Abstract
Trichoderma reesei (syn. Hypocrea jecorina) is the model organism for industrial production of plant cell wall degradating enzymes. The integration of light and nutrient signals for adaptation of enzyme production in T. reesei emerged as an important regulatory mechanism to be tackled for strain improvement. Gene regulation specific for cellulase inducing conditions is different in light and darkness with substantial regulation by photoreceptors. Genes regulated by light are clustered in the genome, with several of the clusters overlapping with CAZyme clusters. Major cellulase transcription factor genes and at least 75% of glycoside hydrolase encoding genes show the potential of light dependent regulation. Accordingly, light dependent protein complex formation occurs within the promoters of cellulases and their regulators. Additionally growth on diverse carbon sources is different between light and darkness and dependent on the presence of photoreceptors in several cases. Thereby, also light intensity plays a regulatory role, with cellulase levels dropping at higher light intensities dependent in the strain background. The heterotrimeric G-protein pathway is the most important nutrient signaling pathway in the connection with light response and triggers posttranscriptional regulation of cellulase expression. All G-protein alpha subunits impact cellulase regulation in a light dependent manner. The downstream cAMP pathway is involved in light dependent regulation as well. Connections between the regulatory pathways are mainly established via the photoreceptor ENV1. The effect of photoreceptors on plant cell wall degradation also occurs in the model filamentous fungus Neurospora crassa. In the currently proposed model, T. reesei senses the presence of plant biomass in its environment by detection of building blocks of cellulose and hemicellulose. Interpretation of the respective signals is subsequently adjusted to the requirements in light and darkness (or on the surface versus within the substrate) by an interconnection of nutrient signaling with light response. This review provides an overview on the importance of light, photoreceptors and related signaling pathways for formation of plant cell wall degrading enzymes in T. reesei. Additionally, the relevance of light dependent gene regulation for industrial fermentations with Trichoderma as well as strategies for exploitation of the observed effects are discussed.
Keywords: Trichoderma reesei, Hypocrea jecorina, CAZymes, Light response, Signal transduction, Surface sensing, Carbon source utilization, EMSA, Genomic clusters
Background
The continuing alteration between light and darkness on earth caused by its rotation resulted in an evolutionary adaptation of the majority of living beings to day and night. The physiological changes connected to day and night are triggered by the circadian clock, which governs preparation to the upcoming day or night. Exposure to light at unexpected times causes phase shifts in the clock controlled cycles and hence alters the connected gene regulation. Nevertheless, control by the circadian clock and response to changing light conditions are distinct processes [1]. This adaptation does not only concern obvious necessities such as the protection against harmful UV light during the day or dealing with higher temperatures and lower humidity levels connected to sunlight versus darkness. Also, the adjustment of metabolic processes to light and darkness has evolved—not only in humans, but also in fungi [2, 3],—which raises the question whether fungi do have an equivalent to the physiological condition of sleep or the physiologically different situation in day and night in humans.
In fungi, light is highly relevant to diverse physiological regulation mechanisms and impacts many signaling pathways that integrate light response with metabolism, stress response and development [4]. Even though many of the fungi currently investigated in academia and industry never experience natural conditions of day and night, their gene expression levels and physiology still follow circadian rhythms [5], which also target metabolism [6–8]. Groundbreaking work in elucidation of the molecular machinery governing circadian rhythmicity and light response has been done in Neurospora crassa [9–11], which involves transcriptional cascades as well as epigenetic and posttranslational modifications [12, 13] and many of the studies on light effects in dthe fungi built on the discoveries from N. crassa thereafter.
Trichoderma reesei represents one of the most important filamentous fungi nowadays used in industry for production of homologous and heterologous enzymes—predominantly for biofuel production [14, 15]. T. reesei expresses diverse carbohydrate active enzymes (CAZymes), the most important being cellulases and hemicellulases [16]. Induction of these enzymes occurs on different carbon sources such as cellulose or lactose or in the presence of sophorose, but also on other carbon sources representing building blocks of plant cell wall material [17, 18]. Repression occurs on easily metabolizable carbon sources like glucose by the function of carbon catabolite repression. Secondary metabolism has not been studied in detail in T. reesei yet, but harmful mycotoxins are not known to be produced by this fungus [19]. T. reesei was the first industrially relevant filamentous fungus for which the method of sexual crossing became available. Sexual development is dependent on specific conditions of light, temperature and carbon source in the medium and numerous regulators, including the T. reesei photoreceptors and several signaling compounds [20, 21].
In this review, I will first give a short overview on composition of plant cell walls and their degradation followed by a general introduction to the light response pathway and known physiological effects of light on fungi in order to familiarize the reader with the two topics connected in this review. Thereafter I briefly describe the discovery of the influence of light on cellulase regulation along with some general findings on the topic later on. Subsequently, I explain the impact of light and the light response machinery on regulatory pathways influencing metabolic functions in Trichoderma with an emphasis on plant cell wall degradation and interconnections between nutrient and light signaling pathways (Fig. 1). Individual regulatory factors as well as their genome/transcriptome wide effects in dependence of light will be discussed including alterations on different carbon sources, non random distribution of light regulated genes and the interplay of light response with carbon catabolite repression. Based on that, light dependent differences at the promoter level and in carbon source utilization are outlined. Finally, the relevance of light for regulation of specific, known factors crucial for highly efficient plant cell wall degradation as well as aspects of light influenced processes for research and industry (Fig. 2) are discussed.
Fig. 1.
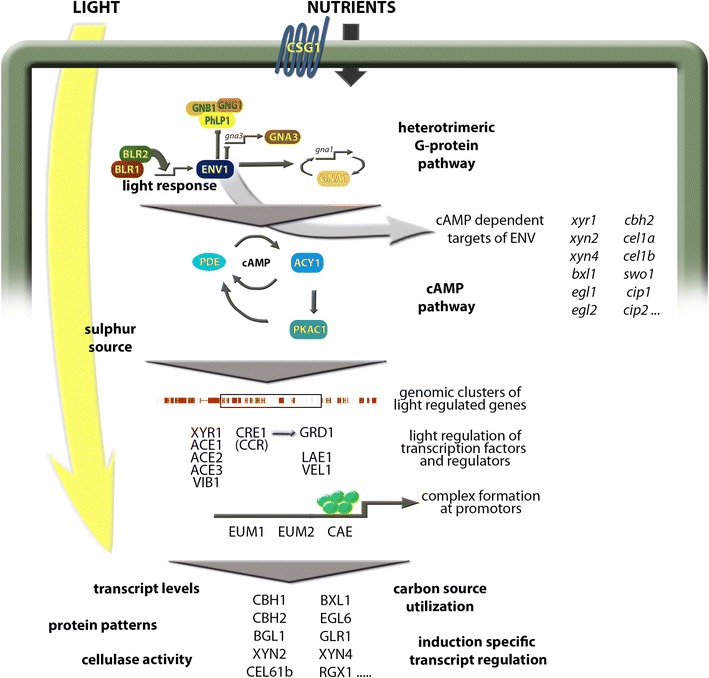
Schematic overview of pathways involved in light regulation of plant cell wall degradation
Fig. 2.
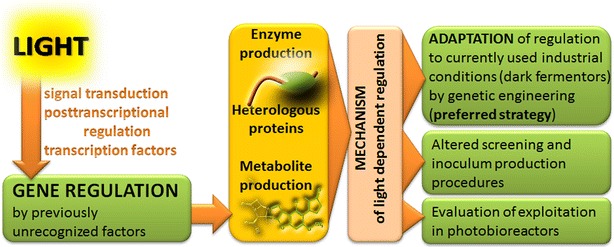
Schematic representation of strategies for strain improvement by exploiting of light dependent effects. Gene regulation in T. reesei is considerably influenced by light, with the light signal coordinated with nutrient signals via the signal transduction pathways of heterotrimeric G-proteins and cAMP signaling. Posttranscriptional regulation of cellulase gene expression is triggered by a G-protein coupled receptor, whose signal is channeled through the G-protein pathway and its subunits. The consequences of light exposure include changes in normal enzyme production, production of heterologous proteins and secondary metabolites expressed from homologous or heterologous gene clusters. Once the mechanisms of light dependent regulation are understood, this information can serve to improve performance under currently applied industrial conditions (dark fermentors) by knowledge based genetic engineering. Additionally, screening procedures are recommended to be performed under controlled light conditions and inoculum production can be improved. For high value products, illumination or specifically applied light regimes in photobioreactors can be evaluated
The plant cell wall and the enzymes degrading it
Lignocellulosic plant biomass represents the most important carbon source on our planet and it is hence of high ecological importance as major component in the global carbon cycle. Plant biomass is composed of several polymers including the recalcitrant cellulose, hemicellulose and lignin, but also pectin and starch [22]. Filamentous fungi are highly efficient degraders of plant biomass and have evolved a complex yet very efficient machinery for degradation of plant cell walls [23, 24]. The involved Carbohydrate Active enZymes (CAZys) act on glycoside linkages in the plant cell wall polysaccharides and can have a broad range of activities [25, 26]. It is assumed that glycoside hydrolases, the most important enzymes for plant cell wall degradation, have evolved from a common ancestor. In the course of the subsequent specialization, saprotropic fungi like T. reesei appear to have lost a significant number of genes including glycoside hydrolases [27]. Since cellulases are usually found coregulated, the cellobiohydrolases CBH1/Cel7a and CBH2/CEL6a are frequently used as representatives for cellulase expression. In recent years, the oxidation of cellulose by polysaccharide monooxygenases (previously assigned to glycoside hydrolase family 61) was shown to significantly contribute to plant cell wall degradation [28, 29]. Interestingly, an involvement of light and photosynthetic pigments in oxidation of polysaccharides was postulated [30].
Regulation of plant cell wall degrading enzymes is complex and occurs at the transcriptional level involving a plethora of transcription factors and regulators, some of the most important being XYR1, ACE1, ACE2, ACE3, VIB1 and the HAP complex [23, 31, 32]. Recently, also a post-transcriptional section of cellulase regulation was shown [33]. Thereby, a considerable number of substrates cause induction of expression of plant cell wall degrading enzymes [16–18]. Importantly, the mechanism of carbon catabolite repression, with its major transcription factor CRE1 serves to avoid biosynthesis of enzymes in the presence of easily metabolizable carbon sources [23, 34]. By application of their sophisticated regulatory machineries, fungi adjust the amount and type of enzymes to produce to their environment—be it a tropical forest or an industrial fermentor.
The light response pathway in Trichoderma spp.
Trichoderma spp. have a long tradition of research towards fungal light responses and their physiological consequences [35, 36]. Eight proteins are considered to be responsible for light perception [36] including BLR1 and BLR2 (blue light regulators 1 and 2): two GATA-type transcription factors predominantly acting as a photoreceptor complex, the photoreceptor ENV1, two photolyases, a cryptochrome, a phytochrome and an opsin, which is however only present in the genome of Trichoderma atroviride, but not in T. reesei [36, 37].
The major blue light photoreceptors BLR1 and BLR2, which are the homologues of N. crassa WC-1 and WC-2 (white collar 1 and 2) [38], were first characterized in T. atroviride, where they are essential for blue light induced conidiation [39]. Thereafter, ENV1, a homologue of N. crassa VVD [40] and the third photoreceptor was detected in T. reesei and an unexpectedly broad range of physiological functions in light response, development, stress response and metabolism was shown for ENV1 that exceed those of BLR1 or BLR2 [41, 42]. In contrast to T. atroviride, deletion of blr1 or blr2 does not abolish conidiation in T. reesei [43, 44], while lack of ENV1 causes a severe growth phenotype in light [42], which does not occur on every carbon source [45, 46]. In both fungi, BLR1 and BLR2 exert common as well as independent functions and also influence gene expression in darkness [46, 47]. Consequently, while there is considerable overlap in functions, there are also some differences in the light response machinery in Trichoderma spp. which may cause altered responses.
BLR1 and BLR2 are GATA type zinc finger transcription factors and are assumed to act in a complex. BLR1 contains three PAS domains, and a PAS/LOV domain, which is responsible for reception of the light signal due to the flavin moiety bound in this domain. BLR2 contains only one PAS domain and ENV1 contains a PAS/LOV domain like BLR1 [36]. Light dependent induction of env1 is strictly dependent on the presence of both BLR1 and BLR2 [43, 48, 49]. Nevertheless, phenotypic effects of deletion of blr1 or blr2 are much less severe than deletion of env1 in light [41–44, 46].
The hierarchy in the light response cascade thereby depends on the function of the respective regulators in T. reesei. In many cases, ENV1 acts via BLR1 and BLR2, for example with respect to growth and sexual fertility [48]. Like BLR1 and BLR2, also ENV1 has individual functions [46], albeit often effects reminiscent of photoadapation occur: If the negative effect of ENV1 is relieved by its deletion, the usually balancing positive effect of BLR1 and BLR2 becomes obvious [48]. This balanced function of ENV1, BLR1 and BLR2 further explains the rescue of the severe growth phenotype of strains lacking ENV1 in light in ∆env1 double or triple mutants with blr1 and/or blr2 deletions [48].
In T. atroviride, enhanced expression of blr2 causes increased photoconidiation and higher transcript levels of light induced genes, while the opposite effect is caused by overexpression of blr1. Blr2 overexpression further causes higher sensitivity to blue light and complex formation of BLR1/BLR2 is assumed to be required for appropriate light perception in T. atroviride [50].
Physiological responses to light in Trichoderma
Life in soil—a mainly dark environment—and on the surface of a substrate are fundamentally different in terms of UV radiation, oxygen, temperature stability, humidity and levels of reactive oxygen species (ROS). Also during day and night many of these environmental properties change and hence, fungi evolved to use light as a fast indicator of the expected changes and prepare for daily alterations by applying circadian rhythms [51].
Blue light responses in T. reesei include enhanced conidiation [43] and sexual reproduction [21, 52], secondary metabolism [46, 53, 54], growth [45] and altered regulation of enzyme gene transcription [46, 53], expression and activity [55]. Genes of the light response machinery including phr1 encoding a photolyase, the photoreceptor gene env1 and frq, which is required for circadian rhythmicity in N. crassa, the MAPkinase gene tmk3 [45] as well as several genes involved in sexual development are light dependently regulated irrespective of the carbon source [33].
The earliest response to light evaluated in T. reesei was after 15 min [45], albeit much shorter light pulses are assumed to be sensed and can cause altered gene regulation. Thereby it has to be considered that not only the duration but also the light intensity (fluence rate) is important [55]. An effect of red light on the regulation of few genes was detected for T. atroviride [56], while in T. reesei no effect of red light was observed so far [43].
In T. atroviride, light stimulates the tolerance to osmotic stress through the Hog1-related MAPkinase TMK3. This stimulation of stress signaling pathways by light is considered a benefit for the cell [57]. Interestingly, T. reesei TMK3 was found to regulate cellulase production [58, 59], albeit an influence of light was not tested in this study. Together these findings indicate that the MAPkinase pathway contribute to light dependent modulation of cellulase gene expression.
In T. reesei an involvement of ENV1 in stress response was observed, which requires the conserved amino acids C96 and T101 [60, 61]. Thereby, C96 is evolutionarily conserved in Hypocreaceae, which in contrast to for example N. crassa, indicating the integration of stress and light responses via ENV1.
An unexpected effect of light and photoreceptors on cellulase gene expression
Traditionally, filamentous fungi were grown under random light conditions, except for investigation of circadian rhythms or photoreceptor functions in N. crassa. Trichoderma spp. also served as models for analysis of light responses in the 1970s and 80s [36], but this did not lead to cultivation under controlled light conditions thereafter because no connection to carbon metabolism was assumed.
A screening assay for genes differentially expressed between the cellulase negative mutant strain QM9978 and the early high production mutant QM9414 surprisingly yielded the photoreceptor gene env1 as differentially regulated between the two strains under inducing conditions (cellulose and sophorose) [62]. ENV1 is a homologue of the N. crassa photoreceptor VIVID (VVD), which negatively acts on the photoreceptor complex established by WC-1 and WC-2, the homologues of BLR1 and BLR2 [63, 64]. However, T. reesei env1 could not complement a N. crassa vvd non-functional mutant and is hence not considered a functional homologue [42]. Still, subsequent research revealed a considerable functional overlap of ENV1 and VVD.
Consequently, cultivation of T. reesei was performed under controlled light conditions and with the cellulase inducing carbon sources cellulose and lactose in order to assess a potential effect of light on enzyme expression [42]. Indeed, striking differences in cellulase transcript levels between growth in light and in darkness were observed upon growth on cellulose, which did not correlate with previous results in a conventional incubator with random light pulses—neither in light nor in darkness. Transcript levels of cbh1 increased by roughly 50% upon growth in constant light on cellulose and an important function of ENV1 in this regulation became obvious, with decreased cbh1 levels on lactose, transient increase on cellulose in darkness and decrease in light in the mutant strain [42]. Upon induction of cellulase gene expression by the natural inducer sophorose, we found that in darkness, cellulase transcript levels increase with time, while in light cellulase induction appears to be accelerated, but transient [65].
Functions of the light response machinery in cellulase regulation
Searching for the basis of light dependent regulation of cellulase genes, comparison of the promoters of env1 and vvd revealed two potential DNA binding motifs, EUM1 (envoy upstream motif; 5′ CTGTGC 3′) and EUM2 (5′ ACCTTGAC 3′). EUM1 is also present in the promoters of the cellulase genes cbh1 and cbh2 as well as in the promoters of blr1 and blr2 suggesting a potential for co-regulation or feedback [42] (see also below). Additionally, the same study revealed that env1 is not expressed in the cellulase negative strain QM9978, which has a mutation in the EUM1 promoter motif in the env1 promoter. However, complementation with the entire env1 gene that included this promoter region did not rescue cellulase expression in QM9978 [42]. EUM1, light responsive elements (LREs) [66] and GATA sites, which all might be predictive for binding of the photoreceptor complex or other light regulatory factors, were found in numerous promoters of known cellulose and hemicellulose degrading enzymes [67].
Later on, investigation of the genome of QM9978 and comparison to that of QM6a recently led to the finding that the PhoG homologue VIB1 is responsible for the defect in cellulase induction of this strain. A translocation of the gene abolished expression of vib1 in QM9978 [68]. Interestingly, vib1 is strongly down-regulated in light upon growth on cellulose in the wild-type strain QM6a (Stappler and Schmoll, unpublished). Downregulation of vib1 in light is further observed upon growth on lactose or sophorose [33]. Transcriptome analysis upon growth on cellulose showed that vib1 is strongly regulated by ENV1 in light and by the adenylate cyclase ACY1 in light [33, 49]. Consequently, vib1 is a cAMP dependent target of the light response pathway [49], which is also in agreement with the finding of the gene encoding ENV1 as a regulator of vib1 to be down-regulated in QM9978 compared to QM9414 under inducing conditions [42] [62].
BLR1 and BLR2 positively regulate transcript levels of cbh1 in T. reesei [43] and in T. atroviride [69]. Interestingly, effects are obvious in light and darkness, indicating that these photoreceptors also have a function in darkness in T. reesei [43, 46]. Accordingly, effects of ENV1, BLR1 and BLR2 were also observed in largely dark fermentor cultivations in T. reesei. Under these conditions, ∆env1 secretes a more efficient enzyme mixture and for BLR2 an effect on secretion capacity was found, which leads to increased cellulase activities as well. ∆blr1 forms more biomass, but its lower secretion capacity counteracts its efficiency in production of cellulose degrading enzymes [67].
Abundance of secreted proteins is considerably altered in light [55] including such important enzymes as CBH1, BGL1, XYN2 or CEL61B. BLR1 and BLR2 have a major impact in this light dependent regulation. While initial analyses on light response of cellulase regulation were performed in the QM9414 strain background, we later found significant differences between light and darkness also for QM6a [55], RutC30 and industrial production strains as well (unpublished results). Thereby, QM6a shows a much lower light tolerance than QM9414 in terms of alteration of cellulase gene expression. In QM9414 cbh1 transcript levels initially increase with increasing light intensities and only drop at 5000 lx, while in QM6a already low light intensities abolish cbh1 transcription. In ∆blr1 and ∆blr2, this drop at high light intensities does not occur [55].
In addition to the photoreceptors, further regulators involved in light dependent signaling were identified as cellulase regulators. In Aspergillus nidulans, VeA and LaeA coordinate the light signal with fungal development and secondary metabolism [70]. Their homologues in T. reesei, VEL1 and LAE1 are important regulators of cellulase gene expression [71, 72]. Unfortunately, the latter studies were done under uncontrolled light conditions and hence a light dependent relevance of this regulation is not known. Since the function of VEL1 in development shows light dependent differences and a connection to photoreceptors [73, 74], this can also be expected for cellulase regulation.
Signaling pathways involved in light dependent modulation of cellulase gene expression
For the presence of cellulose to be sensed in the environment, low molecular weight degradation products liberated from degradable plant material are likely signals that may be sensed by membrane bound receptors. Alternatively, an inducer synthesized outside the cell may be taken up via diffusion or a permease and initiate cellulase formation by an intracellular process. Both hypotheses are likely to describe contributions to the regulation of cellulase gene expression.
The heterotrimeric G-protein pathway
The prime candidate pathway for sensing and transmission of an extracellular cellulose related signals is the heterotrimeric G-protein pathway. Signal transduction via the heterotrimeric G-protein pathway starts with reception of a signaling compound by a G-protein coupled receptor (GPCR). Once such a ligand binds to the GPCR, the heterotrimeric G-protein complex composed of G-protein alpha, beta and gamma subunits, dissociates, GDP is exchanged for GTP on the G-alpha subunit for activation and all subunits then modulate their target pathways [37, 75, 76].
T. reesei has 50 GPCRs, 3 G-alpha subunits (GNA1, GNA2 and GNA3), one G-beta subunit (GNB1) and one G-gamma subunit (GNG1). Additionally, two phosducin like proteins (PhLP1 and PhlP2), which are assumed to act as co-chaperones for G-protein beta and gamma folding are encoded in the genome as well as 7 RGS (regulator of G-protein signaling) domain proteins [37]. The G-protein alpha subunits GNA1 and GNA3 impact cellulase gene expression and this function is dependent on light [77, 78]. Constitutive activation of GNA3 led to a strong increase in cellulase transcripts upon growth on cellulose (around tenfold), but only in light. In darkness, neither constitutive activation nor knock down altered cellulase transcript levels [77]. Interestingly, upregulation of gna3 without constitutive activation caused somewhat increased cellulase levels in light, which however did by far not reach the strong up regulation seen for the constitutive activation of GNA3. It is therefore likely that the rate of inactivation of the alpha subunit by the intrinsic GTPase of GNA3 plays a role. This function is enhanced by RGS (regulator of G-protein signaling) proteins, which are consequently assumed to be involved in cellulase regulation in T. reesei [77]. Of those, rgs1 represents a light independent regulatory target of GNB1, GNG1 and PhLP1 [53].
Lack of GNA1 causes a strong increase of cellulase gene expression in darkness and abolishment in light upon growth on cellulose. Constitutive activation of GNA1 caused—like for GNA3—an increase in cellulase transcript levels in light [78]. For both GNA1 and GNA3, constitutive activation did not enable inducer independent cellulase gene expression. Consequently, the signal for induction of cellulase gene expression is separate from that transmitted by GNA1 and GNA3 and may indeed involve intracellular detection of an inducer.
The G-protein beta and gamma subunits GNB1 and GNG1 as well as the phosducin-like protein PhLP1, which is assumed to act as a chaperone for complex formation of GNB1 and GNG1, influence regulation of several glycoside hydrolases including cbh1 and cbh2 [53]. Cellulase activity increases in all three mutants, while transcript abundance of several genes encoding plant cell wall degrading enzymes decreases, indicating posttranscriptional regulation [53]. However, the light dependent effects of GNB1, GNG1 and PhLP1 are considerably less pronounced than the strongly positive impact of the G-alpha subunits GNA1 and GNA3 [53]. In summary, these findings are in line with a positive effect of phosducin-like proteins on the efficiency of G-protein signaling [79].
Only recently, a new aspect was added to the function of the heterotrimeric G-protein pathway in cellulase regulation. The class XIII of G-protein coupled receptors (GPCRs) was found to have only a minor influence on cellulase transcript levels (up to 50%), but both were required for normal specific cellulase activities on lactose. In the absence of CSG1, representing one of the two class XIII GPCRs of T. reesei, secreted activity levels even decreased to basal levels upon growth on cellulose and lactose [33]. Hence it is assumed that the signal received by CSG1 is responsible for posttranscriptional regulation of cellulase gene expression, a mechanism previously not known to be involved. Based on the light dependent function of GNA1 and GNA3 it can be assumed that these downstream subunits integrate the light dependent relevance with the signal received by CSG1 to a physiological response adapted to the change of day and night.
The cAMP pathway
Cyclic adenosine monophosphate (cAMP) is one of the most important secondary messengers in living organisms. cAMP levels function as coincidence detectors due to their regulation by diverse environmental cues that are integrated to a defined level, which determines the extent of the effect on downstream pathways. Adenylate cyclase synthesizes cAMP and phosphodiestereases, which are activated by the presence of cAMP [80, 81], degrade it [82], forming a feedback cycle for adjustment of levels [80, 81]. Protein kinase A is cAMP dependent and consists of catalytic and regulatory subunits [37]. Light dependent effects on cAMP levels and adenylate cyclase activity have been shown previously for Trichoderma [83–85].
The cAMP dependent protein kinase A plays an important role in regulation of light responses in T. atroviride [86]. In T. reesei, the cAMP pathway is one of the targets of the heterotrimeric G-protein pathway and cAMP levels are impacted by both GNA1 and GNA3 [77, 78]. Investigation of the function of adenylate cyclase (ACY1) and the catalytic subunit 1 of protein kinase A (PKAc1) showed a light dependent effect on cellulase gene expression in both cases upon growth on lactose [87]. Deletion of acy1 and pkac1 caused increased light responsiveness of cellulase genes i.e. strongly increased differences between cellulase levels in light and darkness compared to the light dependent regulation in the wild-type. Regulation of the transcript levels of the transcription factors ACE1 and ACE2 that regulate transcription of cellulase genes show a response to light, but do not correlate well with cellulase levels. Modification of transcript levels of the cellulase regulator XYR1 strongly resembles the pattern of regulation seen for cellulase genes. Hence, it is assumed that the cAMP pathway does not directly target phosphorylation of XYR1, but rather causes phosphorylation of a regulator of XYR1, which by modification of XYR1 acts on cellulase transcription [87].
In this respect it is interesting that deletion of acy1 or pkac1 also causes decreased levels of the photoreceptor gene env1 [87], which regulates cellulase levels [42]. Phosphorylation of the photoreceptor complex by protein kinase A was shown in N. crassa [88] and critically impacts the function of the circadian clock by establishing a negative feedback loop. A similar mechanism may be responsible for regulation of env1 levels in T. reesei. In turn, strains lacking ENV1 show strongly decreased cAMP levels, which is attributed to an impact of ENV1 on the function of phosphodiesterases rather than adenylate cyclase [89]. Such a mechanism would be in agreement with a feedback mechanism for fine-tuning of the integration of light response with nutrient signaling. Only recently we could confirm the involvement of a phosphodiesterase in light dependent cellulase regulation and the connection to ENV1 (E. Stappler et al., manuscript in preparation).
Interconnections between light response machinery and nutrient signaling pathways
The finding that light and its photosensors are involved in regulation of cellulase gene expression, indicated that nutrient utilization is interrelated with light response. The findings of the influence of the light signaling pathway on carbon-compound and carbohydrate metabolism [53] further raised the question as to the interconnections between nutrient signaling and sensing light. More detailed investigation of the heterotrimeric G-protein pathway and the cAMP pathway indicated that the light signal must be integrated with the transmitted nutrient signals.
BLR1 and BLR2 do not have a major impact on the G-protein signaling pathway [49]. ENV1 negatively influences blr1 and blr2 transcript levels, which in turn are required for induction of env1 in light. This mutual regulation causes a steady state level of transcripts for these three genes [49]. ENV1 is required for the positive feedback cycle of GNA1, which increases gna1 transcript levels upon constitutive activation. In turn, GNA1 negatively regulates env1 transcript levels [89]. Concerning the regulation mechanism of GNA3/gna3, ENV1 is not involved in establishing the gna3 feedback cycle, but negatively regulates gna3 transcript levels in light [89]. During investigation of the mutual regulatory effects of BLR1, BLR2 and ENV1 as well as GNA1, GNA3, GNB1, GNG1 and PhLP1, a pair with regulatory interaction at early light response emerged: PhLP1 and ENV1 [49]. With its negative effect on transcript levels of phlp1, gnb1 and gng1, ENV1 dampens G-protein signaling during early light response, presumably to provide resources for protection from light. Thereafter, PhLP1 can exert a positive effect on complex formation of GNB1 and GNG1 and G-protein signaling in general, which is likely to be important for metabolic adaptation to light [49]. The striking overlap in regulatory targets of photoreceptors, G-protein pathway components and ACY1 suggests that this interrelationship constitutes a core function in adaptation beyond transient effects in early light response.
Surprisingly, these interconnection studies revealed that it is not the photoreceptor complex comprising the two BLR proteins, but rather ENV1 that acts in a central position as a checkpoint, which is in line with its strong effect on gene regulation in light [46]. The more prominent role of ENV1 in comparison to BLR1 and BLR2 is also obvious in the role of ENV1 in the regulation of other physiological processes such as sexual development or stress response [41].
The strikingly low cAMP levels in mutants lacking ENV1 indicated that ENV1 could exert at least part of its function via the cAMP pathway adding another link between light response and nutrient signaling (see also above). The ∆acy1 strain showed a striking overlap of regulated genes in light, but not in darkness [49]. These genes have in common their regulation in the presence of strongly decreased or abolished cAMP levels. Among these cAMP targets there are numerous glycoside hydrolases, auxiliary proteins of cellulose degradation as well as the cellulase and hemicellulase regulator gene xyr1 [49]. Functional category analysis of these genes showed a strong enrichment in C-compound and carbohydrate metabolism. Consequently, the regulatory function of ENV1 in targeting regulation of enzyme expression is to a considerable extent mediated by its effect on cAMP levels [49]. Interestingly, many of the genes down-regulated in ∆env1 and ∆acy1 in the light are also down-regulated on soluble carbon sources compared to the insoluble carbon source cellulose [33]. Consequently, the cAMP dependent output of the regulatory cascade triggered by ENV1 could be targeted at sensing and/or reaction to a surface of specifically to cellulose, but not to a soluble carbon source [33].
In summary, ENV1 emerged as a major checkpoint between nutrient and light signaling [41]. However, it still remains to be shown whether this interrelationship is established directly by protein–protein interaction and an influence on activity of the targeted regulators or if the influence is indirect by an impact on regulation of abundance and/or activity of signaling factors, for example kinases, which in turn target regulatory factors. The considerable impact of deletion of env1 on the transcriptome in light [46] makes the latter hypothesis more likely.
Light dependent regulation of cellulose degradation as influenced by the sulphur source
Genes involved in metabolic processes are major targets of light response in T. reesei [46, 53]. Amino acid metabolism and sulphur metabolism are among these targets [33, 46, 53] and these functional categories are further found correlated with altered levels of cellulase production and carbon catabolite repression [90–92].
Studies aimed at identification of regulators binding to CAE within the cbh2 promoter revealed the sulphur related regulator protein LIM1, a homologue of N. crassa SCON2, as a candidate regulator [93]. Investigation of expression of lim1 under cellulase inducing conditions (cellulose, sophorose) in light and darkness supported this hypothesis. Interestingly, in the absence of a sulphur source, the methionine content, which can be used as a sulphur source, was important for cellulase gene expression and high sulphur conditions (5 mM) were even deleterious for growth in light. In the presence of normal sulphur levels in the MA minimal medium on cellulose, the same amount of methionine causes an increase of cellulase gene expression in darkness, but abolishment of cbh1 transcription in light. Therefore, methionine appears to represent a signal molecule with a light dependent impact on cellulase regulation [93].
Additionally, sulphate uptake upon growth on cellulose is regulated by light, while this is not the case with glucose as carbon source [93]. Hence, light dependence of sulfur metabolism is dependent on the carbon source.
Interestingly, we found a significant enrichment (p value > 0.05) of genes involved in amino acid metabolism among those specifically down-regulated under cellulase inducing conditions [33]. Thereby, significant enrichment in methionine metabolism was detected only in darkness [33]. Moreover, significant enrichment in functions in methionine metabolism and sulphur metabolism were detected for down-regulated genes in ∆env1 on cellulose [46]. Only recently, metabolic modelling of T. reesei under chemostat conditions with controlled growth rate revealed sulphur assimilation as a major limiting factor of protein production [94].
Light dependent gene regulation patterns in Trichoderma spp.—a genome wide view
First data on the influence of light on the transcriptome showed regulation of 2.8% of genes in response to light in T. atroviride [56]. In T. reesei, a comparable extent of light dependent gene regulation (2.7%) was detected in the early high cellulase expression mutant QM9414 upon growth on cellulose [53]. The gene set up-regulated upon growth in light on cellulose is significantly enriched in metabolic functions (p-value 1.39e-04), particularly C-compound and carbohydrate metabolism (p-value 7.74e-06) and polysaccharide metabolism (p-value 1.28e-04), but also in nitrogen, sulfur and selenium metabolism, carbohydrate transport and peptide transport. Among down-regulated genes, significant enrichment occurs for metabolism of polyketides and toxin transport [53].
The same study however also showed that in the absence of components of the heterotrimeric G-protein pathway, considerably higher differences in gene regulation between light and darkness occur, with up to 23% of differential regulation in mutants lacking the G-protein beta subunit GNB1. Thereby, a high number of genes with decreased transcript abundance in light compared to darkness was found in strains lacking PhLP1, GNB1 or GNG1. Many of these genes are not simply downregulated in light compared to the wild-type, but rather not up-regulated anymore because the light signal is not transmitted correctly. Accordingly, the highest number of consistently regulated genes in ∆gnb1, ∆gng1 and ∆phlp1 is found in the set of genes downregulated in light (628 genes including 21 glycoside hydrolase genes). This gene set includes a high number of genes associated with metabolism including 89 genes involved in C-compound and carbohydrate metabolism, but significant enrichment was only detected for functions in secondary metabolism (p-value 1.43e-08), photoperception and related functions. The strongest enrichment was observed for unclassified functions, indicating that the major function of GNB1, GNG1 and PhLP1 upon growth on cellulose still remains to be determined [53]. Available data confirm that PhLP1, GNB1 and GNG1 act in the same pathway and suggest that GNB1 and GNG1 may adjust sensitivity of the fungus to environmental signals.
Investigation of the transcriptomes of strains with mutations in the blr1, blr2 or env1 genes showed the same tendency of increased differences between transcript levels in light and darkness compared to wild-type. Light responsiveness of transcript abundances in ∆env1 even exceeds that of ∆gnb1 [46]. This analysis showed that the major function of BLR1 and BLR2 is observed in light, but clearly their absence did not abolish light response as the mutant strains are not blind. The majority of the regulatory targets of BLR1 and BLR2 was found in light, most of them being down-regulated in ∆blr1 and ∆blr2 (769 in ∆blr1 and 873 in ∆blr2) and only few genes being up-regulated. In light and darkness, BLR1 and BLR2 have a considerable number of regulatory targets with major functions in C-compound and carbon metabolism, polysaccharide metabolism and transport [46]. Thereby, BLR1 and BLR2 share many targets, but in addition their function extends to individual targets as well, indicating that BLR1 and BLR2 not only act as a transcription factor complex but have individual functions as well. Independent roles have also been shown for their homologues in T. atroviride and N. crassa [47, 92]. In contrast to BLR1 and BLR2, ENV1 also exerts considerable negative effects on gene regulation in light [46].
In darkness, there are no shared targets between BLR1, BLR2 and ENV1. Only 20 genes show consistent upregulation, while 564 genes are down-regulated in light in ∆blr1, ∆blr2 and ∆env1. These genes are dominated by functions in metabolism, including C-compound and carbohydrate metabolism including 22 glycoside hydrolase encoding genes, sulphur metabolism and with enrichment in secondary metabolism and an unexpectedly high number of genes with no assigned function.
Hence, for the photoreceptors, like for GNB1, GNG1 and PhLP1 their major function lies in up-regulation of their target genes in the presence of light, which is aimed at the light dependent modulation of metabolic functions of primary and secondary metabolism [46, 53].
Considering light dependent regulation patterns in wild-type and the available mutants in BLR1, BLR2, ENV1, GNB1, GNG1 and PhLP1, members of all glycoside hydrolase (GH) families—a remarkably high 75% of all GH encoding genes—show potential light dependent regulation.
cAMP signalling represents one of the most important output pathways of the heterotrimeric G-protein signaling cascade [95] and is hence the prime candidate for a coincidence detector between nutrient and light signaling. Transcriptome analysis of the major components of the cAMP pathway in T. reesei, adenylate cyclase (acy1) and protein kinase A (catalytic subunit 1, pkac1) revealed that their deletion causes an increase in light dependent gene regulation ([49]; unpublished data on ∆pkac1). ACY1 targets a broad array of genes involved in metabolism, Particularly, significant enrichment in metabolic functions was found for its positive regulatory targets in light [49], which is also the condition under which BLR1, BLR2, ENV1, GNB1, GNG1 and PhLP1 show their major function [49]. Comparable with ∆acy1, cAMP levels in ∆env1 are very low [89]. Of the 136 positive targets of ACY1 in light, 114 overlap with those of ENV1 including 25 glycoside hydrolase encoding genes, for example xyn2, xyn4, bxl1, cel3a, cel3b, egl1,egl2, egl6 and cbh2, the lytic polysaccharide monooxygenase encoding cel61a and cel61b as well as swollenin, cip1, cip2 and the major transcription factor gene xyr1. Additionally 16 genes involved in sulphur metabolism are in this gene set [49].
These findings on altered numbers of light responsive genes lead to the conclusion that the signaling pathways for nutrient (heterotrimeric G-protein pathway, cAMP pathway) and light response balance gene expression and this balance is perturbed if single factors are removed. Since a sizable number of the targets of the investigated signaling factors show broad functions in plant cell wall degradation it can be assumed that this balance is aimed at adjustment of substrate utilization to day and night as well as growth within and on the surface of a substrate.
Gene regulation specific for induction of cellulases is light dependent
Cellulases are expressed on diverse carbon sources, mostly representing degradation products of plant cell walls [18, 23, 96, 97]. Cellulase gene expression is not consistently regulated by light on different carbon sources [43, 65, 87, 93], indicating a different relevance of light under different conditions. Therefore, of interestis the a core gene set that is specifically regulated under inducing conditions and whether this gene set is different in light and darkness [33]. This two-dimensional analysis (5 carbon sources including inducing and repressing ones, 2 light conditions) revealed 1324 induction specific genes, comprising a set of 530 genes to be cellulase induction specific in light and in darkness, but also a considerable number of genes, that show regulation specific for cellulase induction only in light or only in darkness. The gene set of cellulase induction specific genes only in darkness includes 16 CAZyme encoding genes and comprises such important genes as egl3 (cel12a), the predicted oxidoreductase TR_56840 (more than eightfold regulation in both cases), bgl1 (cel3a), egl5 (cel45a), cel5b and bgn1 [33]. Genes of the central pathways of primary metabolism, including the pentose phosphate pathway and glycolysis turned out to be targets of light responses, with a particularly important function of ENV1. Generally, photoreceptors clearly contribute to regulation of induction specific genes both in light and darkness [33].
In several cases, specific regulation for cellulase induction [33] and positive regulation by ENV1 [46] overlaps with negative correlation of the specific protein production rate in continuous fermentations controlled for growth rate [98]. Consequently, light dependent processes and their regulation is are to be considered in fermentations as well. Of the 1324 induction specific genes, only 218 were neither light regulated nor photoreceptor targets and this gene set does not include the known cellulase regulators, glycoside hydrolases or proteins involved in oxidative degradation of cellulose [33]. Comparison of gene regulation on the insoluble cellulase inducing carbon source cellulose with the soluble inducing carbon sources lactose and sophorose [33] revealed indications that T. reesei may sense surfaces, particularly in light, due to cellulose specific expression of hydrophobin genes, swollenin, cip1 and cip2, which play a role in the attack on cellulose [17, 99, 100]. Because of overlapping regulation with the cAMP dependent targets of ENV1 [33, 49, 89], surface sensing may be one of the outputs of the function of ENV1.
The pathways described above signal to the cell, among other information inputs, which carbon source is available in the surroundings of the cell. In this respect it is interesting that the number of light regulated genes varies on different carbon sources, like seen with different signaling mutants. In particular, the number of genes increases on lactose, sophorose and glycerol while upon growth on glucose, the number of genes down-regulated in light increases [33]. This finding explains at least in part why perturbing signaling pathways leads to altered light response: the adjustment of gene regulation according to nutrient availability does not work properly anymore.
Genome wide transcription patterns as influenced by light
Genome wide transcriptome patterns are reflect the physiological state of the organism under the tested conditions. Therefore we wanted to assess general effects on deletion of certain signaling genes and how they relate to light response on a given carbon source. For that we re-analyzed transcriptomic data under growth on cellulose in wild-type and signaling mutants in light and darkness for the whole genome and for different gene groups [33]. Generally we found that light dependent gene regulation does not break carbon source dependent regulation [49]. This finding is also in agreement with the higher relevance of the carbon source versus the light status: in other words, light modulates gene expression, but induction or repression is not initiated or abolished by light.
Hierarchical clustering of transcript levels across different carbon sources and in the absence of nutrient and light signaling genes revealed that in mutant strains, the genome wide patterns of carbon source specific regulation are largely retained. Transcripts from cultures grown on cellulose still cluster separately from those grown on soluble carbon sources. Within soluble carbon sources i.e. comparing growth on glucose, glycerol, lactose or sophorose, also patterns on inducing and repressing carbon sources are more similar within the group than with each other. Similarly, transcript profiles from light and dark grown cultures clustered together ([33], our unpublished results). The only exception to these groupings was gene regulation in the ∆env1 strain upon growth in light: although grown on cellulose, gene expression patterns clustered with soluble inducing carbon sources, indicating a certain malfunction in carbon sensing. Accordingly, 77% of the cAMP dependent targets of ENV1 [49] overlap with those potentially involved in cellulose/surface sensing [33].
With respect to environmental sensing, the clustering of genes encoding G-protein coupled receptors indicated perturbed carbon sensing in ∆env1 and also in ∆gnb1 [33]. Although deletion of env1 causes a considerable growth defect in light [42], which is at least in part dependent on altered cAMP levels [49], deletion of gnb1 does not result in strongly altered growth or morphology in light [53]. Consequently, this strong increase in light responsiveness of gene regulation is not simply due to altered growth in light and darkness. Moreover, altered gene regulation in light is correlating with altered growth and reflects a significant physiological modification in response to light and indicates different relevance of the targeted pathways in light and darkness.
Non-random distribution of light regulated genes in the genome
Genes operative in the same pathway are often clustered in a genomic region, facilitating their enhanced regulation or co-regulation for example due to an open chromatin structure in this area as exemplified in secondary metabolite clusters [101]. In the genome of T. reesei, clustering was found for CAZyme encoding genes and in several cases genes associated with secondary metabolism were present in these genomic areas as well [102]. In T. reesei, light regulated genes are clustered in 15 genomic clusters (containing 66 genes) have clusters of light regulated genes when grown on cellulose, but only one cluster for growth on sophorose and 2 for growth on glucose. On lactose and glycerol no significant clusters were found [33]. In part, the light dependent clusters found on cellulose overlap with CAZyme clusters. One cluster contains the lytic polysaccharide monooxygenase encoding cel61a gene along with two polyketide synthase genes [33] and overlaps with a cluster positively correlated with the specific protein production rate of T. reesei under constant cultivation conditions [98]. This cluster represents a secondary metabolite cluster, called the “SOR cluster” which was recently characterized [54, 103]. It is responsible for production of the yellow pigment of T. reesei [103] and regulates biosynthesis of sorbicillin compounds [104] and their derivatives trichodimerol and dihydrotrichotetronine [54]. Interestingly, this cluster overlaps with a CAZyme cluster comprising a candidate chitinase, acetylxylanesterase axe1, the auxiliary protein encoding cip1 in addition to cel61a [102].
The most interesting of the clusters with respect to cellulase gene regulation comprises cbh2/cel6a, one of the two major cellulase encoding genes of T. reesei, cel5a/egl2, a predicted sugar transporter, a putative urea transporter and an FMN dependent oxidoreductase [33]. Normal transcript levels of this cluster require the presence of BLR1, BLR2 and ENV1 in light.
Another cluster contains the mannitol dehydrogenase gene lxr1 [105]. On mannitol, growth of the wild-type in light is slightly slower than in darkness [46]. A cluster of 9 genes around lxr1 is regulated negatively by ENV1 in light on cellulose (up to 40 fold). This cluster overlaps with a light regulated cluster on cellulose [33, 46].
Light influences carbon catabolite repression
Biosynthesis of enzymes is very energy consuming and hence only initiated by the fungus if the environmental conditions require these enzymes for survival [23, 34]. This is mainly the case for insoluble plant material that cannot be taken up by the cell without liberation of low molecular compounds that serve as carbon sources. The most important process for this regulation is carbon catabolite repression, which shuts down enzyme production when easily metabolizable carbon sources are sensed in the environment.
The carbon catabolite repressor CRE1 regulates cellulase gene expression not only upon growth on glucose [106], but also on cellulose [54]. Upon growth on cellulose the regulatory targets of CRE1 are different in light and darkness and few genes were consistently regulated irrespective of the light condition, and targeted functions of CRE1 are different in light and darkness as well [54]. The genes regulated by CRE1 are non-randomly distributed in the genome and form 36 clusters, several of which contain CAZyme encoding genes. Five clusters are found, of which one comprises two CAZyme encoding genes (TR_32243/carbohydrate esterase family 1 and TR_62182/glycosyltransferase family 1) that are consistently upregulated in darkness in ∆cre1 [54]. One of the clusters shows light dependent regulation by CRE1 and photoreceptors i.e. the recently characterized SOR cluster [54, 103, 107].
Of the 1324 genes with cellulase induction specific regulation, 409 are regulated by CRE1. Of those, only 14 are downregulated in ∆cre1 in light and darkness and only 6 (including the hydrophobin gene hfb5) are upregulated in light and darkness [54]. Additionally, genes regulated by CRE1 are in part non-randomly distributed in the genome. Interestingly, 142 genes are upregulated in ∆cre1 only in light, while 251 genes are upregulated in this strain only in darkness. Consequently, the relief from carbon catabolite repression by deletion of cre1 has also light dependent effects [54].
Among the light dependent targets of CRE1, the predicted glucose/ribitol dehydrogenase gene grd1 was analyzed in more detail [65]. Grd1 is positively regulated by CRE1 in darkness (more than eightfold) and its transcript size increases upon onset of inducing conditions, indicating a role of alternative splicing in its regulation. Transcript levels of grd1 are thereby strictly correlated with those of cbh1 and also follow the transient pattern upon sophorose induction in light [65]. GRD1 acts on cellobiose as a substrate with the likely reaction product cellobiitol, which could inhibit beta glucosidase function. With respect to cellulase regulation, GRD1 influences transcript levels, activity and protein abundance of the major cellulases in a light dependent manner. These findings suggest that GRD1 acts in intracellular sensing of cellulase efficiency and adaptation of cellulase levels for optimization of gene regulation [65].
Regulation of plant cell wall degradation by light is conserved in fungi
After the finding that cellulases are regulated by light and photoreceptors in T. reesei, we were interested if this is a conserved phenomenon or specific to Trichoderma. Transcriptome analysis of N. crassa upon growth on cellulose showed that regulation of cellulases by photoreceptors is conserved [92]. Lack of photoreceptors causes initially elevated secreted cellulase activity upon growth on cellulose in N. crassa. After prolonged cultivation, the increased levels are only maintained in ∆vvd, while in the white collar mutants lower levels occur, which is likely due their function in regulation of energy metabolism [92]. Targets of the light response machinery in N. crassa include numerous genes encoding plant cell wall degrading enzymes, including several lytic polysaccharide monooxygenases as well as functions in amino acid metabolism and sulphur metabolism. 55 genes are consistently regulated in one or more of the photoreceptor mutants in N. crassa and T. reesei including the glycoside hydrolase encoding xyn1, xyn3, cbh1 and cbh2. However, the important transcription factor encoding genes xyr1, cre1, clr1 and clr2 are not among these genes [92].
In contrast to T. reesei, gene regulation by the white collar proteins did not overlap with that of VVD, but indications for photoadaptation (contrasting regulation between white collars and VVD) were found. Also the striking difference of gene regulation in ∆env1 compared to wild-type in T. reesei was not observed in ∆vvd compared to wild-type in N. crassa in the same extent. Targets of this photoadaptation include the carbon catabolite repressor CRE1, which is also not the case in T. reesei. It was particularly obvious that ribosomal genes were among those regulated by photoreceptors, which supported the hypothesis of posttranscriptional regulation of cellulase expression.
In general, this study indicates that light dependent regulation of plant cell wall degradation is a conserved process in fungi, albeit some steps of individual regulation of degradation and photoadaptation are not [92]. Exploration of light dependent gene regulation in further fungi will reveal if this conservation is indeed general.
Light alters complex formation in promoters of cellulase genes and their regulators
Regulation of cellulase gene expression at the transcriptional level was shown to involve the CAE (cbh2 activating element) motif in the cbh2 promoter [108] and a similar motif in the cbh1 promoter [109]. This DNA motif is bound by protein complexes both under inducing (sophorose) and repressing (glucose) conditions [108] and induction specific chromatin rearrangement occurs at this site [110]. Thereby, the CAE (cellulase activating element) is crucial for regulation of cbh2 and at least in part similar proteins bind to a comparable motif in cbh1 [111].
The findings that both cbh1 and cbh2 are subject to regulation by light [42, 43] indicated that complex formation within these promoters may be influenced by light. Light dependent promoter binding of the white collar complex (WCC) in N. crassa was shown previously and complex formation of the WCC on its target promoters is influenced by light even in cell free extracts [112].
In T. reesei complex formation at the CAE motif of the cbh2 promoter is clearly different between light and darkness upon growth on lactose [87]. Additionally, one of the formed complexes at CAE is dependent on the presence of the protein kinase A catalytic subunit (PKAC1) [87].
Light dependent alterations in complex formation were also shown at the EUM1 motif, which was found as a conserved motif in the T. reesei env1 promoter and the promoter of the N. crassa homologue vvd [42, 113]. This motif is also present in the cbh1 and cbh2 promoters. Cellulase gene expression is modulated by the G-protein alpha subunit GNA3 in light [77]. The EUM1 was detected in the gna3 promoter as well. In order to investigate a possible connection or feedback between gna3 and env1, we analyzed complex formation at the gna3 promoter in darkness and after illumination upon growth on glycerol [113]. Indeed, we found light dependent changes in complex formation at the EUM1 motif in the promoters of gna3 and env1. The patterns of the protein complexes strongly resembled each other in the two promoters and competition experiments confirmed that similar proteins bind to EUM1 in gna3 and env1 [113]. Consequently, env1 and gna3 are at least in part regulated by similar regulators and due to the regulation of gna3 by ENV1 a feedback mechanism mediated by an as yet unknown transcription factor is likely.
However, the studies on complex formation at EUM1 mentioned above were done under non-inducing conditions. For growth on cellulose, complex formation also occurs within the EUM1 motifs of the cbh2 promoter as well as on the EUM1 motifs in the env1, vvd and gna3 promoters in a light dependent manner. Preliminary data suggest that part of the proteins binding to the EUM1 motif in these promoters also bind to the EUM1 motifs in the env1 and in the gna3 promoter, particularly in light (Schmoll, unpublished results).
Carbon source utilization by Trichoderma is altered by light and regulated by photoreceptors
Early analysis of fungal light responses indicated that the influence of light on growth of a fungus is dependent on the carbon source it grows on [114]. Thereby, a carbon source close to its natural conditions would cause the least growth defect [115].
Using Biolog phenotype microarrays allows for simultaneous testing of growth on 95 different carbon sources [116]. Our analysis showed that there are considerable differences between growth on many of these carbon sources, particularly on substrates associated with plant cell wall degradation such as D-sorbitol, L-arabinose, d-fructose, D-galactose or xylitol [46]. Additionally, the photoreceptor ENV1 caused significantly different growth patterns on many carbon sources [45]. Correlating these results with gene regulation data from transcriptome analyses showed that the pentose-phosphate pathway responsible for utilization of substrates like xylose or lactose, is a target of light response and subject to regulation by photoreceptors. At several of the enzymatic steps in this pathway, modification of growth correlated with light dependent alteration of enzyme gene transcript levels [46].
In addition, the positive regulator of cellulase gene expression, GRD1, influences light dependent growth patterns, especially on several intermediates of lactose and D-galactose carabolism, like D-xylose, D-galactose, d-fructose, D-mannitol and D-xylose. This influence on growth on intermediates of a cellulase inducing carbon source (lactose) is in agreement with the function of GRD1 in cellulase regulation and potentially inducer formation [65].
In T. atroviride, stimulation of growth in light was observed on many carbon sources, particularly 17 carbon sources which are well utilized by this fungus [117]. These carbon sources comprise common building blocks of plant cell walls like fructose, mannose, galactose, glucose or xylose, while growth on the best carbon source for T. atroviride, glycerol, is not influenced by light and growth on sorbitol even decreased in light. The increase in growth rate on several carbon sources in light is dependent on the presence of BLR1 and BLR2. Lack of these factors even causes decreased growth, albeit some exceptions on certain carbon sources for ∆blr1 and ∆blr2 confirm that these strains are not totally blind, but rather show altered and diminished responses [117].
The clear connection between the cAMP pathway and cellulase regulation by light and photoreceptors indicated a role of BLR1 and BLR2 in introducing the light signal into the cAMP pathway upon regulation of utilization of diverse carbon sources. In T. atroviride stimulation of growth by addition of cAMP was observed only on a few carbon sources (glucose, gentiobiose, cellobiose and xylose) and this response was dependent on BLR1 and BLR2. On these carbon sources, stimulation of growth was also achieved by addition of menadione that causes oxidative stress, which should simulate oxidative stress caused by light. Also for this response functional BLR1 and BLR2 are required. However, a general correlation of responses to cAMP and oxidative stress with the presence or absence of BLR1 or BLR2 across all 95 carbon sources tested could not be confirmed [117].
Consequently, light influences growth and carbon utilization of T. atroviride and the cAMP pathway as well as oxidative stress response are involved in this regulation mechanism. Additionally, BLR1 and BLR2 play important roles in this connection particularly on carbon sources representing building blocks of plant cell walls, hence confirming carbohydrate degradation as a major target of light response.
Crucial factors for plant cell wall degradation as targets of light response
The long history of research towards improvement of cellulase gene expression yielded a considerable number of regulators enhancing or decreasing efficiency of plant cell wall degradation [23, 31]. Transcriptome analysis showed that carbon metabolism and degradation of polysaccharides are important targets of light dependent gene regulation in T. reesei and N. crassa [53, 92]. For N. crassa it has been shown that the photoreceptors themselves act on regulation of further transcription factors, constituting a flat hierarchy of regulation [118]. Consequently, the transcription factors involved in plant cell wall degradation are of particular interest as targets of light response.
Therefore we screened the recently published, comprehensive list of transcription factors involved in plant biomass utilization [31] for genes differentially regulated in light and darkness or by photoreceptors. Interestingly, we found that among them, the transcript levels of xyr1, ace1, ace2, ace3, vib1 and amyR are decreased in light compared to darkness in QM6a upon growth on cellulose (Table 1). Also the T. reesei homologues of the important N. crassa transcription factors clr-1 and clr-2 show regulation in QM6a by light, but they do not regulate cellulase expression in T. reesei ([119] and our unpublished results). Xyr1 is regulated by BLR1 and ENV1. Interestingly, the mating type protein MAT1-2-1 interacts with XYR1 and is recruited to the cbh1 promoter in a XYR1 and light dependent manner [120]. The finding that ENV1 considerably regulates mat1-2-1 [44], the hypothesis that this light dependent effect is due to the function of photoreceptors is supported. Ace3, vib1 and amyR are regulated by ENV1 upon growth on cellulose (Table 1; [46]).
Table 1.
Light dependent regulation of transcription factors involved in plant biomass utilization
| Name | Protein ID | Regulatory function (related to plant biomass degradation) in | Cellulase induction specific | Regulation in QM6a in light | Regulated by BLR1 | Regulated by BLR2 | Regulated by ENV1 | ||
|---|---|---|---|---|---|---|---|---|---|
| In light | In darkness | In light and darkness | |||||||
| XYR1 | 122208 | (hemi-)cellulose utilization | x | x | x | -- | x | x | |
| CRE1 | 120117 | Carbon catabolite repression | n | ||||||
| ACE1 | 75418 | Cellulose utilization | - | ||||||
| ACE2 | 78445 | Cellulose utilization | x | -- | |||||
| ACE3 | 77513 | Cellulose utilization | x | x | x | --- | x | ||
| CLR1 | 27600 | (hemi-)cellulose utilization (in N. crassa) | x | - | x | ||||
| CLR2 | 26163 | (hemi-)cellulose utilization (in N. crassa) | -- | x | x | ||||
| XPP1 | 122879 | (hemi-)cellulose utilization | n | ||||||
| VIB1 | 54675 | Cellulose utilization | --- | x | |||||
| HAP2 | 124286 | CAZy regulation | n | ||||||
| HAP3 | 121080 | CAZy regulation | n | ||||||
| HAP5 | 124301 | CAZy regulation | n | ||||||
| AmyR | 55105 | Starch utilization | -- | x | |||||
| MalR | 21997 | Maltose utilization | n | ||||||
| BglR | 52368 | Sugar sensing | n | ||||||
| RhaR1 | 79871 | L-rhamnose utilization | n | ||||||
| RhaR2 | 121107 | L-rhamnose utilization | x | x | x | n | |||
| McmA | 42249 | Cellulase regulation | n | ||||||
| VEL1 | 122284 | Cellulase regulation | ++ | ||||||
| VEL2 | 40551 | Metabolism | n | ||||||
| AreA | 76817 | N-assi mi lation | n | ||||||
| AreB | 120127 | Nitrogen metabolite repression | n | ||||||
| NmrA | 74375 | Nitrogen metabolite repression | n | ||||||
| NIT4 | 76705 | Nitrate pathway | n | ||||||
| PAC1 | 120698 | pH response | n | ||||||
| PACX | 59740 | pH response | - | ||||||
For the regulatory impact of the GATA-type transcription factors BLR1 and BLR2 see text on photoreceptors
n, no regulation; +, upregulation in light; -, downregulation in light; +/-, minor regulation; ++/--, moderate regulation (around 5–10 fold); +++/---, strong regulation
A further important parameter for plant cell wall degradation is the hydrolysis performance as determined by the secretome of T. reesei [121]. This study identified 12 proteins to be limiting for hydrolysis. Ten of them were found to be regulated in a cellulase induction specific manner in light or darkness or both [33] (Table 2). But most importantly, all of the genes encoding these limiting proteins were strongly downregulated in light in QM6a upon growth on cellulose and regulated by ENV1 under the same conditions. Additionally, all those genes except xyn4 are regulated by BLR1 and man1, xyn2 and bgl1 are regulated by BLR2 as well [46].
Table 2.
Light dependent regulation of genes encoding proteins limiting for hydrolysis of pretreated corn stover
| Name | Protein ID | Function | Cellulase induction specific | Regulation in QM6a in light | Regulated by BLR1 | Regulated by BLR2 | Regulated by ENV1 | ||
|---|---|---|---|---|---|---|---|---|---|
| In light | In darkness | In light and darkness | |||||||
| 69276 | Xylanase | x | x | x | --- | x | x | ||
| CIP1 | 73638 | Carbohydrate binding module containing | x | x | x | --- | x | x | |
| CIP2 | 123940 | Glucuronyl esterase | --- | x | x | ||||
| MAN1 | 56996 | Beta-mannanase | x | x | x | --- | x | x | x |
| EGL2 | 120312 | Endoglucanase CEL5A | x | x | x | --- | x | x | |
| XYN2 | 123818 | Xylanase | x | x | x | --- | x | x | x |
| EGL3 | 123232 | Endoglucanase CEL12A | x | --- | x | x | |||
| SWO1 | 123992 | Swollenin | x | x | x | --- | x | x | |
| BGL1 | 76672 | Beta-glucosidase | x | --- | x | x | x | ||
| CBH2 | 72567 | Cellobiohydrolase CEL6A | x | x | x | --- | x | x | |
| EGL6 | 49081 | Xyloglucanase CEL74A | --- | x | x | ||||
| XYN4 | 111849 | Xylanase | x | x | x | --- | x | ||
n, no regulation; +, upregulation in light; -, downregulation in light; +/-, minor regulation; ++/--, moderate regulation (around 5–10 fold); +++/---, strong regulation
These findings are in perfect agreement with polysaccharide degradation being a major target of light response in T. reesei [53]. Moreover, these results indicate that for stable and predictable performance of strains in research and industry, light conditions and random light pulses have to be considered of equal importance as for example pH and oxygen supply and a defined cultivation medium.
Implications for research, industrial application and strain improvement
Fungi sense their environment and adjust growth and enzyme production for efficient use of resources accordingly. They use complex signal transduction pathways to integrate these environmental signals for a defined response to the conditions they perceive—in their natural environment as well as in an industrial setting. Consequently, knowledge on signal transduction pathways is highly relevant for efficient and stable gene expression [122].
In order to draw reliable conclusions on gene regulation, we have to ensure that the growth conditions of the organisms we are testing remain to same. Reproducibility of results requires defined growth media in terms of carbon source, nitrogen source, pH, temperature etc. Often, different standard media and growth conditions for different fungi are discussed as problematic for comparability of data. The research reviewed here clearly shows that light is an important environmental cue not only in T. reesei where light can alter regulation of cellulase genes tenfold and more. Nevertheless, even in T. reesei there is still research towards gene regulation and cellulase gene expression performed under random light conditions. With transcriptional regulation, it is known that even a few seconds of light cause a response in gene regulation. Considering the broad metabolic regulations by light, this compares to using a random mixture of carbon sources in an assay, which may show a beneficial effect first and no effect at all upon repetition. Moreover, we recently showed that cellulase regulation additionally involves a posttranscriptonal section, which is triggered by the light sensitive heterotrimeric G-protein pathway [33]. Consequently, good laboratory practice has to include the use of controlled light conditions in order to draw any reasonable conclusions as to the function of a gene. Especially for transcriptome studies, the effect of random light pulses—for example during harvesting or due to an open shaker—can be dramatic and lead to wrong conclusions on the function of a gene in enzyme production.
The implementation of such data from uncontrolled laboratory experiments into industrial strain improvement may hence lead to costly failures either early in testing or later in upscaling. For example, if constitutive activation of the G-protein alpha subunit GNA3 [77] would be tested for improvement of cellulase fermentations: Shake flask cultivations on table top shakers in the lab (in light) and subsequently in a glass fermentor would lead to beneficial effects on cellulase expression, because GNA3 exerts its function in light. However, upon (cost intensive) upscaling in a dark steel fermentor, the strain would not show benefits anymore, because in darkness constitutive activation of GNA3 does not have an effect on cellulase regulation [77]. Research done under random conditions bears such costly risks for surprises, because based on current knowledge the difference in the function of a given regulator in light and darkness cannot be predicted reliably. Only if part of the pathway is already known, like for the heterotrimeric G-protein pathway, some perspectives can be suggested.
Exploitation of light dependent effects
Available data show that enzyme production is most efficient in darkness in T. reesei. Nevertheless, the strong differences between light and darkness indicate an efficient regulatory mechanism, that—due to previous negligence of light dependent phenomena—bears the chance to identify novel regulators that can be artificially misregulated for dark specific cellulase enhancement in an industrial fermentor (Fig. 2).
Since light does not override the induction process [33, 49], the use of photofermentors for enzyme production in a mass fermentation is unlikely to be cost effective. However, for special high value products like secondary metabolites, the use of light dependent promoters for precisely triggered regulation should be kept in mind. Similarly, secondary metabolism can be altered by application of light which might be beneficial for co-production or production of valuable metabolites. Additionally, the striking differences between light and darkness can be used to select light responsive promoters to enable triggering or abolishing protein production using light pulses. Exemplary studies in yeast [123] and T. reesei [124] have already proven that this approach is successful.
Besides the actual production process, also inoculum production is crucial for stable and high level production. Production of spores is regulated by light in fungi. Moreover, conditions of spore production are crucial for physiology of the next generation of fungi grown from it, particularly with respect to stress response. Accordingly, our own research showed that reproducible results require constant conditions for inoculum generation (precultivation on plates) prior to cultivation in liquid culture and analysis (Schmoll and Tisch, unpublished). Although a circadian rhythmicity has not yet been demonstrated for T. reesei, our experience strongly suggests its operation. Therefore, random light pulses, which reset the molecular circadian clock should be avoided during spore production to ensure reliable production in the cultivation thereafter. It remains to be determined whether constant light or constant darkness or light cycles of defined length during preculture can be used to increase production capacity for fermentations.
Conclusions
Since the discovery that light influences regulation of cellulases and more generally carbohydrate active enzymes, several regulation mechanisms were shown to govern this influence. In some cases, regulators only show an effect under one light status but not another. Growth, enzyme regulation, carbon utilization, carbon catabolite repression and even cross talk with secondary metabolism and sulfur metabolism were shown. Therefore, in order to obtain scientifically meaningful results on gene regulation aimed at investigation of plant cell wall degradation, application of controlled light conditions is mandatory and even short random light pulses are to be avoided.
Authors’ contributions
MS conceived of the review and wrote the manuscript. The author read and approved the final manuscript.
Competing interests
The author declares that she has no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dunlap JC, Loros JJ. Making time: conservation of biological clocks from fungi to animals. Microbiol Spectr. 2017;5(3). 10.1128/microbiolspec.FUNK-0039-2016. [DOI] [PMC free article] [PubMed]
- 2.Hurley JM, Loros JJ, Dunlap JC. The circadian system as an organizer of metabolism. Fungal Genet Biol. 2016;90:39–43. doi: 10.1016/j.fgb.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tisch D, Schmoll M. Light regulation of metabolic pathways in fungi. Appl Microbiol Biotechnol. 2010;85:1259–1277. doi: 10.1007/s00253-009-2320-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmoll M. Assessing the relevance of light for fungi implications and insights into the network of signal transmission. Adv Appl Microbiol. 2011;76:27–78. doi: 10.1016/B978-0-12-387048-3.00002-7. [DOI] [PubMed] [Google Scholar]
- 5.Hurley JM, Loros JJ, Dunlap JC. Circadian oscillators: around the transcription-translation feedback loop and on to output. Trends Biochem Sci. 2016;41:834–846. doi: 10.1016/j.tibs.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hurley JM, Dasgupta A, Emerson JM, Zhou X, Ringelberg CS, Knabe N, Lipzen AM, Lindquist EA, Daum CG, Barry KW, et al. Analysis of clock-regulated genes in Neurospora reveals widespread posttranscriptional control of metabolic potential. Proc Natl Acad Sci USA. 2014;111:16995–17002. doi: 10.1073/pnas.1418963111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montenegro-Montero A, Canessa P, Larrondo LF. Around the fungal clock: recent advances in the molecular study of circadian clocks in Neurospora and other fungi. Adv Genet. 2015;92:107–184. doi: 10.1016/bs.adgen.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Sancar C, Sancar G, Ha N, Cesbron F, Brunner M. Dawn- and dusk-phased circadian transcription rhythms coordinate anabolic and catabolic functions in Neurospora. BMC Biol. 2015;13:17. doi: 10.1186/s12915-015-0126-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker CL, Loros JJ, Dunlap JC. The circadian clock of Neurospora crassa. FEMS Microbiol Rev. 2012;36:95–110. doi: 10.1111/j.1574-6976.2011.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunner M, Kaldi K. Interlocked feedback loops of the circadian clock of Neurospora crassa. Mol Microbiol. 2008;68:255–262. doi: 10.1111/j.1365-2958.2008.06148.x. [DOI] [PubMed] [Google Scholar]
- 11.Schafmeier T, Diernfellner AC. Light input and processing in the circadian clock of Neurospora. FEBS Lett. 2011;585:1467–1473. doi: 10.1016/j.febslet.2011.03.050. [DOI] [PubMed] [Google Scholar]
- 12.Proietto M, Bianchi MM, Ballario P, Brenna A. Epigenetic and posttranslational modifications in light signal transduction and the circadian clock in Neurospora crassa. Int J Mol Sci. 2015;16:15347–15383. doi: 10.3390/ijms160715347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sancar C, Ha N, Yilmaz R, Tesorero R, Fisher T, Brunner M, Sancar G. Combinatorial control of light induced chromatin remodeling and gene activation in Neurospora. PLoS Genet. 2015;11:e1005105. doi: 10.1371/journal.pgen.1005105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bischof RH, Ramoni J, Seiboth B. Cellulases and beyond: the first 70 years of the enzyme producer Trichoderma reesei. Microb Cell Fact. 2016;15:106. doi: 10.1186/s12934-016-0507-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paloheimo M, Haarmann T, Mäkinen S, Vehmaanperä J. Production of industrial enzymes in Trichoderma reesei. In: Schmoll M, Dattenböck C, editors. Gene expression systems in fungi: advancements and applications. Heidelberg: Springer International; 2016. pp. 23–58. [Google Scholar]
- 16.Bazafkan H, Tisch D, Schmoll M. Regulation of glycoside hydrolase expression in Trichoderma. In: Gupta VK, Schmoll M, Herrera-Estrella A, Upadhyay RS, Druzhinina I, Tuohy MG, editors. Biotechnology and biology of Trichoderma. Oxford: Elsevier; 2014. pp. 291–307. [Google Scholar]
- 17.Foreman PK, Brown D, Dankmeyer L, Dean R, Diener S, Dunn-Coleman NS, Goedegebuur F, Houfek TD, England GJ, Kelley AS, et al. Transcriptional regulation of biomass-degrading enzymes in the filamentous fungus Trichoderma reesei. J Biol Chem. 2003;278:31988–31997. doi: 10.1074/jbc.M304750200. [DOI] [PubMed] [Google Scholar]
- 18.Margolles-Clark E, Ilmén M, Penttilä M. Expression patterns of ten hemicellulase genes of the filamentous fungus Trichoderma reesei on various carbon sources. J Biotechnol. 1997;57:167–179. [Google Scholar]
- 19.Blumenthal CZ. Production of toxic metabolites in Aspergillus niger, Aspergillus oryzae, and Trichoderma reesei: justification of mycotoxin testing in food grade enzyme preparations derived from the three fungi. Regul Toxicol Pharmacol. 2004;39:214–228. doi: 10.1016/j.yrtph.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Schmoll M, Wang TF. Sexual development in Trichoderma. In: Wendland J, editor. The Mycota (Vol I): growth, differentiation and sexuality. Cham: Springer; 2016. pp. 457–474. [Google Scholar]
- 21.Seidl V, Seibel C, Kubicek CP, Schmoll M. Sexual development in the industrial workhorse Trichoderma reesei. Proc Natl Acad Sci USA. 2009;106:13909–13914. doi: 10.1073/pnas.0904936106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kowalczyk JE, Benoit I, de Vries RP. Regulation of plant biomass utilization in Aspergillus. Adv Appl Microbiol. 2014;88:31–56. doi: 10.1016/B978-0-12-800260-5.00002-4. [DOI] [PubMed] [Google Scholar]
- 23.Glass NL, Schmoll M, Cate JH, Coradetti S. Plant cell wall deconstruction by ascomycete fungi. Annu Rev Microbiol. 2013;67:477–498. doi: 10.1146/annurev-micro-092611-150044. [DOI] [PubMed] [Google Scholar]
- 24.Kubicek CP, Starr TL, Glass NL. Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu Rev Phytopathol. 2014;52:427–451. doi: 10.1146/annurev-phyto-102313-045831. [DOI] [PubMed] [Google Scholar]
- 25.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucl Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den Brink J, de Vries RP. Fungal enzyme sets for plant polysaccharide degradation. Appl Microbiol Biotechnol. 2011;91:1477–1492. doi: 10.1007/s00253-011-3473-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bech L, Busk PK, Lange L. Cell wall degrading enzymes in Trichoderma asperellum grown on wheat bran. Fungal Genom Biol. 2014;4:116. [Google Scholar]
- 28.Beeson WT, Vu VV, Span EA, Phillips CM, Marletta MA. Cellulose degradation by polysaccharide monooxygenases. Annu Rev Biochem. 2015;84:923–946. doi: 10.1146/annurev-biochem-060614-034439. [DOI] [PubMed] [Google Scholar]
- 29.Kracher D, Scheiblbrandner S, Felice AK, Breslmayr E, Preims M, Ludwicka K, Haltrich D, Eijsink VG, Ludwig R. Extracellular electron transfer systems fuel cellulose oxidative degradation. Science. 2016;352:1098–1101. doi: 10.1126/science.aaf3165. [DOI] [PubMed] [Google Scholar]
- 30.Cannella D, Mollers KB, Frigaard NU, Jensen PE, Bjerrum MJ, Johansen KS, Felby C. Light-driven oxidation of polysaccharides by photosynthetic pigments and a metalloenzyme. Nat Commun. 2016;7:11134. doi: 10.1038/ncomms11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benocci T, Aguilar-Pontes MV, Zhou M, Seiboth B, de Vries RP. Regulators of plant biomass degradation in ascomycetous fungi. Biotechnol Biofuels. 2017;10:152. doi: 10.1186/s13068-017-0841-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stricker AR, Mach RL, de Graaff LH. Regulation of transcription of cellulases- and hemicellulases-encoding genes in Aspergillus niger and Hypocrea jecorina (Trichoderma reesei) Appl Microbiol Biotechnol. 2008;78:211–220. doi: 10.1007/s00253-007-1322-0. [DOI] [PubMed] [Google Scholar]
- 33.Stappler E, Dattenböck C, Tisch D, Schmoll M. Analysis of light- and carbon-specific transcriptomes implicates a class of G-protein-coupled receptors in cellulose sensing. mSphere. 2017;2(3). 10.1128/mSphere.00089-17. [DOI] [PMC free article] [PubMed]
- 34.Kiesenhofer D, Mach-Aigner AR, Mach RL. Understanding the mechanism of carbon catabolite repression to increase protein production in filamentous fungi. In: Schmoll M, Dattenböck D, editors. Gene expression systems in fungi: advancements and applications. Cham: Springer; 2016. pp. 275–288. [Google Scholar]
- 35.Casas-Flores S, Herrera-Estrella A. The influence of light on the biology of Trichoderma. In: Mukherjee P, Horwitz BA, Singh US, Mukherjee M, Schmoll M, editors. Trichoderma—biology and applications. Wallingford: CAB International; 2013. pp. 43–66. [Google Scholar]
- 36.Schmoll M, Esquivel-Naranjo EU, Herrera-Estrella A. Trichoderma in the light of day—physiology and development. Fungal Genet Biol. 2010;47:909–916. doi: 10.1016/j.fgb.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmoll M, Dattenböck C, Carreras-Villasenor N, Mendoza-Mendoza A, Tisch D, Aleman MI, Baker SE, Brown C, Cervantes-Badillo MG, Cetz-Chel J, et al. The genomes of three uneven siblings: footprints of the lifestyles of three Trichoderma species. Microbiol Mol Biol Rev. 2016;80:205–327. doi: 10.1128/MMBR.00040-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ballario P, Macino G. White collar proteins: PASsing the light signal in Neurospora crassa. Trends Microbiol. 1997;5:458–462. doi: 10.1016/S0966-842X(97)01144-X. [DOI] [PubMed] [Google Scholar]
- 39.Casas-Flores S, Rios-Momberg M, Bibbins M, Ponce-Noyola P, Herrera-Estrella A. BLR-1 and BLR-2, key regulatory elements of photoconidiation and mycelial growth in Trichoderma atroviride. Microbiology. 2004;150:3561–3569. doi: 10.1099/mic.0.27346-0. [DOI] [PubMed] [Google Scholar]
- 40.Heintzen C, Loros JJ, Dunlap JC. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell. 2001;104:453–464. doi: 10.1016/s0092-8674(01)00232-x. [DOI] [PubMed] [Google Scholar]
- 41.Schmoll M. Light, stress, sex and carbon—the photoreceptor ENVOY as a central checkpoint in the physiology of Trichoderma reesei. Fungal Biol. 2017. 10.1016/j.funbio.2017.10.007. [DOI] [PubMed]
- 42.Schmoll M, Franchi L, Kubicek CP. Envoy, a PAS/LOV domain protein of Hypocrea jecorina (Anamorph Trichoderma reesei), modulates cellulase gene transcription in response to light. Eukaryot Cell. 2005;4:1998–2007. doi: 10.1128/EC.4.12.1998-2007.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castellanos F, Schmoll M, Martinez P, Tisch D, Kubicek CP, Herrera-Estrella A, Esquivel-Naranjo EU. Crucial factors of the light perception machinery and their impact on growth and cellulase gene transcription in Trichoderma reesei. Fungal Genet Biol. 2010;47:468–476. doi: 10.1016/j.fgb.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 44.Seibel C, Tisch D, Kubicek CP, Schmoll M. ENVOY is a major determinant in regulation of sexual development in Hypocrea jecorina (Trichoderma reesei) Eukaryot Cell. 2012;11:885–890. doi: 10.1128/EC.05321-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuster A, Kubicek CP, Friedl MA, Druzhinina IS, Schmoll M. Impact of light on Hypocrea jecorina and the multiple cellular roles of ENVOY in this process. BMC Genom. 2007;8:449. doi: 10.1186/1471-2164-8-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tisch D, Schmoll M. Targets of light signalling in Trichoderma reesei. BMC Genom. 2013;14:657. doi: 10.1186/1471-2164-14-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanchez-Arreguin A, Perez-Martinez AS, Herrera-Estrella A. Proteomic analysis of Trichoderma atroviride reveals independent roles for transcription factors BLR-1 and BLR-2 in light and darkness. Eukaryot Cell. 2012;11:30–41. doi: 10.1128/EC.05263-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bazafkan H, Beier S, Stappler E, Böhmdorfer S, Oberlerchner JT, Sulyok M, Schmoll M. SUB1 has photoreceptor dependent and independent functions in sexual development and secondary metabolism in Trichoderma reesei. Mol Microbiol. 2017;106(5):742–759. doi: 10.1111/mmi.13842. [DOI] [PubMed] [Google Scholar]
- 49.Tisch D, Schuster A, Schmoll M. Crossroads between light response and nutrient signalling: ENV1 and PhLP1 act as mutual regulatory pair in Trichoderma reesei. BMC Genom. 2014;15:425. doi: 10.1186/1471-2164-15-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Esquivel-Naranjo EU, Herrera-Estrella A. Enhanced responsiveness and sensitivity to blue light by blr-2 overexpression in Trichoderma atroviride. Microbiology. 2007;153:3909–3922. doi: 10.1099/mic.0.2007/007302-0. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez-Romero J, Hedtke M, Kastner C, Muller S, Fischer R. Fungi, hidden in soil or up in the air: light makes a difference. Annu Rev Microbiol. 2010;64:585–610. doi: 10.1146/annurev.micro.112408.134000. [DOI] [PubMed] [Google Scholar]
- 52.Chen CL, Kuo HC, Tung SY, Hsu PW, Wang CL, Seibel C, Schmoll M, Chen RS, Wang TF. Blue light acts as a double-edged sword in regulating sexual development of Hypocrea jecorina (Trichoderma reesei) PLoS ONE. 2012;7:e44969. doi: 10.1371/journal.pone.0044969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tisch D, Kubicek CP, Schmoll M. The phosducin-like protein PhLP1 impacts regulation of glycoside hydrolases and light response in Trichoderma reesei. BMC Genom. 2011;12:613. doi: 10.1186/1471-2164-12-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Monroy AA, Stappler E, Schuster A, Sulyok M, Schmoll M. A CRE1-regulated cluster is responsible for light dependent production of dihydrotrichotetronin in Trichoderma reesei. PLoS One. 2017;12:e0182530. doi: 10.1371/journal.pone.0182530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stappler E, Walton JD, Schmoll M. Abundance of secreted proteins of Trichoderma reesei is regulated by light of different intensities. Front Microbiol. 2017;8:2586. doi: 10.3389/fmicb.2017.02586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosales-Saavedra T, Esquivel-Naranjo EU, Casas-Flores S, Martinez-Hernandez P, Ibarra-Laclette E, Cortes-Penagos C, Herrera-Estrella A. Novel light-regulated genes in Trichoderma atroviride: a dissection by cDNA microarrays. Microbiology. 2006;152:3305–3317. doi: 10.1099/mic.0.29000-0. [DOI] [PubMed] [Google Scholar]
- 57.Esquivel-Naranjo EU, Garcia-Esquivel M, Medina-Castellanos E, Correa-Perez VA, Parra-Arriaga JL, Landeros-Jaime F, Cervantes-Chavez JA, Herrera-Estrella A. A Trichoderma atroviride stress-activated MAPK pathway integrates stress and light signals. Mol Microbiol. 2016;100:860–876. doi: 10.1111/mmi.13355. [DOI] [PubMed] [Google Scholar]
- 58.Wang M, Zhang M, Li L, Dong Y, Jiang Y, Liu K, Zhang R, Jiang B, Niu K, Fang X. Role of Trichoderma reesei mitogen-activated protein kinases (MAPKs) in cellulase formation. Biotechnol Biofuels. 2017;10:99. doi: 10.1186/s13068-017-0789-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang M, Zhao Q, Yang J, Jiang B, Wang F, Liu K, Fang X. A mitogen-activated protein kinase Tmk3 participates in high osmolarity resistance, cell wall integrity maintenance and cellulase production regulation in Trichoderma reesei. PLoS ONE. 2013;8:e72189. doi: 10.1371/journal.pone.0072189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lokhandwala J, Hopkins HC, Rodriguez-Iglesias A, Dattenböck C, Schmoll M, Zoltowski BD. Structural biochemistry of a fungal LOV domain photoreceptor reveals an evolutionarily conserved pathway integrating light and oxidative stress. Structure. 2015;23:116–125. doi: 10.1016/j.str.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 61.Lokhandwala J. Silverman YdlVRI, Hopkins HC, Britton CW, Rodriguez-Iglesias A, Bogomolni R, Schmoll M, Zoltowski BD: a native threonine coordinates ordered water to tune Light-Oxygen-Voltage (LOV) domain photocycle kinetics and osmotic stress signaling in Trichoderma reesei ENVOY. J Biol Chem. 2016;291:14839–14850. doi: 10.1074/jbc.M116.731448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmoll M, Zeilinger S, Mach RL, Kubicek CP. Cloning of genes expressed early during cellulase induction in Hypocrea jecorina by a rapid subtraction hybridization approach. Fungal Genet Biol. 2004;41:877–887. doi: 10.1016/j.fgb.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 63.Malzahn E, Ciprianidis S, Kaldi K, Schafmeier T, Brunner M. Photoadaptation in Neurospora by competitive interaction of activating and inhibitory LOV domains. Cell. 2010;142:762–772. doi: 10.1016/j.cell.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 64.Schwerdtfeger C, Linden H. Blue light adaptation and desensitization of light signal transduction in Neurospora crassa. Mol Microbiol. 2001;39:1080–1087. doi: 10.1046/j.1365-2958.2001.02306.x. [DOI] [PubMed] [Google Scholar]
- 65.Schuster A, Kubicek CP, Schmoll M. Dehydrogenase GRD1 represents a novel component of the cellulase regulon in Trichoderma reesei (Hypocrea jecorina) Appl Environ Microbiol. 2011;77:4553–4563. doi: 10.1128/AEM.00513-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He Q, Liu Y. Molecular mechanism of light responses in Neurospora: from light-induced transcription to photoadaptation. Genes Dev. 2005;19:2888–2899. doi: 10.1101/gad.1369605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gyalai-Korpos M, Nagy G, Mareczky Z, Schuster A, Reczey K, Schmoll M. Relevance of the light signaling machinery for cellulase expression in Trichoderma reesei (Hypocrea jecorina) BMC Res Notes. 2010;3:330. doi: 10.1186/1756-0500-3-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ivanova C, Ramoni J, Aouam T, Frischmann A, Seiboth B, Baker SE, Le Crom S, Lemoine S, Margeot A, Bidard F. Genome sequencing and transcriptome analysis of Trichoderma reesei QM9978 strain reveals a distal chromosome translocation to be responsible for loss of vib1 expression and loss of cellulase induction. Biotechnol Biofuels. 2017;10:209. doi: 10.1186/s13068-017-0897-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Friedl M. Global and specific effects of light on Hypocrea/Trichoderma (master thesis). Vienna University of Technology, Vienna; 2006.
- 70.Bayram O, Krappmann S, Ni M, Bok JW, Helmstaedt K, Valerius O, Braus-Stromeyer S, Kwon NJ, Keller NP, Yu JH, Braus GH. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science. 2008;320:1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]
- 71.Karimi Aghcheh R, Nemeth Z, Atanasova L, Fekete E, Paholcsek M, Sandor E, Aquino B, Druzhinina IS, Karaffa L, Kubicek CP. The VELVET A orthologue VEL1 of Trichoderma reesei regulates fungal development and is essential for cellulase gene expression. PLoS ONE. 2014;9:e112799. doi: 10.1371/journal.pone.0112799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seiboth B, Karimi RA, Phatale PA, Linke R, Hartl L, Sauer DG, Smith KM, Baker SE, Freitag M, Kubicek CP. The putative protein methyltransferase LAE1 controls cellulase gene expression in Trichoderma reesei. Mol Microbiol. 2012;84:1150–1164. doi: 10.1111/j.1365-2958.2012.08083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bazafkan H, Dattenböck C, Böhmdorfer S, Tisch D, Stappler E, Schmoll M. Mating type dependent partner sensing as mediated by VEL1 in Trichoderma reesei. Mol Microbiol. 2015;96:1103–1118. doi: 10.1111/mmi.12993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bazafkan H, Dattenbock C, Stappler E, Beier S, Schmoll M. Interrelationships of VEL1 and ENV1 in light response and development in Trichoderma reesei. PLoS ONE. 2017;12:e0175946. doi: 10.1371/journal.pone.0175946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cabrera-Vera TM, Vanhauwe J, Thomas TO, Medkova M, Preininger A, Mazzoni MR, Hamm HE. Insights into G protein structure, function, and regulation. Endocr Rev. 2003;24:765–781. doi: 10.1210/er.2000-0026. [DOI] [PubMed] [Google Scholar]
- 76.Li L, Borkovich KA. GPR-4 is a predicted G-protein-coupled receptor required for carbon source-dependent asexual growth and development in Neurospora crassa. Eukaryot Cell. 2006;5:1287–1300. doi: 10.1128/EC.00109-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schmoll M, Schuster A, do Nascimento Silva R, Kubicek CP. The G-alpha protein GNA3 of Hypocrea jecorina (anamorph Trichoderma reesei) regulates cellulase gene expression in the presence of light. Eukaryot Cell. 2009;8:410–420. doi: 10.1128/EC.00256-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seibel C, Gremel G, Silva RD, Schuster A, Kubicek CP, Schmoll M. Light-dependent roles of the G-protein subunit GNA1 of Hypocrea jecorina (anamorph Trichoderma reesei) BMC Biol. 2009;7:58. doi: 10.1186/1741-7007-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Willardson BM, Howlett AC. Function of phosducin-like proteins in G protein signaling and chaperone-assisted protein folding. Cell Signal. 2007;19:2417–2427. doi: 10.1016/j.cellsig.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hicks JK, Bahn YS, Heitman J. Pde1 phosphodiesterase modulates cyclic AMP levels through a protein kinase A-mediated negative feedback loop in Cryptococcus neoformans. Eukaryot Cell. 2005;4:1971–1981. doi: 10.1128/EC.4.12.1971-1981.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang L, Griffiths K, Jr, Zhang YH, Ivey FD, Hoffman CS. Schizosaccharomyces pombe adenylate cyclase suppressor mutations suggest a role for cAMP phosphodiesterase regulation in feedback control of glucose/cAMP signaling. Genetics. 2005;171:1523–1533. doi: 10.1534/genetics.105.047233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Houslay MD, Adams DR. PDE4 cAMP phosphodiesterases: modular enzymes that orchestrate signalling cross-talk, desensitization and compartmentalization. Biochem J. 2003;370:1–18. doi: 10.1042/BJ20021698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kolarova N, Haplova J, Gresik M. Light-activated adenyl cyclase from Trichoderma viride. FEMS Microbiol Lett. 1992;72:275–278. doi: 10.1016/0378-1097(92)90474-3. [DOI] [PubMed] [Google Scholar]
- 84.Sestak S, Farkas V. Metabolic regulation of endoglucanase synthesis in Trichoderma reesei: participation of cyclic AMP and glucose-6-phosphate. Can J Microbiol. 1993;39:342–347. doi: 10.1139/m93-048. [DOI] [PubMed] [Google Scholar]
- 85.Gresik M, Kolarova N, Farkas V. Membrane potential, ATP, and cyclic AMP changes induced by light in Trichoderma viride. Exp Mycol. 1988;12:295–301. [Google Scholar]
- 86.Casas-Flores S, Rios-Momberg M, Rosales-Saavedra T, Martinez-Hernandez P, Olmedo-Monfil V, Herrera-Estrella A. Cross talk between a fungal blue-light perception system and the cyclic AMP signaling pathway. Eukaryot Cell. 2006;5:499–506. doi: 10.1128/EC.5.3.499-506.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schuster A, Tisch D, Seidl-Seiboth V, Kubicek CP, Schmoll M. Roles of protein kinase A and adenylate cyclase in light-modulated cellulase regulation in Trichoderma reesei. Appl Environ Microbiol. 2012;78:2168–2178. doi: 10.1128/AEM.06959-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang G, Chen S, Li S, Cha J, Long C, Li L, He Q, Liu Y. Protein kinase A and casein kinases mediate sequential phosphorylation events in the circadian negative feedback loop. Genes Dev. 2007;21:3283–3295. doi: 10.1101/gad.1610207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tisch D, Kubicek CP, Schmoll M. New insights into the mechanism of light modulated signaling by heterotrimeric G-proteins: ENVOY acts on gna1 and gna3 and adjusts cAMP levels in Trichoderma reesei (Hypocrea jecorina) Fungal Genet Biol. 2011;48:631–640. doi: 10.1016/j.fgb.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Peter GJ, Düring L, Ahmed A. Carbon catabolite repression regulates amino acid permeases in Saccharomyces cerevisiae via the TOR signaling pathway. J Biol Chem. 2006;281:5546–5552. doi: 10.1074/jbc.M513842200. [DOI] [PubMed] [Google Scholar]
- 91.Portnoy T, Margeot A, Linke R, Atanasova L, Fekete E, Sandor E, Hartl L, Karaffa L, Druzhinina IS, Seiboth B, et al. The CRE1 carbon catabolite repressor of the fungus Trichoderma reesei: a master regulator of carbon assimilation. BMC Genom. 2011;12:269. doi: 10.1186/1471-2164-12-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schmoll M, Tian C, Sun J, Tisch D, Glass NL. Unravelling the molecular basis for light modulated cellulase gene expression—the role of photoreceptors in Neurospora crassa. BMC Genom. 2012;13:127. doi: 10.1186/1471-2164-13-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gremel G, Dorrer M, Schmoll M. Sulphur metabolism and cellulase gene expression are connected processes in the filamentous fungus Hypocrea jecorina (anamorph Trichoderma reesei) BMC Microbiol. 2008;8:174. doi: 10.1186/1471-2180-8-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pakula TM, Nygren H, Barth D, Heinonen M, Castillo S, Penttilä M, Arvas M. Genome wide analysis of protein production load in Trichoderma reesei. Biotechnol Biofuels. 2016;9:132. doi: 10.1186/s13068-016-0547-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lengeler KB, Davidson RC, D’Souza C, Harashima T, Shen WC, Wang P, Pan X, Waugh M, Heitman J. Signal transduction cascades regulating fungal development and virulence. Microbiol Mol Biol Rev. 2000;64:746–785. doi: 10.1128/mmbr.64.4.746-785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Aro N, Pakula T, Penttilä M. Transcriptional regulation of plant cell wall degradation by filamentous fungi. FEMS Microbiol Rev. 2005;29:719–739. doi: 10.1016/j.femsre.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 97.Znameroski EA, Glass NL. Using a model filamentous fungus to unravel mechanisms of lignocellulose deconstruction. Biotechnol Biofuels. 2013;6:6. doi: 10.1186/1754-6834-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Arvas M, Pakula T, Smit B, Rautio J, Koivistoinen H, Jouhten P, Lindfors E, Wiebe M, Penttila M, Saloheimo M. Correlation of gene expression and protein production rate—a system wide study. BMC Genom. 2011;12:616. doi: 10.1186/1471-2164-12-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Saloheimo M, Paloheimo M, Hakola S, Pere J, Swanson B, Nyyssonen E, Bhatia A, Ward M, Penttilä M. Swollenin, a Trichoderma reesei protein with sequence similarity to the plant expansins, exhibits disruption activity on cellulosic materials. Eur J Biochem. 2002;269:4202–4211. doi: 10.1046/j.1432-1033.2002.03095.x. [DOI] [PubMed] [Google Scholar]
- 100.Li XL, Spanikova S, de Vries RP, Biely P. Identification of genes encoding microbial glucuronoyl esterases. FEBS Lett. 2007;581:4029–4035. doi: 10.1016/j.febslet.2007.07.041. [DOI] [PubMed] [Google Scholar]
- 101.Gacek A, Strauss J. The chromatin code of fungal secondary metabolite gene clusters. Appl Microbiol Biotechnol. 2012;95:1389–1404. doi: 10.1007/s00253-012-4208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, Chapman J, Chertkov O, Coutinho PM, Cullen D, et al. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina) Nat Biotechnol. 2008;26:553–560. doi: 10.1038/nbt1403. [DOI] [PubMed] [Google Scholar]
- 103.Derntl C, Rassinger A, Srebotnik E, Mach RL, Mach-Aigner AR. Identification of the main regulator responsible for synthesis of the typical yellow pigment produced by Trichoderma reesei. Appl Environ Microbiol. 2016;82:6247–6257. doi: 10.1128/AEM.01408-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Derntl C, Kluger B, Bueschl C, Schuhmacher R, Mach RL, Mach-Aigner AR. Transcription factor Xpp1 is a switch between primary and secondary fungal metabolism. Proc Natl Acad Sci USA. 2017;114:E560–E569. doi: 10.1073/pnas.1609348114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Metz B, de Vries RP, Polak S, Seidl V, Seiboth B. The Hypocrea jecorina (syn. Trichoderma reesei) lxr1 gene encodes a D-mannitol dehydrogenase and is not involved in L-arabinose catabolism. FEBS Lett. 2009;583:1309–1313. doi: 10.1016/j.febslet.2009.03.027. [DOI] [PubMed] [Google Scholar]
- 106.Antonieto AC, dos Santos Castro L, Silva-Rocha R, Persinoti GF, Silva RN. Defining the genome-wide role of CRE1 during carbon catabolite repression in Trichoderma reesei using RNA-Seq analysis. Fungal Genet Biol. 2014;73:93–103. doi: 10.1016/j.fgb.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 107.Salo O, Guzman-Chavez F, Ries MI, Lankhorst PP, Bovenberg RA, Vreeken RJ, Driessen AJ. Identification of a polyketide synthase involved in sorbicillin biosynthesis by Penicillium chrysogenum. Appl Environ Microbiol. 2016;82:3971–3978. doi: 10.1128/AEM.00350-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zeilinger S, Mach RL, Kubicek CP. Two adjacent protein binding motifs in the cbh2 (cellobiohydrolase II- encoding) promoter of the fungus Hypocrea jecorina (Trichoderma reesei) cooperate in the induction by cellulose. J Biol Chem. 1998;273:34463–34471. doi: 10.1074/jbc.273.51.34463. [DOI] [PubMed] [Google Scholar]
- 109.Saloheimo A, Aro N, Ilmen M, Penttila M. Isolation of the ace1 gene encoding a Cys(2)-His(2) transcription factor involved in regulation of activity of the cellulase promoter cbh1 of Trichoderma reesei. J Biol Chem. 2000;275:5817–5825. doi: 10.1074/jbc.275.8.5817. [DOI] [PubMed] [Google Scholar]
- 110.Zeilinger S, Schmoll M, Pail M, Mach RL, Kubicek CP. Nucleosome transactions on the Hypocrea jecorina (Trichoderma reesei) cellulase promoter cbh2 associated with cellulase induction. Mol Genet Genomics. 2003;270:46–55. doi: 10.1007/s00438-003-0895-2. [DOI] [PubMed] [Google Scholar]
- 111.Leonhartsberger M. Untersuchungen an einem cis-wirkenden Element der Cellulaseinduktion (master thesis, german). Vienna University of Technology, Vienna; 1999.
- 112.Froehlich AC, Liu Y, Loros JJ, Dunlap JC. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science. 2002;297:815–819. doi: 10.1126/science.1073681. [DOI] [PubMed] [Google Scholar]
- 113.Schuster A, Schmoll M. Heterotrimeric G-protein signaling and light response: two signaling pathways coordinated for optimal adjustment to nature. Commun Integr Biol. 2009;2:308–310. doi: 10.4161/cib.2.4.8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chovanec P, Hudecova D, Varecka L. Vegetative growth, aging- and light-induced conidiation of Trichoderma viride cultivated on different carbon sources. Folia Microbiol (Praha) 2001;46:417–422. doi: 10.1007/BF02814432. [DOI] [PubMed] [Google Scholar]
- 115.Carlile MJ. The photobiology of fungi. Ann Rev Plant Physiol. 1965;16:175–202. [Google Scholar]
- 116.Druzhinina IS, Schmoll M, Seiboth B, Kubicek CP. Global carbon utilization profiles of wild-type, mutant, and transformant strains of Hypocrea jecorina. Appl Environ Microbiol. 2006;72:2126–2133. doi: 10.1128/AEM.72.3.2126-2133.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Friedl MA, Schmoll M, Kubicek CP, Druzhinina IS. Photostimulation of Hypocrea atroviridis growth occurs due to a cross-talk of carbon metabolism, blue light receptors and response to oxidative stress. Microbiology. 2008;154:1229–1241. doi: 10.1099/mic.0.2007/014175-0. [DOI] [PubMed] [Google Scholar]
- 118.Smith KM, Sancar G, Dekhang R, Sullivan CM, Li S, Tag AG, Sancar C, Bredeweg EL, Priest HD, McCormick RF, et al. Transcription factors in light and circadian clock signaling networks revealed by genome wide mapping of direct targets for Neurospora white collar complex. Eukaryot Cell. 2010;9:1549–1556. doi: 10.1128/EC.00154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Häkkinen M, Valkonen MJ, Westerholm-Parvinen A, Aro N, Arvas M, Vitikainen M, Penttila M, Saloheimo M, Pakula TM. Screening of candidate regulators for cellulase and hemicellulase production in Trichoderma reesei and identification of a factor essential for cellulase production. Biotechnol Biofuels. 2014;7:14. doi: 10.1186/1754-6834-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zheng F, Cao Y, Wang L, Lv X, Meng X, Zhang W, Chen G, Liu W. The mating type locus protein MAT1-2-1 of Trichoderma reesei interacts with Xyr1 and regulates cellulase gene expression in response to light. Sci Rep. 2017;7:17346. doi: 10.1038/s41598-017-17439-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lehmann L, Ronnest NP, Jorgensen CI, Olsson L, Stocks SM, Jorgensen HS, Hobley T. Linking hydrolysis performance to Trichoderma reesei cellulolytic enzyme profile. Biotechnol Bioeng. 2016;113:1001–1010. doi: 10.1002/bit.25871. [DOI] [PubMed] [Google Scholar]
- 122.Stappler E, Rodriguez-Iglesias A, Bazafkan H, Li G, Schmoll M. Relevance of signal transduction pathways for efficient gene expression in fungi. In: Schmoll M, Dattenböck C, editors. Gene expression systems in fungi: advancements and applications. Cham: Springer; 2016. pp. 309–334. [Google Scholar]
- 123.Salinas F, Rojas V, Delgado V, Agosin E, Larrondo LF. Optogenetic switches for light-controlled gene expression in yeast. Appl Microbiol Biotechnol. 2017;101:2629–2640. doi: 10.1007/s00253-017-8178-8. [DOI] [PubMed] [Google Scholar]
- 124.Zhang G, Liu P, Wei W, Wang X, Wei D, Wang W. A light-switchable bidirectional expression system in filamentous fungus Trichoderma reesei. J Biotechnol. 2016;240:85–93. doi: 10.1016/j.jbiotec.2016.11.003. [DOI] [PubMed] [Google Scholar]


