ABSTRACT
Ethanolamine is a ubiquitous and essential molecule within a host. Significantly, bacterial pathogens exploit ethanolamine during infection to promote growth and regulate virulence. The ethanolamine permease EutH is dispensable for growth in vitro under standard conditions, whereas EutH is required for ethanolamine utilization at low pH. These findings suggested a model in which EutH facilitates diffusion of ethanolamine into the bacterial cell in acidic environments. To date, the ecological significance of this model has not been thoroughly investigated, and the importance of EutH to bacterial growth under physiologically relevant conditions is not known. During infection, immune cells internalize invading bacteria within an acidic, nutrient-depleted vacuole called the phagosome. Here, we investigated the hypothesis that EutH promotes bacterial survival following phagocytosis. Our findings indicate that EutH is important for survival and replication of the facultative intracellular pathogens Salmonella enterica serovar Typhimurium and Listeria monocytogenes during prolonged or transient exposure to the phagosome, respectively. Furthermore, in agreement with EutH being important in the acidic environment, neutralization of the vacuole abolished the requirement for EutH. Significantly, consistent with a role for EutH in promoting intramacrophage survival, EutH was not required during S. Typhimurium local intestinal infection but specifically conferred an advantage upon dissemination to peripheral organs. These findings reveal a physiologically relevant and conserved role for EutH in spatiotemporal niche adaptation during infection.
KEYWORDS: Listeria, Salmonella, ethanolamine, macrophage, pathogenesis, vacuole
INTRODUCTION
Ethanolamine (EA) is a ubiquitous molecule within a host as a base constituent of phosphatidylethanolamine (PE), an abundant lipid in mammalian and bacterial cell membranes. Additionally, free EA is present within cells and in bodily fluids (1–5). In mammals, EA-containing lipids as well as EA itself are essential for health by modulating immune processes, energy balance, cell growth, regulated cell death pathways, and cardioprotection (4, 6–12). In bacteria, EA plays a dynamic role as a metabolite that promotes growth of pathogens during infection as well as a signal that modulates virulence (13–20). Genes encoding EA metabolism and signaling are carried in the EA utilization (eut) locus. eut loci have been identified in nearly 100 fully sequenced bacterial genomes and can be generally classified according to the number of genes within a respective locus (short versus long) (21). The short loci may contain only the genes encoding the EA-ammonia lyase EutBC, which catalyzes the first step in the breakdown of EA. In contrast, the long loci may also encode autoregulatory components, auxiliary proteins involved in EA catabolism, as well as genes encoding a microcompartment (22). Significantly, most short and long eut loci encode an EA permease—EutH or Eat (21). In vitro studies have shown that EutH and Eat are dispensable for bacterial growth in vitro under standard conditions (23–27), whereas EutH is required for EA utilization at low pH, suggesting a model in which EutH facilitates EA diffusion into the bacterial cell in acidic environments (25). To date, an ecologically relevant role for EutH in EA utilization and/or signaling has not been established.
Salmonella enterica serovar Typhimurium and Listeria monocytogenes are facultative intracellular pathogens that cause acute or mild gastroenteritis, respectively, and can cause invasive systemic disease in susceptible individuals. Importantly, both S. Typhimurium and L. monocytogenes exploit EA to promote pathogenesis (13, 18, 20, 28). In S. Typhimurium, the eut locus contains 17 genes, including the transcription factor EutR (23, 29, 30) (Fig. 1A). In response to EA and vitamin B12, EutR directly activates eut expression to promote EA utilization (16, 29). In the intestine, S. Typhimurium exploits EA as a noncompetitive metabolite to sidestep nutritional competition from the microbiota and establish infection (13, 20). Subsequently, S. Typhimurium invades the epithelial barrier and penetrates to the lamina propria, where S. Typhimurium is internalized by macrophages. In the intramacrophage environment, EutR promotes S. Typhimurium survival and replication by directly activating expression of ssrB (13), which encodes SsrB, the master regulator of Salmonella pathogenicity island 2 (SPI-2) (31–33). SPI-2 encodes the type 3 secretion system 2 (T3SS-2) and effectors that convert the phagosome into a replicative niche called the Salmonella-containing vacuole (SCV) (34–37). Significantly, EutR-mediated regulation of SPI-2 during systemic infection is independent of EA metabolism (13), suggesting a dynamic role for EutR in S. Typhimurium host colonization. In L. monocytogenes, the eut locus contains at least 19 genes (Fig. 1B) that are regulated by a two-component system, EutVW, and a riboswitch-containing small RNA (sRNA), Rli55/EutX, that functions as an antiterminator (18, 38, 39). Similar to S. Typhimurium, EA metabolism also promotes L. monocytogenes host colonization, as disruption of eutB impacts growth of L. monocytogenes in HeLa cells as well as during systemic infection (18, 28). Although the disease progressions of S. Typhimurium and L. monocytogenes share common features, L. monocytogenes uses a distinct strategy for replicating within host cells. L. monocytogenes secretes the pore-forming toxin listeriolysin O (LLO) and the phospholipases C to escape the phagosome and replicate in the cytosol (40–44). Regardless of prolonged or transient exposure to the vacuole, S. Typhimurium and L. monocytogenes must be able to withstand the acidic, nutrient-limiting environment of the phagosome to cause disease. Here, we provide evidence that the EA permease EutH contributes to fitness of these pathogens during macrophage infection. Furthermore, in agreement with EutH being important in the acidic environment, neutralization of the vacuole abolished the requirement for EutH. Additionally, although EutH was dispensable during S. Typhimurium local intestinal infection, EutH specifically conferred an advantage during dissemination to peripheral organs. These findings reveal a conserved and physiologically relevant role for EutH in spatiotemporal niche adaptation during infection.
FIG 1.
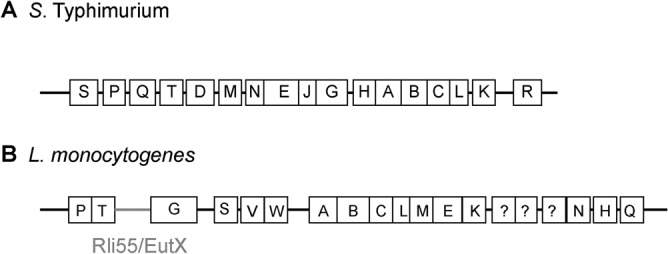
Schematic of the eut locus in (A) S. Typhimurium or (B) L. monocytogenes.
RESULTS AND DISCUSSION
EutH promotes S. Typhimurium survival with macrophages.
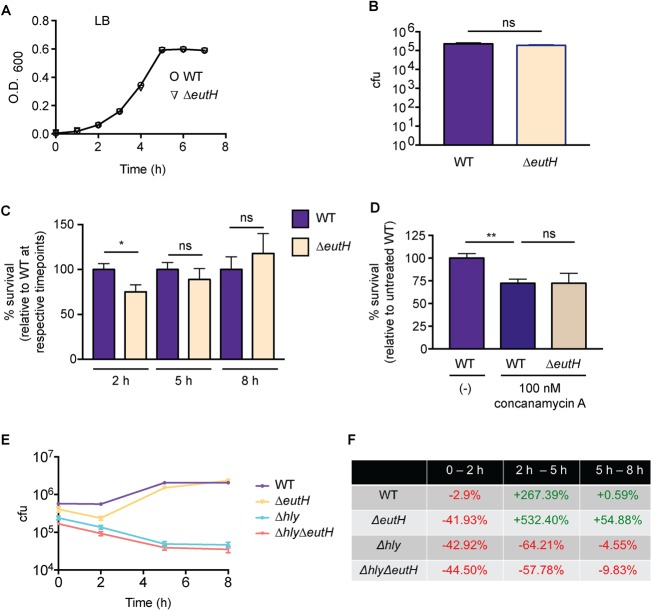
eutH was originally identified as an open reading frame within the eut locus (27). Sequence analysis and structure predictions indicated that EutH was a hydrophobic protein containing at least six transmembrane helices, and thus EutH was hypothesized to function as a permease (27). Although deletion of eutH did not render S. Typhimurium unable to utilize EA in vitro, this deletion resulted in mild attenuation during intraperitoneal (i.p.) mouse infection, suggesting that EutH and EA were important for S. Typhimurium replication in vivo (27). Subsequent studies confirmed that eutH was dispensable for bacterial growth in vitro under standard conditions (23, 25) (Fig. 2A) but also showed that EA enters cells in a charge-dependent manner (25). These latter findings suggested that pH may influence the requirement for EutH in EA utilization (25). Consistent with this idea, a ΔeutH S. Typhimurium strain was unable to grow on EA at low pH (with the addition of 20 or 41 mM EA to the culture medium) (25). These data suggested a model in which EutH facilitates EA diffusion into the bacterial cell in acidic environments (25), which we hypothesize includes the phagosome of macrophages. EA signaling through the transcription factor EutR promotes S. Typhimurium fitness within macrophages (Fig. 2B) (13); therefore, we examined the importance of EutH to S. Typhimurium survival during macrophage infection. In accordance with the proposed model, the ΔeutH S. Typhimurium strain was similarly attenuated compared to the ΔeutR or ΔeutH ΔeutR strains following infection of RAW macrophages or bone marrow-derived macrophages (BMDMs), respectively (Fig. 2C and D). Furthermore, trans-complementation with eutS-H expressed from the native eut P1 promoter restored survival of the ΔeutH strain within BMDMs to near-wild-type (WT) levels (Fig. 2E). In agreement with our previous findings (13), the macrophage survival phenotype was dependent on the T3SS-2 (Fig. 2F). Importantly, there were no differences in phagocytic uptake of the ΔeutH or ΔeutH ΔeutR strains compared to the WT (Fig. 2G) or during in vitro growth in a macrophage-like medium (SPI-2 inducing medium) (Fig. 2H).
FIG 2.
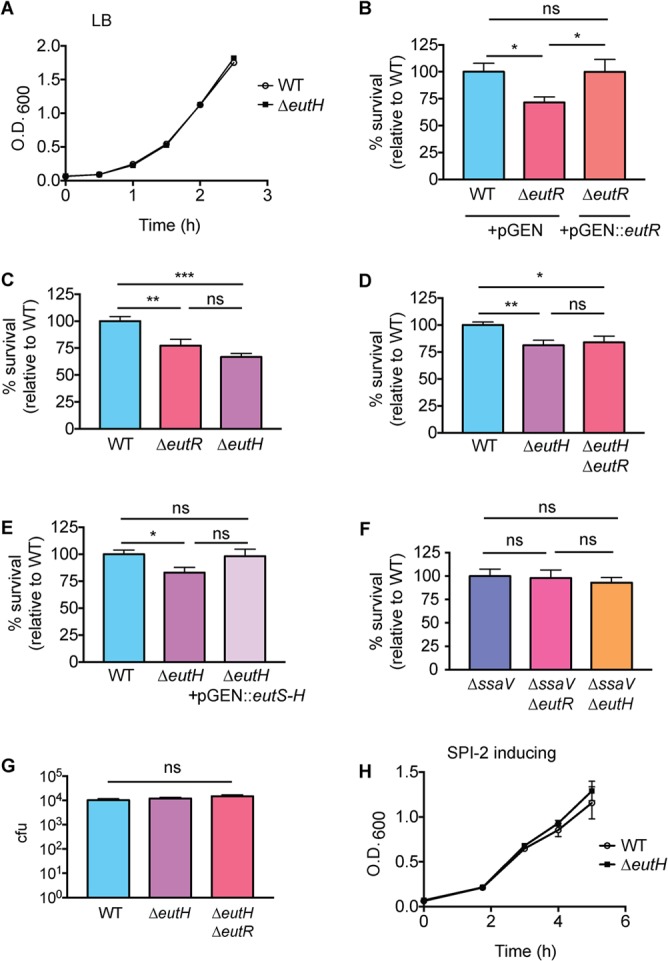
EutH promotes S. Typhimurium survival within macrophages. (A) In vitro growth curve of the WT S. Typhimurium (SL1344) and the ΔeutH (CJA052) strains grown in LB broth. Each data point shows the average from three biological replicates. Error bars represent the mean ± standard deviation (SD). (B) Intramacrophage survival and replication of WT (CJA034), ΔeutR (CJA032), or eutR complemented (CJA033) strains in peritoneal exudate macrophages after 7 h postphagocytosis. n = 9 replicates per strain. (C) Intramacrophage survival and replication of WT (AJK61), ΔeutR (CJA023), or ΔeutH (CJA043) S. Typhimurium strains in RAW macrophages after 5 h postphagocytosis. n = 18 replicates per strain. (D) Intramacrophage survival and replication of WT S. Typhimurium (AJK61), ΔeutH (CJA043), and ΔeutH ΔeutR (CJA168) strains after 5 h postphagocytosis in BMDMs. n = 15 replicates per strain. (E) Intramacrophage survival and replication of WT S. Typhimurium (CJA034), ΔeutH (CJA087), and eutH (CJA184) complemented strains after 5 h postphagocytosis in BMDMs. n = 18 replicates per strain. (F) Intramacrophage survival and replication of ΔssaV, ΔssaV ΔeutR (CJA064), and ΔssaV ΔeutH (CJA172) S. Typhimurium strains after 5 h postphagocytosis in BMDMs. ssaV encodes an essential component of the T3SS-2, and the ΔssaV strain is completely defective for T3SS-2-mediated secretion (48). n = 9 replicates per strain. (G) Initial phagocytosis of S. Typhimurium WT (AJK61), ΔeutH (CJA043), and ΔeutH ΔeutR (CJA168) strains at time zero (see Materials and Methods). n = 24 replicates per strain. (H) In vitro growth curve of the S. Typhimurium WT (SL1344) and ΔeutH (CJA052) strains grown in SPI-2 inducing medium. Each data point shows the average from three biological replicates. Error bars represent the mean ± SD. For panels B to G, the mean and standard error of the mean (SEM) are shown. *, P ≤ 0.05; **, P ≤ 0.005; ***, P ≤ 0.0005. P values of >0.05 are not significant (ns).
As an alternative method to demonstrate the importance of EutH to EA-enhanced survival, we infected RAW macrophages and then supplemented the medium with EA following gentamicin treatment. The latter step ensured that the added EA was restricted to internalized bacteria. EA supplementation augmented WT S. Typhimurium survival within macrophages in a concentration-dependent manner (Fig. 3A). In contrast, EA addition did not affect survival of the ΔeutR and ΔeutH strains (Fig. 3B and C), and EA enhanced survival of the complemented ΔeutH strain (Fig. 3D). Collectively, these data indicate that EutH is required for EA-dependent survival and replication within macrophages.
FIG 3.
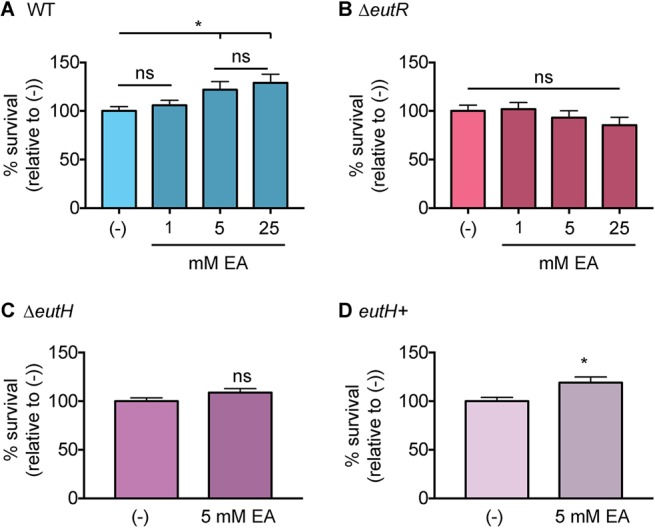
Bacterial survival following EA supplementation. (A) Intramacrophage survival and replication of WT S. Typhimurium (AJK61) after 5 h postphagocytosis in RAW macrophages without or with EA supplementation. n = 9. (B) Intramacrophage survival and replication of ΔeutR S. Typhimurium (CJA023) after 5 h postphagocytosis in RAW macrophages without or with EA supplementation. n = 9. (C) Intramacrophage survival and replication of ΔeutH S. Typhimurium (CJA043) after 5 h postphagocytosis in RAW macrophages without or with EA supplementation. n = 9. (D) Intramacrophage survival and replication of the eutH complemented strain (eutH+) of S. Typhimurium (CJA184) after 5 h postphagocytosis in RAW macrophages without or with EA supplementation. n = 12. For all values, the mean and SEM are shown. *, P ≤ 0.05. P values of >0.05 are not significant (ns).
Vacuole acidification drives the requirement for EutH.
Uncharged EA freely diffuses across the bacterial membrane, whereas protonated EA cannot. The pH of the environment influences the ratio of uncharged to charged EA. At low pH, the protonated form of EA predominates, and EutH facilitates diffusion of EA into the bacterial cell (25). Therefore, to further test the proposed model that EutH mediates EA diffusion specifically at low pH, we examined how neutralization of the vacuole affected the requirement for EutH in enhancing S. Typhimurium survival within macrophages. For the initial experiments, BMDMs were treated with concanamycin A, an inhibitor of vacuolar ATPases that prevents acidification. Consistent with our previous data, in the untreated BMDMs, the ΔeutH strain was significantly less fit than the WT. However, after concanamycin A treatment, both strains were significantly attenuated compared to infection of untreated BMDMs and survived similarly to each other (Fig. 4A). Acidification of the SCV is required for expression of SPI-2 and formation of the T3SS-2 (45–50), and complete ablation of vacuole acidification with concanamycin A renders S. Typhimurium susceptible to macrophage killing (47). Thus, these data further underscore a role for EA signaling in enhancing T3SS-2-mediated survival and replication. Next, we repeated these experiments using NH4Cl, a mild base. Treatment with NH4Cl did not impact replication of WT S. Typhimurium during BMDM infection; however, neutralization using 10 or 50 mM NH4Cl restored survival of the ΔeutH strain to nearly WT levels (Fig. 4B). These findings reveal that EutH contributes to S. Typhimurium intramacrophage survival and replication specifically in response to vacuole acidification.
FIG 4.
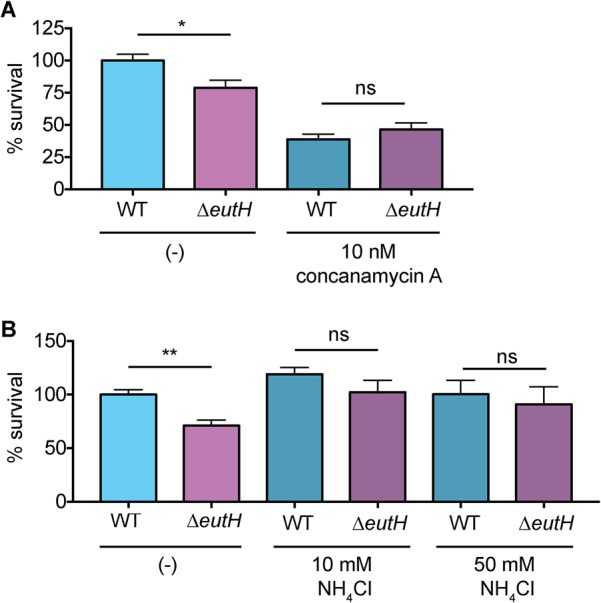
EutH enhances S. Typhimurium fitness in the acidified SCV. (A) Intramacrophage survival and replication of the WT (AJK61) or ΔeutH (CJA043) strain in BMDMs without or with concanamycin A treatment. n = 9 replicates per strain per condition. (B) Intramacrophage survival and replication of the WT (AJK61) or ΔeutH (CJA043) strain in BMDMs without or with NH4Cl. n = 9 replicates per strain per condition. The mean and SEM are shown. *, P ≤ 0.05; **, P ≤ 0.005. P values of >0.05 are not significant (ns).
EutH promotes L. monocytogenes vacuole adaptation.
To examine whether EutH plays a more general role in bacterial fitness within the acidic environment, we assessed the requirement for EutH in L. monocytogenes during in vitro growth as well as during BMDM infection. Similar to S. Typhimurium, EutH did not impact L. monocytogenes growth in vitro (Fig. 5A) or uptake by macrophages (Fig. 5B). Although L. monocytogenes can begin to escape the vacuole within 30 min following internalization (51, 52), the majority of L. monocytogenes bacteria are contained within the vacuole during the first 2 to 3 h following uptake (53, 54) and then escape to the cytosol within 5 to 10 h (54–57). Therefore, we assessed the contribution of EutH to L. monocytogenes survival and replication at 2, 5, and 8 h postinfection (hpi) as time points reflective of vacuolar containment (2 h) and cytosolic replication (5 and 8 h). At 2 hpi, the ΔeutH L. monocytogenes strain was significantly attenuated compared to the WT (Fig. 5C). Correlating with increased access to the neutral, nutrient-replete cytosol (58), similar numbers of WT and ΔeutH cells were recovered as infection progressed (Fig. 5C). Consistent with the S. Typhimurium neutralization assays, treatment of macrophages with concanamycin A ablated attenuation of the eutH L. monocytogenes strain in comparison to the WT strain (Fig. 5D).
FIG 5.
EutH contributes to survival of WT L. monocytogenes during macrophage infection. (A) In vitro growth curve of WT (10403S) and ΔeutH (VK01) L. monocytogenes strains grown in LB broth. Each data point shows the average from three biological replicates. Error bars represent the mean ± SD. (B) Initial phagocytosis of WT (10403S) and ΔeutH (VK01) L. monocytogenes strains at time zero (see Materials and Methods). (C) Intramacrophage survival and replication of WT (10403S) or ΔeutH (VK01) L. monocytogenes strains in BMDMs at the indicated times postphagocytosis. 2 hpi, n = 18 replicates per strain; 5 hpi, n = 12 replicates per strain; 8 hpi, n = 9 replicates per strain. For each time point, WT survival and replication were set to 100%. (D) Intramacrophage survival and replication of WT (10403S) or ΔeutH (VK01) L. monocytogenes strains in BMDMs without or with concanamycin A treatment at 2 h postphagocytosis. n = 9 replicates per strain per condition. (E) Intramacrophage CFU per milliliter of the WT (10403S), ΔeutH (VK01), Δhly (DP-2161), or Δhly ΔeutH (VK06) strains at time zero and 2, 5, and 8 h postphagocytosis. n = 12 to 18 replicates per strain per time point. (F) The percentage of change in intramacrophage survival and replication over time is calculated from data shown in panel E. Each percentage is indicative of the change between the two time points listed and was calculated as (later time point – earlier time point)/(earlier time point). For panels B to E, the mean and SEM are shown. *, P ≤ 0.05; **, P ≤ 0.005. P values of >0.05 are not significant (ns).
LLO is essential for L. monocytogenes escape from the vacuole (43, 59). Therefore, to further interrogate the importance of EutH to L. monocytogenes fitness within the vacuole, we assessed time-dependent survival and replication of L. monocytogenes carrying a deletion of hly (that encodes LLO) in the WT or ΔeutH background strains. At 2 hpi, we again measured a significant decrease in bacterial recovery (CFU) of the ΔeutH strain compared to the WT, and no significant differences in CFU were measured at 5 or 8 hpi (Fig. 5E). Notably, the ΔeutH, Δhly, and Δhly ΔeutH strains displayed similar survival defects within the first 2 hpi (Fig. 5E and F). In agreement with previous studies (43, 60), our data show that the Δhly strain did not replicate within the macrophages, and we did not measure any further attenuation in the context of the eutH deletion (Fig. 5E and F). A caveat to this experiment is that LLO functions in many aspects of L. monocytogenes pathogenesis (61), such as intracellular growth and survival within the phagosome (43, 60); therefore, further attenuation may not be detectable. Alternatively, EutH- and LLO-dependent survival may be functionally linked. Additional studies are needed to fully understand how EutH impacts L. monocytogenes survival within the phagosome. Nevertheless, these findings reveal a conserved role for EutH in pH-dependent vacuole adaptation in distinct intracellular pathogens.
EutH plays a spatiotemporal role in EA signaling during mammalian infection.
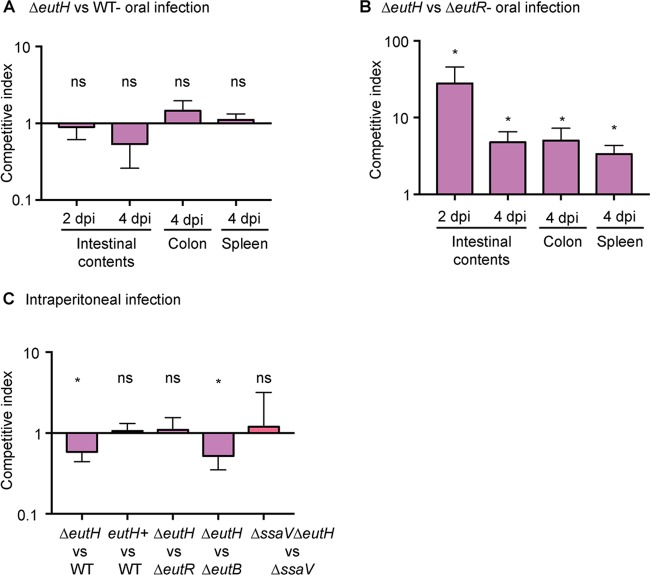
The gastrointestinal (GI) tract and intramacrophage environments are primary niches for S. Typhimurium during host infection (62). The GI tract is characterized by neutral pH and millimolar concentrations of EA (20, 63, 64), indicating that during infection, EutH may play a niche-specific role in host adaptation. To investigate this idea, we performed competition experiments using S. Typhimurium-infected murine models of colitis and systemic infection (65). To examine the importance of EutH to S. Typhimurium intestinal colonization, we infected streptomycin-treated mice with equal numbers of cells of the WT and ΔeutH strains or with equal numbers of cells of the ΔeutR and ΔeutH strains and assessed bacterial burden in the intestinal contents as well as in the colon and spleen. The ΔeutR strain cannot metabolize EA, and this strain is attenuated for intestinal colonization (13). Equal numbers of cells of the WT and ΔeutH strains were recovered from intestinal contents at 2 and 4 days postinfection (dpi) as well as from the colon and spleen (4 dpi) (Fig. 6A), whereas the ΔeutH strain significantly outcompeted the ΔeutR strain under all conditions (Fig. 6B). These findings indicate that EutH is dispensable for EutR-dependent regulation of EA utilization during colitis.
FIG 6.
Contribution of EutH to S. Typhimurium fitness in vivo. (A) Competitive indices of the ΔeutH (CJA046) versus WT (SL1344) strain during colitis. n = 7 or 8 mice. (B) Competitive indices of the ΔeutH (CJA052) versus ΔeutR (CJA007) strain during colitis. n = 8 mice. (C) Competitive indices in the spleen of the ΔeutH (CJA046) versus WT (SL1344) strain, the ΔeutH complemented (CJA192) (eutH+, which contains pGEN::eutS-eutH) versus WT (CJA182, which contains pGEN empty vector) strain, the ΔeutH (CJA052) versus ΔeutR (CJA007) strain, the ΔeutH (CJA043) versus ΔeutB (CJA018) strain, and the ΔssaV ΔeutH (CJA172) versus ΔssaV strain at 6 hpi following intraperitoneal injection. n = 8 to 11 mice per competition. The median and interquartile range are shown *, P ≤ 0.05. P values of >0.05 are not significant (ns).
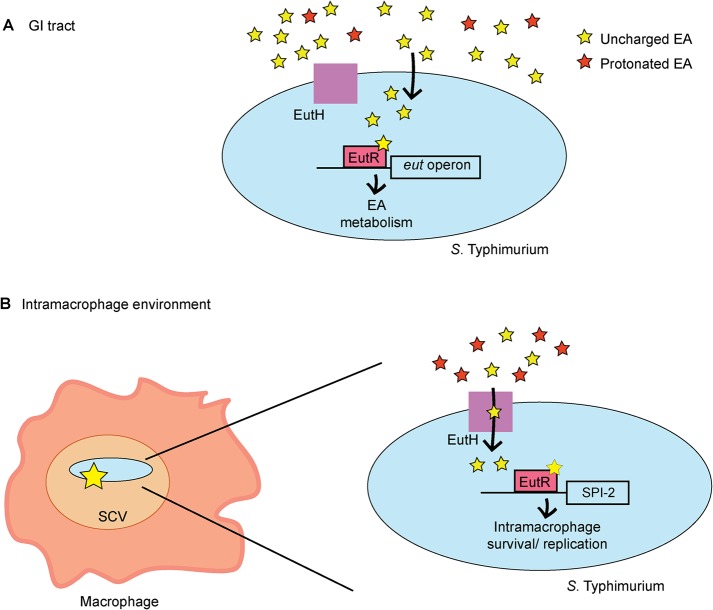
To evaluate the role of EutH in S. Typhimurium dissemination, we intraperitoneally infected mice and assessed bacterial numbers recovered from the spleen at 6 hpi. Macrophages play a major role in S. Typhimurium dissemination, and in agreement with data shown in Fig. 2 to 4, the ΔeutH strain was recovered in significantly lower numbers compared to the WT, and this defect could be complemented when eutH was expressed in trans (Fig. 6C). A previous study reported that an eutH deletion strain did not significantly impact dissemination (27); however, the experimental details were not described and may contribute to differences between these studies. Additionally, because the difference in recovery of the WT and ΔeutH strains was ∼2-fold, we substantiated the importance of EutH to systemic infection by performing competition infections between the ΔeutH and ΔeutR strains as well as between the ΔeutH and ΔeutB strains. We previously reported that the ΔeutR strain was attenuated during early systemic infection compared to the WT and the ΔeutB strains as EutR regulates SPI-2 independently of EA metabolism (13). In agreement with a role for EutH in EutR signaling and macrophage survival, the ΔeutH strain was recovered in similar numbers compared to the ΔeutR strain and was significantly outcompeted by the ΔeutB strain (Fig. 6C). Moreover, equal numbers of the T3SS-2-deficient ΔssaV and ΔssaV ΔeutH strains were recovered, which further underscores the requirement of the T3SS-2 in EA-dependent dissemination (Fig. 6C). Collectively, these data demonstrate that EutH contributes to S. Typhimurium dissemination during infection. Significantly, the initial study that focused on EutH concluded with the idea that EutH is selectively maintained within bacterial genomes because S. Typhimurium (and presumably other bacteria) frequently encounters EA at concentrations or under pH conditions that limit the external unprotonated EA—conditions under which EutH is required to enhance the ability to utilize EA (25). Our findings are consistent with the original in vitro model proposed nearly 15 years ago and reveal a physiologically relevant role for EutH in spatiotemporal niche adaptation during infection (Fig. 7A and B).
FIG 7.
EutH plays a spatiotemporal role in S. Typhimurium niche adaptation. (A) EutH is dispensable for EA utilization in the intestine. (B) EutH contributes to EutR-dependent signaling and S. Typhimurium survival and replication during systemic infection.
Conclusions
Following entry into host cells, the majority of intracellular pathogens are at least initially trapped within a host vacuole. Although bacterial strategies to withstand or evade immune defenses within the phagosome may be redundant, many are specific to a particular pathogen (66). In contrast, host-specific signals that promote virulence programs and/or metabolic processes may be shared among diverse pathogens. Indeed, EA is a seemingly ubiquitous molecule within the host, and diverse pathogens rely on EA as a metabolite to support growth during host infection as well as co-opt EA as a signal to modulate virulence. Although a deletion of eutH does not ablate bacterial virulence, EutH consistently and reproducibly enhanced bacterial survival and replication within the phagosome and during S. Typhimurium systemic infection. Altogether, our findings demonstrate that the EA permease EutH contributes to the ability of distinct bacterial pathogens to survive within the acidified vacuole of macrophages.
MATERIALS AND METHODS
Strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. S. Typhimurium cultures were routinely grown overnight in LB broth with antibiotics when appropriate. Antibiotics were used in the following concentrations: ampicillin (100 μg/ml), streptomycin (100 μg/ml), chloramphenicol (20 μg/ml), and kanamycin (50 μg/ml). SPI-2-inducing medium was prepared as described: 100 mM Bis/Tris-HCl (pH 7.0), 5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 1 mM KH2PO4, 38 mM glycerol, 0.1% Casamino Acids, and 8 μM MgCl2 (67). Strains of L. monocytogenes were routinely grown overnight in brain heart infusion (BHI) broth without antibiotics.
Deletions of eutR and eutH were constructed in the WT S. Typhimurium SL1344, invG mutant, or ssaV mutant backgrounds using λ-red mutagenesis (68) using primers eutR_λ-red F/R or eutH_λ-red F/R listed in Table S2 in the supplemental material. To create the nonpolar deletions, the chloramphenicol or kanamycin cassettes were resolved using pCP20 (68). Unresolved strains were used as indicated for competition experiments in vivo. The invG eutH eutR mutant strain (CJA168) was left unresolved, as subsequent use of pCP20 removed all genomic DNA between eutH and eutR. The eutH mutant was complemented with pCJA031. pCJA031 was constructed by amplifying S. Typhimurium genomic DNA with primers eutH_complement F/R, which are specific to the eut operon to include the native P1 promoter through eutH (listed in Table S2). Amplified PCR product was digested with NdeI and NotI and inserted into pGEN-MCS (69) (Addgene MTA). When appropriate, WT and deletion strains were transformed with empty pGEN-MCS vectors as controls.
An in-frame L. monocytogenes 10403S eutH deletion mutant strain and listeriolysin O mutant hly eutH deletion strain were generated as described previously (70) with modifications. Briefly, primer pairs ΔeutH.PA/ΔeutH.PB and ΔeutH.PC/ΔeutH.PD (Table S2) were used to amplify two 400-bp fragments upstream and downstream of eutH. Subsequently, an 800-bp deletion fragment was constructed via joining these 400-bp DNA fragments by overlap extension PCR with the ΔeutH.PA/ΔeutH.PD pair. The 800-bp deletion fragment was cloned into the pMAD vector after restriction digestion with EcoRI and BamHI (New England BioLabs) and ligation reactions with T4 ligase (New England BioLabs). The resulting pMAD::ΔeutH deletion construct was introduced into the L. monocytogenes 10403S and Δhly strains via electroporation (71). Following electroporation, transformants were selected on BHI plates supplemented with 0.5 M sucrose, 10 μg/ml erythromycin, and 20 μg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) after 48 h of growth at 30°C. After reisolation on a BHI-erythromycin plate, blue colonies were picked for growth in BHI containing erythromycin at 39°C for 24 to 48 h. This culture was plated on BHI containing erythromycin and X-Gal and incubated at 39°C to isolate transformants that harbor the chromosomally integrated deletion construct. Blue colonies were used to start serial passages (1 per day) in BHI at 30°C to mediate excision of pMAD plasmid from the chromosome. After 3 to 5 passages, the temperature was shifted to 39°C and incubation carried out for an additional 5 h. Final cultures were plated on BHI–X-Gal plates and incubated at 39°C for 24 h. White colonies, which are cured of pMAD plasmid, were screened for erythromycin sensitivity. In-frame deletion mutants were identified from these white erythromycin-sensitive clones by PCR. Final verification of all deletions was performed by sequencing.
Animal experiments.
All animal experiments were approved by the Animal Care and Use Committee at the University of Virginia. For all infections, 10- to 12-week-old, female C57BL/6 mice were used (Envigo). For the mouse colitis model, 24 h prior to infection, mice received a single dose via oral gavage of 20 mg streptomycin (72). Mice were infected via oral gavage with an equal mixture of 5 × 108 CFU of each of the indicated S. Typhimurium strains. Fresh fecal samples were collected daily, and mice were euthanized at 4 dpi to assess bacterial burden in the indicated tissues. Tissue samples were weighed and then homogenized in 1 ml phosphate-buffered saline (PBS), and bacterial burden was quantified by plating serial dilutions of homogenized tissue on MacConkey agar with streptomycin to obtain total bacterial CFU and MacConkey agar with chloramphenicol to obtain unresolved mutant CFU. The competitive index for each tissue was calculated as the ratio of indicated strains recovered from tissue normalized to the ratio in the inoculum.
For i.p. infections, mice were infected with an equal mixture of 5 × 104 CFU of each of the indicated S. Typhimurium strains. Spleens were collected at 6 h postinfection. Spleens were homogenized and processed and competitive infections calculated as described above. For complementation studies, samples were plated on LB agar with ampicillin to obtain total bacterial CFU and LB agar with ampicillin and chloramphenicol to obtain unresolved mutant CFU.
Tissue culture.
RAW cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and 1× penicillin-streptomycin-glutamine. Peritoneal exudate macrophages (PEMs) from 8- to 12-week-old C57BL/6 mice were isolated as described previously (13, 73). Bone marrow-derived macrophages (BMDMs) were isolated and cultured as described previously (73, 74). Briefly, 8- to 12-week-old C57BL/6 (S. Typhimurium infections) or BALB/c (L. monocytogenes infections) mice were euthanized, and bone marrow was harvested from femurs. The extracted bone marrow was incubated with 0.84% ammonium chloride solution at room temperature for 10 min to lyse red blood cells. Bone marrow cells were centrifuged at 1,500 rpm for 5 min and resuspended in the indicated medium. Additional medium was provided to cell cultures 3 days after harvest, and medium was replaced after 6 days of culture before cells were used in assays. PEMs and BMDMs were cultured in RPMI 1640 supplemented with 10% FBS, 20% L-929 conditioned medium, and 1× penicillin-streptomycin-glutamine. All cells were seeded into 12-well plates at 5 × 105 cells/well for experiments.
Gentamicin protection assays were performed as described previously (13, 35, 73–77). We used the invG mutant and indicated eut mutants for macrophage infections as invasive S. Typhimurium strains kill macrophages in vitro (76). Furthermore, expression of invasion-associated genes is decreased upon phagocytosis, and thus these strains more closely mimic S. Typhimurium encountering macrophages during systemic infection (78). Overnight cultures of S. Typhimurium or L. monocytogenes were washed and resuspended in PBS before incubation with macrophages at a multiplicity of infection (MOI) of 10. RAW cells were infected using DMEM supplemented with 10% heat-inactivated FBS without antibiotics. PEMs and BMDMs were infected using RPMI 1640 supplemented with 10% heat-inactivated FBS without antibiotics. After 30 min of incubation, extracellular bacteria were killed with 100-μg/ml gentamicin treatment for 30 min before replacement with medium containing 10 μg/ml gentamicin for the remainder of the assay. Cells were lysed at the indicated time points in 1% Triton X-100, and CFU were determined by serial dilutions and plating onto LB agar (S. Typhimurium) or BHI agar (L. monocytogenes). Percentage of survival was calculated as viable CFU at indicated time points as a percentage of CFU following 30 min of incubation with 100 μg/ml gentamicin (time zero) and normalized such that the wild type was equal to 100%.
For vacuole neutralization, BMDMs were incubated for 1 h prior to infection with the indicated concentration of concanamycin A (Sigma), dimethyl sulfoxide (DMSO) vehicle, or ammonium chloride (79). Following a 1-h pretreatment, cells were washed twice to remove concanamycin A or DMSO vehicle before continuing with a standard gentamicin protection assay as described above. Ammonium chloride was maintained in the medium throughout infection.
For ethanolamine supplementation to RAW cell infection, ethanolamine was added to culture medium at the indicated concentration following the 30-min incubation with 100 μg/ml gentamicin (time zero) until cells were lysed at 5 h postinfection.
Statistical analysis.
Statistical significance of in vivo competitive indexes was determined by Wilcoxon's signed-rank test with a theoretical median of 1. Student's t test was used for the comparison of viable CFU in tissue culture experiments.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jim Casanova for providing the ssaV S. Typhimurium strain and members of the Kendall lab for critical feedback on the project.
This work was supported by National Institutes of Health (NIH) grants R01AI118732 and R21AI130439 to M.M.K. and R01AI073904 to H.A. C.J.A. received support through the NIH training grant 5T32AI007046 and University of Virginia School of Medicine Wagner Fellowship. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00172-18.
REFERENCES
- 1.Lipton BA, Davidson EP, Ginsberg BH, Yorek MA. 1990. Ethanolamine metabolism in cultured bovine aortic endothelial cells. J Biol Chem 265:7195–7201. [PubMed] [Google Scholar]
- 2.Lipton BA, Yorek MA, Ginsberg BH. 1988. Ethanolamine and choline transport in cultured bovine aortic endothelial cells. J Cell Physiol 137:571–576. doi: 10.1002/jcp.1041370325. [DOI] [PubMed] [Google Scholar]
- 3.Nikawa J, Tsukagoshi Y, Yamashita S. 1986. Cloning of a gene encoding choline transport in Saccharomyces cerevisiae. J Bacteriol 166:328–330. doi: 10.1128/jb.166.1.328-330.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel D, Witt SN. 2017. Ethanolamine and phosphatidylethanolamine: partners in health and disease. Oxid Med Cell Longev 2017:4829180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandra A, Cai J. 1991. Plasma membrane appearance of phosphatidylethanolamine in stimulated macrophages. J Leukoc Biol 50:19–27. doi: 10.1002/jlb.50.1.19. [DOI] [PubMed] [Google Scholar]
- 6.Bakovic M, Fullerton MD, Michel V. 2007. Metabolic and molecular aspects of ethanolamine phospholipid biosynthesis: the role of CTP:phosphoethanolamine cytidylyl-transferase (Pcyt2). Biochem Cell Biol 85:283–300. doi: 10.1139/O07-006. [DOI] [PubMed] [Google Scholar]
- 7.Meijerink J, Plastina P, Vincken JP, Poland M, Attya M, Balvers M, Gruppen H, Gabriele B, Witkamp RF. 2011. The ethanolamide metabolite of DHA, docosahexaenoylethanolamine, shows immunomodulating effects in mouse peritoneal and RAW264.7 macrophages: evidence for a new link between fish oil and inflammation. Br J Nutr 105:1798–1807. doi: 10.1017/S0007114510005635. [DOI] [PubMed] [Google Scholar]
- 8.Sugiura T, Kobayashi Y, Oka S, Waku K. 2002. Biosynthesis and degradation of anandamide and 2-arachidonoylglycerol and their possible physiological significance. Prostaglandins Leukot Essent Fatty Acids 66:173–192. doi: 10.1054/plef.2001.0356. [DOI] [PubMed] [Google Scholar]
- 9.Kelly RF, Lamont KT, Somers S, Hacking D, Lacerda L, Thomas P, Opie LH, Lecour S. 2010. Ethanolamine is a novel STAT-3 dependent cardioprotective agent. Basic Res Cardiol 105:763–770. doi: 10.1007/s00395-010-0125-0. [DOI] [PubMed] [Google Scholar]
- 10.Sasaki H, Kume H, Nemoto A, Narisawa S, Takahashi N. 1997. Ethanolamine modulates the rate of rat hepatocyte proliferation in vitro and in vivo. Proc Natl Acad Sci U S A 94:7320–7325. doi: 10.1073/pnas.94.14.7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murakami H, Masui H, Sato GH, Sueoka N, Chow TP, Kano-Sueoka T. 1982. Growth of hybridoma cells in serum-free medium: ethanolamine is an essential component. Proc Natl Acad Sci U S A 79:1158–1162. doi: 10.1073/pnas.79.4.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kano-Sueoka T, Oda T, Kawamoto JK. 2001. Phosphatidylethanolamine deficiency in membrane lipids inhibits keratinocyte intercellular networks formation. In Vitro Cell Dev Biol Anim 37:691–697. doi:. [DOI] [PubMed] [Google Scholar]
- 13.Anderson CJ, Clark DE, Adli M, Kendall MM. 2015. Ethanolamine signaling promotes Salmonella niche recognition and adaptation during infection. PLoS Pathog 11:e1005278. doi: 10.1371/journal.ppat.1005278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonyar LA, Kendall MM. 2014. Ethanolamine and choline promote expression of putative and characterized fimbriae in enterohemorrhagic Escherichia coli O157:H7. Infect Immun 82:193–201. doi: 10.1128/IAI.00980-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kendall MM, Gruber CC, Parker CT, Sperandio V. 2012. Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7. mBio 3:e00050-12. doi: 10.1128/mBio.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luzader DH, Clark DE, Gonyar LA, Kendall MM. 2013. EutR is a direct regulator of genes that contribute to metabolism and virulence in enterohemorrhagic Escherichia coli O157:H7. J Bacteriol 195:4947–4953. doi: 10.1128/JB.00937-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maadani A, Fox KA, Mylonakis E, Garsin DA. 2007. Enterococcus faecalis mutations affecting virulence in Caenorhabditis elegans model host. Infect Immun 75:2634–2637. doi: 10.1128/IAI.01372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mellin JR, Koutero M, Dar D, Nahori MA, Sorek R, Cossart P. 2014. Riboswitches. Sequestration of a two-component response regulator by a riboswitch-regulated noncoding RNA. Science 345:940–943. doi: 10.1126/science.1255083. [DOI] [PubMed] [Google Scholar]
- 19.Subashchandrabose S, Hazen TH, Brumbaugh AR, Himpsl SD, Smith SN, Ernst RD, Rasko DA, Mobley HLT. 2014. Host-specific induction of Escherichia coli fitness genes during human urinary tract infection. Proc Natl Acad Sci U S A 111:18327–18332. doi: 10.1073/pnas.1415959112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Bäumler AJ. 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A 108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsoy O, Ravcheev D, Mushegian A. 2009. Comparative genomics of ethanolamine utilization. J Bacteriol 191:7157–7164. doi: 10.1128/JB.00838-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garsin DA. 2010. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat Rev Microbiol 8:290–295. doi: 10.1038/nrmicro2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kofoid E, Rappleye C, Stojiljkovic I, Roth J. 1999. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J Bacteriol 181:5317–5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lundgren BR, Sarwar Z, Pinto A, Ganley JG, Nomura CT. 2016. Ethanolamine catabolism in Pseudomonas aeruginosa PAO1 is regulated by the enhancer-binding protein EatR (PA4021) and the alternative sigma factor RpoN. J Bacteriol 198:2318–2329. doi: 10.1128/JB.00357-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penrod JT, Mace CC, Roth JR. 2004. A pH-sensitive function and phenotype: evidence that EutH facilitates diffusion of uncharged ethanolamine in Salmonella enterica. J Bacteriol 186:6885–6890. doi: 10.1128/JB.186.20.6885-6890.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roof DM, Roth JR. 1989. Functions required for vitamin B12-dependent ethanolamine utilization in Salmonella typhimurium. J Bacteriol 171:3316–3323. doi: 10.1128/jb.171.6.3316-3323.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stojiljkovic I, Bäumler AJ, Heffron F. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol 177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joseph B, Przybilla K, Stuhler C, Schauer K, Slaghis J, Fuchs TM, Goebel W. 2006. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol 188:556–568. doi: 10.1128/JB.188.2.556-568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roof DM, Roth JR. 1992. Autogenous regulation of ethanolamine utilization by a transcriptional activator of the eut operon in Salmonella typhimurium. J Bacteriol 174:6634–6643. doi: 10.1128/jb.174.20.6634-6643.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheppard DE, Roth JR. 1994. A rationale for autoinduction of a transcriptional activator: ethanolamine ammonia-lyase (EutBC) and the operon activator (EutR) compete for adenosyl-cobalamin in Salmonella typhimurium. J Bacteriol 176:1287–1296. doi: 10.1128/jb.176.5.1287-1296.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng X, Walthers D, Oropeza R, Kenney LJ. 2004. The response regulator SsrB activates transcription and binds to a region overlapping OmpR binding sites at Salmonella pathogenicity island 2. Mol Microbiol 54:823–835. doi: 10.1111/j.1365-2958.2004.04317.x. [DOI] [PubMed] [Google Scholar]
- 32.Walthers D, Carroll RK, Navarre WW, Libby SJ, Fang FC, Kenney LJ. 2007. The response regulator SsrB activates expression of diverse Salmonella pathogenicity island 2 promoters and counters silencing by the nucleoid-associated protein H-NS. Mol Microbiol 65:477–493. doi: 10.1111/j.1365-2958.2007.05800.x. [DOI] [PubMed] [Google Scholar]
- 33.Worley MJ, Ching KH, Heffron F. 2000. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol Microbiol 36:749–761. doi: 10.1046/j.1365-2958.2000.01902.x. [DOI] [PubMed] [Google Scholar]
- 34.Cirillo DM, Valdivia RH, Monack DM, Falkow S. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol 30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 35.Hensel M, Shea JE, Waterman SR, Mundy R, Nikolaus T, Banks G, Vazquez-Torres A, Gleeson C, Fang FC, Holden DW. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol Microbiol 30:163–174. doi: 10.1046/j.1365-2958.1998.01047.x. [DOI] [PubMed] [Google Scholar]
- 36.Ochman H, Soncini FC, Solomon F, Groisman EA. 1996. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci U S A 93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shea JE, Hensel M, Gleeson C, Holden DW. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci U S A 93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DebRoy S, Gebbie M, Ramesh A, Goodson JR, Cruz MR, vanHoof A, Winkler WC, Garsin DA. 2014. Riboswitches. A riboswitch-containing sRNA controls gene expression by sequestration of a response regulator. Science 345:937–940. doi: 10.1126/science.1255091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fox KA, Ramesh A, Stearns JE, Bourgogne A, Reyes-Jara A, Winkler WC, Garsin DA. 2009. Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proc Natl Acad Sci U S A 106:4435–4440. doi: 10.1073/pnas.0812194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camilli A, Paynton C, Portnoy DA. 1989. Intracellular methicillin selection of Listeria monocytogenes mutants unable to replicate in a macrophage cell line. Proc Natl Acad Sci U S A 86:5522–5526. doi: 10.1073/pnas.86.14.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marquis H, Doshi V, Portnoy DA. 1995. The broad-range phospholipase C and a metalloprotease mediate listeriolysin O-independent escape of Listeria monocytogenes from a primary vacuole in human epithelial cells. Infect Immun 63:4531–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mengaud J, Braun-Breton C, Cossart P. 1991. Identification of phosphatidylinositol-specific phospholipase C activity in Listeria monocytogenes: a novel type of virulence factor? Mol Microbiol 5:367–372. doi: 10.1111/j.1365-2958.1991.tb02118.x. [DOI] [PubMed] [Google Scholar]
- 43.Portnoy DA, Jacks PS, Hinrichs DJ. 1988. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J Exp Med 167:1459–1471. doi: 10.1084/jem.167.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vazquez-Boland JA, Kocks C, Dramsi S, Ohayon H, Geoffroy C, Mengaud J, Cossart P. 1992. Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect Immun 60:219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alpuche-Aranda CM, Swanson JA, Loomis WP, Miller SI. 1992. Salmonella typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc Natl Acad Sci U S A 89:10079–10083. doi: 10.1073/pnas.89.21.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chakraborty S, Mizusaki H, Kenney LJ. 2015. A FRET-based DNA biosensor tracks OmpR-dependent acidification of Salmonella during macrophage infection. PLoS Biol 13:e1002116. doi: 10.1371/journal.pbio.1002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rathman M, Sjaastad MD, Falkow S. 1996. Acidification of phagosomes containing Salmonella typhimurium in murine macrophages. Infect Immun 64:2765–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beuzon CR, Banks G, Deiwick J, Hensel M, Holden DW. 1999. pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol Microbiol 33:806–816. doi: 10.1046/j.1365-2958.1999.01527.x. [DOI] [PubMed] [Google Scholar]
- 49.Rappl C, Deiwick J, Hensel M. 2003. Acidic pH is required for the functional assembly of the type III secretion system encoded by Salmonella pathogenicity island 2. FEMS Microbiol Lett 226:363–372. doi: 10.1016/S0378-1097(03)00638-4. [DOI] [PubMed] [Google Scholar]
- 50.Hansen-Wester I, Stecher B, Hensel M. 2002. Type III secretion of Salmonella enterica serovar Typhimurium translocated effectors and SseFG. Infect Immun 70:403–409. doi: 10.1128/IAI.70.1.403-406.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Myers JT, Tsang AW, Swanson JA. 2003. Localized reactive oxygen and nitrogen intermediates inhibit escape of Listeria monocytogenes from vacuoles in activated macrophages. J Immunol 171:5447–5453. doi: 10.4049/jimmunol.171.10.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tilney LG, Portnoy DA. 1989. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J Cell Biol 109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Chastellier C, Berche P. 1994. Fate of Listeria monocytogenes in murine macrophages: evidence for simultaneous killing and survival of intracellular bacteria. Infect Immun 62:543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh R, Jamieson A, Cresswell P. 2008. GILT is a critical host factor for Listeria monocytogenes infection. Nature 455:1244–1247. doi: 10.1038/nature07344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahmed JK, Freitag NE. 2016. Secretion chaperones PrsA2 and HtrA are required for Listeria monocytogenes replication following intracellular induction of virulence factor secretion. Infect Immun 84:3034–3046. doi: 10.1128/IAI.00312-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Czuczman MA, Fattouh R, vanRijn JM, Canadien V, Osborne S, Muise AM, Kuchroo VK, Higgins DE, Brumell JH. 2014. Listeria monocytogenes exploits efferocytosis to promote cell-to-cell spread. Nature 509:230–234. doi: 10.1038/nature13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gedde MM, Higgins DE, Tilney LG, Portnoy DA. 2000. Role of listeriolysin O in cell-to-cell spread of Listeria monocytogenes. Infect Immun 66:999–1003. doi: 10.1128/IAI.68.2.999-1003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Casey JR, Grinstein S, Orlowski J. 2010. Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol 11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- 59.Gaillard JL, Berche P, Mounier J, Richard S, Sansonetti P. 1987. In vitro model of penetration and intracellular growth of Listeria monocytogenes in the human enterocyte-like cell line Caco-2. Infect Immun 55:2822–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Birmingham CL, Canadien V, Kaniuk NA, Steinberg BE, Higgins DE, Brumell JH. 2008. Listeriolysin O allows Listeria monocytogenes replication in macrophage vacuoles. Nature 451:350–354. doi: 10.1038/nature06479. [DOI] [PubMed] [Google Scholar]
- 61.Osborne SE, Brumell JH. 2017. Listeriolysin O: from bazooka to Swiss army knife. Philos Trans R Soc Lond B Biol Sci 372:20160222. doi: 10.1098/rstb.2016.0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anderson CJ, Kendall MM. 2017. Salmonella enterica serovar Typhimurium strategies for host adaptation. Front Microbiol 8:1983. doi: 10.3389/fmicb.2017.01983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Evans DF, Pye G, Bramley R, Clark AG, Dyson TJ, Hardcastle JD. 1988. Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut 29:1035–1041. doi: 10.1136/gut.29.8.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khutoryanskiy VV. 2015. Longer and safer gastric residence. Nat Mater 15:963–964. doi: 10.1038/nmat4432. [DOI] [PubMed] [Google Scholar]
- 65.Santos RL, Zhang S, Tsolis RM, Kingsley RM, Kingsley RA, Adams LG, Baumler AJ. 2001. Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect 3:1335–1344. doi: 10.1016/S1286-4579(01)01495-2. [DOI] [PubMed] [Google Scholar]
- 66.da Silva CV, Cruz L, da Silva Araujo N, Angeloni MB, Fonseca BB, de Oliveira Gomes A, dos Reis Carvalho F, Goncalves ALR, de Freitas Barbosa B. 2012. A glance at Listeria and Salmonella cell invasion: different strategies to promote host actin polymerization. Int J Med Microbiol 302:19–32. doi: 10.1016/j.ijmm.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 67.Deiwick J, Nikolaus T, Erdogan S, Hensel M. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol Microbiol 31:1759–1773. doi: 10.1046/j.1365-2958.1999.01312.x. [DOI] [PubMed] [Google Scholar]
- 68.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lane MC, Alteri CJ, Smith SN, Mobley HLT. 2007. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc Natl Acad Sci U S A 104:16669–16674. doi: 10.1073/pnas.0607898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arnaud M, Chastanet A, Débarbouillé M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, Gram-positive bacteria. Appl Environ Microbiol 70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park SF, Stewart G. 1990. High-efficiency transformation of Listeria monocytogenes by electroporation of penicillin-treated cells. Gene 94:129–132. doi: 10.1016/0378-1119(90)90479-B. [DOI] [PubMed] [Google Scholar]
- 72.Barthel M, Hapfelmeier S, Quintanilla-Martínez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Rüssmann H, Hardt W-D. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang X, Goncalves R, Mosser DM. 2008. The isolation and characterization of murine macrophages. Curr Protoc Immunol Chapter 14:Unit 14.1. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kadl A, Meher AK, Sharma PR, Lee MY, Doran AC, Johnstone SR, Elliott MR, Gruber F, Han J, Chen W, Kensler T, Ravichandran KS, Isakson BE, Wamhoff BR, Leitinger N. 2010. Identification of a novel macrophage phenotype that develops in response to atherogenic phospholipids via Nrf2. Circ Res 107:737–746. doi: 10.1161/CIRCRESAHA.109.215715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Buchmeier NA, Heffron F. 1989. Intracellular survival of wild-type Salmonella typhimurium and macrophage-sensitive mutants in diverse populations of macrophages. Infect Immun 57:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen LM, Kaniga K, Galan JE. 1996. Salmonella spp. are cytotoxic for cultured macrophages. Mol Microbiol 21:1101–1115. doi: 10.1046/j.1365-2958.1996.471410.x. [DOI] [PubMed] [Google Scholar]
- 77.Owen K, Meyer CB, Bouton AH, Casanova JE. 2014. Activation of focal adhesion kinase by Salmonella suppresses autophagy via an Akt/mTOR signaling pathway and promotes bacterial survival in macrophages. PLoS Pathog 10:e1004159. doi: 10.1371/journal.ppat.1004159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Laughlin RC, Knodler LA, Barhoumi R, Payne HR, Wu J, Gomez G, Pugh R, Lawhon S, Baumler AJ, Steele-Mortimer O, Adams LG. 2014. Spatial segregation of virulence gene expression during acute enteric infection with Salmonella enterica serovar Typhimurium. mBio 5:e00946-13. doi: 10.1128/mBio.00946-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gilmartin AA, Ralston KS, Petri WAJ. 2017. Inhibition of amebic lysosomal acidification blocks amebic trogocytosis and cell killing. mBio 8:e01187-17. doi: 10.1128/mBio.01187-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.