HW, Zhang YP. Molecular phylogeny of the Drosophila virilis section (Diptera: Drosophilidae) based on mitochondrial and nuclear sequences. Mol Phylogenet Evol. 2006;40:484–500. doi: 10.1016/j.ympev.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Woolley S, Johnson J, Smith MJ, Crandall KA, McClellan DA. TreeSAAP: Selection on amino acid properties using phylogenetic trees. Bioinformatics. 2003;19:671–672. doi: 10.1093/bioinformatics/btg043. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Bestor TH. A candidate mammalian DNA methyltransferase related to pmt1p of fission yeast. Hum Mol Genet. 1998;7:279–284. doi: 10.1093/hmg/7.2.279. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
Internet Resources Section
- Bächli G. TaxoDros: The database on taxonomy of Drosophilidae, v.1.04, Database 2017/2. 2016. [(accessed 26 February 2016)]. Available via http://taxodros.unizh.ch/
- Flybase - http://flybase.bio.indiana.edu/blast/
- Drosophila buzzatii Genome Project. - http://dbuz.uab.cat/welcome.php.
- SpottedWingFlyBase - http://spottedwingflybase.oregonstate.edu/
- Evolutionary Rate Covariation (ERC) Analysis - http://csb.pitt.edu/erc_analysis/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.