Abstract
Diverse chromatin modifiers are involved in regulation of gene expression at the level of transcriptional regulation. Among these modifiers are ATP-dependent chromatin remodelers, where the SWI/SNF complex is the founding member. It has been observed that High Mobility Group (HMG) proteins can influence the activity of a number of these chromatin remodelers. In this context, we have previously demonstrated that the yeast HMG proteins Nhp6 and Hmo1 can stimulate SWI/SNF activity. Here, we studied the genome-wide binding patterns of Nhp6, Hmo1 and the SWI/SNF complex, finding that most of gene promoters presenting high occupancy of this complex also display high enrichment of these HMG proteins. Using deletion mutant strains we demonstrate that binding of SWI/SNF is significantly reduced at numerous genomic locations by deletion of NHP6 and/or deletion of HMO1. Moreover, alterations in the nucleosome landscape take place at gene promoters undergoing reduced SWI/SNF binding. Additional analyses show that these effects also correlate with alterations in transcriptional activity. Our results suggest that, besides the ability to stimulate SWI/SNF activity, these HMG proteins are able to assist the loading of this complex onto gene regulatory regions.
Keywords: Chromatin remodeling, HMG, Nhp6, Hmo1, SWI/SNF
1. Introduction
Chromatin dynamics have a deep impact on gene expression at the level of regulation of transcriptional activity. Several proteins and protein complexes, collectively called chromatin modifiers, are involved in this critical aspect of transcriptional regulation. Among these modifiers are ATP-dependent chromatin remodelers, which use the energy of ATP hydrolysis for mobilizing nucleosomes or altering their composition [1,2]. Within this family of chromatin remodelers, the SWI/SNF complex is the founding member. Its action at gene regulatory regions is involved in either transcriptional activation or repression. In both cases, the complex needs to be recruited to these regulatory regions in order to exert its action, mainly through physical interactions of one or more of its subunits with transcription factors [1].
High Mobility Group (HMG) proteins are abundant proteins involved in several nuclear activities, including transcription. HMG proteins are subdivided into three families: HMGA, HMGB and HMGN [3]. Among the functions described for these proteins is their ability to assist binding of transcription factors to their cognate sites at gene regulatory regions [4]. HMGA and HMGB proteins have the ability to bend DNA, a property related to this functional association with transcription factors [5] and also to other nuclear functions (see below). Seven genes encode HMGB proteins in the yeast Saccharomyces cerevisiae, including NHP6A, NHP6B and HMO1 [6]. Several studies have reported the involvement of these three HMG proteins in different aspects of transcriptional regulation [7–14]. Among the properties ascribed to HMGB proteins is their ability to stimulate ATP-dependent nucleosome remodeling activity. HMGB1 is able to stimulate the remodeling activities of the ACF, CHRAC [15] and SWI/SNF complexes [16,17]. In yeast, physical and functional interactions were demonstrated between Nhp6 and RSC, a complex of the SWI/SNF subfamily of chromatin remodelers [18]. Subsequently, with the use of several in vitro analyses, we have recently demonstrated the ability of Nhp6A, Nhp6B and Hmo1 proteins to stimulate SWI/SNF nucleosome remodeling activity. Interestingly, Hmo1 and Nhp6 exert a differential stimulatory effect on SWI/SNF activity. Among other differences, only Hmo1 is able to stimulate SWI/SNF binding to the nucleosome [19]. Consistent with our findings, a number of studies point to a functional connection between Nhp6 and SWI/SNF. A subset of yeast genes whose expression is affected by both Nhp6A/B and SWI/SNF has been observed in early high throughput gene expression analyses [12]. Consistently, from a genome-wide analysis performed by Venters and colleagues for 200 transcription regulatory proteins, numerous genes with a relative high co-occupancy of both Nhp6A and SWI/SNF can be identified [20]. Furthermore, the triple mutant nhp6a nhp6b swi2 (SWI2 = catalytic subunit of ySWI/SNF) is lethal [7]. To date, there are no in vivo studies focusing on functional relationships between Hmo1 and SWI/SNF. In our current study, with the use of ChIP-chip analyses we found that the S. cerevisiae HMGB proteins Nhp6A/B and Hmo1 are required for binding of the SWI/SNF complex to the promoters of numerous genes. Further analyses in a large fraction of these genes indicate that this dependence correlates with variations in mRNA levels and in the nucleosome landscape at the promoter region of the genes analyzed. Additional analyses suggest that this Hmo1 and Nhp6-dependent binding of SWI/SNF to gene promoters may rely, at least in part, on a stimulatory effect on SWI/SNF recruitment by transcription factors.
2. Materials and methods
2.1. Yeast strains
The strains used in this work are listed in Supplementary Table S1. All the strains are derived from the S288C strain. Their identity was confirmed by PCR and/or western blot (Supplementary Fig. S1; primer sets listed in Supplementary Table S2).
2.2. Antibodies
ChIP assays were performed using the following antibodies: anti-Snf5 (Upstate #07-320); Histone H3 antibody (Active Motif #39163); normal IgG antibody (SantaCruz #2015). IgG Sepharose 6FF resin (GE Lifescience #17-0969) was used in the case of ChIPs for Nhp6 and Hmo1 (TAP-tag strains). Western blots were performed using anti-CBP (TAP-HRP, Sigma #P1291) and the anti-Snf5 and anti-H3 antibodies described above.
2.3. Chromatin immunoprecipitation assays
Yeast strains were grown in 200 ml of YPD medium at 30 °C, crosslinked and processed for ChIP as described [21,22]. Cells were crosslinked with 1% formaldehyde at room temperature (25–28 °C) under continuous shaking. The ChIP lysates were used to immunoprecipitate Snf5 and H3, and to affinity-purify (ChAP, chromatin affinity purification) the HMG proteins. Immunoprecipitation and further processing were carried out essentially as previously described [21], using 50 µg of chromatin (DNA content). The immunocomplexes and TAP-tagged proteins were pulled down with Protein G-Sepharose 4FF (GE Healthcare Life Sciences) or IgG Sepharose 6FF resin (GE Healthcare Life Sciences), respectively. Three biological replicates were grown for each strain and used for the ChIP-on-chip assays. In the case of ChIP-qPCR, DNA was analyzed using Brilliant III QPCR Master Mix (Stratagene, 600882), according to manufacturer's instructions. Real-time PCRs were performed in a Mx3000p thermocycler (Stratagene), using specific primer sets whose sequences are given in the Supplementary Table S2.
2.4. Microarray analysis
ChIP-on-chip assays for the genome-wide distribution of the TAP-HMG proteins and Snf5 were performed using 8 × 60k yeast genome DNA arrays (Agilent, cat. 031697) with an average probe spacing of ~200 bp. We used 50 ng of input and immunoprecipitation (or affinity purification) samples for double T7 linear amplification (Epicentre) and labeling as described [23,24]. Inputs were labeled with Cy3 dye and immunoprecipitations (or affinity purifications) with Cy5 dye (GE Healthcare). We combined 4 µg of each sample (input and immunoprecipitation/affinity purification) for hybridization. Three biological repeats were done for all microarray-based experiments. Microarrays were scanned with an Agilent DNA Microarray Scanner (model no. G2505B; Agilent) and the information treated with Feature Extraction software (Agilent). The data was normalized with GeneSpringGX software (Agilent). R software was used to analyze, normalize and plot all microarray data. The microarray data from this publication have been submitted to the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) and is available as a super series under GEO accession number GSE86279.
2.5. Data analysis
Normalized ChIP-on-chip data were analyzed using a modified average gene analysis, as previously described [25]. ORFs were subdivided into 14 equal-sized bins, irrespective of gene length. Intergenic regions (480 bp upstream and downstream of genes) were allocated into three bins each. A matrix of twenty columns was generated, representing coding and intergenic regions, and each row representing a gene (Supplementary Fig. S2). The columns were averaged to generate the whole-genome average plots. To generate enrichment profiles of TAP-tagged proteins (Nhp6A, Nhp6B and Hmo1), background values obtained from a ChAP performed with the wild-type strain (BY4741) were subtracted from values obtained from the TAP-tag strains. Gene clustering criterion to group genes presenting occupancy values higher than a defined log2 value consisted in selecting those genes presenting a value ≥ a defined threshold in at least one bin, in a delimited gene section (promoter/TSS or ORF). Clustering criterion to group genes presenting occupancy values lower than a defined log2 value consisted in selecting those genes displaying all bin values below a defined threshold, in a delimited gene section (promoter/TSS or ORF). To determine genes presenting significant reduction in SWI/SNF occupancy, we used as criterion a reduction ≥1 at least in the bin displaying the highest occupancy value in the wild-type strain.
2.6. RNA isolation and RT-qPCR
Yeast strains were grown in YPD medium at 30 °C until reaching an optical density at 600 nm (OD600) of 0.8. Total RNA was isolated using acid phenol extraction as previously described [26]. RNA quality and quantity were assessed by agarose gel electrophoresis and UV spectroscopy, respectively. In each case, 1 µg of RNA was treated with DNase and then reverse transcribed using 0.25 µg of Anchored Oligo(dT)20 Primer (Invitrogen, 12577-011) and M-MLV reverse transcriptase (Promega, M170A), in a final volume of 20 µl; 0.5 µl of this sample was analyzed by real-time PCR using Brilliant III QPCR Master Mix (Stratagene, 600882), according to the manufacturer's instructions. Real-time PCRs were performed in a Mx3000p thermocycler (Stratagene) and mRNA levels were determined using the Standard Curve Method using actin (ACT1) as a reference gene.
2.7. Recombinant proteins and protein complexes
Recombinant proteins Nhp6A, Nhp6B, Hmo1 and HMGB1 were purified as previously described [19]. Gal4-VP16 was purified as described previously [27]. The ySWI/SNF complex was obtained by tandem affinity purification, concentrated and quantified as described [28].
2.8. DNA probes, nucleosome reconstitution and EMSA
A 216 bp DNA segment was generated by PCR, using the pGEM-3Z/601-Gal4 plasmid as template [28]. Prior to PCR, one of the PCR primers was end-labeled with [γ-32P] dATP. Nucleosome reconstitution was carried out as described previously [28]. Electrophoretic mobility shift assays (EMSAs) were performed essentially as previously reported [28]. In each assay, reaction mixes contained 7.4 µl remodeling buffer (70 mM KCl, 20 mM Hepes-KOH pH 7.9, 2 mM DTT, 0.5 mM PMSF, 10% glycerol, 0.05% NP-40, 10 mM MgCl2 and 100 µg/ml BSA), 0.6 µl deionized water, 0.5 µl of short oligonucleosomes (400 ng/µl), 3 µl of SWI/SNF or SWI/SNF buffer (150 mM NaCl, 10 mM Tris–Cl pH 8.0, 1 mM Mg(CH3COO)2, 1 mM imidazole, 2 mM EGTA, 0.1% NP-40, 10% glycerol, 1 mM DTT and 0.5 mM PMSF), 0.5 µl 300 nM Gal4-VP16 (or Gal4 buffer), 0.5 µl of one of the HMG proteins (3 or 6 µM stock concentration) or HMG buffer (100 mM KCl, 10 mM Hepes-KOH pH 7.9 and 15% glycerol) and 2.5 µl of probe. All these components, with the exception of SWI/SNF (or SWI/SNF buffer), were mixed and then the reactions were incubated for 20 min at 30 °C, followed by addition of the complex (or SWI/SNF buffer) and a 40 min incubation at 30 °C. The samples were then subjected to electrophoresis in a non-denaturing polyacrylamide gel (0.3× TBE, 3.5% acrylamide, 60:1 AA:Bis proportion). Afterwards, the gel was dried and subjected to autoradiography and also scanned using phosphor screen and Molecular Imager FX (BioRad). None of the reaction mixes included ATP.
2.9. Mapping of chromatin landscape at specific promoters
MNase digestion of chromatin was performed on spheroplasts by using a procedure that minimizes the time after the isolation of intact cells [29]. Promoter mapping was carried out following the protocol described by Lam and colleagues [30], with the following modifications: the analysis was performed for wild-type (BY4741) and both nhp6a/b and hmo1 deletion mutant strains; 25 ml of exponentially growing cells, cultured in YPD at 30 °C, were used in each case; primer pairs were designed to tile ~500 bp of a promoter region (see sequence information in Supplementary Table S2), yielding 90–110 bp PCR products centered every 40 bp. An analysis of chromatin digestion by MNase is presented in Supplementary Fig. S3.
3. Results
3.1. The binding profile of SWI/SNF and the HMG proteins Nhp6 and Hmo1 are highly correlated
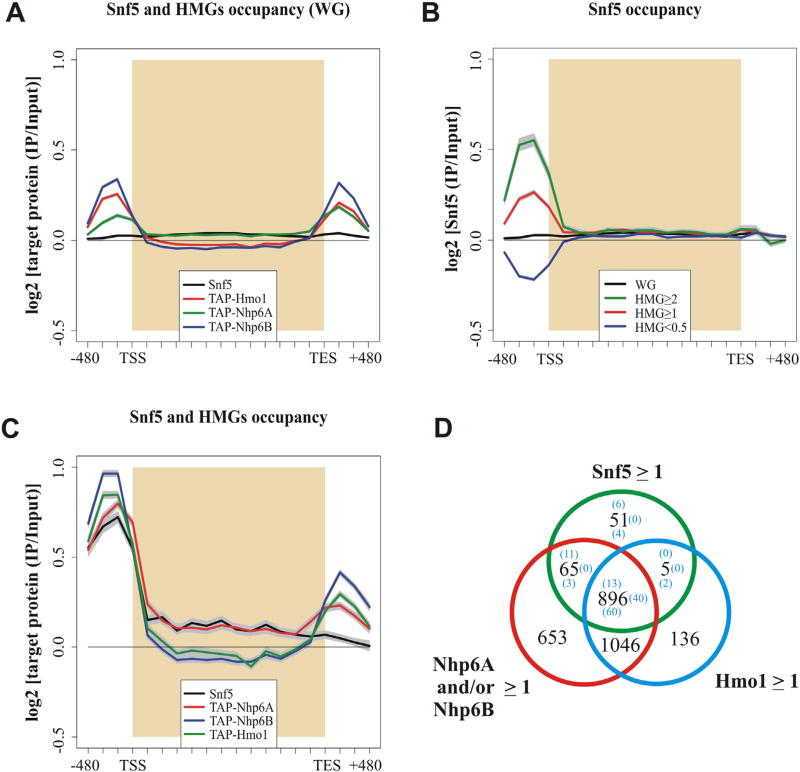
We have recently demonstrated the ability of Nhp6 and Hmo1 proteins to stimulate SWI/SNF nucleosome remodeling activity, by performing different in vitro remodeling assays [19]. In light of these results, we decided to analyze the in vivo characteristics of this connection between the afore-mentioned HMG proteins and the SWI/SNF complex in S. cerevisiae. To pursue this goal, we first performed ChIP-chip assays in order to analyze the occupancy profile of SWI/SNF and these HMG proteins. In the case of the SWI/SNF complex, the immunoprecipitation was directed against one of its core subunits, Snf5 [31,32]. In the case of the HMG proteins analyzed, strains expressing TAP epitope-tagged proteins were used. The enrichment profiles obtained were analyzed using average gene analysis [25]. When looking at the genome-wide occupancy profile (ca. 6600 ORFs), no significant preference of SWI/SNF for any particular gene region is observed (Fig. 1A), which is consistent with previous studies [33]. In the case of Nhp6A, Nhp6B and Hmo1 proteins, preferential occupancy at promoter regions can be observed (Fig. 1A). We then looked at the SWI/SNF occupancy profile in different subsets of genes generated according to the enrichment levels of Nhp6 and Hmo1 proteins at gene promoters [including the transcription start site (TSS), see Fig. 1 legend for details]. Strikingly, SWI/SNF occupancy at gene promoters displaying high Nhp6 and/or Hmo1 enrichment is considerably higher than that observed for the whole genome. Conversely, the occupancy of this complex falls down at gene promoters presenting low enrichment of these HMG proteins (Fig. 1B). Consistent with these observations, when looking at genes enriched in SWI/SNF occupancy at their promoter region [genes with an occupancy value of Snf5 ≥ 1 (log2 ratio) in at least one of the bins spanning the promoter/TSS region, 1017 genes] a higher enrichment of these HMG proteins at the promoter is observed, as compared to their genome-wide occupancy at this gene region (Fig. 1C). Interestingly, in this group of genes, the HMG proteins display the same enrichment profile shown by the SWI/SNF complex, characterized by a high enrichment in the promoter relative to the gene body (Fig. 1C). Considering that a role in transcription elongation has also been ascribed to the SWI/SNF complex [34], we asked how the enrichment profile of Nhp6 and Hmo1 proteins would be in genes with a high SWI/SNF occupancy at the gene body region (Snf5 ≥ 1 in at least one of the bins spanning the gene body, 845 genes). In this set of genes the occupancy profile of the complex is consistent with previous reports, characterized by a higher enrichment in the first half of the gene body [33]. Notably, the enrichment profile of Nhp6 and Hmo1 follows the same profile displayed by the SWI/SNF complex in this set of genes (Supplementary Fig. S4A), which is remarkably different to the enrichment profile observed for these HMG proteins in the set of genes that possess high SWI/SNF occupancy at their promoter region (Fig. 1C). Taken together, these enrichment profiles unveil a strong correlation between SWI/SNF and Nhp6/Hmo1 binding to different gene regions at a genome-wide scale. In the same line of evidence, the grouping of genes according to high occupancy of SWI/SNF, Nhp6A/B or Hmo1 at their promoter region (occupancy value ≥1 in at least one of the bins spanning the promoter/TSS region) reveals that 95% of the genes with high SWI/SNF occupancy also have high Nhp6 and/or Hmo1 occupancy (966 out of 1017 genes). In fact, most of these genes (896) display high occupancy of SWI/SNF, Nhp6 and Hmo1 (Fig. 1D).
Fig. 1.
SWI/SNF binding to gene promoters correlates with Nhp6 and Hmo1 binding. ChIP-chip assays were performed using yeast genomic tiling arrays. The log2 enrichment ratios were subjected to average gene analysis. In figures A–C, whole-genome average data were calculated and plotted as mean ± s.e.m. (gray) and represent three independent experiments. The transcription start site (TSS) and termination site (TES) are indicated. A) Whole-genome (6575 genes) enrichment profiles of Snf5, TAP-Nhp6A, TAP-Nhp6B and TAP-Hmo1. B) Snf5 enrichment profile for the whole genome and for subsets of genes displaying occupancy levels of both Nhp6 and Hmo1 (collectively termed HMG) which are over or below defined thresholds at the promoter/TSS: HMG ≥ 2 = 776 genes (green); HMG ≥ 1 = 2801 genes (red); HMG < 0.5 = 1942 genes (blue). C) Enrichment profiles of Snf5, TAP-Nhp6A, TAPNhp6B, and TAP-Hmo1 for the subset of genes displaying high occupancy of Snf5 (≥1) at the promoter/TSS region (1017 genes). D) Venn diagram depicting correlation between occupancy levels of SWI/SNF (Snf5), Nhp6A/B, and Hmo1 at the promoter/TSS region. Numbers in brackets correspond to genes displaying a significant reduction (≥1) of SWI/SNF occupancy at this gene region in deletion mutants nhp6a/b (top), hmo1 (bottom) or in both deletion mutants (right). A full list of these genes is given in Supplementary Table S3. The global effect on SWI/SNF occupancy generated by these deletion mutants is depicted in Supplementary Fig. S5.
3.2. SWI/SNF binding to numerous gene promoters is negatively affected by the absence of Nhp6 and/or the absence of Hmo1
With the aim of assessing the effect of Nhp6 and Hmo1 proteins on the residence of SWI/SNF at gene promoters, our genome-wide analyses included ChIP-chip for Snf5 comparing a wild-type strain to nhp6a/b and hmo1 deletion mutants. Average gene analysis at the whole-genome level showed a reduction in SWI/SNF occupancy at gene promoters in both deletion mutants. Importantly, in genes displaying high enrichment of Nhp6 or Hmo1 at this region the reduction in SWI/SNF occupancy upon deletion of NHP6A/B or HMO1 is more pronounced. Conversely, the low SWI/SNF occupancy observed at gene promoters also displaying low enrichment of these HMG proteins remains unaltered in these deletion mutants (Supplementary Fig. S5).
Among the gene promoters presenting high enrichment of SWI/SNF and the HMG proteins under study (966 genes), we determined those with a significant occupancy reduction of SWI/SNF in the deletion mutants, using as criterion a reduction of log2 ≥ 1 in SWI/SNF occupancy. According to this criterion, a total of 129 out of these 966 genes show a significant reduction in SWI/SNF enrichment, most of them belonging to the group of genes displaying high occupancy of SWI/SNF and both HMG proteins (896 genes). Within this group, 60 showed this reduction in the hmo1 deletion mutant, 13 in the nhp6a/b mutant and 40 in both mutant strains (Fig. 1D and Supplementary Table S3). In this analysis we additionally found reduction in SWI/SNF occupancy in genes displaying only high occupancy of the complex (51 genes, Fig. 1D). However, as detailed above, at the genome-wide level the occupancy of SWI/SNF is reduced essentially in genes displaying high enrichment of Nhp6 and/or Hmo1. We also found genes showing an increase in SWI/SNF occupancy upon deletion of NHP6A/B and/or deletion of HMO1 (Supplementary Fig. S6). Nevertheless, the overall trend corresponds to reduction in SWI/SNF occupancy at the genome-wide level, in the set of gene promoters displaying high occupancy of this complex and in the set of gene promoters displaying high Nhp6 and/or Hmo1 occupancy (Supplementary Fig. S5).
3.3. Reduced SWI/SNF binding is accompanied by alterations in transcriptional activity
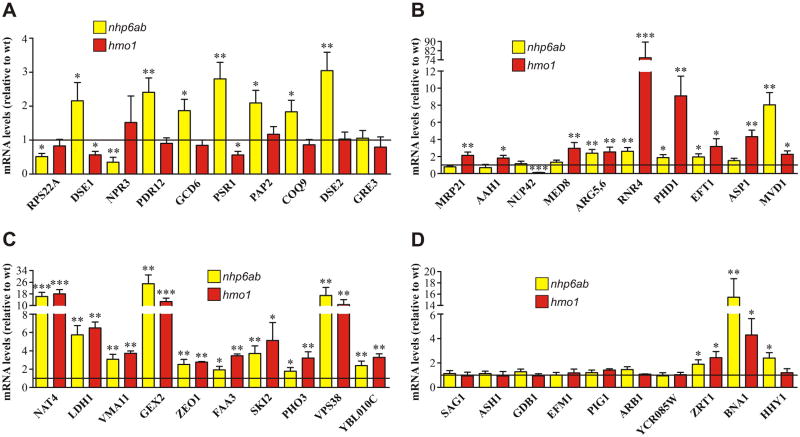
In order to validate the results observed in our genome-wide analyses and to further study the roles of Nhp6 and Hmo1 proteins in chromatin dynamics at gene regulatory regions, we selected those genes showing the highest reduction in SWI/SNF occupancy in the deletion mutants for further analyses. This selection was performed only from genes displaying both high SWI/SNF occupancy in the wild-type strain and high HMG occupancy in the TAP-tag strains. Our further analyses were focused on this group of genes considering that within this set we would find the most relevant effects derived from a reduction of SWI/SNF occupancy. A total of 40 genes were selected, divided into four equal groups: affected in nhp6a/b deletion mutant; affected in hmo1; affected in both nhp6a/b and hmo1 deletion mutants; and genes not affected in any of these deletion mutants (control group). The term “affected” stands for reduction in SWI/SNF occupancy. To ease data presentation, these groups are defined as groups A, B, C and D, respectively, in all future figures (Table 1, an expanded version of this table, containing numerical data, is provided as Supplementary Table S4). The group of non-affected genes is composed of genes that possess high occupancy of SWI/SNF and high occupancy of Nhp6 and/or Hmo1, showing no reduction in SWI/SNF occupancy in any deletion mutant, defined as a reduction ≤0.263 in log2 ratio (i.e., a SWI/SNF occupancy value at most 1.2 times higher in the wild-type strain than in the deletion mutant strains). We first determined whether changes in SWI/SNF enrichment correlate with alterations in mRNA levels of the selected genes. To do this, we performed RT-qPCR for all 40 selected genes in the wild-type and both deletion mutant strains. Significant differences in mRNA levels (reduction or increment) were observed in most of the genes affected by deletion of NHP6 and/or HMO1, relative to mRNA levels measured in the wild-type strain (Fig. 2A–C). In each set of genes the effect appears to be mostly linked to the HMG protein responsible for SWI/SNF occupancy. Thus, in group A (genes with SWI/SNF occupancy affected in the nhp6a/b deletion mutant) 90% of the genes show significant variations in mRNA levels in the nhp6a/b deletion mutant, while only 20% show significant variations in the hmo1 deletion mutant (Fig. 2A). On the other hand, in group B (genes affected in the hmo1 deletion mutant) all 10 genes show significant variations in mRNA levels in the hmo1 mutant, while 5 genes display significant changes in the nhp6a/b deletion mutant (Fig. 2B). In most of these 5 genes deletion of HMO1 derives in a larger variation in mRNA levels than that observed upon deletion of NHP6A/B. Consistently, in the group of genes affected in both deletion mutants (group C), a significant change in mRNA levels for every gene is observed in the absence of Nhp6 and also in the absence of Hmo1 (Fig. 2C). Importantly, changes in mRNA levels occur in 29 out of the 30 selected genes where SWI/SNF occupancy is affected by the absence of Nhp6 and/or Hmo1 proteins. Conversely, significant variations in mRNA levels occur in only 3 out of the 10 genes that constitute the group of genes where SWI/SNF binding is not affected (group D, Fig. 2D), despite that these genes possess high occupancy levels of Nhp6 and/or Hmo1 proteins, as determined by the ChIP-chip analyses performed in the wild-type (TAP-tag) strains. Taken together, these results suggest a general trend where the absence of Nhp6 and Hmo1 affects transcriptional activity of genes where their presence favors SWI/SNF binding.
Table 1.
Genes selected for validation analyses and further studies.
| Reduced SWI/SNF occupancy in nhp6ab (group A) |
Reduced SWI/SNF occupancy in hmo1 (group B) |
Reduced SWI/SNF occupancy in nhp6ab and hmo1 (group C) |
No reduction of SWI/SNF occupancy in deletion mutants (group D) |
|---|---|---|---|
| RPS22A1, 2 | MRP211, 2 | NAT41, 2 | SAG11, 2 |
| DSE11, 2 | AAH11, 2 | LDH11, 2 | ASH11, 2 |
| NPR31 | NUP421 | VMA111 | GDB11 |
| PDR121 | MED81 | GEX21 | EFM11 |
| GCD6 | ARG5,6 | ZEO1 | PIG1 |
| PSR1 | RNR4 | FAA3 | ARB1 |
| PAP2 | PHD1 | SKI2 | YCR085W |
| COQ9 | EFT1 | PHO3 | ZRT1 |
| DSE2 | ASP1 | VPS38 | BNA1 |
| GRE3 | MVD1 | YBL010C | HHY1 |
RT-qPCR analyses were performed for all genes listed in the table. Colour key: yellow = high Snf5 and Nhp6 occupancy; light-blue = high Snf5 and Hmo1 occupancy; light-green = high Snf5, Nhp6 and Hmo1 occupancy.
Genes selected for ChIP-qPCR analyses.
Genes selected for analyses of nucleosome landscape at their promoter regions.
Fig. 2.
The absence of Nhp6 and/or Hmo1 affects transcriptional activity of genes where these proteins are required for SWI/SNF binding to their promoters. RT-qPCR analyses were performed to determine changes in mRNA levels generated by deletion of NHP6A/B (yellow bars) or HMO1 (red bars) for all genes listed in Table 1. The groups of genes depicted in figures A, B, C, and D are in correspondence with the groups of genes listed in Table 1. All measures of mRNA levels were normalized using ACT1 as reference gene. In the graphs, these normalized mRNA levels of each gene in each deletion mutant strain are given relative to the normalized mRNA levels of the corresponding gene determined in the wild-type strain. Thus, the horizontal black line is representative of mRNA levels in the wild-type strain. Data in each graph correspond to an assay representative of two independent assays, each performed in triplicate. Error bars represent one standard deviation. Asterisks denote a statistically significant difference (*p < 0.05; **p < 0.01; ***p < 0.001), as deducted from the t-test. Supplementary Fig. S7 is an expanded version of this figure. A) Analysis for genes undergoing reduction of SWI/SNF occupancy at their promoter/TSS upon deletion of NHP6A/B. B) Analysis for genes undergoing reduction of SWI/SNF occupancy at their promoter/TSS upon deletion of HMO1. C) Analysis for genes undergoing reduction of SWI/SNF occupancy at their promoter/TSS in both deletion mutants. D) Analysis for genes where deletion of neither NHP6A/B nor HMO1 affects SWI/SNF binding to the promoter/TSS.
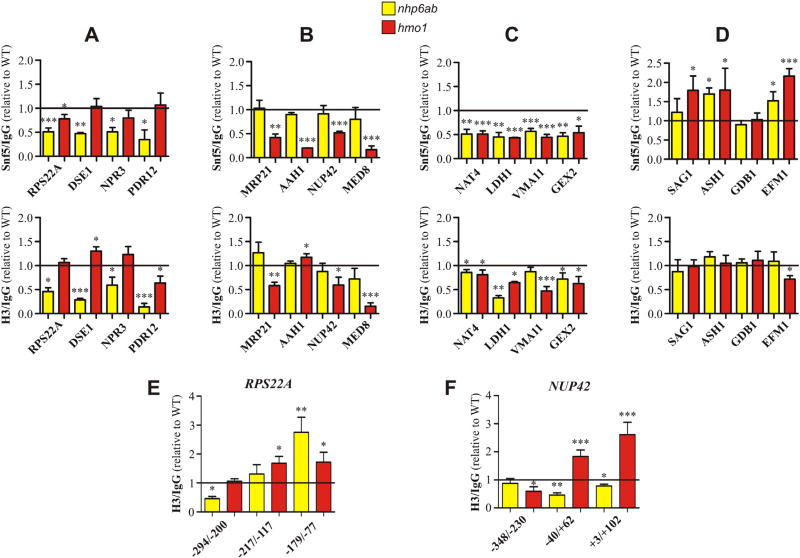
We next selected 16 genes (4 genes from each group) to validate our ChIP-chip studies by performing ChIP-qPCR. PCR reactions for these analyses were directed to the same promoter regions where reduction in SWI/SNF occupancy was detected in the genome-wide studies. Immunoprecipitation reactions were carried out for Snf5 and also for histone H3, in the latter case with the aim of additionally exploring changes in nucleosome occupancy at these regions. With no exceptions, the analysis of SWI/SNF occupancy (ChIP directed against Snf5) confirmed the results obtained in the ChIP-chip studies for all these 16 genes (Fig. 3A–D, top panel). Reduction of SWI/SNF occupancy in the nhp6a/b deletion mutant occurs only in the group of genes defined by the genome-wide analyses as affected in this deletion mutant (group A) and in the group of genes determined as affected in both deletion mutants (group C). On the other hand, SWI/SNF occupancy is reduced by the absence of Hmo1 only in the group of genes determined as affected in the hmo1 deletion mutant (group B) and in the group of genes defined as affected in both deletion mutants (group C). Consistently, this approach confirmed no reduction in SWI/SNF enrichment within the group of non-affected genes (group D). In the case of RPS22A, a reduction of SWI/SNF occupancy was also observed upon deletion of HMO1, although at a lower extent than that observed upon deletion of NHP6A/B (Fig. 3A, top panel). Interestingly, ChIP-qPCR analyses performed for histone H3 exhibited changes in its occupancy at those gene promoters where deletion of NHP6A/B and/or HMO1 results in reduction of SWI/SNF occupancy. Histone H3 occupancy is not altered in any gene within the group of genes defined as non-affected in deletion mutants, with the exception of a minor reduction observed for EFM1 in the hmo1 deletion mutant (group D, Fig. 3D, bottom panel). On the other hand, H3 enrichment is altered in all genes belonging to the groups defined as affected in the nhp6a/b and/or hmo1 deletion mutants, with the exception of VMA11, for which no change of H3 occupancy occurs upon deletion of NHP6A/B (Figs. 3A–C, bottom panels). Moreover, within group A (genes affected by deletion of NHP6A/B), major changes in H3 occupancy occur in the nhp6a/b deletion mutant but not in the hmo1 deletion mutant (Fig. 3A, bottom panel). Conversely, within group B (genes affected in the hmo1 deletion mutant), changes in H3 enrichment occur only by deletion of HMO1 (Fig. 3B, bottom panel).
Fig. 3.
Histone H3 occupancy is affected at gene promoters undergoing reduction of SWI/SNF occupancy upon deletion of NHP6A/B and/or deletion of HMO1. ChIP-qPCR analyses were performed to determine changes in Snf5 and H3 occupancy generated by deletion of NHP6A/B (yellow bars) or HMO1 (red bars) for selected genes listed in Table 1. The groups of genes depicted in figures A, B, C, and D are in correspondence with the groups of genes listed in Table 1. Values obtained from the qPCR reactions for all genes in wild-type and deletion mutant strains were expressed as “times over IgG” and the resulting values obtained for the deletion mutants are expressed in the graphs relative to the corresponding values obtained for the wild-type strain. Thus, the horizontal black line is representative of occupancy levels in the wild-type strain. Data in each graph correspond to an assay representative of two independent assays, each performed in triplicate. Error bars represent one standard deviation. Asterisks denote a statistically significant difference (*p < 0.05; **p < 0.01; ***p < 0.001), as deducted from the t-test. Top (figures A–D): ChIP-qPCR analysis for Snf5. Bottom (figures A–D): ChIP-qPCR analysis for histone H3. Positions given here and in the figures are relative to the translation start site. Supplementary Fig. S8 is an expanded version of figures A–D. A) Analysis for genes undergoing reduction of SWI/SNF occupancy at their promoter/TSS upon deletion of NHP6A/B, as determined by the ChIP-chip assays. Regions spanned by PCR reactions: RPS22A, −294/−200; DSE1, −97/+4; NPR3, −222/−97; PDR12, −356/−227. B) Analysis for genes undergoing reduction of SWI/SNF occupancy at their promoter/TSS upon deletion of HMO1, as determined by the ChIP-chip assays. Regions spanned by PCR reactions: MRP21, −115/−15; AAH1, −213/−119; NUP42, −348/−230; MED8, −175/−44. C) Analysis for genes undergoing reduction of SWI/SNF occupancy at their promoter/TSS in both deletion mutants, as determined by the ChIP-chip assays. Regions spanned by PCR reactions: NAT4, −162/−62; LDH1, −94/+8; VMA11, −210/−96; GEX2, −356/−223. D) Analysis for genes where deletion of neither NHP6A/B nor HMO1 affects SWI/SNF binding to the promoter/TSS, as determined by the ChIP-chip assays. Regions spanned by PCR reactions: SAG1, −101/+2; ASH1, −145/−43; GDB1, −201/−73; EFM1, −226/−115. E) ChIP-qPCR analysis of histone H3 occupancy at different stretches of the RPS22A promoter. F) ChIP-qPCR analysis of histone H3 occupancy at different stretches of the NUP42 promoter.
Although changes in H3 enrichment observed in our ChIP-qPCR analyses were mostly consistent with variations in mRNA levels determined by our RT-qPCR assays, we found unexpected results for a few number of genes. Considering that transcriptional repression is commonly linked to an increase in nucleosome occupancy at gene regulatory regions [35], the reduction in H3 occupancy observed for the RPS22A, NPR3 and NUP42 genes (Fig. 3A and B, bottom panels) was not consistent with the repressive effect observed for these genes by deletion of NHP6A/B or HMO1 (Fig. 2A and B). However, when testing other promoter regions of the RPS22A and NUP42 genes, an increase of H3 occupancy can be observed (Fig. 3E and F, respectively), now consistent with the variations in mRNA levels observed for these genes. This result pointed to the need of further analyses of changes in the nucleosome landscape at the promoters of affected genes, rather than limiting the analysis to nucleosome occupancy at fixed stretches of the promoters (see below).
3.4. Nhp6 and Hmo1 stimulate targeting of the SWI/SNF complex by Gal4-VP16
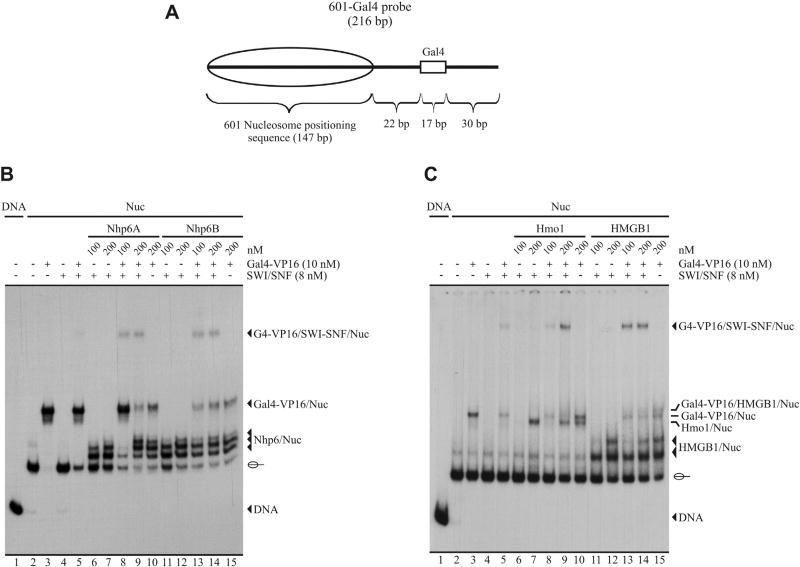
The results of our ChIP-chip analyses, validated by the ChIP-qPCR assays performed for Snf5, raised an issue in relation to the effect observed by deletion of NHP6A/B. We have previously shown that Hmo1, but not Nhp6, has the property of stimulating SWI/SNF binding to the nucleosome [19]. However, our ChIP-chip analyses show that in >60 genes SWI/SNF occupancy is significantly diminished at their promoter region in the absence of Nhp6A/B (see above). This result suggests that Nhp6 is required for SWI/SNF binding to defined gene promoters in vivo. Hence, in a cellular context, additional factors may play a role for the occurrence of this effect of Nhp6 on SWI/SNF binding to gene promoters. We reasoned that among these additional factors could be transcription factors able to recruit this complex to gene regulatory regions [1,36]. Considering this reasoning, we decided to test in vitro whether Nhp6 is able to stimulate recruitment of SWI/SNF to a mononucleosome driven by the chimeric transcription factor Gal4-VP16, using EMSA. In these assays we used as probe a mononucleosome reconstituted onto a radiolabeled DNA segment containing a single Gal4 binding site in the region of extranucleosomal DNA (Fig. 4A; [28]). Importantly, a large amount of unlabeled oligonucleosomes was added to the binding reactions in order to establish conditions where SWI/SNF is unable to interact with the nucleosome probe in the absence of Gal4-VP16. In addition, SWI/SNF and Gal4-VP16 concentrations were adjusted to obtain a weak recruitment signal, in order to be able to detect any potential stimulatory effect given by the HMG proteins. ATP was not included in the reactions, with the aim of detecting only binding patterns of the SWI/SNF complex (see Materials and methods for details). As observed in Fig. 4B, the Gal4-VP16-mediated recruitment of SWI/SNF is stimulated by both Nhp6A and Nhp6B, evidenced by a higher intensity of the slowest migrating band (Fig. 4B, compare lane 5 to lanes 8–9 and to lanes 13–14). This result suggests that the requirement of Nhp6 for SWI/SNF binding to defined gene promoters, observed in our in vivo analyses, would rely on the ability of this HMG protein to stimulate transcription factor-mediated recruitment of the complex. EMSA analyses performed for Hmo1 and the human HMGB1 proteins show that they also have the ability of stabilizing targeting of the SWI/SNF complex by Gal4-VP16 (Fig. 4C, compare lane 5 to lane 9 and to lanes 13–14).
Fig. 4.
Nhp6 and Hmo1 stimulate targeting of the SWI/SNF complex mediated by Gal4-VP16. EMSA analyses comparing the effect of different concentrations of each HMG protein on Gal4-VP16-mediated recruitment of SWI/SNF to a mononucleosome probe. A 216 bp DNA segment containing a single Gal4 binding site and the 601 nucleosome positioning sequence was reconstituted into a mononucleosome. The resulting probe contains the Gal4 binding site in the extranucleosomal DNA region, as depicted in figure A. After binding incubations, the samples were analyzed by electrophoresis in non-denaturing polyacrylamide gels (3.5%, AA:Bis 60:1). Migration of naked DNA, mononucleosome, and the different complexes is indicated at right of the pictures. Migration of the mononucleosome is indicated schematically. The components of each reaction are depicted on top of the pictures. Pictures in figures B and C are representative of three independent assays performed for each set of HMG proteins. These proteins may also be present in the Gal4-VP16/SWI-SNF/Nuc complex, which cannot be confirmed or discarded through this analysis. A) Schematic representation of the probe used in the assays. B) EMSA analysis for Nhp6A and Nhp6B proteins. C) EMSA analysis for Hmo1 and HMGB1 proteins.
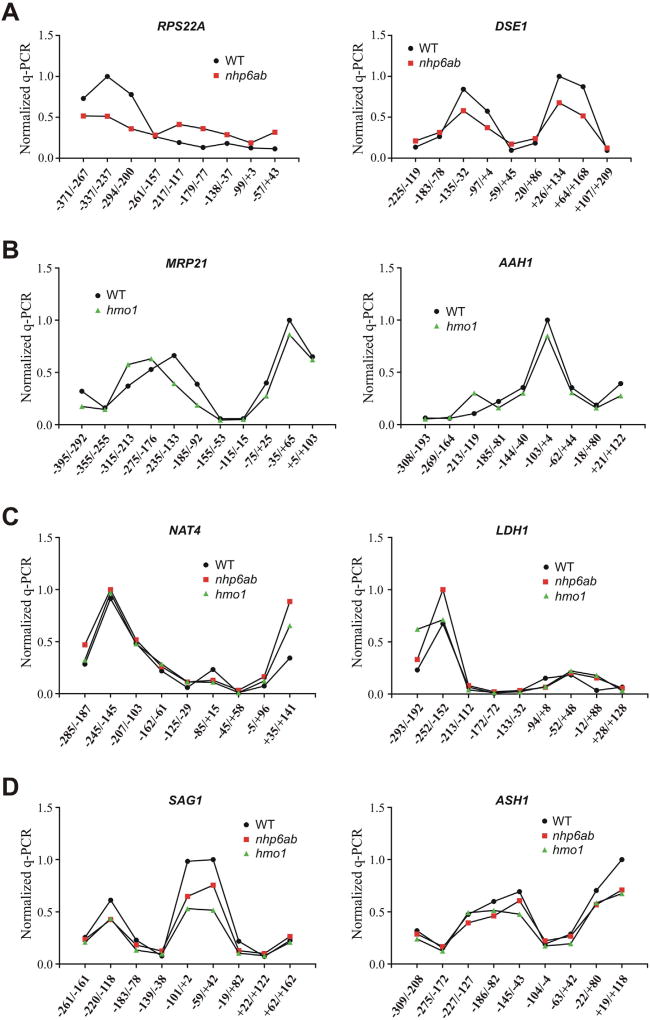
3.5. Chromatin landscape is altered at gene promoters undergoing reduction of SWI/SNF binding
To further study the impact of Nhp6 and Hmo1 on chromatin dynamics at gene promoters where, according to our analyses, their presence contributes to SWI/SNF binding, we decided to analyze whether deletion of NHP6A/B or HMO1 has an impact on the nucleosome landscape at the promoter region of genes selected from groups A to D (see Table 1). The need for this type of experimental approach was emphasized after obtaining variations in mRNA levels which, in some cases, were not consistent with variations in H3 occupancy, depending on the promoter stretches tested for genes such as RPS22A and NUP42 (see above). Nucleosome positions were analyzed using micrococcal nuclease digestion of chromatin in yeast spheroplasts followed by qPCR analysis of digestion patterns using overlapping primer pairs tiling 400 to 500 base pairs (bp) of selected gene promoters (see Materials and methods for details). Two genes from each group were tested using this approach. Among genes tested from group A (genes where SWI/SNF occupancy is reduced by deletion of NHP6A/B), a manifest change was observed for the RPS22A promoter. The change observed suggests the movement of a nucleosome from a region upstream the TSS of RPS22A to a position closer to the TSS (Fig. 5A, left panel). This result is consistent with the repressive effect observed for this gene upon deletion of NHP6A/B (Fig. 2A) and explains the opposite ChIP-qPCR patterns found in the analysis of H3 occupancy performed for this gene promoter, where increase or reduction of its occupancy was observed depending on the promoter region tested by qPCR (Fig. 3A and E). Within the same group A, a more subtle effect is observed in the case of the DSE1 promoter, pointing to a reduction in the positioning of the nucleosomes spanning this region. That is, deletion of NHP6A/B does not result in a marked change in nucleosome positioning or occupancy, although a reduction in nucleosome occupancy is observed in all positions displaying high occupancy in the wild-type and, conversely, an increase in nucleosome occupancy is observed in all the low occupancy positions present in the wild-type (Fig. 5A, right panel). In the case of gene promoters tested from group B (genes where SWI/SNF enrichment is reduced by deletion of HMO1) the absence of Hmo1 results in alterations of the nucleosome landscape in both gene promoters. At the MRP21 promoter, a nucleosome located around −185 (middle point of the −235/−133 primer pair, Fig. 5B, left panel) is shifted nearly 50 bp upstream upon HMO1 deletion. At the AAH1 promoter, the assay suggests the appearance of a nucleosome around −165 (middle point of the −213/−119 primer pair, Fig. 5B, right panel) in the absence of Hmo1. Similarly, both genes within group C (genes where SWI/SNF occupancy is reduced by deletion of NHP6A/B and by deletion of HMO1) show alterations in nucleosome landscape derived from the absence of Nhp6 and the absence of Hmo1 (Fig. 5C). For both genes, the changes in nucleosome landscape point to the establishment or expansion of a nucleosome-depleted region near the TSS [37], which is consistent with the increase in mRNA levels observed for these two genes in the deletion mutant strains (Fig. 2C). As expected, no effect on nucleosome positioning at the promoter region of the genes within group D (gene promoters where SWI/SNF occupancy is not affected in deletion mutants) were observed upon deletion of NHP6A/B or HMO1 (Fig. 5D), which is consistent with the unaffected mRNA levels observed for these genes (Fig. 2D). Taken together, these analyses demonstrate that the absence of Nhp6 or Hmo1 has an impact on nucleosome landscape at the promoters where this absence negatively affects SWI/SNF binding.
Fig. 5.
Deletion of NHP6A/B or HMO1 impacts nucleosome landscape at gene promoters where SWI/SNF occupancy is also impacted. Analysis of nucleosome positioning on the promoter region of selected genes, performed for wild-type and deletion mutant strains. Chromatin in yeast spheroplasts was subjected to MNase digestion and purified nucleosome-length DNA was used for tiling qPCR covering ~500 bp of each gene promoter analyzed. The heterochromatic REC104 locus was used as a normalization control in each assay and values given in each graph are relative to the highest value obtained in the corresponding assay. Data in each graph correspond to an assay representative of two independent assays, each performed in triplicate. The positions covered by each primer pair are given in the figures relative to the translation start site. A) Analysis for genes undergoing reduction of SWI/SNF occupancy at their promoter/TSS upon deletion of NHP6A/B. B) Analysis for genes undergoing reduction of SWI/SNF occupancy at their promoter/TSS upon deletion of HMO1. C) Analysis for genes undergoing reduction of SWI/SNF occupancy at their promoter/TSS in both deletion mutants. D) Analysis for genes where deletion of neither NHP6A/B nor HMO1 affects SWI/SNF binding to the promoter/TSS.
4. Discussion
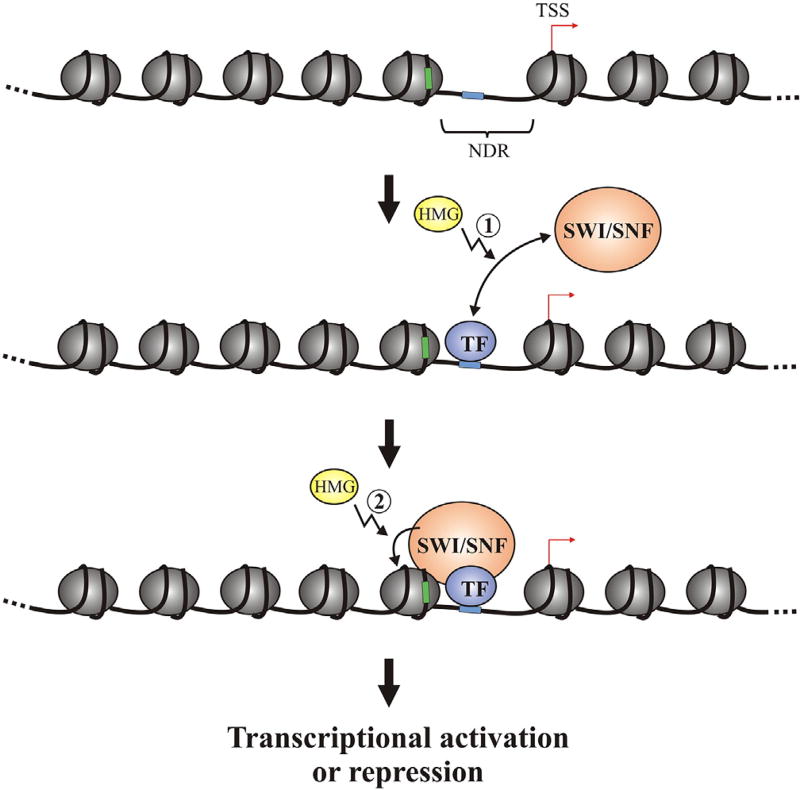
In this work we have demonstrated that the yeast HMG proteins Nhp6 and Hmo1 contribute to SWI/SNF enrichment at numerous gene promoters in vivo. Moreover, we have shown that, in genes where this binding depends on Nhp6 and/or Hmo1, their absence also affects transcriptional activity and nucleosome landscape at the promoter region. In addition to allowing determination of gene promoters where NHP6A/B and/or HMO1 deletion affects SWI/SNF occupancy, our ChIP-chip analyses unveiled that most SWI/SNF target genes are also targets of these HMG proteins, and that the enrichment profile of the complex at its target genes is highly correlated to the enrichment profile of these proteins. The results of the in vivo and in vitro assays performed in this work, together with our previously published studies [19], strongly support a model consisting in involvement of these HMG proteins in the recruitment as well as in stimulating the ATP-dependent remodeling activity of the SWI/SNF complex (Fig. 6).
Fig. 6.
Role of the HMG proteins Nhp6 and Hmo1 in SWI/SNF-mediated chromatin dynamics at gene regulatory regions. The model highlights the processes where the HMG proteins Nhp6 and Hmo1 could play a stimulatory role, which are: 1) Stimulation of SWI/SNF recruitment to gene promoters mediated by specific transcription factors (TF). Hmo1 stimulates direct binding of SWI/SNF to nucleosomes [19], implicating that this protein could also assist SWI/SNF loading onto gene regulatory regions mediated by other promoter features, such as histone modifications. 2) Stimulation of the ATP-dependent chromatin remodeling activity of SWI/SNF. TSS = transcription start site. NDR = nucleosome-depleted region. The blue box represents a specific binding site for a TF able to recruit SWI/SNF to gene regulatory regions. The green box represents a cis-regulatory element embedded in a nucleosome, which may become accessible upon SWI/SNF action.
In the context of our results involving Nhp6, a study performed by Celona and colleagues [38] has shown that S. cerevisiae cells lacking Nhp6A/B contain a lower number of histones, leading to a global reduction in nucleosome occupancy of about 30%, but not to a pronounced change in nucleosome positioning at the genome-wide level. Regarding the latter observation, the authors found, however, that around 30% of the nucleosomes shift their positions in >20 bp. Taking into account this observation and focusing on gene regulatory regions where SWI/SNF binds, our results suggest that the absence of Nhp6 proteins affects nucleosome positioning at those gene promoters where their presence is required for SWI/SNF binding, having no major impact on this feature at gene promoters where SWI/SNF remains bound despite the absence of these HMG proteins. As demonstrated in our RT-qPCR analyses, this observation also extends to transcriptional activity of genes showing or not showing reduction in SWI/SNF occupancy by deletion of NHP6A/B, where an impact was observed essentially in genes undergoing reduction in SWI/SNF occupancy. However, changes in mRNA levels were also observed for a small fraction of genes in which deletion of NHP6A/B does not result in reduction in SWI/SNF binding. In these genes, other effects derived from the absence of Nhp6 would be acting, with reduction in nucleosome occupancy being one of the possibilities.
Roles as transcriptional activators as well as repressors have been found for both Hmo1 and Nhp6 proteins. Hall and colleagues determined that deletion of the HMO1 gene results in up-regulation of 261 genes and down-regulation of 570 [9]. Similarly, Celona and colleagues observed that deletion of the NHP6 genes results in the up-regulation of 219 genes and down-regulation of 251 [38]. Consistently, we observed genes with increased mRNA levels and genes with reduced mRNA levels upon deletion of HMO1 or NHP6A/B. More importantly, changes in mRNA levels were essentially found in genes where deletion of HMO1 or NHP6A/B results in a reduction of SWI/SNF occupancy at their promoter region.
Our in vitro analyses suggest that the requirement of Nhp6 and/or Hmo1 for in vivo binding of SWI/SNF to defined gene promoters could rely on stimulation of SWI/SNF recruitment by transcription factors that physically interact with this complex. In the case of Hmo1, we have additionally shown that this protein is able to directly stimulate binding of SWI/SNF to nucleosomes [19], suggesting the existence of more means by which this HMG protein could stimulate SWI/SNF loading onto gene promoters. According to our results, this stimulation of SWI/SNF recruitment would not necessarily rely on enhancement of transcription factor binding to its cognate sequence. In this context, it would be interesting to address whether the large number of genes presenting high occupancy of Nhp6 and/or Hmo1 at their promoter region, but not SWI/SNF, corresponds to genes lacking binding sites for transcription factors displaying physical interaction with the complex.
Our ChIP-chip analyses allowed us to determine that most of the genes displaying high SWI/SNF occupancy at their promoter region also present high occupancy of both Nhp6 and Hmo1. Within this group of genes about 13% display a significant reduction of SWI/SNF enrichment in the absence of these HMG proteins. The percentage of gene promoters undergoing reduction in SWI/SNF occupancy is higher in those groups presenting high enrichment of this complex and either Nhp6 or Hmo1. Although the total number of genes comprising these groups is considerably lower, the relatively low percentage of affected genes in the group of genes displaying high occupancy of SWI/SNF plus both Nhp6 andHmo1 suggests that, within this group, the presence of one of these HMG proteins might compensate for the absence of the other. In this context, previous studies have proposed that Hmo1 and Nhp6 do not have overlapping functions [39]. Our results suggest that Nhp6 and Hmo1 might play redundant roles in assisting SWI/SNF binding, especially in genes displaying high occupancy of all these players and where occupancy of the complex is unaffected by deletion of NHP6A/B or HMO1. It has to be pointed out, however, that there are several gene promoters where only one of these HMG proteins is required for SWI/SNF binding, even though in many of these promoters both HMG proteins are present. Also in the context of our genome-wide analyses, we found gene promoters displaying high occupancy of SWI/SNF plus either Nhp6 or Hmo1, in which binding of the complex is not reduced by deleting the corresponding HMG gene. Such a phenomenon has been previously reported for the GAL1 promoter, where SWI/SNF binding is independent of the presence of Nhp6 [40]. The occurrence of genes presenting high occupancy of SWI/SNF and either Nhp6 or Hmo1 at their promoters, in which SWI/SNF binding is not affected by the absence of the corresponding HMG protein, clearly indicates the existence of other factors relevant for binding of the complex to these promoters. In this regard, it is likely that transcription factors are among these other factors. The ability of SWI/SNF recruitment to gene promoters has been described for several transcription factors [35,36,41]. We speculate that these HMG proteins would be more necessary for SWI/SNF binding at gene promoters containing weak binding sites for SWI/SNF-interacting transcription factors or at gene promoters regulated by transcription factors whose protein-protein interactions with SWI/SNF might be enhanced by Nhp6 or Hmo1 proteins.
In our analysis of the effect on nucleosome landscape derived from deletion of NHP6A/B or HMO1, shifts in the preferential position of particular nucleosomes were found for the MRP21 and LDH1 genes. Changes in positioning roughly ranged from 30 to 50 bp. In agreement with these observations, Reja and colleagues found an average shift of ~20 bp in the position of the +1 nucleosome in the Hmo1-enriched ribosomal protein genes, upon deletion of HMO1 [14].
In our present study, we focused our analyses in gene promoters. However, data mining of our ChIP-chip assays also uncovered a strong correlation between the enrichment profiles of SWI/SNF and both Nhp6 and Hmo1 over the gene body, in those genes displaying high occupancy of the complex at this region. Similar to what we found at gene promoters, most of the genes presenting high SWI/SNF occupancy over their gene body (845 genes) also display high occupancy of both Nhp6 and Hmo1 at this region (713 genes). Deletion of NHP6A/B or HMO1 results in a less marked effect on SWI/SNF occupancy in this group of genes, as compared to the effect observed in the group of genes presenting high occupancy of the complex at the promoter region. Nevertheless, several of these genes undergo a significant reduction in SWI/SNF occupancy upon deletion of NHP6A/B or HMO1, mostly by deletion of HMO1 (Supplementary Fig. S4). The fact that a role in transcription elongation has also been ascribed to SWI/SNF [33,34] makes it interesting to perform further studies focusing on the combined role that SWI/SNF and these HMG proteins might play at this gene region.
Supplementary Material
Acknowledgments
We thank Dr. David Stillman (University of Utah Health Sciences Center) for providing yeast strains.
Funding
This work was supported by grants CONICYT, FONDECYT/Regular 1130818 to JLG and 1140394 to MT, and private funding from the Stowers Institute for Medical Research to JLW. MIH was supported by a CONICYT scholarship for Ph.D. students and CONICYT scholarships AT-24100076 and P-75110059.
Abbreviations
- SWI/SNF
switching defective/sucrose non-fermenting
- HMG
high mobility group
- Nhp6A/B
non-histone protein 6A/B
- Hmo1
high mobility group family 1
- MNase
micrococcal nuclease
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbagrm.2017.01.002.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
MIH, MS, CG, NV, AA, AM, and MMG performed laboratory studies. MIH, MS, RA, and MMG performed bioinformatic analyses. MIH, MS, MT, JLW, and JLG designed the experiments and wrote the manuscript.
Transparency document
The Transparency document associated with this article can be found, in online version.
References
- 1.Becker PB, Workman JL. Nucleosome remodeling and epigenetics. Cold Spring Harb. Perspect. Biol. 2013;5:a017905. doi: 10.1101/cshperspect.a017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Narlikar GJ, Sundaramoorthy R, Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154:490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reeves R. Nuclear functions of the HMG proteins. Biochim. Biophys. Acta. 2010;1799:3–14. doi: 10.1016/j.bbagrm.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr. Opin. Genet. Dev. 2005;15:496–506. doi: 10.1016/j.gde.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Hock R, Furusawa T, Ueda T, Bustin M. HMG chromosomal proteins in development and disease. Trends Cell Biol. 2007;17:72–79. doi: 10.1016/j.tcb.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stillman DJ. Nhp6: a small but powerful effector of chromatin structure in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2010;1799:175–180. doi: 10.1016/j.bbagrm.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biswas D, Imbalzano AN, Eriksson P, Yu Y, Stillman DJ. Role for Nhp6, Gcn5, and the Swi/Snf complex in stimulating formation of the TATA-binding protein-TFIIA-DNA complex. Mol. Cell. Biol. 2004;24:8312–8321. doi: 10.1128/MCB.24.18.8312-8321.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dowell NL, Sperling AS, Mason MJ, Johnson RC. Chromatin-dependent binding of the S. cerevisiae HMGB protein Nhp6A affects nucleosome dynamics and transcription. Genes Dev. 2010;24:2031–2042. doi: 10.1101/gad.1948910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall DB, Wade JT, Struhl K. An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26:3672–3679. doi: 10.1128/MCB.26.9.3672-3679.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasahara K, Ohyama Y, Kokubo T. Hmo1 directs pre-initiation complex assembly to an appropriate site on its target gene promoters by masking a nucleosome-free region. Nucleic Acids Res. 2011;39:4136–4150. doi: 10.1093/nar/gkq1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kassavetis GA, Steiner DF. Nhp6 is a transcriptional initiation fidelity factor for RNA polymerase III transcription in vitro and in vivo. J. Biol. Chem. 2006;281:7445–7451. doi: 10.1074/jbc.M512810200. [DOI] [PubMed] [Google Scholar]
- 12.Moreira JM, Holmberg S. Chromatin-mediated transcriptional regulation by the yeast architectural factors NHP6A and NHP6B. EMBO J. 2000;19:6804–6813. doi: 10.1093/emboj/19.24.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paull TT, Carey M, Johnson RC. Yeast HMG proteins NHP6A/B potentiate promoter-specific transcriptional activation in vivo and assembly of preinitiation complexes in vitro. Genes Dev. 1996;10:2769–2781. doi: 10.1101/gad.10.21.2769. [DOI] [PubMed] [Google Scholar]
- 14.Reja R, Vinayachandran V, Ghosh S, Pugh BF. Molecular mechanisms of ribosomal protein gene coregulation. Genes Dev. 2015;29:1942–1954. doi: 10.1101/gad.268896.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonaldi T, Längst G, Strohner R, Becker PB, Bianchi ME. The DNA chaperone HMGB1 facilitates ACF/CHRAC-dependent nucleosome sliding. EMBO J. 2002;21:6865–6873. doi: 10.1093/emboj/cdf692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patenge N, Elkin SK, Oettinger MA. ATP-dependent remodeling by SWI/SNF and ISWI proteins stimulates V(D)J cleavage of 5 S arrays. J. Biol. Chem. 2004;279:35360–35367. doi: 10.1074/jbc.M405790200. [DOI] [PubMed] [Google Scholar]
- 17.Ugrinova I, Pashev IG, Pasheva EA. Nucleosome binding properties and Co-remodeling activities of native and in vivo acetylated HMGB-1 and HMGB-2 proteins. Biochemistry. 2009;48:6502–6507. doi: 10.1021/bi9004304. [DOI] [PubMed] [Google Scholar]
- 18.Szerlong H, Saha A, Cairns BR. The nuclear actin-related proteins Arp7 and Arp9: a dimeric module that cooperates with architectural proteins for chromatin remodeling. EMBO J. 2003;22:3175–3187. doi: 10.1093/emboj/cdg296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hepp MI, Alarcon V, Dutta A, Workman JL, Gutiérrez JL. Nucleosome remodeling by the SWI/SNF complex is enhanced by yeast High Mobility Group Box (HMGB) proteins. Biochim. Biophys. Acta. 2014;1839:764–772. doi: 10.1016/j.bbagrm.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, et al. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol. Cell. 2011;41:480–492. doi: 10.1016/j.molcel.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B, Reese JC. Ssn6-Tup1 regulates RNR3 by positioning nucleosomes and affecting the chromatin structure at the upstream repression sequence. J. Biol. Chem. 2001;276:33788–33797. doi: 10.1074/jbc.M104220200. [DOI] [PubMed] [Google Scholar]
- 22.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 23.Venkatesh S, Smolle M, Li H, Gogol MM, Saint M, Kumar S, Natarajan K, Workman JL. Set2 methylation of histone H3 lysine36 suppresses histone exchange on transcribed genes. Nature. 2012;489:452–455. doi: 10.1038/nature11326. [DOI] [PubMed] [Google Scholar]
- 24.van Bakel H, van Werven FJ, Radonjic M, Brok MO, van Leenen D, Holstege FCP, Timmers HTM. Improved genome-wide localization by ChIP-chip using double-round T7 RNA polymerase-based amplification. Nucleic Acids Res. 2008;36:e21. doi: 10.1093/nar/gkm1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smolle M, Venkatesh S, Gogol MM, Li H, Zhang Y, Florens L, Washburn MP, Workman JL. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat. Struct. Mol. Biol. 2012;19:884–892. doi: 10.1038/nsmb.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev. 2007;21:1422–1430. doi: 10.1101/gad.1539307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Utley RT, Ikeda K, Grant PA, Côté J, Steger DJ, Eberharter A, John S, Workman JL. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 28.Gutiérrez JL, Chandy M, Carrozza MJ, Workman JL. Activation domains drive nucleosome eviction by SWI/SNF. EMBO J. 2007;26:730–740. doi: 10.1038/sj.emboj.7601524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kent NA, Mellor J. Chromatin structure snap-shots: rapid nuclease digestion of chromatin in yeast. Nucleic Acids Res. 1995;23:3786–3787. doi: 10.1093/nar/23.18.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam FH, Steger DJ, O'Shea EK. Chromatin decouples promoter threshold from dynamic range. Nature. 2008;453:246–250. doi: 10.1038/nature06867.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martens JA, Winston F. Evidence that Swi/Snf directly represses transcription in S. cerevisiae. Genes Dev. 2002;16:2231–2236. doi: 10.1101/gad.1009902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 33.Yen K, Vinayachandran V, Batta K, Koerber RT, Pugh BF. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell. 2012;149:1461–1473. doi: 10.1016/j.cell.2012.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwabish MA, Struhl K. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol. Cell. Biol. 2007;27:6987–6995. doi: 10.1128/MCB.00717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 36.Yudkovsky N, Logie C, Hahn S, Peterson CL. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 1999;13:2369–2374. doi: 10.1101/gad.13.18.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Celona B, Weiner A, Di Felice F, Mancuso FM, Cesarini E, Rossi RL, Gregory L, Baban D, Rossetti G, et al. Substantial histone reduction modulates genomewide nucleosomal occupancy and global transcriptional output. PLoS Biol. 2011;9:e1001086. doi: 10.1371/journal.pbio.1001086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu J, Kobayashi R, Brill SJ. Characterization of a high mobility group 1/2 homolog in yeast. J. Biol. Chem. 1996;271:33678–33685. doi: 10.1074/jbc.271.52.33678. [DOI] [PubMed] [Google Scholar]
- 40.Lemieux K, Larochelle M, Gaudreau L. Variant histone H2A.Z, but not the HMG proteins Nhp6a/b, is essential for the recruitment of Swi/Snf, Mediator, and SAGA to the yeast GAL1 UAS(G) Biochem. Biophys. Res. Commun. 2008;369:1103–1107. doi: 10.1016/j.bbrc.2008.02.144. [DOI] [PubMed] [Google Scholar]
- 41.Boyer LA, Logie C, Bonte E, Becker PB, Wade PA, Wolffe AP, Wu C, Imbalzano AN, Peterson CL. Functional delineation of three groups of the ATP-dependent family of chromatin remodeling enzymes. J. Biol. Chem. 2000;275:18864–18870. doi: 10.1074/jbc.M002810200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.