ABSTRACT
Phytoplasmas are plant-pathogenic bacteria transmitted by hemipteran insects. The leafhopper Euscelidius variegatus is a natural vector of chrysanthemum yellows phytoplasma (CYp) and a laboratory vector of flavescence dorée phytoplasma (FDp). The two phytoplasmas induce different effects on this species: CYp slightly improves whereas FDp negatively affects insect fitness. To investigate the molecular bases of these different responses, transcriptome sequencing (RNA-seq) analysis of E. variegatus infected with either CYp or FDp was performed. The sequencing provided the first de novo transcriptome assembly for a phytoplasma vector and a starting point for further analyses on differentially regulated genes, mainly related to immune system and energy metabolism. Insect phenoloxidase activity, immunocompetence, and body pigmentation were measured to investigate the immune response, while respiration and movement rates were quantified to confirm the effects on energy metabolism. The activation of the insect immune response upon infection with FDp, which is not naturally transmitted by E. variegatus, confirmed that this bacterium is mostly perceived as a potential pathogen. Conversely, the acquisition of CYp, which is naturally transmitted by E. variegatus, seems to increase the insect fitness by inducing a prompt response to stress. This long-term relationship is likely to improve survival and dispersal of the infected insect, thus enhancing the opportunity of phytoplasma transmission.
KEYWORDS: “Candidatus Phytoplasma asteris”, flavescence dorée phytoplasma, energy metabolism, immune response, leafhopper
INTRODUCTION
Phytoplasmas are wall-less plant-pathogenic bacteria of the class Mollicutes that cause yield losses in many crops worldwide. They colonize plant phloem tissues and are transmitted by phloem-feeding hemipterans (1). In insects, ingested phytoplasmas cross the gut, multiply in the hemocoel, and invade salivary glands before being transmitted during feeding on a new plant (2). Currently, genomes of four phytoplasmas are fully sequenced and annotated and a few others are available as drafts (1). Phytoplasmas have small (∼750-kb) genomes due to a genome reduction that resulted in the loss of important metabolic pathways; as a consequence, these intracellular bacteria depend on their hosts for many essential metabolites (3). Due to these strict interactions with hosts and to the difficulty of their axenic cultivation, phytoplasmas need to be studied directly in their hosts. Little is known about pathogenicity mechanisms, even though some pathogen-secreted virulence factors have been identified, mainly in strains of “Candidatus Phytoplasma asteris” (2). Studies on this species suggest that phytoplasmas are able to modulate their gene expression during host switching between plant and insect (4, 5).
Flavescence dorée (FD) is an important grapevine disease caused by a 16SrV phytoplasma, mainly transmitted under field conditions by the hemipteran cicadellid Scaphoideus titanus. FD phytoplasma (FDp) is a plant quarantine pathogen in the European Union and represents one of the major threats to southern European viticulture. Vitis vinifera and S. titanus do not represent ideal experimental organisms for laboratory tests: grapevine is a perennial woody plant, and the leafhopper is a monovoltine species. Thus, a laboratory model has been established to manage the FDp infection cycle with the herbaceous Vicia faba as the plant and the polivoltine leafhopper Euscelidius variegatus as the vector (6).
Chrysanthemum yellows phytoplasma (CYp), 16SrI-B “Ca. P. asteris,” is associated with a disease of ornamental plants in northwestern Italy, where E. variegatus is one of the most important natural vectors (7). Like FDp infections, CYp infections can be obtained under controlled conditions with Chrysanthemum carinatum as the host plant and E. variegatus as the vector. The two phytoplasmas have opposite effects on the vector fitness: FDp significantly reduces insect longevity and fecundity (8), whereas CYp induces a slight fitness increase (9). CYp shows greater ability than FDp to colonize the salivary glands of the vector and therefore is more efficiently transmitted by E. variegatus (7). So far, physical maps of FDp and CYp genomes, drafts of their genome sequences, and an FDp transcriptome analysis (10) are available. In contrast, neither the genome nor the transcriptome of E. variegatus is available. Few proteins of the insect have been identified, such as in vitro interacting partners of CYp antigenic membrane protein (Amp) (11), which is necessary for CYp acquisition by insect vectors (12). The specificity of FDp transmission is presumably mediated by variable membrane protein A (VmpA), a phytoplasma protein that is supposed to interact with insect tissues and shows high sequence variability in different strains transmitted by different vector species (13).
In a few studies, the plant response to phytoplasma infection was addressed with next-generation-sequencing (NGS) transcriptomic approaches (1), but to our knowledge, this technique has never been used to investigate either the transcriptomic profile of an insect vector infected by these pathogens or the alterations induced by different phytoplasmas in the same host. Here we investigated the effects of two genetically different phytoplasmas on the same insect vector and describe (i) de novo assembly of E. variegatus transcriptome, (ii) differential expression analysis of E. variegatus transcripts under infection with CY or FD phytoplasmas, (iii) validation of the differential expression profiles, and (iv) biological experiments to support the transcript profiling results.
RESULTS
Transcriptome sequencing (RNA-seq) and differential gene expression.
Diagnostic reverse transcription-quantitative PCR (RT-qPCR) assays confirmed the presence of CY and FD phytoplasmas in E. variegatus samples (Eva_CY and Eva_FD). The phytoplasma populations, expressed as mean phytoplasma 16S/insect 18S ratios, were 4.67 E−03 (standard error of the mean [SEM], ±9.84 E−04) and 2.08 E−04 (SEM, ±6.54 E−05) for insects infected with CYp and FDp, respectively. Analyses of the cDNA libraries obtained from Eva_CY and Eva_FD resulted in a combined de novo assembly comprising around 135,000 transcripts, with an average GC content of 40%, a median contig length of 433 bp, and an average contig length of 833 bp. Due to the lack of E. variegatus genomic sequence information, the functional annotation of transcripts was conducted using blastx against the NCBI nonredundant (nr) database. The Blast2GO platform was then used to assign the Gene Ontology (GO) terms to the predicted proteins with known function. The results of the de novo assembly and the following transcript annotations are summarized in Table S1 in the supplemental material. The species distributions of the best blastx matches for each sequence are shown in Table S2.
Transcriptome profiles of E. variegatus infected by the beneficial CY or the pathogenic FD phytoplasmas were compared to elucidate the differential vector responses.
Differential expression analysis revealed that 84 transcripts were upregulated and 13 downregulated in Eva_CY in comparison with Eva_FD (Tables 1 and 2; see also Tables S3 and S4). The upregulated genes could be classified into a few main functional categories: immune response (11 transcripts), movement and energy metabolism (34 transcripts), proteases (9 transcripts), extracellular matrix (20 transcripts), nucleic acid binding (6 transcripts), and detoxification (4 transcripts). Downregulated genes could be ascribed to immune response (10 transcripts), movement and energy metabolism (1 transcript), proteases (1 transcript), and detoxification (1 transcript) functional categories. Some of these putative metabolic functions were further investigated to explore the phenotypes correlated with the altered gene regulation.
TABLE 1.
Overview of Euscelidius variegatus transcripts upregulated during chrysanthemum yellows phytoplasma infection compared with insects infected by flavescence dorée phytoplasmasa
| Contig accession no. | FPKM (avg ±SD) |
Ln fold change | P value | Sequence description | |
|---|---|---|---|---|---|
| Eva_CY | Eva_FD | ||||
| Immune response | |||||
| GFTU01010641.1 | 69.0 ± 1.4 | 39.9 ± 5.6 | +0.802 | 2E−14 | Predicted: serpin B3-like |
| GFTU01009442.1 | 63.5 ± 3.1 | 39.9 ± 2.4 | +0.729 | 2E−23 | hypothetical protein g.45731 (protein disulfide isomerase [PDIa] family, redox active TRX domains) |
| GFTU01009443.1 | 34.9 ± 1.4 | 22.0 ± 1.8 | +0.723 | 7E−22 | Predicted: uncharacterized protein LOC109042410 isoform X4 (protein disulfide isomerase [PDIa] family, redox active TRX domains) |
| GFTU01009445.1 | 35.4 ± 1.5 | 22.4 ± 1.8 | +0.721 | 6E−22 | Predicted: uncharacterized protein LOC106678838 isoform X8 (protein disulfide isomerase [PDIa] family, redox active TRX domains) |
| GFTU01009444.1 | 59.5 ± 3.0 | 37.8 ± 1.9 | +0.717 | 6E−24 | Hypothetical protein g.45731 (protein disulfide isomerase [PDIa] family, redox active TRX domains) |
| GFTU01012880.1 | 111.7 ± 6.6 | 80.8 ± 5.5 | +0.518 | 6E−07 | Predicted: protein disulfide-isomerase A6 |
| GFTU01010415.1 | 40.3 ± 0.4 | 26.3 ± 4.8 | +0.592 | 4E−05 | Predicted: gamma-interferon-inducible lysosomal thiol reductase-like |
| GFTU01003389.1 | 30.9 ± 2.2 | 20.9 ± 2.9 | +0.559 | 5E−05 | Hypothetical protein g.13589 (single domain von Willebrand factor type C) |
| GFTU01000362.1 | 1136.7 ± 171.8 | 589.7 ± 137.6 | +0.556 | 8E−3 | Predicted: hexamerin 4 isoform X1 |
| GFTU01005409.1 | 3.9 ± 0.5 | 2.6 ± 0.5 | +0.548 | 2E−03 | chitinase |
| GFTU01006368.1 | 14.7 ± 0.3 | 10.8 ± 0.6 | +0.512 | 2E−09 | Predicted: E3 ubiquitin-protein ligase HUWE1 isoform X5 |
| Movement and energy metabolism | |||||
| GFTU01004258.1 | 57.3 ± 3.3 | 27.7 ± 0.2 | +1.082 | 1E−43 | Predicted: troponin I-like isoform X2 |
| GFTU01000669.1 | 44.4 ± 4.2 | 27.0 ± 0.7 | +0.719 | 8E−10 | Predicted: maltase A1-like |
| GFTU01001629.1 | 47.1 ± 0.9 | 28.5 ± 0.9 | +0.791 | 2E−32 | Predicted: twitchin isoform X25 |
| GFTU01012455.1 | 47.2 ± 0.9 | 28.6 ± 0.9 | +0.791 | 2E−32 | Predicted: twitchin isoform X19 |
| GFTU01012453.1 | 47.5 ± 0.9 | 28.7 ± 0.9 | +0.790 | 2E−32 | Predicted: twitchin isoform X23 |
| GFTU01001631.1 | 47.0 ± 0.9 | 28.4 ± 0.9 | +0.790 | 2E−32 | Predicted: twitchin isoform X1 |
| GFTU01001630.1 | 47.4 ± 0.9 | 28.7 ± 0.9 | +0.790 | 2E−32 | Predicted: twitchin isoform X1 |
| GFTU01001638.1 | 47.3 ± 0.9 | 28.6 ± 0.9 | +0.790 | 2E−32 | Predicted: twitchin isoform X1 |
| GFTU01001632.1 | 47.3 ± 0.9 | 28.7 ± 0.9 | +0.789 | 2E−32 | Predicted: twitchin isoform X1 |
| GFTU01001635.1 | 47.2 ± 0.9 | 28.6 ± 0.9 | +0.789 | 2E−32 | Predicted: twitchin isoform X1 |
| GFTU01001634.1 | 47.7 ± 0.9 | 28.9 ± 0.9 | +0.789 | 2E−32 | Predicted: twitchin isoform X23 |
| GFTU01001636.1 | 47.5 ± 0.9 | 28.8 ± 0.9 | +0.789 | 2E−32 | Predicted: twitchin isoform X25 |
| GFTU01001637.1 | 47.3 ± 1.0 | 28.7 ± 0.9 | +0.789 | 2E−32 | Predicted: twitchin isoform X25 |
| GFTU01001633.1 | 47.3 ± 0.9 | 28.6 ± 0.9 | +0.789 | 2E−32 | Predicted: twitchin isoform X24 |
| GFTU01012458.1 | 54.6 ± 0.3 | 34.6 ± 0.3 | +0.723 | 1E−26 | Predicted: twitchin isoform X16 |
| GFTU01012456.1 | 46.6 ± 3.3 | 32.8 ± 1.2 | +0.567 | 2E−11 | Predicted: twitchin isoform X13 |
| GFTU01001910.1 | 1319.6 ± 93.2 | 893.8 ± 69.3 | +0.633 | 3E−21 | Predicted: arginine kinase |
| GFTU01009195.1 | 633.2 ± 64.8 | 428.9 ± 54.4 | +0.618 | 2E−13 | Predicted: myosin light chain alkali |
| GFTU01001677.1 | 1054.6 ± 59.2 | 786.0 ± 22.8 | +0.506 | 6E−18 | Predicted: myosin heavy chain, muscle isoform X30 |
| GFTU01002981.1 | 76.1 ± 1.5 | 51.4 ± 0.3 | +0.633 | 1E−17 | Predicted: PDZ and LIM domain protein 3 isoform X3 |
| GFTU01000140.1 | 79.3 ± 1.4 | 53.6 ± 0.7 | +0.629 | 3E−17 | Predicted: PDZ and LIM domain protein 3 isoform X4 |
| GFTU01012488.1 | 35.2 ± 1.0 | 24.1 ± 0.1 | +0.609 | 5E−16 | Predicted: sarcalumenin isoform X2 |
| GFTU01006510.1 | 1983.5 ± 71.2 | 1262.8 ± 211.9 | +0.598 | 1E−4 | Actin muscle protein |
| GFTU01009078.1 | 9.6 ± 0.0 | 6.5 ± 0.6 | +0.601 | 2E−07 | Predicted: muscle M-line assembly protein unc-89 isoform X1 |
| GFTU01012457.1 | 40.3 ± 2.3 | 27.9 ± 0.5 | +0.595 | 2E−14 | Predicted: muscle M-line assembly protein unc-89-like |
| GFTU01012454.1 | 37.0 ± 2.0 | 25.8 ± 0.8 | +0.588 | 2E−13 | Predicted: muscle M-line assembly protein unc-89-like |
| GFTU01001628.1 | 38.2 ± 2.2 | 26.7 ± 0.8 | +0.582 | 3E−13 | Predicted: muscle M-line assembly protein unc-89-like |
| GFTU01007383.1 | 33.9 ± 0.0 | 23.2 ± 3.4 | +0.588 | 2E−08 | Predicted: titin isoform X2 |
| GFTU01010045.1 | 28.3 ± 2.7 | 20.2 ± 1.2 | +0.533 | 5E−08 | Predicted: titin-like, partial |
| GFTU01008365.1 | 28.6 ± 1.1 | 20.2 ± 2.0 | +0.566 | 2E−12 | Predicted: ryanodine receptor |
| GFTU01008363.1 | 28.3 ± 1.0 | 20.0 ± 1.9 | +0.565 | 1E−12 | Predicted: ryanodine receptor |
| GFTU01002369.1 | 19.8 ± 1.0 | 13.3 ± 3.0 | +0.547 | 3E−04 | d-beta-hydroxybutyrate dehydrogenase, putative |
| GFTU01014213.1 | 1182.4 ± 91.5 | 871.7 ± 9.5 | +0.519 | 6E−18 | Predicted: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type isoform X3 (SERCA) |
| GFTU01010911.1 | 8.2 ± 1.4 | 5.9 ± 0.6 | +0.505 | 1E−4 | Predicted: dynein beta chain, ciliary-like |
| Proteases | |||||
| GFTU01004471.1 | 134.2 ± 14.5 | 64.3 ± 13.3 | +0.972 | 5E−14 | Predicted: cathepsin L1 |
| GFTU01003425.1 | 135.3 ± 7.5 | 62.6 ± 11.3 | +0.871 | 2E−07 | Hypothetical protein g.35645 (peptidase M1 aminopeptidase N) |
| GFTU01001733.1 | 23.5 ± 0.4 | 12.8 ± 2.2 | +0.872 | 6E−16 | Hypothetical protein g.22606 (peptidase m1 aminopeptidase N family) |
| GFTU01012473.1 | 16.5 ± 2.1 | 9.6 ± 0.8 | +0.716 | 1E−06 | Predicted: prostatic acid phosphatase-like |
| GFTU01013458.1 | 172.1 ± 18.2 | 92.9 ± 17.8 | +0.638 | 9E−04 | Hypothetical protein g.31954 (chitin-binding domain type 2, peptidase M1 aminopeptidase N family) |
| GFTU01002473.1 | 27.0 ± 0.7 | 18.3 ± 1.2 | +0.593 | 2E−08 | Predicted: neprilysin isoform X1 (membrane metallo-endopeptidase) |
| GFTU01016619.1 | 59.8 ± 2.1 | 42.2 ± 3.8 | +0.563 | 2E−10 | Predicted: leucyl-cystinyl aminopeptidase-like |
| GFTU01003281.1 | 76.7 ± 4.9 | 54.7 ± 3.4 | +0.553 | 2E−12 | Predicted: membrane metallo-endopeptidase-like 1 isoform X3 |
| GFTU01012719.1 | 44.2 ± 2.6 | 32.5 ± 0.8 | +0.515 | 2E−11 | Hypothetical protein g.32075 (invasion associated secreted endopeptidase; provisional) |
| Extracellular matrix | |||||
| GFTU01000571.1 | 6.3 ± 0.3 | 3.7 ± 0.1 | +0.793 | 2E−16 | Predicted: neurogenic locus notch homolog protein 3-like, partial |
| GFTU01005879.1 | 28.4 ± 1.7 | 18.2 ± 0.5 | +0.686 | 5E−14 | Predicted: neurogenic locus notch homolog protein 1 |
| GFTU01007832.1 | 24.3 ± 2.2 | 15.7 ± 2.3 | +0.658 | 1E−9 | Predicted: membrane-associated guanylate kinase, WW and PDZ domain-containing protein 3-like |
| GFTU01007834.1 | 23.9 ± 2.2 | 15.4 ± 2.5 | +0.657 | 3E−09 | Predicted: membrane-associated guanylate kinase, WW and PDZ domain-containing protein 3-like |
| GFTU01007833.1 | 25.1 ± 2.4 | 16.2 ± 2.0 | +0.655 | 9E−10 | Predicted: membrane-associated guanylate kinase, WW and PDZ domain-containing protein 3-like |
| GFTU01007831.1 | 25.5 ± 2.4 | 16.5 ± 1.7 | +0.655 | 3E−10 | Predicted: membrane-associated guanylate kinase, WW and PDZ domain-containing protein 3-like |
| GFTU01011451.1 | 8.3 ± 0.5 | 5.1 ± 0.0 | +0.709 | 4E−11 | Hypothetical protein g.48125 (calcium-binding EGF-like domain) |
| GFTU01001449.1 | 7.1 ± 0.2 | 4.8 ± 0.0 | +0.622 | 1E−13 | Predicted: uncharacterized protein LOC658528 (calcium-binding EGF-like domain) |
| GFTU01001450.1 | 6.6 ± 0.7 | 4.5 ± 0.1 | +0.583 | 2E−06 | Predicted: uncharacterized protein LOC106669909 (calcium-binding EGF-like domain) |
| GFTU01001448.1 | 6.4 ± 0.2 | 4.6 ± 0.1 | +0.540 | 4E−09 | Predicted: uncharacterized protein LOC658528 (calcium-binding EGF-like domain) |
| GFTU01011448.1 | 5.4 ± 0.5 | 3.7 ± 0.4 | +0.509 | 1E−03 | Predicted: uncharacterized protein LOC106669909 (calcium-binding EGF-like domain) |
| GFTU01009271.1 | 32.3 ± 0.6 | 23.1 ± 0.6 | +0.560 | 6E−17 | Predicted: basement membrane-specific heparan sulfate proteoglycan core protein isoform X19 |
| GFTU01009275.1 | 30.4 ± 0.6 | 21.9 ± 0.6 | +0.551 | 2E−16 | Predicted: basement membrane-specific heparan sulfate proteoglycan core protein isoform X7 |
| GFTU01001084.1 | 31.0 ± 0.6 | 22.4 ± 0.6 | +0.549 | 3E−16 | Predicted: basement membrane-specific heparan sulfate proteoglycan core protein isoform X8 |
| GFTU01009272.1 | 31.2 ± 0.4 | 22.8 ± 0.5 | +0.532 | 7E−16 | Predicted: basement membrane-specific heparan sulfate proteoglycan core protein isoform X6 |
| GFTU01009276.1 | 30.1 ± 0.5 | 22.0 ± 0.5 | +0.531 | 7E−16 | Predicted: basement membrane-specific heparan sulfate proteoglycan core protein isoform X13 |
| GFTU01009270.1 | 30.3 ± 0.4 | 22.2 ± 0.5 | +0.529 | 9E−16 | Predicted: basement membrane-specific heparan sulfate proteoglycan core protein isoform X21 |
| GFTU01009274.1 | 30.2 ± 0.4 | 22.1 ± 0.5 | +0.528 | 1E−15 | Predicted: basement membrane-specific heparan sulfate proteoglycan core protein isoform X15 |
| GFTU01009267.1 | 30.7 ± 0.4 | 22.5 ± 0.5 | +0.526 | 2E−15 | Predicted: basement membrane-specific heparan sulfate proteoglycan core protein |
| GFTU01003839.1 | 31.1 ± 2.1 | 22.2 ± 0.6 | +0.543 | 5E−09 | Predicted: protein mesh isoform X2 |
| Nucleic acid binding | |||||
| GFTU01010183.1 | 10.0 ± 0.1 | 5.2 ± 0.0 | +0.905 | 1E−14 | Predicted: piggyBac transposable element-derived protein 4-like |
| GFTU01001044.1 | 19.5 ± 2.3 | 11.8 ± 2.5 | +0.532 | 8E−03 | Predicted: RNA-directed DNA polymerase from mobile element jockey-like |
| GFTU01010403.1 | 11.5 ± 0.1 | 7.6 ± 0.6 | +0.592 | 4E−06 | Predicted: retrovirus-related polymerase (Pol) polyprotein from transposon 17.6 |
| GFTU01004281.1 | 5.7 ± 0.3 | 3.8 ± 0.9 | +0.554 | 4E−04 | Predicted: uncharacterized protein K02A2.6-like (2 integrases) |
| GFTU01004309.1 | 39.6 ± 1.7 | 28.3 ± 4.9 | +0.522 | 6E−06 | Hypothetical protein g.15643 (integrase) |
| GFTU01007993.1 | 11.2 ± 1.1 | 7.8 ± 0.6 | +0.521 | 2E−04 | Predicted: nuclear factor interleukin-3-regulated protein |
| Detoxification | |||||
| GFTU01009741.1 | 23.5 ± 0.6 | 13.8 ± 0.3 | +0.784 | 2E−14 | Predicted: venom carboxylesterase-6 |
| GFTU01002688.1 | 17.8 ± 5.7 | 6.4 ± 1.4 | +0.691 | 5E−04 | Predicted: cytochrome P450 4C1-like |
| GFTU01002689.1 | 13.1 ± 4.6 | 4.9 ± 1.2 | +0.593 | 3E−03 | Predicted: cytochrome P450 4C1-like |
| GFTU01000057.1 | 7.8 ± 0.4 | 5.1 ± 0.4 | +0.605 | 1E−05 | Predicted: gamma-aminobutyric acid receptor subunit alpha-6-like |
Transcripts were classified into functional categories according to the putative identification assigned by a blastx search. Contig accession numbers correspond to BioProject PRJNA393620. CYp, chrysanthemum yellows phytoplasmas; FDp, flavescence dorée phytoplasmas; FPKM, fragments per kilobase million.
TABLE 2.
Overview of Euscelidius variegatus transcripts downregulated during chrysanthemum yellows phytoplasma infection compared with insects infected by flavescence dorée phytoplasmasa
| Contig accession no. | FPKM (avg ±SD) |
Ln fold change | P value | Sequence description | |
|---|---|---|---|---|---|
| Eva_CY | Eva_FD | ||||
| Immune response | |||||
| GFTU01001177.1 | 34.4 ± 7.3 | 70.6 ± 2.9 | −0.719 | 7E−06 | Predicted: mitogen-activated protein kinase kinase kinase 12 isoform X2 |
| GFTU01016523.1 | 40.5 ± 6.2 | 70.1 ± 1.9 | −0.654 | 2E−14 | Predicted: serine protease snake-like isoform X2 |
| GFTU01005213.1 | 16.7 ± 4.3 | 30.7 ± 0.4 | −0.635 | 2E−05 | Hypothetical protein g.9121 (Kazal-type 1 serine protease inhibitor-like protein type gamma) |
| GFTU01006779.1 | 195.0 ± 11.6 | 307.4 ± 5.2 | −0.536 | 3E−14 | Hypothetical protein g.7830 (Kazal-type 1 serine protease inhibitor-like protein type gamma) |
| GFTU01007903.1 | 6.5 ± 0.3 | 11.3 ± 0.3 | −0.587 | 4E−05 | Predicted: protein farnesyltransferase/ geranylgeranyltransferase type-1 subunit alpha |
| GFTU01003663.1 | 8.0 ± 0.1 | 13.2 ± 1.2 | −0.558 | 3E−06 | Predicted: Kruppel-like factor 10 |
| GFTU01003662.1 | 5.7 ± 0.1 | 9.4 ± 0.9 | −0.534 | 2E−05 | Predicted: Kruppel-like factor 10 |
| GFTU01005275.1 | 16.7 ± 2.1 | 27.4 ± 0.2 | −0.529 | 1E−04 | Predicted: circadian clock-controlled protein-like |
| GFTU01002709.1 | 11.6 ± 0.9 | 18.5 ± 1.4 | −0.519 | 4E−06 | Predicted: heat shock protein 68-like |
| GFTU01007012.1 | 8.2 ± 2.1 | 14.0 ± 2.3 | −0.512 | 4E−34 | Predicted: pancreatic triacylglycerol lipase-like |
| Movement and energy metabolism | |||||
| GFTU01009515.1 | 4.7 ± 0.0 | 8.3 ± 0.0 | −0.620 | 1E−06 | Dihydropyridine-sensitive l-type calcium channel (DHPR) |
| Proteases | |||||
| GFTU01013038.1 | 16.0 ± 3.0 | 27.5 ± 2.4 | −0.571 | 3E−05 | Predicted: cathepsin D-like |
| Detoxification | |||||
| GFTU01002121.1 | 15.0 ± 1.2 | 24.2 ± 0.7 | −0.541 | 4E−07 | Predicted: probable cytochrome P4506a14 |
Transcripts were classified into functional categories according to the putative identification assigned by a blastx search. Contig accession numbers correspond to BioProject PRJNA393620. CYp, chrysanthemum yellows phytoplasmas; FDp, flavescence dorée phytoplasmas; FPKM, fragments per kilobase million.
Cellular and humoral immunity in response to phytoplasma infections.
To investigate the effects of phytoplasma presence on immune response, gene expression analysis of selected transcripts and enzymatic activity and biological assays were performed (Fig. 1A; see also Tables S5, S6, and S7). Healthy controls (Eva_H), not exposed to phytoplasmas and PCR negative, were included in the following experiments to highlight the mechanisms underpinning the differential effects on E. variegatus immune response upon infection with the two phytoplasma species. Gender was taken into account, but whenever no sex-related differences were recorded within the same category, data were pooled. RT-qPCR validation was run on 42 samples (each made up of five pooled insects), phenoloxidase (PO) activity was measured for 36 samples (each made up of hemolymph collected from five insects), and pigmentation and immunocompetence assays were tested on 170 and 46 specimens, respectively.
FIG 1.
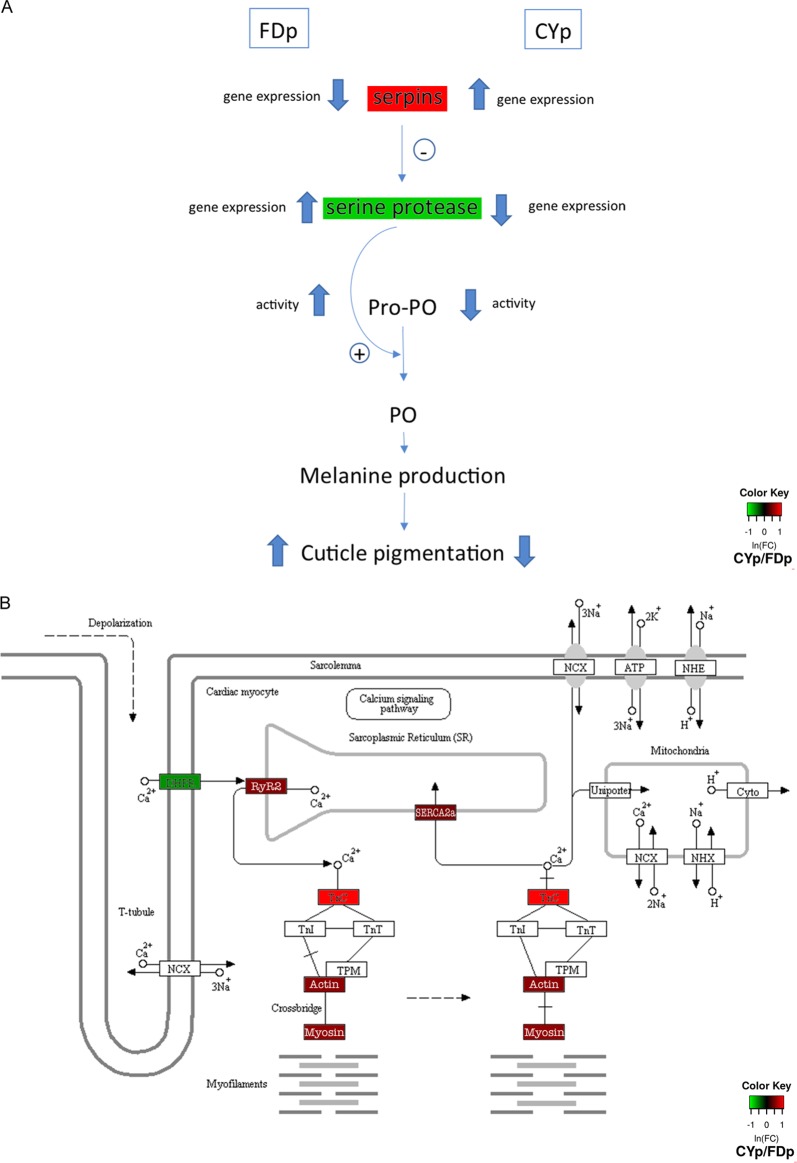
Regulation of prophenoloxidase cascade and muscle contraction pathway. (A) During Euscelidius variegatus infection with flavescence dorée (FDp) or chrysanthemum yellows (CYp) phytoplasmas, the prophenoloxidase (ProPO) cascade, which produces melanine as innate immunity response, is regulated in reverse manners, being activated in FDp and inhibited in CYp-infected insects. (B) The expression of most of genes involved in muscle contraction was altered upon CYp infection (indicated in colored boxes). DHPR, dihydropyridine receptor; RyR2, ryanodine receptor; TnC, troponin C; SERCA2a, calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum. Data in the heat map of expression are indicated as follows: red and green correspond to up- and downregulation levels, respectively, during CYp infection compared with infection by FDp. (Republished from reference 70 with permission.)
(i) Gene expression.
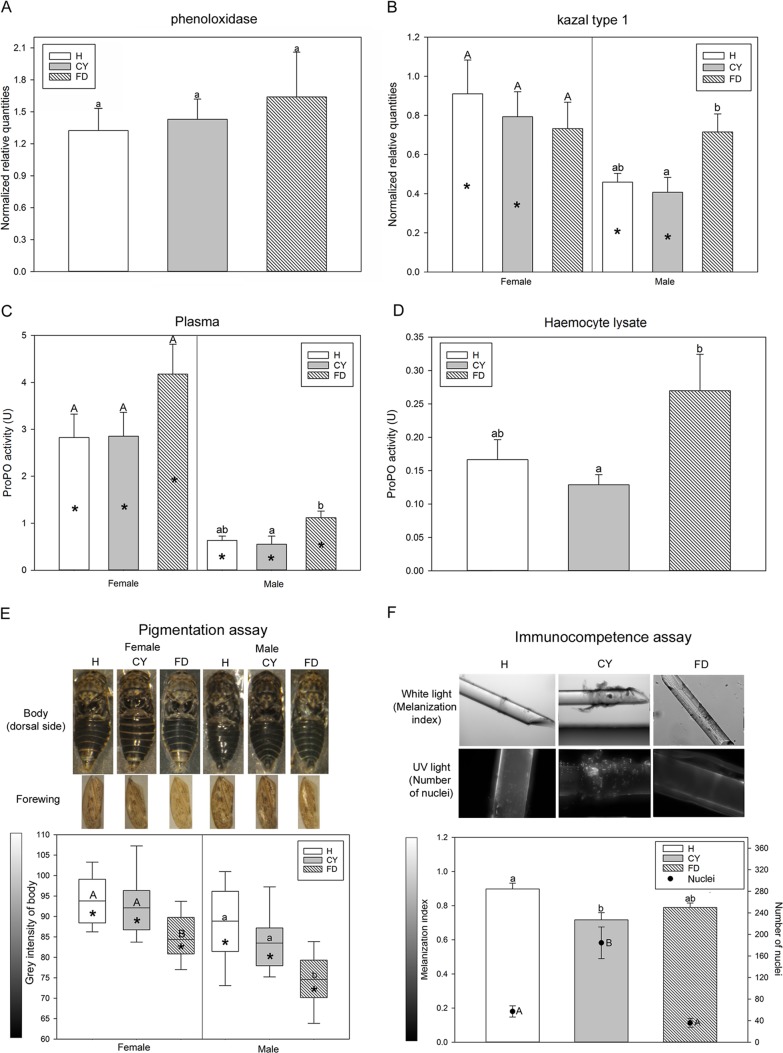
Kazal-type 1 serine protease inhibitor and phenoloxidase genes were selected from the RNA-seq results and a literature search (14), respectively, and analyzed by RT-qPCR in CYp-infected, FDp-infected, and healthy insects (Eva_H, Eva_CY, and Eva_FD). Similar levels of phenoloxidase transcripts were recorded in the insects, regardless of the sex and the infection status (Fig. 2A), whereas Kazal-type 1 transcripts were significantly more abundant in Eva_FD males than in Eva_CY ones (P = 0.022) (Fig. 2B). Transcripts of the same gene were significantly more abundant in healthy females than in healthy males (P = 0.002) and in CYp-infected females than in CYp-infected males (P = 0.023) (Table S5). Upregulation of Kazal-type 1 serine protease inhibitor in Eva_FD confirmed the differential expression results obtained by RNA-seq analysis (Table 2).
FIG 2.
Phytoplasma infection modulates insect immune response. (A and B) The gene expression profile data are indicated as mean normalized relative quantities ± standard errors of results from analysis of phenoloxidase (A) and Kazal-type 1 serine protease inhibitor (B) in healthy (H; open columns) Euscelidius variegatus insects or insects infected by chrysanthemum yellows (CYp [CY; gray columns]) or flavescence dorée (FDp [FD; diagonally striped columns]) phytoplasmas. (C and D) Mean enzymatic activities (U) ± standard errors of data corresponding to prophenoloxidase (ProPO) measured in plasma (C) and hemocyte lysate (D) fractions of H, CY, and FD insects are indicated. (E) Box plot of intensity data (gray) calculated for bodies (dorsal side) of H, CY, and FD insects. Intensity data (gray) range from 0 (black) to 255 (white). Within the same category (H, CY, or FD), asterisks indicate significant differences between females and males of each category. Within the same gender, different letters indicate significant differences among the categories (uppercase letters for females and lowercase letters for males). In all cases in which no sex-related differences were recorded within the same category, female and male data were pooled. (F) Mean melanization index (MI; bars) and mean numbers of adherent cells (dots) (± standard error) measured on nylon threads implanted into H, CY, and FD insects. MI data range from 0 (black) to 1 (white). Different letters indicate significant differences among the categories (lowercase letters for MI and uppercase letters for adherent cells).
(ii) Enzymatic activity.
Phenoloxidase (PO) and prophenoloxidase (ProPO) activities associated with hemocytes and the plasma fraction of hemolymph were quantified. The specific inhibitor phenylthiourea inhibited PO and ProPO activities in all assays with the exception of PO in hemocytes (data not further analyzed). The PO activities of the plasma fractions were similar irrespective of sex and infection status (Table S6). The plasma ProPO activities were significantly lower in males than in females, irrespective of the infection status (P < 0.001, P < 0.001, and P = 0.018, for H, CY, and FD, respectively); this activity was higher in FDp-infected insects than in CYp-infected insects (2-fold increase), although the difference was significant only for males (P = 0.046) (Fig. 2C). ProPO activities were similar in hemocytes from females and males of each infection status, whereas the enzymatic activity of Eva_FD was double that of Eva_CY (P = 0.017) (Fig. 2D). Steeper slopes of the linear phases of data from Eva_FD assays further confirmed faster enzymatic reactions in FDp-infected insects (Table S6).
(iii) Pigmentation assay.
Since PO activity may be correlated to cuticular color (15), pigmentation of bodies (dorsal side) and forewings of healthy and phytoplasma-infected E. variegatus was further explored. Pigmentation was expressed as gray intensity values: 0 for black to 255 for white (Fig. 2E). Considering the three experimental conditions (Eva_H, Eva_CY, and Eva_FD), males always showed significantly darker bodies and wings than females (P = 0.005 for healthy bodies and P < 0.001 for all other comparisons) (Table S7). Wing pigmentation did not show any significant variation in the presence of phytoplasmas, while bodies of FDp-infected female and male insects were significantly darker than those of healthy and CYp-infected ones (for females, P < 0.001 for both comparisons; for males, P < 0.001 and P = 0.003 for H versus FD and CY versus FD, respectively) (Fig. 2E).
(iv) Immunocompetence assay.
To determine whether the phytoplasma presence could influence insect immune responses, nylon threads were implanted in insect abdomens to measure melanization as well as the number of hemocytes that adhered to the threads (Fig. 2F; see also Table S7). There was no influence of sex on melanization index (MI) or on the number of cells that adhered to the nylon threads, irrespective of the infection status, but there was a significant difference in the MI values determined for nylon threads implanted in CYp-infected E. variegatus compared to those implanted in healthy insects (Fig. 2F, bars). Up to five times more hemocytes adhered to nylon threads implanted into CYp-infected E. variegatus than adhered to those implanted into H and FDp-infected insects, and this difference was significant (P < 0.001) in both cases (Fig. 2F, dots).
Movement and energy metabolism.
Genes involved in muscle contraction and synthesis of intermediate metabolites for energy production were selected among those differentially regulated according to the RNA-seq and literature search results (16) (Fig. 1B) (Table S5). To investigate the effects of phytoplasma infection on insect mobility and respiration rate, several parameters were measured (Fig. 3E, F, G, and H) (Table S8). Healthy controls (Eva_H) were included in the following experiments to better describe the metabolic response of E. variegatus challenged by the two phytoplasmas, and whenever no sex-related differences were recorded within the same category, data were pooled. RT-qPCR validation was run on 42 samples (each made up of five pooled insects), movement assays were performed on 138 specimens, and CO2 production was measured in 24 insect groups (each made up of three specimens).
FIG 3.
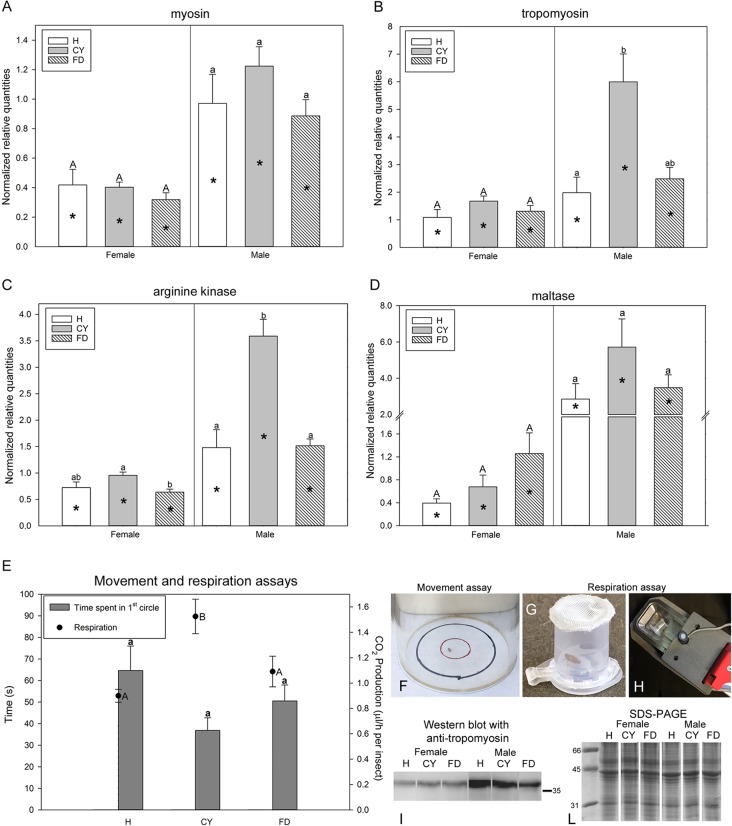
Chrysanthemum yellows phytoplasma infection increases energy metabolism. (A to D) Gene expression profiles are indicated as mean normalized relative quantities ± standard errors for myosin light chain (A), tropomyosin (B), arginine kinase (C), and maltase (D) in Euscelidius variegatus insects that were healthy (H) and in those that were infected by chrysanthemum yellows (CYp) or by flavescence dorée (FDp) phytoplasmas. (E) Mean time (in seconds) required to leave the first circle (bars [“Time spent in 1st circle”]) and mean CO2 production (dots [“Respiration”]) ± standard errors measured in H, CY, and FD insects. (F to H) Concentric circles used for movement assay (F), insect minicage (G), and leaf minichamber (H) used for respiration assay. (I and L) Western blot with anti-tropomyosin antisera (I) and SDS-PAGE for internal loading control (L) of total proteins extracted from H, CY, or FD insects. Within the same category (H, CY, or FD), asterisks indicate significant differences between females and males of each category. Within the same gender, different letters indicate significant differences among the categories (uppercase letters for females and lowercase letters for males). In all cases in which no sex-related differences were recorded within the same category, female and male data were pooled.
(i) Gene expression.
Levels of myosin light chain, tropomyosin, arginine kinase (AK), and maltase were analyzed by RT-qPCR in insects under the three experimental conditions (Eva_H, Eva_CY, and Eva_FD). The four analyzed transcripts were significantly more abundant in males than in females, regardless of the infection status, with the exception of tropomyosin and arginine kinase in healthy insects (Fig. 3A to D; see also Table S5). In general, CYp-infected males showed higher transcript levels than healthy and FDp-infected males. These differences were significant only for tropomyosin, with about three times more transcripts in CYp-infected males than in healthy males (P = 0.018), and for arginine kinase, with about twice as many transcripts in CYp-infected males as in healthy and FDp-infected ones (P < 0.001 for both comparisons). Arginine kinase transcripts were also significantly more abundant in CYp-infected females than in FDp-infected ones (P = 0.024) (Fig. 3C). Upregulation of arginine kinase and maltase in Eva_CY confirmed the differential expression results of the RNA-seq analysis (Table 1).
(ii) Protein expression.
To further characterize the differential expression levels of proteins involved in movement, Western blot analysis was performed on healthy and phytoplasma-infected insects with antitropomyosin commercial antisera (Fig. 3I; see also Fig. S1 in the supplemental material). Tropomyosin was more highly expressed in males than in females, regardless of the infection status, confirming the transcriptional analyses. Nevertheless, there were no evident differences in protein expression levels among the Eva_H, Eva_CY, and Eva_FD categories.
(iii) Movement and respiration functional assays.
Movement parameters (permanence time in a two-circle arena and numbers of jumps) showed no significant variation either between males and females or among the three experimental conditions, although a trend of faster movement was observed for CYp-infected males than for healthy and FDp-infected ones (Fig. 3E, bars; see also Table S8). Consistently, measurements performed with the gas analyzer showed that Eva_CY produced a significantly larger amount of CO2 than Eva_H and Eva_FD, irrespective of sex (P = 0.002 and P = 0.027 for H versus CY and CY versus FD, respectively) (Fig. 3E, dots; see also Table S8).
Protease regulation.
Cathepsin L was selected to investigate the effect of phytoplasma infection on protease regulation. This protein is the major component of the gut digestive enzymes in many invertebrates (17) and, among other functions, is involved in controlling symbiont populations (18). Healthy controls (Eva_H) were included in the following experiments, to dissect the effects of the different phytoplasmas on E. variegatus protease regulation.
(i) Gene expression.
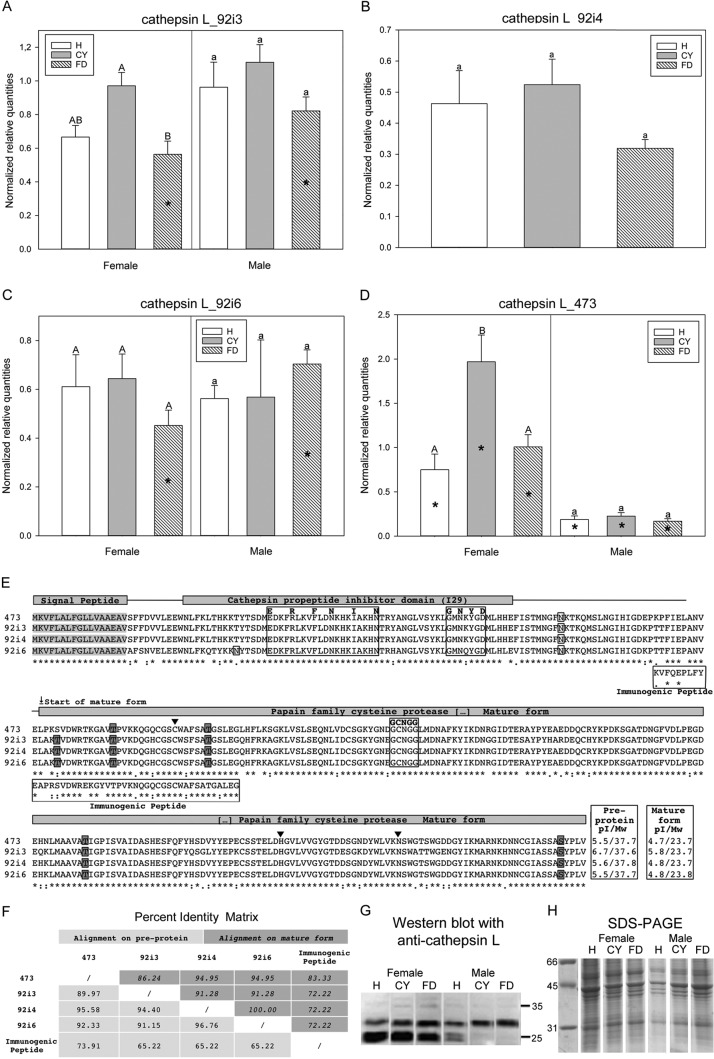
The expression profile of four isoforms (namely, 92i3, 92i4, 92i6, and 473) of cathepsin L was analyzed by RT-qPCR in insects under the three experimental conditions (Eva_H, Eva_CY, and Eva_FD). Isoform 473 was the most highly upregulated in Eva_CY within the protease category (Table 1), whereas isoforms 92i3, 92i4, and 92i6 were selected since, among all the E. variegatus transcripts annotated as “cathepsin L,” they showed the highest identity with the immunogenic peptide recognized by the commercial anti-cathepsin L antibody used for the Western blotting. Significant differences between female and male insects were present for isoform 473, irrespective of the infection status (upregulated in females; P = 0.007, P < 0.001, and P < 0.001 for Eva_H, Eva_CY, and Eva_FD, respectively), and for isoforms 92i3 and 92i6 specifically for FDp-infected E. variegatus (upregulated in males; P = 0.043 and P = 0.017 for 92i3 and 92i6, respectively) (Fig. 4A to D; see also Table S5). Transcripts of isoform 92i3 were significantly upregulated in CYp-infected females versus FDp-infected ones (P = 0.012) (Fig. 4A). Those of isoform 473 were twice as abundant in CYp-infected females as in healthy and FDp-infected ones, and these differences were significant (P = 0.003 and P = 0.012 for H versus CY and CY versus FD, respectively) (Fig. 4D). Upregulation of cathepsin L_473 in Eva_CY compared to Eva_FD confirmed the results of the RNA-seq analysis (Table 1).
FIG 4.
Phytoplasma infection alters protease regulation. (A to D) Gene expression profiles are indicated as mean normalized relative quantities ± standard errors of cathepsin L isoforms 92i3 (A), 92i4 (B), 92i6 (C), and 473 (D) in Euscelidius variegatus insects that were healthy (H) and in those that were infected by chrysanthemum yellows (CYp) or by flavescence dorée (FDp) phytoplasmas. Within the same category (H, CY, or FD), asterisks indicate significant differences between females and males of each category. Within the same gender, different letters indicate significant differences among the categories (uppercase letters for females and lowercase letters for males). In all cases in which no sex-related differences were recorded within the same category, female and male data were pooled. (E) Alignment of predicted amino acid sequences for cathepsin L isoforms. Positions of signal peptide (gray highlighting), cathepsin propeptide inhibitor domains (I29), and mature protease domains are indicated above the alignment. Predicted glycosylation sites are indicated as single boxed amino acid data (white, N-glycosylation; gray, O-glycosylation). Highly conserved regions containing the three typical cathepsin L consensus sequences are boxed and indicated in bold, and required conserved amino acids within each consensus sequence are located above the sequences. The immunogenic peptide sequence is boxed and aligned below the sequences. Three predicted active sites are indicated by black full arrowheads. Data corresponding to the predicted isoelectric point and molecular weight of preproteins and mature forms are indicated at the end of each sequence. (F) Percent identity matrix of the four cathepsin L isoforms and immunogenic peptide. Values for preproteins are indicated at bottom left in the matrix and mature forms in italics at upper right in the matrix. (G and H) Western blot with anti-cathepsin L antisera (G) and SDS-PAGE for internal loading control (H) of total proteins extracted from H, CY, or FD insects.
(ii) Putative protein characterization.
The predicted amino acid sequences of the four cathepsin L isoforms were analyzed (Fig. 4E and F). All four isoforms showed putative signal peptide from amino acid (aa) 1 to aa 16, a proregion containing the propeptide inhibitor domain and a predicted mature protein, including the cysteine protease domain. Putative glycosylation sites were predicted on different isoforms, namely, 2 nitrogen-linked and 5 oxygen-linked isoforms. Isoform 92i6 showed a unique putative N-linked site at position 37, and isoform 473 was missing an O-linked site at position 125. Isoforms 92i4 and 92i6 showed highly similar preproteins and identical mature forms, whereas isoforms 473 and 92i3 were the most diverse ones (Fig. 4F). The immunogenic peptide, recognized by the commercial antibody used for Western blotting, was more similar to isoform 473 than to the other ones (Fig. 4F).
(iii) Protein expression.
To further characterize the effect of phytoplasma infection on the expression of cathepsin L protein, Western blotting was performed on healthy and phytoplasma-infected insects with anti-cathepsin L commercial antisera (Fig. 4G; see also Fig. S1). The antibody was raised to detect both the preprotein and the mature form, as it reacts to the immunogenic peptide indicated in Fig. 4E. Indeed, the Western blot analyses performed with anti-cathepsin L antibody (Fig. 4G) showed a complex pattern: two faint bands with high molecular mass (around 37 to 35 kDa) possibly corresponding to preproteins and two intense bands with low molecular mass (around 27 to 25 kDa) possibly corresponding to the mature forms. Similar band patterns were observed for total proteins of E. variegatus females, irrespective of the infection status. A similar profile, although less intense, was evident for healthy males. Surprisingly, almost no signal from the bands corresponding to the lowest molecular mass (25 kDa) was detected for the phytoplasma-infected males.
DISCUSSION
Relationships between phytoplasmas and their vectors may be pathogenic, neutral, or mutualistic (9). CYp and FDp establish different types of relationships with their vector E. variegatus: the former slightly improves vector fitness, whereas the latter is pathogenic. The transcriptional landscape of E. variegatus infected with the two different phytoplasma species was analyzed, focusing on long-lasting modifications occurring in insects during the response to chronic phytoplasma infection and thus avoiding any possible differences related to CYp and FDp multiplication dynamics occurring during early stages of infection. Sex-specific effects were recorded for several of the tested parameters, as already described for immune response (19, 20) as well as for insect movement and dispersal (21).
Among insects of the Cicadellidae family, few de novo transcriptome assemblies are available; some were obtained from specific insect tissues, namely, salivary glands of Nephotettix cincticeps (22) and Empoasca fabae (23) and intestinal tract of Empoasca vitis (24), and others from whole bodies, such as those of Graminella nigrifrons (25), Homalodisca vitripennis (26), and Zyginidia pullula (27). Interestingly, the transcriptomic response of G. nigrifrons vector to different plant virus infections reveals that the expression of cytoskeleton and immunity genes increases in the presence of the persistent propagative rhabdovirus Maize fine streak virus (28).
Phytoplasma infection modulates insect immune responses.
Molecular and biological analyses indicate that the modulation of the E. variegatus immune response that occurred following FDp infection was different from that seen following CYp infection.
The altered regulation of the immune system was revealed by RNA-seq analysis during infection with both phytoplasmas. Among these, transcripts of the Kazal type 1 serine protease inhibitor were more abundant in FDp-infected insects than in CY-infected insects. Similar serine protease inhibitors have antibacterial activity against bacteria (29) as well as antifungal activity against both plant-pathogenic and entomopathogenic fungi, as inhibitors of microbial serine proteases (30). Serpin was the most highly upregulated transcript in CYp-infected insects, whereas the snake-like serine protease was among the most highly downregulated ones. Clip domain serine proteases, such as the snake-like ones, are involved in the activation of the ProPO proteolytic cascade in invertebrate immune systems (31, 32), while serpins from different arthropod species inhibit clip domain serine proteases by blocking the activation of ProPO melanization pathway (33, 34). The ProPO cascade is involved in melanization and encapsulation processes and provides arthropod immunity to bacteria, fungi, protozoan, and parasites (34). The opposing forms of regulation of these two transcripts correlate with the lower prophenoloxidase activity and with the less intense cuticular pigmentation observed in CYp-infected than in FDp-infected insects. Cuticular color is related to immune response in insects (15), and the darker body pigmentation of FDp-infected E. variegatus suggests stimulation of melanization pathway due to stronger activation of the immune response. The presence of FDp is perceived by the insect as a stress status and therefore elicits intense production of melanin. Indeed, the prophenoloxidase activities of both plasma and hemocyte lysates were more intense in Eva_FD than in Eva_CY and Eva_H. On the other hand, the infection with the two phytoplasmas had no effect either on naturally activated phenoloxidase (PO) activity, representing a good estimation of invertebrate immunocompetence (35), or on the abundance of the corresponding transcripts. This could have been due to the fact that the analyses were performed at late, chronic stages of phytoplasma infection, when colonization of the insect body was complete (12). A burst of activated phenoloxidase is, in fact, expected at the onset of the infectious event, as a defense reaction to the immunological challenge (35), as reported for Micrococcus luteus infection of the leafhopper Circulifer haematoceps (36). Surprisingly, when insects were challenged by an additional stress (wounding through nylon thread), the scenario changed. In the immunocompetence assay, insertion of a nylon thread into the insect body mimicked a parasite invader and induced encapsulation. The response of FDp-infected insects was similar to that of the healthy ones. In contrast, CYp-infected insects showed higher MI levels and higher numbers of hemocytes, indicating a better capacity of these insects to react to and isolate an invader. E. variegatus is a natural vector of CYp and shares the same ecological niches; these factors could have shaped the insect immune system to fight more promptly against incoming pathogenic organisms.
The Kruppel-like factor, a zinc finger DNA-binding protein, is crucial for mediation of white spot syndrome virus (WSSV) infection in two different shrimp species (37), and two transcripts of this gene were regulated in reverse manners in E. variegatus (more abundant in the FDp-infected category), suggesting a role for this protein in the response to phytoplasma infection. On the other hand, hexamerin transcripts were upregulated upon CYp infection. Members of this protein family are effector proteins involved in insect immunity that are inducible upon ingestion of bacteria and have a putative role in gut repair (38). Moreover, in the closely related mollicute-leafhopper association (C. haematoceps/Spiroplasma citri), hexamerin is upregulated following infection and is required for vector survival after spiroplasma inoculation (36). Besides their role in energy metabolism (see below), arthropod arginine kinases (AK) are also involved in stress response and innate immunity; Apis cerana AK is induced by abiotic and biotic stresses (39), pacific oyster AK modulates bactericidal immune response in hemolymph (40), and AK from Fenneropenaeus chinensis shrimp has been hypothesized to be the putative receptor of the WSSV virus envelope protein (41). AK transcript was upregulated upon CYp infection, suggesting different potential roles for this protein during infection with the two phytoplasmas.
Disulfide bonds are redox-controlled switches for pathogen invasion and are involved in regulating pathogen entry into the endocytic pathway of vertebrates (42) and some invertebrates (43, 44). Recently, vesicle-mediated colonization of salivary glands has been suggested for CYp infection of E. variegatus (12). Consistently, the protein disulfide-isomerase (five transcripts) and the gamma interferon-inducible lysosomal thiol genes were upregulated upon CYp infection, supporting the involvement of the endocytic pathway in phytoplasma colonization of the host, as described for Leishmania, Listeria, and Chlamydia spp. (42). Other bacterial pathogens have developed strategies to interfere with host lipidation mechanisms (45). For example, Salmonella enterica and Legionella pneumophila exploit host prenylation to direct effector proteins to the pathogen-containing vacuole of the host cell (46). Interestingly, transcripts of the farnesyl-geranylgeranyl transferases, the key enzymes of the prenylation pathway, were regulated in a reverse manner in E. variegatus (downregulated in CYp-infected insects), suggesting different alterations of vesicular trafficking upon infection with CYp and FDp. Phytoplasma may modulate the host metabolism through active secretion of effector molecules, and the diversity of the effector arsenals among phytoplasmas (2, 47) may explain the contrasting transcription profiles of this gene.
Chrysanthemum yellows phytoplasma infection increases energy metabolism.
Molecular and biological analyses indicate activation of E. variegatus energy production metabolism and increased locomotion activity upon CYp infection. According with RNA-seq results, movement and energy production metabolism was the functional category with the highest number of gene transcripts altered upon phytoplasma infection.
Titin, twitchin, and protein unc-89 are members of the giant cytoskeletal kinase family that mediate sensing and transduction of mechanical signals in the myofibril. These big proteins display elastic conformational deformation and regulate muscle tissue in adaptation to external stimuli (48). Several isoforms of these gene transcripts were upregulated in CYp-infected E. variegatus. These kinases participate in regulating protein turnover in muscle, and, in particular, unc-89 regulates ubiquitin-mediated protein degradation, through recruitment of E3 ubiquitin ligases (48), which indeed was also upregulated upon CYp infection.
Two isoforms of PDZ and LIM domain protein 3 were upregulated in CYp-infected insects. PDZ/LIM genes encode a large group of proteins that play important and diverse biological roles but that can all functionally influence or be associated with the actin cytoskeleton (49). Ryanodine receptor (RYR) is the main calcium release complex of the sarcoplasmic reticulum involved in the excitation-contraction coupling of muscle cells (50), and the dihydropyridine receptor (DHPR) is the plasma membrane L-type calcium channel involved in opening of the RYR by a calcium-induced calcium release mechanism (51). Transcripts of these genes were inversely regulated (RYR upregulated and DHPR downregulated) upon CYp infection, suggesting altered Ca2+ regulation in the cytosol of muscle cells in response to phytoplasmas. Indeed, transcripts of the calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum (SERCA) and of the sarcalumenin were upregulated in CYp-infected insects. The former is a pump involved in translocation of cytosolic calcium into the sarcoplasmic reticulum to allow relaxation of muscle fibers (16); the latter is a calcium-binding protein involved in fine regulation of cellular calcium storage (52). Moreover, transcripts of the main proteins involved in contraction and cytoskeletal motion, namely, tropomyosin, troponin, myosin, actin, and dynein (16), were all upregulated upon CYp infection. The analysis of tropomyosin confirmed the stronger expression in males than in females, as revealed by RT-qPCR and Western blotting, but no differences were evident among the different infection categories (H, CY, and FD), possibly due to the presence of several isoforms derived from alternative splicing, a well-known phenomenon for this gene (53). Increased insect movement has been observed in some pathogen-vector associations, such as Diaphorina citri infected with “Candidatus Liberibacter asiaticus” (54) and Bombyx mori with Bombyx mori nuclear polyhedrosis virus (BmNPV) (55). We tested the intriguing hypothesis that CY phytoplasma, which is naturally transmitted by the insect host, can manipulate the vector movement to increase its transmission, but the parameters recorded during the movement assays did not clearly support this hypothesis. Despite that, muscle contraction is also involved in active insect respiration, so the higher expression of the genes in CYp-infected insects mentioned above could have been related to an increase in the respiration rate. This was indeed the case, as higher CO2 levels were produced in the respiration assay by CYp-infected E. variegatus than by FDp-infected and healthy insects. Additionally, the upregulation of maltase, hydroxybutyrate dehydrogenase, and arginine kinase (AK) transcripts in CYp-infected E. variegatus indicates a stronger activation of the energy production metabolism. Altogether, these data point to augmented movement in CYp-infected insects which may positively influence phytoplasma transmission.
Phytoplasma infection alters protease regulation.
Cathepsins are proteases generally stored in lysosomes and are involved in several processes such as development, apoptosis, and immunity of arthropods (17, 56). Upon CYp infection, transcripts of cathepsin L were upregulated and those of cathepsin D downregulated, suggesting different roles of these enzymes in response to phytoplasma infection. To confirm the RNA-seq analysis results, four isoforms were chosen among all the E. variegatus “cathepsin L” transcripts: 473, derived from differential expression analysis, and 92i3, 92i4, and 92i6, showing the highest identity with the immunogenic peptide recognized by the anti-cathepsin L antibody used in Western blotting. The upregulation of cathepsin L was confirmed by RT-qPCR for three of the four isoforms upon CYp infection in comparison with FDp infection. The same upregulation was not present at the protein level. Molecular weights of cathepsin L mature proteins differed from the theoretical ones, and this may be explained by differences in glycosylation, one of the posttranslational processes occurring during cathepsin maturation (57). Indeed, the anti-cathepsin L antibody detected proteins of different sizes, and, intriguingly, the smallest ones were poorly present in healthy males and nearly absent in phytoplasma-infected males. As transcripts of isoform 473 were significant upregulated in females, these smallest protein bands presumably represented its mature forms. Indeed, isoform 473 showed one fewer glycosylation site than other isoforms and could migrate faster. The absence of the mature form of this isoform in infected males might indicate that phytoplasma presence could prevent cathepsin L maturation. Expression of this gene is altered upon microbial infection, either in pathogenic combinations, such as Serratia marcescens in the pea aphid Acyrthosiphon pisum (18), or in symbiotic associations, such as Burkholderia symbionts ingested by the bean bug Riptortus pedestris (58). Indeed, in both pathogenic and mutualistic associations, bacteria need to avoid lysosomal degradation to establish an intracellular association, and this could be true also for CY and FD phytoplasmas.
In conclusion, transcriptomic and phenotypic results shed some light on the molecular mechanisms underlying the different effects of the two phytoplasmas on the insect vector E. variegatus. Our data show that E. variegatus perceives FD as a pathogen, since it activates an immune response. Lack of natural interactions between FD phytoplasma, mainly restricted to Vitis spp., and the laboratory vector E. variegatus, which does not feed on grapevine, may explain the perception of this phytoplasma as non-self. On the other hand, the long-lasting interactions between CY phytoplasma and E. variegatus (which are sympatric) might have driven the organisms toward a mutualistic relationship.
The prompt and aggressive response to the menace of an external pathogen, mimicked by the nylon thread, may have been due to an immune priming activated by CYp and, together with the increased energy metabolism, is likely to provide an ecological advantage to both the vector and the phytoplasma.
MATERIALS AND METHODS
Insect and phytoplasmas.
E. variegatus isolate to-1 was collected in Piedmont (Italy) and reared on oat, Avena sativa (L.) (7). Chrysanthemum yellows phytoplasma (CYp) was isolated in Italy and maintained by insect transmission on daisy, Chrysanthemum carinatum Schousboe (7). Flavescence dorée phytoplasma (FDp) was isolated in Italy and maintained by insect transmission on broad bean, Vicia faba L. Daisies, broad beans, and oats were all grown from seed in greenhouses (59). For each acquisition access period (AAP), the sanitary status of source plants was confirmed by symptom observation and PCR as detailed in reference 4.
Three experimental conditions were set up. Fifth-instar healthy nymphs were separately fed on (i) healthy daisies and broad beans (Eva_H), (ii) CYp-infected daisies (Eva_CY), or (iii) FDp-infected broad beans (Eva_FD) for an AAP of 7 days and then transferred to oat for a 28-day latency period (LP). At 35 days postacquisition, the surviving insects were sexed, analyzed for phytoplasma infection, and used in experiments.
RNA extraction.
Total RNA was extracted from 64 samples (each made of 5 insects), including nearly 20 (usually 10 female and 10 male) for each experimental condition, using a Direct-zol RNA Mini Prep kit (Zymo Research). RNA was analyzed in a Nanodrop spectophotometer and in an Agilent Technologies model 2100 Expert Bioanalyzer to evaluate the concentration, purity, and quality of the samples.
Phytoplasma detection and quantification.
Total RNA was treated with Turbo RNase-free DNase I (Applied Biosystems). For CYp and FDp diagnosis, cDNA was synthesized from total RNA (800 ng) using a High Capacity cDNA reverse transcription kit (Applied Biosystems). A 2-μl volume of cDNA was used as the template in qPCR analyses performed with iTaq Universal Probes Supermix (Bio-Rad) and primers CYS2Fw/CYS2Rv and TaqMan CYS2 probe (4). The same primers and probe, targeting phytoplasma 16S rRNA, and primers MqFw/MqRv with TaqMan Mq probe, targeting insect 18S rRNA (4), were used to quantify phytoplasma loads. Four serial 100-fold dilutions of pGem-T Easy (Promega) plasmids, harboring portions of ribosomal genes from phytoplasma and insect, were included to calculate phytoplasma 16S/insect 18S ratios.
RNA-seq, differential gene expression, and sequence analyses.
Six micrograms of RNA extracted from insects fed on phytoplasma-infected plants (Eva_CY and Eva_FD) and showing similar phytoplasma amounts were sent to Macrogen (South Korea) for construction and sequencing of cDNA libraries, as detailed in reference 59. Each library was obtained from a pooled sample of 20 males and 20 females. To generate a comprehensive landscape of the E. variegatus transcriptome, the data sets generated by the cDNA libraries (two biological replicates for each condition) were pooled, trimmed by Trimmomatic v0.32 (60), quality checked by FastQC v0.9 (61), de novo assembled using Trinity v2.0.6 (62), and clustered by cd-hit-est (63), with a sequence identity cutoff value of 0.98. Each transcript was analyzed by blastx against the NCBI nr database with a cutoff expected value of 1 E−04. Only transcripts with arthropods corresponding to species representing the best top hits were retained for further analysis. Functional annotation was obtained for each of the selected transcripts by loading the corresponding XML output files in Blast2GO and running the mapping and annotation options with default parameters to retrieve GO terms and assign reliable functions, respectively. In addition, sequences were analyzed for orthology predictions with eggnog (64) using the DIAMOND mapping mode. Open reading frames (ORF) were predicted by TransDecoder (65) using the “–single_best_orf” and “–retain_pfam_hits” options, which allow retention of only the single best ORF for each transcript according to the presence of a significant Pfam hit.
For differentially expressed gene (DEG) identification, DESeq2 package (66) v. 1.14.1 was run on a 60 core and 256-GB RAM local machine, using Ubuntu server 12.04 LTS. DEG selection was based on an adjusted P value of ≤0.01 and log2FC (fold change) values of ≥0.5 for upregulated genes and ≤−0.5 for downregulated genes.
SignalP 4.1 (67), Prosite (68), and GlycoEP (69) were used to predict putative signal peptide sites, active sites, and glycosylation sites on cathepsin L isoforms, respectively. The KEGG pathway database was used for preparation of Fig. 1B (70).
qPCR validation.
Some genes were selected from the RNA-seq results and the literature search and analyzed by RT-qPCR in CYp-infected, FDp-infected, and healthy insects (Eva_H, Eva_CY, and Eva_FD). Reverse transcriptase reactions were performed on the RNA extracted from 42 samples (each made up of five pooled insects): seven samples of males and seven of females under each of the three conditions. These samples included those used for library construction as well as new ones. cDNA was used as the template for qPCR with primers listed in Table S9 in the supplemental material and iTaq Universal SYBR green Supermix (Bio-Rad) with an annealing/extension temperature of 60°C. Primers were checked to target unique isoforms in the whole E. variegatus transcriptome. Among the six putative reference genes tested, those encoding insect elongation factor-1α, glutathione S-transferase, and heat shock protein 70-1 were selected as the most stable under the three conditions (Eva_H, Eva_FD, and Eva_CY) (Table S10) and used for qPCR gene expression analysis, according to methods described in reference 71. Normalized relative quantities for each condition were compared.
Phenoloxidase activity.
The enzymatic activity of naturally activated phenoloxidase (PO) and the proenzyme prophenoloxidase (ProPO) were measured in plasma and hemocyte lysate supernatant (HLS) as described previously (35, 36). About 5 hemolymph samples were tested for each sex and condition. The optical density at 490 nm (OD490) was determined immediately, after 30 min, and then every hour for 15 h using a Bio-Rad microplate spectrophotometer. One unit of activity was defined as an OD490 change of 0.001 per min in the linear phase of reaction. Specificity was tested using phenylthiourea (Sigma) (4 mg/ml) to inhibit enzyme activity.
Pigmentation assay.
The pigmentation of forewing and body (dorsal side) devoid of appendices was calculated through image analysis for about 50 insects for each condition. Images were taken under a D5000 Nikon stereo microscope controlled by Camera Control Pro 2 software and analyzed with Fiji software (72). The outline of the object to be measured was marked by the freehand selection tool. Light conditions and camera and software settings were not changed throughout the process of image acquisition for the whole set of samples. Nevertheless, measurement data for each object were normalized against a white area used as an internal standard. The mean degree of gray intensity was expressed as a numerical reading ranging from 0 for black to 255 for white.
Immunocompetence assay.
A nylon thread (length, 2 to 4 mm; diameter, 80 μm) was implanted in abdomen of CO2-anesthetized insects under a stereoscope. About 50 insects were treated for each condition. Insects were transferred to oat for 72 h, collected, and dissected to recover the nylon thread in 900 μl 10% phosphate-buffered saline (PBS). Following overnight fixation at 4°C (4% paraformaldehyde–0.1% Triton X-100–10% PBS), the threads were washed, stained with DAPI (4′,6-diamidino-2-phenylindole), and photographed under a light and UV microscope. To ensure the best count of nuclei of cells adherent to the thread, three images were taken at different z axes. Image analyses was performed with Fiji software (72), and the melanization index (MI) was calculated as the ratio between the measured integrated densities per surface unit of nylon portions inside and outside the body of each insect. About 15 insects were analyzed for each condition. Numbers of adherent cells were calculated by summing the data from the DAPI-stained nuclei in the three pictures of each thread.
Western blotting.
For each category, proteins were extracted from four samples (each made up of five pooled insects), quantified by Bradford reagent (Bio-Rad), and loaded onto 12% polyacrylamide gels (12 μg/lane), together with prestained and unstained broad-range standards (Bio-Rad) (11). Gels were either stained with colloidal Coomassie stain or blotted on a polyvinylidene difluoride (PVDF) membrane. Membranes were blocked for 1 h (3% bovine serum albumin–Tris-buffered saline with 0.1% Tween [BSA-TBST]), incubated overnight at 4°C with primary antibodies (ab50567 rat-developed anti-tropomyosin and ab200738 rabbit-developed anti-cathepsin L [both diluted 1:1,000 in BSA-TBST]; Abcam plc), washed, incubated for 2 h with corresponding horseradish peroxidase (HRP)-conjugated secondary antibodies (A4416 GAM-HRP and A0545 GAR-HRP [both diluted 1:10,000 in BSA-TBST] [Sigma], respectively), washed, and developed with West Pico SuperSignal chemiluminescent substrate (Pierce) in a VersaDoc 4000 MP system (Bio-Rad). Each experiment was repeated three times.
Movement and respiration assays.
To evaluate insect movement under the three conditions, insects were anesthetized for 30 s and put one at a time into the middle of two concentric circles (2 cm and 6 cm in diameter) drawn on a paper, covered with a glass cylinder (height 20 cm), and continuously observed for 5 min. The time required to leave the two circles and the numbers of jumps were recorded. About 20 insects were tested for each sex and condition.
To evaluate insect respiration, CO2 production was monitored within the standard “broad leaf chamber” of a LCpro+ (ADC BioScientific) gas analyzer, as described for Drosophila (73). To measure gas exchange, groups of three adults (same sex and same category) were put in a minicage (1.5-ml tube; deprived of the bottom and sealed with a net). To allow better survival, 200 μl of feeding solution (12) was put in the minicage cap and covered by a Parafilm layer. For each sex and category, 4 groups were analyzed. Each minicage was left in the chamber for 30 min before measurements (5 reading replicates) were performed. The level of CO2 production, expressed by the analyzer in micromoles per second per square meter, was transformed into microliters per hour per insect in accordance with previously published methods (73).
Statistical analyses.
Depending on whether or not the data were normally distributed, t tests or Mann-Whitney tests were used for sex comparisons and analysis of variance (ANOVA) or Kruskal-Wallis tests for category comparisons (H versus CY versus FD) (Table S11). Tukey or Dunn post hoc tests were used following the ANOVA or Kruskal-Wallis tests, respectively. Whenever no sex-related differences were recorded within the same category, female and male data were pooled. SIGMAPLOT 11 (Systat Software) was used.
Accession number(s).
Reads were loaded into NCBI's Sequence Read Archive (SRA) database with the following accession numbers: SRR5816888, SRR5816889, SRR5816890, and SRR5816891.
Supplementary Material
ACKNOWLEDGMENTS
We thank Brigitte Batailler for helping with immunocompetence assay and Francesco Pennacchio and Gennaro Di Prisco, University of Naples Federico II, for helpful discussion and suggestions.
This work was part of the FitoDigIt Project funded by Fondazione Cassa di Risparmio di Torino, Turin (Italy), within the Richieste Ordinarie 2014 and Richieste Ordinarie 2015 calls. M. Rossi and M. Pegoraro were supported by a fellowship funded by the following grant-making foundations: Fondazione Cassa di Risparmio di Cuneo, Fondazione Cassa di Risparmio di Torino, and Fondazione Cassa di Risparmio di Asti in the framework of the INTEFLAVI project. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
L. Galetto, S. Abbà, C. Marzachì, M. Rossi, M. Vallino, and D. Bosco designed the experiments. S. Abbà performed RNA-seq analysis, bioinformatic analysis, and differential gene expression profiling. L. Galetto performed RT-qPCR validation, statistical analysis, and protein expression experiments. L. Galetto, C. Marzachì, and M. Pesando analyzed phenoloxidase enzymatic activity. M. Rossi, M. Vallino, and S. Abbà performed pigmentation assays. C. Marzachì, N. Arricau-Bouvery, and M.-P. Dubrana performed immunocompetence assays. S. Abbà, M. Rossi, M. Vallino, M. Pesando, and L. Galetto performed movement assays. L. Galetto and W. Chitarra performed respiration assays. M. Pegoraro performed insect rearing and plant production. L. Galetto, S. Abbà, M. Rossi, M. Vallino, and C. Marzachì wrote the paper, and all of us reviewed the manuscript.
We declare that we have no conflicts of interest with the contents of this article.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00042-18.
REFERENCES
- 1.Marcone C. 2014. Molecular biology and pathogenicity of phytoplasmas. Ann Appl Biol 165:199–221. doi: 10.1111/aab.12151. [DOI] [Google Scholar]
- 2.Maejima K, Oshima K, Namba S. 2014. Exploring the phytoplasmas, plant pathogenic bacteria. J Gen Plant Pathol 80:210–221. doi: 10.1007/s10327-014-0512-8. [DOI] [Google Scholar]
- 3.Oshima K, Maejima K, Namba S. 2013. Genomic and evolutionary aspects of phytoplasmas. Front Microbiol 4:230. doi: 10.3389/fmicb.2013.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pacifico D, Galetto L, Rashidi M, Abbà S, Palmano S, Firrao G, Bosco D, Marzachì C. 2015. Decreasing global transcript levels over time suggest that phytoplasma cells enter stationary phase during plant and insect colonization. Appl Environ Microbiol 81:2591–2602. doi: 10.1128/AEM.03096-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oshima K, Ishii Y, Kakizawa S, Sugawara K, Neriya Y, Himeno M, Minato N, Miura C, Shiraishi T, Yamaji Y, Namba S. 2011. Dramatic transcriptional changes in an intracellular parasite enable host switching between plant and insect. PLoS One 6:e23242. doi: 10.1371/journal.pone.0023242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caudwell A, Kuszala C, Larrue J, Bachelier J. 1972. Transmission de la flavescence dorée de la fève à la fève par des cicadelles des genres Euscelis et Euscelidius. Ann Phytopathol 1972:181–189. [Google Scholar]
- 7.Rashidi M, D'Amelio R, Galetto L, Marzachì C, Bosco D. 2014. Interactive transmission of two phytoplasmas by the vector insect. Ann Appl Biol 165:404–413. doi: 10.1111/aab.12146. [DOI] [Google Scholar]
- 8.Bressan A, Clair D, Sémétey O, Boudon-Padieu É. 2005. Effect of two strains of flavescence dorée phytoplasma on the survival and fecundity of the experimental leafhopper vector Euscelidius variegatus Kirschbaum. J Invertebr Pathol 89:144–149. doi: 10.1016/j.jip.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Bosco D, Marzachì C. 2016. Insect transmission of phytoplasmas, p 319–327. In Brown JK. (ed), Vector-mediated transmission of plant pathogens. APS Press, St. Paul, MN. [Google Scholar]
- 10.Abbà S, Galetto L, Carle P, Carrère S, Delledonne M, Foissac X, Palmano S, Veratti F, Marzachì C. 2014. RNA-Seq profile of flavescence dorée phytoplasma in grapevine. BMC Genomics 15:1088. doi: 10.1186/1471-2164-15-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galetto L, Bosco D, Balestrini R, Genre A, Fletcher J, Marzachì C. 2011. The major antigenic membrane protein of ‘Candidatus Phytoplasma asteris’ selectively interacts with ATP synthase and actin of leafhopper vectors. PLoS One 6:e22571. doi: 10.1371/journal.pone.0022571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rashidi M, Galetto L, Bosco D, Bulgarelli A, Vallino M, Veratti F, Marzachì C. 2015. Role of the major antigenic membrane protein in phytoplasma transmission by two insect vector species. BMC Microbiol 15:193. doi: 10.1186/s12866-015-0522-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arricau-Bouvery N, Duret S, Dubrana M-P, Batailler B, Desqué D, Béven L, Danet J-L, Monticone M, Bosco D, Malembic-Maher S, Foissac X. 9 February 2018. Variable membrane protein A of flavescence dorée phytoplasma binds the midgut perimicrovillar membrane of Euscelidius variegatus and promotes adhesion to its epithelial cells. Appl Environ Microbiol doi: 10.1128/AEM.02487-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howell L, Sampson CJ, Xavier MJ, Bolukbasi E, Heck MMS, Williams MJ. 2012. A directed miniscreen for genes involved in the Drosophila anti-parasitoid immune response. Immunogenetics 64:155–161. doi: 10.1007/s00251-011-0571-3. [DOI] [PubMed] [Google Scholar]
- 15.Armitage SAO, Siva-Jothy MT. 2005. Immune function responds to selection for cuticular colour in Tenebrio molitor. Heredity 94:650–656. doi: 10.1038/sj.hdy.6800675. [DOI] [PubMed] [Google Scholar]
- 16.Morano I. 2013. Muscles and motility, p 461–478. In Galizia CG, Lledo P-M (ed), Neurosciences - from molecule to behavior: a university textbook. Springer, Berlin, Germany. [Google Scholar]
- 17.Waniek PJ, Pacheco Costa JE, Jansen AM, Costa J, Araújo CAC. 2012. Cathepsin L of Triatoma brasiliensis (Reduviidae, Triatominae): sequence characterization, expression pattern and zymography. J Insect Physiol 58:178–187. doi: 10.1016/j.jinsphys.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Renoz F, Noël C, Errachid A, Foray V, Hance T. 2015. Infection dynamic of symbiotic bacteria in the pea aphid Acyrthosiphon pisum gut and host immune response at the early steps in the infection process. PLoS One 10:e0122099. doi: 10.1371/journal.pone.0122099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rolff J. 2002. Bateman's principle and immunity. Proc Biol Sci 269:867–872. doi: 10.1098/rspb.2002.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheridan LAD, Poulin R, Ward DF, Zuk M. 2000. Sex differences in parasitic infections among arthropod hosts: is there a male bias? Oikos 88:327–334. doi: 10.1034/j.1600-0706.2000.880211.x. [DOI] [Google Scholar]
- 21.Blaauw BR, Jones VP, Nielsen AL. 2016. Utilizing immunomarking techniques to track Halyomorpha halys (Hemiptera: Pentatomidae) movement and distribution within a peach orchard. PeerJ 4:e1997. doi: 10.7717/peerj.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsumoto Y, Suetsugu Y, Nakamura M, Hattori M. 2014. Transcriptome analysis of the salivary glands of Nephotettix cincticeps (Uhler). J Insect Physiol 71:170–176. doi: 10.1016/j.jinsphys.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 23.DeLay B, Mamidala P, Wijeratne A, Wijeratne S, Mittapalli O, Wang J, Lamp W. 2012. Transcriptome analysis of the salivary glands of potato leafhopper, Empoasca fabae. J Insect Physiol 58:1626–1634. doi: 10.1016/j.jinsphys.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 24.Shao E, Lin G, Liu S, Ma X, Chen M, Lin L, Wu S, Sha L, Liu Z, Hu X, Guan X, Zhang L. 2017. Identification of transcripts involved in digestion, detoxification and immune response from transcriptome of Empoasca vitis (Hemiptera: Cicadellidae) nymphs. Genomics 109:58–66. doi: 10.1016/j.ygeno.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Cassone BJ, Bai X, Redinbaugh MG, Michel AP. 2012. Transcriptome of the plant virus vector Graminella nigrifrons, and the molecular interactions of Maize fine streak rhabdovirus transmission. PLoS One 7:e40613. doi: 10.1371/journal.pone.0040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nandety RS, Kamita SG, Hammock BD, Falk BW. 2013. Sequencing and de novo assembly of the transcriptome of the glassy-winged sharpshooter (Homalodisca vitripennis). PLoS One 8:e81681. doi: 10.1371/journal.pone.0081681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asgharian H, Chang PL, Mazzoglio PJ, Negri I. 2014. Wolbachia is not all about sex: male-feminizing Wolbachia alters the leafhopper Zyginidia pullula transcriptome in a mainly sex-independent manner. Front Microbiol 5:430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cassone BJ, Cisneros Carter FM, Michel AP, Stewart LR, Redinbaugh MG. 2014. Genetic insights into Graminella nigrifrons competence for Maize fine streak virus infection and transmission. PLoS One 9:e113529. doi: 10.1371/journal.pone.0113529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumaresan V, Harikrishnan R, Arockiaraj J. 2015. A potential Kazal-type serine protease inhibitor involves in kinetics of protease inhibition and bacteriostatic activity. Fish Shellfish Immunol 42:430–438. doi: 10.1016/j.fsi.2014.11.027. [DOI] [PubMed] [Google Scholar]
- 30.Kim BY, Lee KS, Zou FM, Wan H, Choi YS, Yoon HJ, Kwon HW, Je YH, Jin BR. 2013. Antimicrobial activity of a honeybee (Apis cerana) venom Kazal-type serine protease inhibitor. Toxicon 76:110–117. doi: 10.1016/j.toxicon.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 31.Monwan W, Amparyup P, Tassanakajon A. 2017. A snake-like serine proteinase (Pm Snake) activates prophenoloxidase-activating system in black tiger shrimp Penaeus monodon. Dev Comp Immunol 67:229–238. doi: 10.1016/j.dci.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 32.Barillas-Mury C. 2007. CLIP proteases and Plasmodium melanization in Anopheles gambiae. Trends Parasitol 23:297–299. doi: 10.1016/j.pt.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Hou F, He S, Qian Z, Wang X, Mao A, Sun C, Liu X. 2014. Identification, characterization and functional analysis of a serine protease inhibitor (Lvserpin) from the Pacific white shrimp, Litopenaeus vannamei. Dev Comp Immunol 43:35–46. doi: 10.1016/j.dci.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 34.Dubovskiy I, Kryukova N, Glupov V, Ratcliffe N. 2016. Encapsulation and nodulation in insects. Invertebr Surviv J 13:229–246. [Google Scholar]
- 35.Cornet S, Biard C, Moret Y. 2009. Variation in immune defence among populations of Gammarus pulex (Crustacea: Amphipoda). Oecologia 159:257–269. doi: 10.1007/s00442-008-1211-y. [DOI] [PubMed] [Google Scholar]
- 36.Eliautout R, Dubrana M-P, Vincent-Monégat C, Vallier A, Braquart-Varnier C, Poirié M, Saillard C, Heddi A, Arricau-Bouvery N. 2016. Immune response and survival of Circulifer haematoceps to Spiroplasma citri infection requires expression of the gene hexamerin. Dev Comp Immunol 54:7–19. doi: 10.1016/j.dci.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 37.Huang P-H, Lu S-C, Yang S-H, Cai P-S, Lo C-F, Chang L-K. 2014. Regulation of the immediate-early genes of white spot syndrome virus by Litopenaeus vannamei Kruppel-like factor (LvKLF). Dev Comp Immunol 46:364–372. doi: 10.1016/j.dci.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 38.Castagnola A, Jurat-Fuentes JL. 2016. Intestinal regeneration as an insect resistance mechanism to entomopathogenic bacteria. Curr Opin Insect Sci 15:104–110. doi: 10.1016/j.cois.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X, Yao P, Chu X, Hao L, Guo X, Xu B. 2015. Isolation of arginine kinase from Apis cerana cerana and its possible involvement in response to adverse stress. Cell Stress Chaperones 20:169–183. doi: 10.1007/s12192-014-0535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang S, Jia Z, Chen H, Wang L, Song L. 2016. The modulation of haemolymph arginine kinase on the extracellular ATP induced bactericidal immune responses in the Pacific oyster Crassostrea gigas. Fish Shellfish Immunol 54:282–293. doi: 10.1016/j.fsi.2016.03.153. [DOI] [PubMed] [Google Scholar]
- 41.Ma C, Gao Q, Liang Y, Li C, Liu C, Huang J. 2016. Shrimp arginine kinase being a binding protein of WSSV envelope protein VP31. Chin J Oceanol Limnol 34:1287–1296. doi: 10.1007/s00343-016-5198-7. [DOI] [Google Scholar]
- 42.Sun J. 2012. Roles of cellular redox factors in pathogen and toxin entry in the endocytic pathways, p 61–90. In Ceresa B. (ed), Molecular regulation of endocytosis. InTech https://www.intechopen.com/books/molecular-regulation-of-endocytosis/roles-of-cellular-redox-factors-in-pathogen-and-toxin-entry-in-the-endocytic-pathways.
- 43.Kongton K, McCall K, Phongdara A. 2014. Identification of gamma-interferon-inducible lysosomal thiol reductase (GILT) homologues in the fruit fly Drosophila melanogaster. Dev Comp Immunol 44:389–396. doi: 10.1016/j.dci.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 44.Ren C, Chen T, Jiang X, Luo X, Wang Y, Hu C. 2015. The first echinoderm gamma-interferon-inducible lysosomal thiol reductase (GILT) identified from sea cucumber (Stichopus monotuberculatus). Fish Shellfish Immunol 42:41–49. doi: 10.1016/j.fsi.2014.10.024. [DOI] [PubMed] [Google Scholar]
- 45.Al-Quadan T, Price CT, London N, Schueler-Furman O, AbuKwaik Y. 2011. Anchoring of bacterial effectors to host membranes through host-mediated lipidation by prenylation: a common paradigm. Trends Microbiol 19:573–579. doi: 10.1016/j.tim.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 46.Hicks SW, Galán JE. 2013. Exploitation of eukaryotic subcellular targeting mechanisms by bacterial effectors. Nat Rev Microbiol 11:316–326. doi: 10.1038/nrmicro3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anabestani A, Izadpanah K, Abbà S, Galetto L, Ghorbani A, Palmano S, Siampour M, Veratti F, Marzachì C. 2017. Identification of putative effector genes and their transcripts in three strains related to ‘Candidatus Phytoplasma aurantifolia’. Microbiol Res 199:57–66. doi: 10.1016/j.micres.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 48.Mayans O, Benian GM, Simkovic F, Rigden DJ. 2013. Mechanistic and functional diversity in the mechanosensory kinases of the titin-like family. Biochem Soc Trans 41:1066–1071. doi: 10.1042/BST20130085. [DOI] [PubMed] [Google Scholar]
- 49.Velthuis AJW, Bagowski CP. 2007. PDZ and LIM domain-encoding genes: molecular interactions and their role in development. Sci World J 7:1470–1492. doi: 10.1100/tsw.2007.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rossi D, Sorrentino V. 2002. Molecular genetics of ryanodine receptors Ca2+-release channels. Cell Calcium 32:307–319. doi: 10.1016/S0143416002001987. [DOI] [PubMed] [Google Scholar]
- 51.Takekura H, Franzini-Armstrong C. 2002. The structure of Ca2+ release units in arthropod body muscle indicates an indirect mechanism for excitation-contraction coupling. Biophys J 83:2742–2753. doi: 10.1016/S0006-3495(02)75284-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ji T, Yin L, Liu Z, Shen F, Shen J. 2014. High-throughput sequencing identification of genes involved with Varroa destructor resistance in the eastern honeybee, Apis cerana. Genet Mol Res 13:9086–9096. doi: 10.4238/2014.October.31.24. [DOI] [PubMed] [Google Scholar]
- 53.Ayme-Southgate A, Feldman S, Fulmer D. 2015. Myofilament proteins in the synchronous flight muscles of Manduca sexta show both similarities and differences to Drosophila melanogaster. Insect Biochem Mol Biol 62:174–182. doi: 10.1016/j.ibmb.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 54.Martini X, Hoffmann M, Coy MR, Stelinski LL, Pelz-Stelinski KS. 2015. Infection of an insect vector with a bacterial plant pathogen increases its propensity for dispersal. PLoS One 10:e0129373. doi: 10.1371/journal.pone.0129373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang G, Zhang J, Shen Y, Zheng Q, Feng M, Xiang X, Wu X. 2015. Transcriptome analysis of the brain of the silkworm Bombyx mori infected with Bombyx mori nucleopolyhedrovirus: a new insight into the molecular mechanism of enhanced locomotor activity induced by viral infection. J Invertebr Pathol 128:37–43. doi: 10.1016/j.jip.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 56.Saikhedkar N, Summanwar A, Joshi R, Giri A. 2015. Cathepsins of lepidopteran insects: aspects and prospects. Insect Biochem Mol Biol 64:51–59. doi: 10.1016/j.ibmb.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 57.Katunuma N. 2010. Posttranslational processing and modification of cathepsins and cystatins. J Signal Transduct 2010:375345. doi: 10.1155/2010/375345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Futahashi R, Tanaka K, Tanahashi M, Nikoh N, Kikuchi Y, Lee BL, Fukatsu T. 2013. Gene expression in gut symbiotic organ of stinkbug affected by extracellular bacterial symbiont. PLoS One 8:e64557. doi: 10.1371/journal.pone.0064557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abbà S, Galetto L, Vallino M, Rossi M, Turina M, Sicard A, Marzachì C. 2017. Genome sequence, prevalence and quantification of the first iflavirus identified in a phytoplasma insect vector. Arch Virol 162:799–809. doi: 10.1007/s00705-016-3158-3. [DOI] [PubMed] [Google Scholar]
- 60.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.FastQC. 2016. FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 62.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, LeDuc RD, Friedman N, Regev A. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 64.Powell S, Szklarczyk D, Trachana K, Roth A, Kuhn M, Muller J, Arnold R, Rattei T, Letunic I, Doerks T, Jensen LJ, von Mering C, Bork P. 2012. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res 40:D284–D289. doi: 10.1093/nar/gkr1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.GitHub Inc. 2016. TransDecoder (find coding regions within transcripts). https://transdecoder.github.io/.
- 66.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 68.Sigrist CJA, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, Bougueleret L, Xenarios I. 2013. New and continuing developments at PROSITE. Nucleic Acids Res 41:D344–D347. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chauhan JS, Rao A, Raghava GPS. 2013. In silico platform for prediction of N-, O- and C-glycosites in eukaryotic protein sequences. PLoS One 8:e67008. doi: 10.1371/journal.pone.0067008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. 2017. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:research0034.1. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cooper R, McLetchie D. 2004. Monitoring carbon dioxide production by Drosophila larvae. Drosoph Infect Serv 87:88–91. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.