ABSTRACT
Coccidiosis, caused by different species of Eimeria parasites, is an economically important disease of poultry and livestock worldwide. Here we report previously unknown alterations in the gut microbes and metabolism of BALB/c mice infected with Eimeria falciformis. Specifically, we observed a significant shift in the abundance of cecal bacteria and disrupted metabolism in parasitized animals. The relative abundances of Lachnospiraceae bacterium NK4A136, Ruminiclostridium, Alistipes, and Lactobacillus declined in response to E. falciformis infection, whereas Escherichia, Shigella, Helicobacter, Klebsiella, and Bacteroides were increased. Carbohydrate and amino acid metabolites in the serum samples of infected mice were significantly altered compared to naïve controls. Levels of amino acids, including asparagine, histidine, l-cysteine, tryptophan, lysine, glycine, serine, alanine, proline, ornithine, methionine, and valine, decreased on day 7 postinfection before returning to baseline on day 14. In addition, increased levels of indolelactate and mannitol and a reduced amount of oxalic acid indicated impaired carbon metabolism upon parasitic infection. These data demonstrate that intestinal coccidial infection perturbs the microbiota and disrupts carbon and nitrogen metabolism.
KEYWORDS: host-parasite interaction, gut microbiota, metabolic pathway, Eimeria falciformis, mouse
INTRODUCTION
The phylum Apicomplexa consists of single-celled parasites, including those belonging to the genera Toxoplasma, Plasmodium, Eimeria, Cryptosporidium, and Theileria, which collectively cause severe morbidity and mortality in humans and livestock (1–4). Protozoan infection involves a complex interplay between host and parasite, since these pathogens divert host resources for their own survival and reproduction (5). Thus, understanding host-parasite interactions may facilitate the development of improved diagnostics and therapeutics.
Gut microbiota (referred to as bacteria herein) may alter the pathophysiology of parasite infections, and changes in microbiota can confer resistance to enteric protozoa or promote protozoan infection. For example, germfree mice developed Cryptosporidium parvum infections, whereas normal intestinal microbiota decreased host susceptibility to this parasite (6). In contrast, previous studies suggest that Eimeria spp. cannot infect germfree animals (7, 8). Recent reports indicate that the commensal intestinal microflora can modulate various physiologic processes, including a protective immune response, nutrition and metabolism, and pathogen exclusion (9–11). Commensal gut bacteria can also serve as molecular adjuvants, providing indirect immunostimulation that protects mice from Toxoplasma gondii and Giardia infections (12, 13). In brief, the gut microbiota is important for intestinal homeostasis and host responses to enteric pathogens, and understanding how bacteria respond to infection should broaden our view of infectious diseases.
Metabolomics enables the analysis of global metabolite perturbations in response to biological stimuli (14). It has been reported that host metabolism is altered by protozoan infection during intracellular development. These studies provide novel insights into disease pathogenesis and infection responses (15, 16). Many significant biochemical alterations in Trypanosoma cruzi-infected mice have been reported: glucose import, the sorbitol pathway, and fatty acid and phospholipid synthesis decreased in plasma but increased in heart tissue (17). Urine and plasma samples of Plasmodium berghei-infected mice showed glycolytic upregulation, increased energy demand, and disrupted gut microbiota (5, 18–20). Similarly, many intermediates of amino acid and energy metabolism were altered in mouse serum and brain samples after infection with Toxoplasma gondii (21–24).
The genus Eimeria of the phylum Apicomplexa comprises more than 1,800 species, each infecting a distinct host. These parasites usually infect intestinal tissues, completing their entire life cycle in a single host (25–27). Little is known about the influence of Eimeria infection on host homeostasis. In this study, we used Eimeria falciformis, a natural mouse pathogen (4, 28) and an established model for the study of in vivo parasite-host interactions. Using 16S rRNA gene sequences and gas chromatography-mass spectrometry (GC-MS)-based metabolomic analyses, we investigated the effect of E. falciformis infection on the intestinal microbiota and serum metabolism of infected animals.
RESULTS
Infection of the mouse cecum with E. falciformis.
Cecal contents and blood samples of animals infected with oocysts were collected at various time points (Fig. 1A). Oocysts were first detected at day 7 postinfection (p.i.) and peaked at day 8 p.i. A few oocysts were still detected in feces at day 11 p.i. (Fig. 1B). Infected animals had decreased body weight at day 7 p.i. (Fig. 1C) concurrently with bloody feces during early oocyst shedding (Fig. 1D). The body weight recovered at day 14 p.i. (Fig. 1C). Histological analysis showed immune cell infiltration, crypt elongation, and blunting of villi in cecal sections in E. falciformis-infected mice at day 7 (Fig. 1D). Our results are consistent with previous data on the E. falciformis life cycle in mice (28).
FIG 1.
Characteristic signs of coccidiosis in E. falciformis-infected mice. (A) Timeline for administration of E. falciformis oocysts and collection of samples from infected and control BALB/c mice. (B) The oocyst yield was tested daily from day 6 until day 14 p.i. Arrow a indicates that approximately 2,000 oocysts were detected at day 7 p.i. Arrow b indicates that 13,400 to 48,000 oocysts were detected at day 11 p.i. (C) Body weight changes in infected mice over the course of the experiment. (D) Histopathological images of cecal tissues of infected mice at day 7 p.i. Red arrows indicate gametocytes. EC, epithelial cell; LP, lamina propria; LU, lumen; SM, submucosa. Bars, 50 μm.
The composition of the gut microbiota was significantly altered after Eimeria infection.
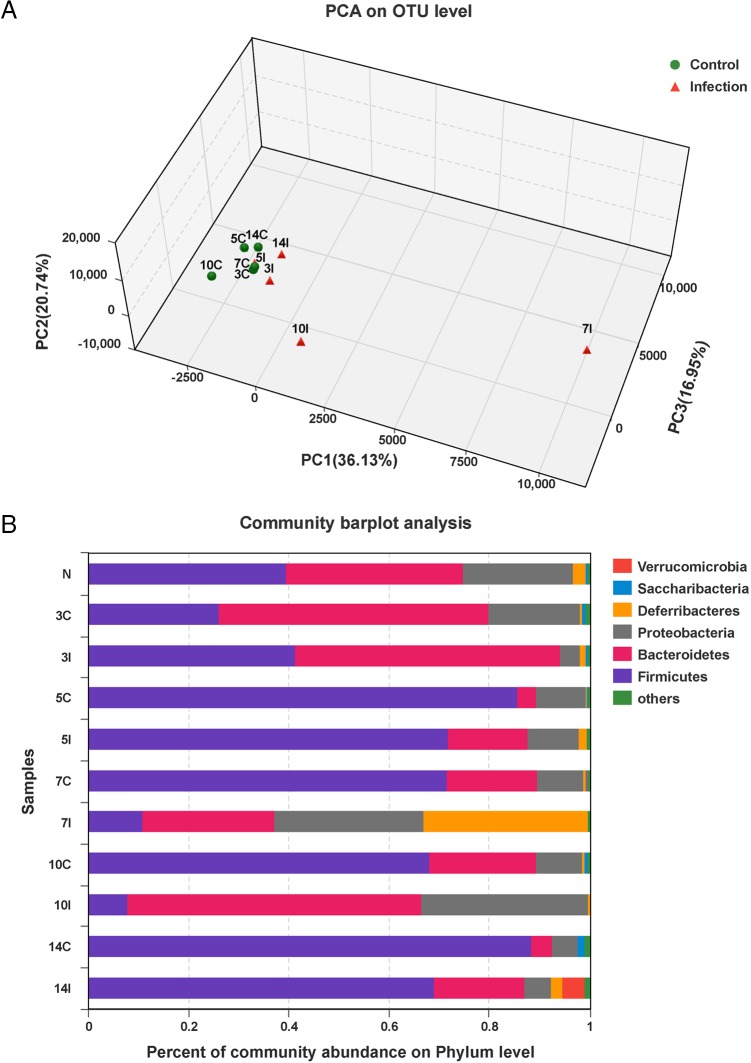
We used 16S rRNA gene sequencing to investigate cecal microbes. Principal-component analysis (PCA) of operational taxonomic units (OTUs) for bacteria (97% identity) showed that the gut microbiota from infected samples differed significantly from control samples at days 7 and 10 p.i. (Fig. 2A). The gut microbiota identified consisted of six phyla. Members of the Firmicutes were the most abundant, followed by Bacteroidales, Proteobacteria, Deferribacteres, Verrucomicrobia, and “Candidatus Saccharibacteria” (Fig. 2B). Eimeria infection reduced the abundance of Firmicutes and increased the abundances of Bacteroidales and Proteobacteria at days 7 and 10 p.i. (Fig. 2B). We also observed a predominance of Deferribacteres at day 7 p.i. (Fig. 2B).
FIG 2.
Alterations of gut microbiota caused by E. falciformis infection. (A) PCA of unweighted UniFrac values for samples from infected and control mice. Each point represents a sample collected on the day indicated by the number before the letter C (control) or I (infected). (B) Relative abundances of bacterial phyla in the microbiota of infected and control mice over the course of the experiment. Cecal contents from mice (n = 8) at each time were analyzed. N, uninfected samples (day 0).
Eimeria infection was associated with increased abundances of pathogenic bacteria.
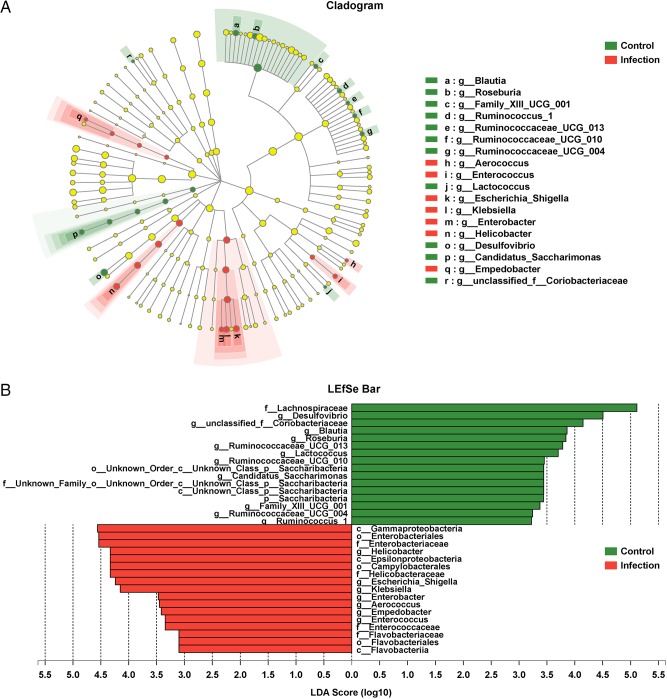
Taxon-specific differences in the gut microbiota associated with Eimeria infection were evaluated by utilizing the linear discriminant analysis (LDA) effect size (LEfSe) algorithm (Fig. 3A). A logarithmic LDA cutoff score of 3.0 was used to identify important taxonomic differences between infected animals and uninfected controls. Scores in parasitized animals had increased for Gammaproteobacteria, Enterobacteriales, Enterobacteriaceae, Epsilonproteobacteria, and Campylobacterales. Also, pathogenic taxa, including the genera Helicobacter, Escherichia-Shigella, Klebsiella, Enterobacter, Aerococcus, Empedobacter, and Enterococcus, were more abundant in parasite-infected mice than in controls. In contrast, the abundances of probiotics, including Lachnospiraceae, Desulfovibrio, Blautia, Roseburia, Ruminococcus, “Candidatus Saccharibacteria,” Alistipes, and Lactobacillus, were reduced in Eimeria-infected mice (Fig. 3B; see also Fig. S1 in the supplemental material).
FIG 3.
Increases in the abundances of opportunistic pathogenic bacteria in infected mice. (A) Cladogram of bacterial taxa differentially abundant in the gut microbiota of E. falciformis-infected and control mice generated by LDA and LEfSe analyses. (B) Histogram of LDA scores for differentially abundant bacterial clades in fecal samples of infected and control mice.
The abundance of microbiota contributing to carbohydrate and amino acid metabolism declined after infection.
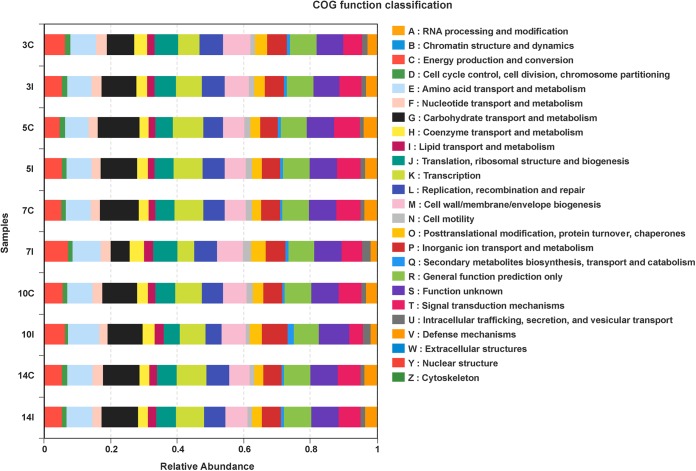
COG (clusters of orthologous groups) ontology (http://www.ncbi.nlm.nih.gov/COG/) analysis identified 25 different functional categories in data sets for infected mice. These included carbohydrate transport and metabolism; amino acid transport and metabolism; translation, ribosomal structure, and biogenesis; energy production and conversion; cell wall, membrane, and envelope biogenesis; replication, recombination, and repair; and signal transduction mechanisms (Fig. 4). The relative abundance of the microbiota contributing to carbohydrate transport and metabolism decreased in the infected group at day 7 p.i. and returned to baseline by day 10 p.i. Conversely, the abundance of the microbiota contributing to energy production and conversion; lipid transport and metabolism; posttranslational modification, protein turnover, and chaperones; and intracellular trafficking, secretion, and vesicular transport was increased over that for the control group at day 7 p.i (Fig. 4).
FIG 4.
COG category analysis of the cecal microbiota for infected and control mice. COG analysis revealed the relative abundances of metabolic functions that differed between infected and control mice. Each sample is designated by the number representing the day of collection (day 3, 5, 7, 10, or 14), followed by the letter C (control) or I (infected).
Eimeria-infected mice exhibit a significantly altered metabolic profile.
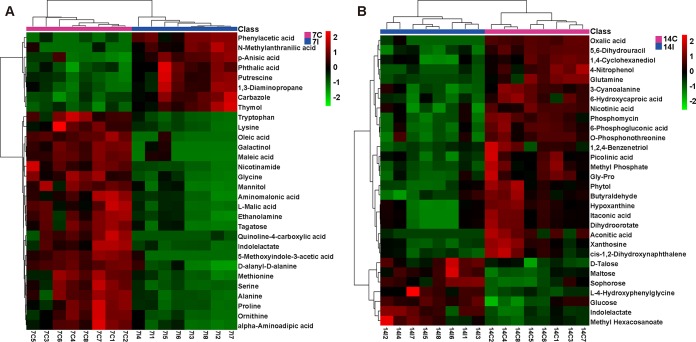
Next, we used GC-MS techniques to analyze metabolic differences between the sera isolated from Eimeria-infected and control animals. In total, 159 and 104 metabolites were identified at days 7 and 14 p.i., respectively. PCA showed notable differences between the serum metabolites of infected and control mice on both days (see Fig. S2 in the supplemental material). The levels of amino acids, including asparagine (fold change [FC], 2.54), histidine (FC, 5.15), l-cysteine (FC, 2.30), tryptophan (FC, 2.04), alanine (FC, 2.02), ornithine (FC, 2.04), and valine (FC, 2.09), were significantly decreased at day 7 p.i. (P < 0.05) (Fig. 5A; see also Table S1 in the supplemental material) and then returned to normal at day 14 p.i. (Fig. 5B; see also Table S2). In addition, glucose (FC, 2.20 and 0.64 on days 7 and 14 p.i., respectively), indolelactate (FC, 2.16 and 0.56), and mannitol (FC, 1.59 and 0.49) levels were altered (Tables S1 and S2). Also, oxalic acid levels (FC, 0.76 and 9.71) increased on day 7 p.i. (P < 0.05) but decreased on day 14 p.i. (P < 0.05). Thus, the glycolytic pathway in the host was impaired at day 14 after Eimeria infection (Fig. 5; also Tables S1 and S2). Heat maps constructed with the 30 most affected metabolites confirmed a significant difference between the infected group and the corresponding control group (Fig. 5). In particular, tryptophan levels in sera decreased, a finding consistent with the upregulation of the kynurenine pathway and the amino acid degradation reported for mice infected with E. falciformis (4, 28).
FIG 5.
Heat maps of the 30 most affected metabolites in E. falciformis-infected mice. Profiles of the top 30 serum metabolites from samples on days 7 (A) and 14 (B) were used to construct heat maps. Eight samples from infected mice at day 7 (7I1 to 7I8) or 14 (14I1 to 14I8) and eight samples from control mice at day 7 (7C1 to 7C8) or 14 (14C1 to 14C8) were analyzed.
ROC curve analysis identified three potential biomarkers of E. falciformis infection.
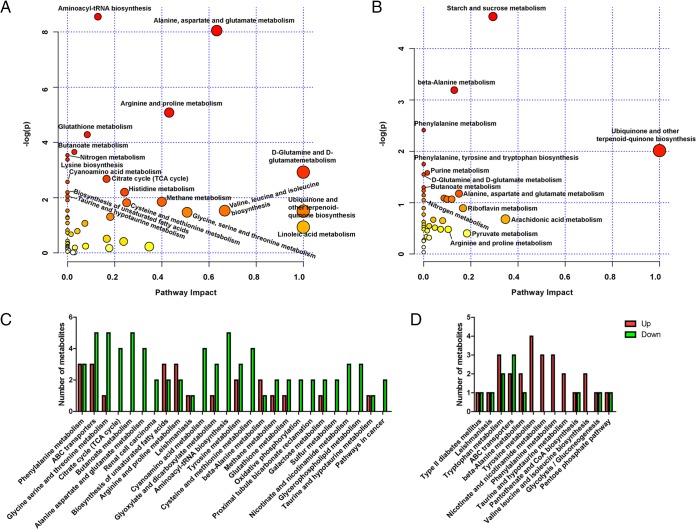
MetaboAnalyst 3.0 identified the metabolic pathways of Eimeria-infected mice that were altered, including the tricarboxylic acid (TCA) cycle; glycolysis and gluconeogenesis; nitrogen metabolism; lysine biosynthesis; glycine, serine, and threonine metabolism; aminoacyl-tRNA biosynthesis; alanine, aspartate, and glutamate metabolism; methane metabolism; and beta-alanine metabolism. In total, 23 and 13 pathways were found to be significantly affected at days 7 (Fig. 6A; see also Table S3 in the supplemental material) and 14 (Fig. 6B; see also Table S4 in the supplemental material) p.i., respectively (P < 0.05 in both cases). The corresponding metabolites were up- or downregulated in infected mice (Fig. 6C and D). Receiver operator characteristic (ROC) analysis showed mannitol, oxalic acid, and indolelactate to have the highest sensitivity and specificity to E. falciformis infection on day 7 as well as on day 14 (Fig. 7A and B), suggesting that these three metabolites could serve as diagnostic markers of E. falciformis infection in mice.
FIG 6.
Metabolic pathway analyses for infected and control mice. (A and B) MetaboAnalyst was used to identify the metabolic pathways that are important for the host response to E. falciformis infection on day 7 (A) or 14 (B). A darker circle color indicates more-significant changes in the metabolites in the corresponding pathway, while the size of each circle corresponds to the pathway impact score: larger circles reflect higher centrality of the metabolites involved. (C and D) Up- and downregulated metabolites of all major pathways enriched in E. falciformis-infected mice on day 7 (C) or 14 (D).
FIG 7.
ROC analyses of potential biomarkers for E. falciformis infection. ROC analyses were performed to measure the sensitivities and specificities of mannitol, oxalic acid, and indolelactate for Eimeria infection at days 7 (A) and 14 (B) p.i. AUC, area under the ROC curve.
DISCUSSION
Understanding the pathophysiology of coccidiosis may help identify abnormal metabolic pathways that could be potential biomarkers and therapeutic targets (9, 29, 30). Here we report significant changes in the abundances of gut microbiota and plasma metabolites in E. falciformis-infected mice. Eimeria infection increased the abundances of Escherichia, Shigella, and Helicobacter bacteria, most of which have been described as opportunistic pathogens (31). In contrast, parasitic infection decreased levels of Lachnospiraceae bacterium NK4A136, Ruminiclostridium, Alistipes, and Lactobacillus organisms, which are considered beneficial for gut health (32–34). In agreement with previous work, Firmicutes was the most abundant phylum in control mice, followed by Bacteroidetes and Proteobacteria (35, 36). In contrast, decreased Firmicutes levels and more Bacteroidetes and Proteobacteria were found in cecal samples from parasitized animals. The microbiota did not change in the early phase of infection, a finding consistent with an earlier report on the sister taxon to Eimeria, Cryptosporidium (37). However, we could not rule out the possibility that a gradual change in bacterial abundance is driven by earlier life cycle stages.
Metabolic analysis showed that Eimeria infection induced widespread effects on metabolic pathways in the host. In particular, carbohydrate- and amino acid-related pathways were perturbed during oocyst shedding. Perturbation of amino acids on day 7 is consistent with the dominance of Bacteroides spp. in infected mice. Bacteroides spp. have been reported to ferment aromatic amino acids to produce potentially bioactive products (38, 39). Previous studies have suggested that a high concentration of mannitol is present in unsporulated oocysts and that mannitol functions as an endogenous carbon source enabling oocysts to sporulate outside the host (40, 41). We noted a depletion of mannitol at day 7 p.i. during the sexual stages (zygote formation) of the life cycle. Mannitol biosynthesis depends on plasma glucose, which disrupts the glycolytic pathway at fructose-6-phosphate (41). The alteration of the glycolytic, indolelactate, and mannitol pathways may be important for host repair and parasite reproduction (41). On a different note, mannitol, oxalic acid, and indolelactate may prove to be diagnostic biomarkers of coccidiosis.
In conclusion, these findings enhance our understanding of the gut microbiota and metabolic profiles of E. falciformis-infected mice, which could be useful for studies in other host-parasite models.
MATERIALS AND METHODS
Ethics statement.
All experiments complied with the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of China (42) and were approved by the Institutional Animal Care and Use Committee of China Agricultural University (Beijing Laboratory Animal employee certificate 1114120800083). All efforts were made to minimize animal suffering.
Parasites.
E. falciformis oocysts were preserved in potassium dichromate and were maintained by serial propagation in 4- to 6-week-old BALB/c mice. Oocyst collection, purification, and sporulation were performed according to standard procedures as described previously (4, 43). To ensure that the gastrointestinal microbiota was not influenced by existing bacteria, oocysts were surface-sterilized with sodium hypochlorite (6.25% available chlorine).
Mice.
Female BALB/c mice (n = 96), 6 weeks of age, were randomized to two treatments (n, 48/group): infection and control (no infection). Infected mice were orally inoculated with 200 purified sporulated oocysts suspended in sterile phosphate-buffered saline (PBS), and controls were mock infected with the same volume of PBS. All mice were housed in a biosafety level 2 (BSL-2) experimental facility at China Agricultural University, where they had ad libitum access to sterilized feed and water. Oocyst yields were evaluated by collecting feces on a daily basis. All animals were weighed at days 0, 7, and 14 p.i. to monitor weight loss during infection.
Sample collection.
Eight animals from each group were euthanized for the collection of cecal contents at day 0 (preinfection) and days 3, 5, 7, 10, and 14 p.i., as well as serum samples at days 7 and 14 postinfection (Fig. 1A). Serum samples were derived from blood collected into Vacutainer serum tubes (BD, Franklin Lakes, NJ) and centrifuged at 3,500 × g for 25 min. Sera were stored at −80°C until use. Cecal contents were collected and were stored at −80°C for bacterial DNA extraction. Cecal tissues were removed and were immediately fixed in 4% formaldehyde for histopathological examination.
H&E staining.
After immersion, cecal tissues were dehydrated by gradual soaking in alcohol and xylene, embedded in paraffin, and then sliced into 4-μm-thick sections, which were stained with hematoxylin and eosin (H&E) to visualize intestinal morphology (44). Slides were examined under a digital optical microscope (Olympus, Tokyo, Japan).
DNA extraction and 16S rRNA gene sequencing.
To assess the cecal microbiota by use of 16S rRNA gene sequencing, total genomic DNA from samples was extracted using cetyltrimethylammonium bromide (CTAB)/sodium dodecyl sulfate (SDS) methods as described previously (45). DNA concentration and purity were assayed with 1% agarose gels. The V3-V4 hypervariable region of the 16S rRNA gene was amplified using forward primer 338F (5′-ACTCCTRCGGGAGGCAGCAG-3′) and reverse primer 806R (5′-GGACTACCVGGGTATCTAAT-3′) (46). Sequencing libraries were generated using the NEBNext Ultra DNA Library Prep kit (New England BioLabs, Ipswich, MA) according to the manufacturer's instructions. Library quality was assessed and sequenced on an Illumina MiSeq platform as described previously (47). Raw FASTQ files were demultiplexed, quality-filtered by Trimmomatic, and merged by fast length adjustment of short reads (FLASH) with the following criteria. (i) The reads were truncated at any site receiving an average quality score of <20 over a 50-bp sliding window. (ii) Primers were exactly matched allowing 2-nucleotide mismatching, and reads containing ambiguous bases were removed. (iii) Sequences with overlaps longer than 10 bp were merged according to the overlap sequence.
Sample processing for metabolomics analysis.
Serum samples stored at −80°C were thawed at room temperature. Metabolites were extracted as described elsewhere with minor modifications (48). Briefly, 80 μl serum was added to a 1.5-ml Eppendorf tube with 10 μl of 2-chloro-l-phenylalanine (0.3 mg/liter) dissolved in methanol as an internal standard. Subsequently, 240 μl of an ice-cold mixture of methanol and acetonitrile (2:1, vol/vol) was added, and the mixtures were vortexed and centrifuged at 12,000 × g and 4°C for 10 min. Supernatants were dried in a glass vial using a centrifugal freeze concentration dryer, and 80 μl of 15-mg/ml methoxyamine hydrochloride in pyridine was subsequently added. Then 80 μl of N,O-bis(trimethylsilyl) trifluoroacetamide (with 1% trimethylchlorosilane) and 20 μl n-hexane were added to the mixture, followed by incubation at 37°C for 90 min. Samples were kept at ambient temperature (25°C to 28°C) for 30 min before GC-MS analysis. Derivatized serum samples were analyzed on an Agilent 7890B gas chromatography system coupled to an Agilent 5977A MSD system (Agilent, CA).
Data preprocessing and statistical analysis.
OTUs were clustered with a 97% similarity cutoff using UPARSE (version 7.1). The taxonomy of each 16S rRNA gene sequence was analyzed by the Ribosomal Database Project Classifier (http://rdp.cme.msu.edu/) against the Silva (SSU128) 16S rRNA database with a confidence threshold of 70% (49). Metastats software (http://metastats.cbcb.umd.edu/) was used to confirm differences in taxonomic abundance between the two groups. LEfSe was used for quantitative analysis of biomarkers within different groups (50). This method was designed to analyze data for which the number of bacterial species was greater than the number of samples and to provide biological class explanations to establish statistical significance.
Acquired MS data from GC-MS were analyzed using ChromaTOF software (version 4.34; LECO, St Joseph, MI). Metabolites were quantified using the Fiehn database linked with ChromaTOF software. Unique metabolites were selected based on the combination of a statistically significant threshold of variable influence on projection (VIP) and P values. VIP values were obtained from the orthogonal partial least squares discriminant analysis (OPLS-DA) model, while P values were from a two-tailed Student t test of normalized peak areas for metabolites. VIP values of >1.0 and P values of <0.05 were considered statistically significant (51).
Accession number(s).
The raw reads for the 16S rRNA genes determined in this study were deposited in the NCBI Sequence Read Archive (SRA) database (accession number SRP127344).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Key Research and Development Program of China (grant 2017YFD0500400), the National Natural Science Foundation of China (grants 31572507, 31330076, 31472180, and 31772728), the National Transgenic Major Program of China (grant 2014ZX0800603B), and the Beijing Municipal Natural Science Foundation (grant 6152011).
We thank Majorbio Bio-Pharm Technology Co., Ltd., Shanghai, China, for technical assistance. We thank LetPub for providing linguistic assistance during the preparation of the manuscript.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00073-18.
REFERENCES
- 1.Dubey JP. 1977. Toxoplasma, Hammondia, Besnoitia, Sarcocystis, and other tissue cyst-forming coccidia of man and animals, p 101–237. In Kreier JP. (ed), Parasitic protozoa, vol 3 Academic Press, San Diego, CA. [Google Scholar]
- 2.Rothwell L, Gramzinski RA, Rose ME, Kaiser P. 1995. Avian coccidiosis—changes in intestinal lymphocyte populations associated with the development of immunity to Eimeria maxima. Parasite Immunol 17:525–533. doi: 10.1111/j.1365-3024.1995.tb00883.x. [DOI] [PubMed] [Google Scholar]
- 3.Chapman HD, Barta JR, Blake D, Gruber A, Jenkins M, Smith NC, Suo X, Tomley FM. 2013. A selective review of advances in coccidiosis research. Adv Parasitol 83:93–171. doi: 10.1016/B978-0-12-407705-8.00002-1. [DOI] [PubMed] [Google Scholar]
- 4.Schmid M, Heitlinger E, Spork S, Mollenkopf H, Lucius R, Gupta N. 2014. Eimeria falciformis infection of the mouse caecum identifies opposing roles of IFN gamma-regulated host pathways for the parasite development. Mucosal Immunol 7:969–982. doi: 10.1038/mi.2013.115. [DOI] [PubMed] [Google Scholar]
- 5.Olszewski KL, Morrisey JM, Wilinski D, Burns JM, Vaidya AB, Rabinowitz JD, Llinas M. 2009. Host-parasite interactions revealed by Plasmodium falciparum metabolomics. Cell Host Microbe 5:191–199. doi: 10.1016/j.chom.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harp JA, Chen WX, Harmsen AG. 1992. Resistance of severe combined immunodeficient mice to infection with Cryptosporidium parvum—the importance of intestinal microflora. Infect Immun 60:3509–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Owen D. 1975. Eimeria falciformis (Eimer, 1870) in specific pathogen free and gnotobiotic mice. Parasitology 71:293–303. doi: 10.1017/S0031182000046734. [DOI] [PubMed] [Google Scholar]
- 8.Gouet P, Yvore P, Naciri M, Contrepois M. 1984. Influence of digestive microflora on parasite development and the pathogenic effect of Eimeria ovinoidalis in the axenic, gnotoxenic and conventional lamb. Res Vet Sci 36:21–23. [PubMed] [Google Scholar]
- 9.Brestoff JR, Artis D. 2013. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol 14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamada N, Chen GY, Inohara N, Nunez G. 2013. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knaus UG, Hertzberger R, Pircalabioru GG, Yousefi SP, Branco Dos Santos F. 2017. Pathogen control at the intestinal mucosa—H2O2 to the rescue. Gut Microbes 8:67–74. doi: 10.1080/19490976.2017.1279378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benson A, Pifer R, Behrendt CL, Hooper LV, Yarovinsky F. 2009. Gut commensal bacteria direct a protective immune response against Toxoplasma gondii. Cell Host Microbe 6:187–196. doi: 10.1016/j.chom.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barash NR, Maloney JG, Singer SM, Dawson SC. 2017. Giardia alters commensal microbial diversity throughout the murine gut. Infect Immun 85:e00948-. doi: 10.1128/IAI.00948-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kafsack BF, Llinas M. 2010. Eating at the table of another: metabolomics of host-parasite interactions. Cell Host Microbe 7:90–99. doi: 10.1016/j.chom.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen XQ, Zhou CX, Elsheikha HM, He S, Hu GX, Zhu XQ. 2017. Profiling of the perturbed metabolomic state of mouse spleen during acute and chronic toxoplasmosis. Parasite Vector 10:339. doi: 10.1186/s13071-017-2282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacot D, Waller RF, Soldati-Favre D, MacPherson DA, MacRae JI. 2016. Apicomplexan energy metabolism: carbon source promiscuity and the quiescence hyperbole. Trends Parasitol 32:56–70. doi: 10.1016/j.pt.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Gironès N, Carbajosa S, Guerrero NA, Poveda C, Chillon-Marinas C, Fresno M. 2014. Global metabolomic profiling of acute myocarditis caused by Trypanosoma cruzi infection. PLoS Negl Trop Dis 8:e3337. doi: 10.1371/journal.pntd.0003337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li JV, Wang Y, Saric J, Nicholson JK, Dirnhofer S, Singer BH, Tanner M, Wittlin S, Holmes E, Utzinger J. 2008. Global metabolic responses of NMRI mice to an experimental Plasmodium berghei infection. J Proteome Res 7:3948. doi: 10.1021/pr800209d. [DOI] [PubMed] [Google Scholar]
- 19.Xu T, Ping J, Yu Y, Yu F, Yu Y, Hao P, Li X. 2010. Revealing parasite influence in metabolic pathways in Apicomplexa infected patients. BMC Bioinformatics 11(Suppl 11):S13. doi: 10.1186/1471-2105-11-S11-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sengupta A, Ghosh S, Basant A, Malusare S, Johri P, Pathak S, Sharma S, Sonawat HM. 2011. Global host metabolic response to Plasmodium vivax infection: a 1H NMR based urinary metabonomic study. Malar J 10:384. doi: 10.1186/1475-2875-10-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charron AJ, Sibley LD. 2002. Host cells: mobilizable lipid resources for the intracellular parasite Toxoplasma gondii. J Cell Sci 115:3049–3059. [DOI] [PubMed] [Google Scholar]
- 22.Nelson MM, Jones AR, Carmen JC, Sinai AP, Burchmore R, Wastling JM. 2008. Modulation of the host cell proteome by the intracellular apicomplexan parasite Toxoplasma gondii. Infect Immun 76:828–844. doi: 10.1128/IAI.01115-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prandovszky E, Gaskell E, Martin H, Dubey JP, Webster JP, McConkey GA. 2011. The neurotropic parasite Toxoplasma gondii increases dopamine metabolism. PLoS One 6:e23866. doi: 10.1371/journal.pone.0023866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou CX, Zhou DH, Elsheikha HM, Zhao Y, Suo X, Zhu XQ. 2016. Metabolomic profiling of mice serum during toxoplasmosis progression using liquid chromatography-mass spectrometry. Sci Rep 6:19557. doi: 10.1038/srep19557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long PL, Rose ME, Pierce AE. 1963. Effects of fowl sera on some stages in life cycle of Eimeria tenella. Exp Parasitol 14:210–217. doi: 10.1016/0014-4894(63)90025-0. [DOI] [PubMed] [Google Scholar]
- 26.Suo X, Zhang JX, Li ZG, Yang CT, Min QR, Xu LT, Liu Q, Zhu XQ. 2006. The efficacy and economic benefits of Supercox, a live anticoccidial vaccine in a commercial trial in broiler chickens in China. Vet Parasitol 142:63–70. doi: 10.1016/j.vetpar.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 27.McDonald V, Shirley MW. 2009. Past and future: vaccination against Eimeria. Parasitology 136:1477–1489. doi: 10.1017/S0031182009006349. [DOI] [PubMed] [Google Scholar]
- 28.Schmid M, Lehmann MJ, Lucius R, Gupta N. 2012. Apicomplexan parasite, Eimeria falciformis, co-opts host tryptophan catabolism for life cycle progression in mouse. J Biol Chem 287:20197–20207. doi: 10.1074/jbc.M112.351999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allen PC, Fetterer RH. 2002. Recent advances in biology and immunobiology of Eimeria species and in diagnosis and control of infection with these coccidian parasites of poultry. Clin Microbiol Rev 15:58–65. doi: 10.1128/CMR.15.1.58-65.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quiroz-Castañeda RE, Dantan-Gonzalez E. 2015. Control of avian coccidiosis: future and present natural alternatives. Biomed Res Int 2015:430610. doi: 10.1155/2015/430610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macdonald SE, Nolan MJ, Harman K, Boulton K, Hume DA, Tomley FM, Stabler RA, Blake DP. 2017. Effects of Eimeria tenella infection on chicken caecal microbiome diversity, exploring variation associated with severity of pathology. PLoS One 12:e0184890. doi: 10.1371/journal.pone.0184890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walter J. 2008. Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl Environ Microbiol 74:4985–4996. doi: 10.1128/AEM.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meehan CJ, Beiko RG. 2014. A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol Evol 6:703–713. doi: 10.1093/gbe/evu050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ravachol J, de Philip P, Borne R, Mansuelle P, Maté MJ, Perret S, Fierobe HP. 2016. Mechanisms involved in xyloglucan catabolism by the cellulosome-producing bacterium Ruminiclostridium cellulolyticum. Sci Rep 6:22770. doi: 10.1038/srep22770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, Burcelin R. 2008. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 36.Murphy EF, Cotter PD, Healy S, Marques TM, O'Sullivan O, Fouhy F, Clarke SF, O'Toole PW, Quigley EM, Stanton C, Ross PR, O'Doherty RM, Shanahan F. 2010. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut 59:1635–1642. doi: 10.1136/gut.2010.215665. [DOI] [PubMed] [Google Scholar]
- 37.Ras R, Huynh K, Desoky E, Badawy A, Widmer G. 2015. Perturbation of the intestinal microbiota of mice infected with Cryptosporidium parvum. Int J Parasitol 45:567–573. doi: 10.1016/j.ijpara.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 38.Louis P, Hold GL, Flint HJ. 2014. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol 12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 39.Mariño E, Richards JL, McLeod KH, Stanley D, Yap YA, Knight J, McKenzie C, Kranich J, Oliveira AC, Rossello FJ, Krishnamurthy B, Nefzger CM, Macia L, Thorburn A, Baxter AG, Morahan G, Wong LH, Polo JM, Moore RJ, Lockett TJ, Clarke JM, Topping DL, Harrison LC, Mackay CR. 2017. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat Immunol 18:552. doi: 10.1038/ni.3713. [DOI] [PubMed] [Google Scholar]
- 40.Schmatz DM. 1997. The mannitol cycle in Eimeria. Parasitology 114S:S81–S89. [PubMed] [Google Scholar]
- 41.Michalski WP, Edgar JA, Prowse SJ. 1992. Mannitol metabolism in Eimeria tenella. Int J Parasitol 22:1157–1163. doi: 10.1016/0020-7519(92)90035-J. [DOI] [PubMed] [Google Scholar]
- 42.National Research Council. 1996. Guide for the care and use of laboratory animals—Chinese version. National Academies Press, Washington, DC. [Google Scholar]
- 43.Ryley JF, Meade R, Hazelhurst J, Robinson TE. 1976. Methods in coccidiosis research: separation of oocysts from feces. Parasitology 73:311–326. doi: 10.1017/S0031182000046990. [DOI] [PubMed] [Google Scholar]
- 44.Fischer AH, Jacobson KA, Rose J, Zeller R. 2008. Hematoxylin and eosin staining of tissue and cell sections. CSH Protoc 2008:pdb.prot4986. doi: 10.1101/pdb.prot4986. [DOI] [PubMed] [Google Scholar]
- 45.Cruaud P, Vigneron A, Lucchetti-Miganeh C, Ciron PE, Godfroy A, Cambon-Bonavita M-A. 2014. Influence of DNA extraction method, 16S rRNA targeted hypervariable regions, and sample origin on microbial diversity detected by 454 pyrosequencing in marine chemosynthetic ecosystems. Appl Environ Microbiol 80:4626–4639. doi: 10.1128/AEM.00592-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baker GC, Smith JJ, Cowan DA. 2003. Review and re-analysis of domain-specific 16S primers. J Microbiol Methods 55:541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 47.Heijs SK, Haese RR, van der Wielen PW, Forney LJ, van Elsas JD. 2007. Use of 16S rRNA gene based clone libraries to assess microbial communities potentially involved in anaerobic methane oxidation in a Mediterranean cold seep. Microb Ecol 53:384–398. doi: 10.1007/s00248-006-9172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.A J, Trygg J, Gullberg J, Johansson AI, Jonsson P, Antti H, Marklund SL, Moritz T. 2005. Extraction and GC/MS analysis of the human blood plasma metabolome. Anal Chem 77:8086–8094. doi: 10.1021/ac051211v. [DOI] [PubMed] [Google Scholar]
- 49.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J Bacteriol 192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia J, Wishart DS. 2016. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr Protoc Bioinformatics 55:14.10.1–14.10.91. doi: 10.1002/cpbi.11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.