ABSTRACT
Combinatorial chemotherapy is necessary for the treatment of malaria. However, finding a suitable partner drug for a new candidate is challenging. Here we develop an algorithm that identifies all of the gene pairs of Plasmodium falciparum that possess orthologues in yeast that have a synthetic lethal interaction but are absent in humans. This suggests new options for drug combinations, particularly for inhibitors of targets such as P. falciparum calcineurin, cation ATPase 4, or phosphatidylinositol 4-kinase.
KEYWORDS: antimalarials, combinatorial chemotherapy, gene orthology, synthetic lethality, yeast genetics
TEXT
There is a persistent need for new antimalarials due to the evolution of drug-resistant parasites. Under the auspices of the Medicines for Malaria Venture (MMV), new drug candidates that are active against artemisinin-resistant isolates of Plasmodium falciparum are being developed; the frontrunners are artefenomel, KAF156, cipargamin, DSM265, MMV390048, ferroquine, and tafenoquine (1, 2). However, the choice of the right partner drug will be critical for the success of these new molecules, as the WHO enforces the application of antimalarials in combination therapy (3). In addition to protecting each other from drug resistance, two molecules to be combined need to be compatible for coformulation, should have matching pharmacokinetic profiles, and must not have unfavorable polypharmacology (4–6). Ideally, the two molecules would potentiate each other, thereby decreasing the duration of treatment and the required doses. Thus, combinatorial chemotherapy not only can reduce the risk of drug resistance but also can enhance drug safety and drug efficacy, enabling the ambitious goal of a “single-exposure radical cure” (7, 8).
Here we propose to support the matchmaking of antimalarial candidates by learning from yeast reverse genetics. Saccharomyces cerevisiae is probably the best studied of all eukaryotes. Only about 20% of its protein-coding genes are essential for growth on rich medium (9). High-throughput crossing experiments have shown that many viable S. cerevisiae gene deletion mutants possess synthetic phenotypes, i.e., growth defects that become apparent only in the absence of another nonessential gene. The concept of genetic synthetic lethality can be adopted to combination chemotherapy (8, 10–12). The principal idea is to extrapolate from synthetic lethal gene pairs in S. cerevisiae to orthologous pairs of genes in P. falciparum, assuming that the combined inhibition of the respective gene products will produce a synergistic effect. However, this seemingly straightforward approach is complicated by the fact that S. cerevisiae is more closely related to Homo sapiens than to P. falciparum (13). Thus, a drug combination inferred from yeast synthetic genetic lethality might enhance the toxicity to humans rather than enhancing the antimalarial efficacy. To avoid such a scenario, we developed an algorithm to exclude gene pairs that are conserved in H. sapiens.
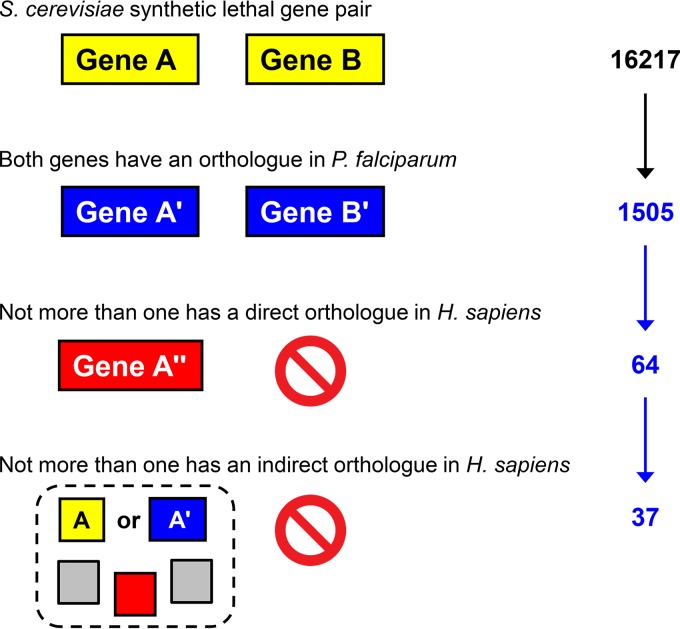
Yeast synthetic lethal gene pairs were obtained from BioGRID 3.4 (14) and pairs and groups of orthologous genes from the OrthoMCL 5 database, based on the similarity of the derived protein sequences (15, 16). Mining the OrthoMCL database with the 16,217 synthetic lethal gene pairs of S. cerevisiae identified in BioGRID, we found that only 1,505 pairs (9.3%) had direct orthologues in P. falciparum for both gene products (Fig. 1). From this set, we tested all of the proteins for the presence of an orthologue in the human proteome, again referring to the downloaded OrthoMCL database. This assessment included direct pairwise orthology between the P. falciparum or S. cerevisiae protein and a H. sapiens protein or indirect orthology in which either the malaria protein or its yeast orthologue belonged to an OrthoMCL group that also contained a human protein (Fig. 1). All of the P. falciparum gene pairs for which both gene products tested positive for direct or indirect human orthology were eliminated. This process yielded 37 pairs composed of 55 unique P. falciparum proteins that fulfilled the conditions that (i) their direct orthologues in S. cerevisiae exhibit synthetic lethality and (ii) at least one of the two proteins has neither a direct nor an indirect orthologue in the human proteome. Therefore, we suggest these pairs as targets for combinatorial chemotherapy. The comparative genomics pipeline (Fig. 1) is built with self-developed Python scripts that are available for download at the GitHub repository (https://github.com/suvi-subra/SynthLeth).
FIG 1.
Graphic representation of the algorithm, with the numbers of P. falciparum gene pairs that passed the filters; the final 37 are shown in Table 1. Yellow, S. cerevisiae; blue, P. falciparum; red, H. sapiens.
The final set of 37 pairs was enriched in druggable proteins (Table 1). Of the 55 proteins in the set, 30 either had been validated as drug targets or had a positive “druggability index,” as predicted by TDR Targets (17). Some of the suggested combinations affected the same pathway, e.g., the pyridoxal kinase-like protein and pyridoxine biosynthesis protein involved in vitamin B6 metabolism or NAD(P)H-dependent glutamate synthase and NADP-specific glutamate dehydrogenase; the latter is selectively inhibited by isophthalic acid (18), while glutamate synthase had been suggested as a target based on comparative genomics (19). Calcineurin subunit B paired with the P. falciparum cation/H+ antiporter (PfCHA), which is sensitive to known inhibitors such as KB-R7943 (20). Hubs of inferred interactions were P. falciparum apurinic/apyrimidinic endonuclease 1 (PfAPN1) and the P. falciparum U5 small nuclear ribonucleoprotein (PfSNU114) of the spliceosome, both of which are involved in the processing of nucleic acids. Two proteins in the target set were of particular pharmacological interest, namely, P. falciparum Ca2+-ATPase 4 (PfATP4) and P. falciparum phosphatidylinositol 4-kinase (PfPI4K). Either protein is targeted by new antimalarial candidates (21–27). PfATP4 is the target of cipargamin and paired with PfCHA (Table 1), suggesting testing for potential synergy between cipargamin and KB-R7943. PfPI4K, the target of imidazolopiperazines and MMV390048, paired with ubiquitin-conjugating enzyme E2 (Table 1). An inhibitor of Atg8-Atg3 interactions was identified from the MMV Malaria Box (28), and ubiquitin-protein ligase E3 was proposed as an antimalarial target (29). The inferred link between phosphatidylinositol 4-kinase and ubiquitination suggests testing for potential synergy between PfPI4K inhibitors and P. falciparum proteasome inhibitors (30–32).
TABLE 1.
Pairs of P. falciparum proteins suggested as targets for combinatorial chemotherapy, based on synthetic lethal genetic interactions in S. cerevisiae
| Gene 1 identification | Gene 1 producta | Gene 2 identification | Gene 2 product |
|---|---|---|---|
| PF14_0492 | Calcineurin subunit B | PFF0170w | Cation/H+ antiporter |
| PFL0590c | Non-SERCA-type Ca2+-transporting P-ATPase 4 | PFF0170w | Cation/H+ antiporter |
| PFE0485w | Phosphatidylinositol 4-kinase | PFF0305c | Ubiquitin-conjugating enzyme E2 |
| PF08_0031 | Dicarboxylate/tricarboxylate carrier | mal_mito_2 | Cytochrome c oxidase subunit 1 |
| PFF1105c | Chorismate synthase | PF14_0511 | Glucose-6-phosphate dehydrogenase |
| PFL2465c | Thymidylate kinase | PF13_0176 | Apurinic/apyrimidinic endonuclease |
| MAL13P1.346 | DNA repair endonuclease | PF13_0176 | Apurinic/apyrimidinic endonuclease |
| PFB0160w | ERCC1 nucleotide excision repair protein | PF13_0176 | Apurinic/apyrimidinic endonuclease |
| PFF0715c | Endonuclease III homologue | PF13_0176 | Apurinic/apyrimidinic endonuclease |
| PFD0865c | Cdc2-related protein kinase 1 | PFF0165c | Conserved Plasmodium protein, unknown function |
| PFL1635w | Sentrin-specific protease 1 | PF10_0092 | Metallopeptidase |
| PF13_0251 | DNA topoisomerase 3 | PF10_0092 | Metallopeptidase |
| PFF0775w | Pyridoxal kinase-like protein | PFF1025c | Pyridoxine biosynthesis protein |
| PF11_0192 | Histone acetyltransferase | PFF1180w | Anaphase-promoting complex subunit |
| PFL2440w | DNA repair protein | MAL7P1.94 | Prefoldin subunit 3 |
| PF11_0087 | DNA repair protein | PF10_0041 | U5 small nuclear ribonucleoprotein |
| PFB0445c | ATP-dependent RNA helicase | PF10_0041 | U5 small nuclear ribonucleoprotein |
| PFE0925c | ATP-dependent RNA helicase | PF10_0041 | U5 small nuclear ribonucleoprotein |
| PF10_0294 | Pre-mRNA-splicing factor ATP-dependent RNA helicase | PF10_0041 | U5 small nuclear ribonucleoprotein |
| PFC1060c | U4/U6.U5 tri-small-nuclear-ribonucleoprotein-associated protein 1 | PF10_0041 | U5 small nuclear ribonucleoprotein |
| PF13_0096 | U4/U6.U5 tri-small-nuclear-ribonucleoprotein-associated protein 2 | PF10_0041 | U5 small nuclear ribonucleoprotein |
| PFC0365w | Pre-mRNA-processing factor 19 | PF10_0041 | U5 small nuclear ribonucleoprotein |
| PFD0685c | Structural maintenance of chromosomes protein 3 | PF10_0041 | U5 small nuclear ribonucleoprotein |
| MAL13P1.214 | Phosphoethanolamine N-methyltransferase | PFA0455c | Fatty acid elongation protein, GNS1/SUR4 family |
| MAL13P1.214 | Phosphoethanolamine N-methyltransferase | PFL0950c | Aminophospholipid-transporting P-ATPase |
| MAL8P1.17 | Protein disulfide isomerase | PF10_0092 | Metallopeptidase |
| MAL8P1.17 | Protein disulfide isomerase | PFB0920w | DnaJ protein |
| PF07_0029 | Heat shock protein 86 | MAL13P1.139 | Mitochondrial fission 1 protein |
| PF07_0029 | Heat shock protein 86 | PFI0300w | Vacuolar protein sorting-associated protein 46 |
| PF14_0068 | rRNA 2′-O-methyltransferase fibrillarin | PFF1180w | Anaphase-promoting complex subunit |
| PF14_0261 | Proliferation-associated protein 2g4 | PF14_0612 | Zinc finger protein |
| PF14_0286 | NADP-specific glutamate dehydrogenase | PF14_0334 | NAD(P)H-dependent glutamate synthase |
| PF14_0401 | tRNA import protein | PF13_0257 | Glutamate-tRNA ligase |
| PFC0510w | E3 ubiquitin-protein ligase | PFI0300w | Vacuolar protein sorting-associated protein 46 |
| PFE0750c | Pre-mRNA-splicing factor | PF14_0688 | Pre-mRNA-splicing factor ISY1 |
| PFF1385c | Conserved Plasmodium protein | PFB0920w | DnaJ protein |
| PFL1140w | Vacuolar iron transporter | PFL0725w | Thioredoxin peroxidase 2 |
SERCA, sarcoendoplasmic reticulum calcium transport ATPase; ERCC1, excision repair cross-complementation group 1.
The present approach critically depends on the existence of S. cerevisiae genes that (i) possess synthetic lethal phenotypes and (ii) are orthologous to known P. falciparum drug target genes. This seems contradictory; by definition, drug targets are essential and genes with synthetic phenotypes are nonessential. However, we show here that several validated drug targets of P. falciparum possess orthologues in S. cerevisiae that are nonessential (Table 1). Phosphoethanolamine methyltransferase and phosphatidylinositol 4-kinase (Table 1), for instance, have been demonstrated to be essential enzymes in P. falciparum (24, 33). Most of the genes that are conserved between S. cerevisiae and P. falciparum also have an orthologue in H. sapiens (the OrthoMCL database contains only 80 yeast genes with an orthologue in P. falciparum but not in H. sapiens). We speculate that, of the conserved genes that are essential in yeast, many may also be essential in H. sapiens and their products not suitable as drug targets. On the other hand, conserved genes that are devoid of synthetic phenotypes in yeast might also be disposable in P. falciparum and thus not suitable either. The conserved genes that have synthetic lethal phenotypes in yeast might be the most interesting pharmacologically.
The proposed algorithm strongly narrows the target space for antimalarial drug combinations by including potentially synergistic interactions involving efficacy against P. falciparum as well as toxicity against H. sapiens. The fact that it relies on genome-scale experimental data from S. cerevisiae rather than P. falciparum makes the algorithm straightforward and unbiased but also difficult to validate experimentally. Presently, experimental testing of the identified target pairs in Table 1 is precluded by the lack of inhibitors for most of the proposed targets. However, we think that the presence of targets such as PfPI4K and PfATP4 in Table 1 validates the algorithm, and we hope that the algorithm will help identify future combinations of antimalarial molecules that potentiate each other.
ACKNOWLEDGMENTS
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
We are grateful to Andrew M. Waterhouse for help with Python programming and to Matthias Rottmann for critical reading of the manuscript.
REFERENCES
- 1.Medicines for Malaria Venture. 2017. MMV drug development portfolio. https://www.mmv.org/research-development/mmv-supported-projects.
- 2.White NJ. 2016. Can new treatment developments combat resistance in malaria? Expert Opin Pharmacother 17:1303–1307. doi: 10.1080/14656566.2016.1187134. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization. 2010. Guidelines for the treatment of malaria. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 4.Wells TN, Hooft van Huijsduijnen R, Van Voorhis WC. 2015. Malaria medicines: a glass half full? Nat Rev Drug Discov 14:424–442. doi: 10.1038/nrd4573. [DOI] [PubMed] [Google Scholar]
- 5.Kremsner PG, Krishna S. 2004. Antimalarial combinations. Lancet 364:285–294. doi: 10.1016/S0140-6736(04)16680-4. [DOI] [PubMed] [Google Scholar]
- 6.malERA Consultative Group on Drugs. 2011. A research agenda for malaria eradication: drugs. PLoS Med 8:e1000402. doi: 10.1371/journal.pmed.1000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diagana TT. 2015. Supporting malaria elimination with 21st century antimalarial agent drug discovery. Drug Discov Today 20:1265–1270. doi: 10.1016/j.drudis.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 8.Conde-Pueyo N, Munteanu A, Sole RV, Rodriguez-Caso C. 2009. Human synthetic lethal inference as potential anti-cancer target gene detection. BMC Syst Biol 3:116. doi: 10.1186/1752-0509-3-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang C, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 10.Fügi MA, Kaiser M, Tanner M, Schneiter R, Mäser P, Guan XL. 2015. Match-making for posaconazole through systems thinking. Trends Parasitol 31:46–51. doi: 10.1016/j.pt.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Kaelin WG., Jr 2005. The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer 5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 12.Roemer T, Boone C. 2013. Systems-level antimicrobial drug and drug synergy discovery. Nat Chem Biol 9:222–231. doi: 10.1038/nchembio.1205. [DOI] [PubMed] [Google Scholar]
- 13.Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S, James TY, Karpov S, Kugrens P, Krug J, Lane CE, Lewis LA, Lodge J, Lynn DH, Mann DG, McCourt RM, Mendoza L, Moestrup O, Mozley-Standridge SE, Nerad TA, Shearer CA, Smirnov AV, Spiegel FW, Taylor MF. 2005. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol 52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- 14.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. 2006. BioGRID: a general repository for interaction datasets. Nucleic Acids Res 34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen F, Mackey AJ, Stoeckert CJ Jr, Roos DS. 2006. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res 34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer S, Brunk BP, Chen F, Gao X, Harb OS, Iodice JB, Shanmugam D, Roos DS, Stoeckert CJ Jr. 2011. Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr Protoc Bioinformatics Chapter 6:Unit 6.12.1–6.12.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aguero F, Al-Lazikani B, Aslett M, Berriman M, Buckner FS, Campbell RK, Carmona S, Carruthers IM, Chan AW, Chen F, Crowther GJ, Doyle MA, Hertz-Fowler C, Hopkins AL, McAllister G, Nwaka S, Overington JP, Pain A, Paolini GV, Pieper U, Ralph SA, Riechers A, Roos DS, Sali A, Shanmugam D, Suzuki T, Van Voorhis WC, Verlinde CL. 2008. Genomic-scale prioritization of drug targets: the TDR Targets database. Nat Rev Drug Discov 7:900–907. doi: 10.1038/nrd2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aparicio IM, Marin-Menendez A, Bell A, Engel PC. 2010. Susceptibility of Plasmodium falciparum to glutamate dehydrogenase inhibitors: a possible new antimalarial target. Mol Biochem Parasitol 172:152–155. doi: 10.1016/j.molbiopara.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Ludin P, Woodcroft B, Ralph SA, Maser P. 2012. In silico prediction of antimalarial drug target candidates. Int J Parasitol Drugs Drug Resist 2:191–199. doi: 10.1016/j.ijpddr.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salcedo-Sora JE, Ward SA, Biagini GA. 2012. A yeast expression system for functional and pharmacological studies of the malaria parasite Ca2+/H+ antiporter. Malar J 11:254. doi: 10.1186/1475-2875-11-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rottmann M, McNamara C, Yeung BK, Lee MC, Zou B, Russell B, Seitz P, Plouffe DM, Dharia NV, Tan J, Cohen SB, Spencer KR, Gonzalez-Paez GE, Lakshminarayana SB, Goh A, Suwanarusk R, Jegla T, Schmitt EK, Beck HP, Brun R, Nosten F, Renia L, Dartois V, Keller TH, Fidock DA, Winzeler EA, Diagana TT. 2010. Spiroindolones, a potent compound class for the treatment of malaria. Science 329:1175–1180. doi: 10.1126/science.1193225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldgof GM, Durrant JD, Ottilie S, Vigil E, Allen KE, Gunawan F, Kostylev M, Henderson KA, Yang J, Schenken J, LaMonte GM, Manary MJ, Murao A, Nachon M, Stanhope R, Prescott M, McNamara CW, Slayman CW, Amaro RE, Suzuki Y, Winzeler EA. 2016. Comparative chemical genomics reveal that the spiroindolone antimalarial KAE609 (cipargamin) is a P-type ATPase inhibitor. Sci Rep 6:27806. doi: 10.1038/srep27806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jimenez-Diaz MB, Ebert D, Salinas Y, Pradhan A, Lehane AM, Myrand-Lapierre ME, O'Loughlin KG, Shackleford DM, Justino de Almeida M, Carrillo AK, Clark JA, Dennis AS, Diep J, Deng X, Duffy S, Endsley AN, Fedewa G, Guiguemde WA, Gomez MG, Holbrook G, Horst J, Kim CC, Liu J, Lee MC, Matheny A, Martinez MS, Miller G, Rodriguez-Alejandre A, Sanz L, Sigal M, Spillman NJ, Stein PD, Wang Z, Zhu F, Waterson D, Knapp S, Shelat A, Avery VM, Fidock DA, Gamo FJ, Charman SA, Mirsalis JC, Ma H, Ferrer S, Kirk K, Angulo-Barturen I, Kyle DE, DeRisi JL, Floyd DM, Guy RK. 2014. (+)-SJ733, a clinical candidate for malaria that acts through ATP4 to induce rapid host-mediated clearance of Plasmodium. Proc Natl Acad Sci U S A 111:E5455–E5462. doi: 10.1073/pnas.1414221111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McNamara CW, Lee MC, Lim CS, Lim SH, Roland J, Simon O, Yeung BK, Chatterjee AK, McCormack SL, Manary MJ, Zeeman AM, Dechering KJ, Kumar TS, Henrich PP, Gagaring K, Ibanez M, Kato N, Kuhen KL, Fischli C, Nagle A, Rottmann M, Plouffe DM, Bursulaya B, Meister S, Rameh L, Trappe J, Haasen D, Timmerman M, Sauerwein RW, Suwanarusk R, Russell B, Renia L, Nosten F, Tully DC, Kocken CH, Glynne RJ, Bodenreider C, Fidock DA, Diagana TT, Winzeler EA. 2013. Targeting Plasmodium PI(4)K to eliminate malaria. Nature 504:248–253. doi: 10.1038/nature12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spillman NJ, Allen RJ, McNamara CW, Yeung BK, Winzeler EA, Diagana TT, Kirk K. 2013. Na+ regulation in the malaria parasite Plasmodium falciparum involves the cation ATPase PfATP4 and is a target of the spiroindolone antimalarials. Cell Host Microbe 13:227–237. doi: 10.1016/j.chom.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dembele L, Ang X, Chavchich M, Bonamy GMC, Selva JJ, Lim MY, Bodenreider C, Yeung BKS, Nosten F, Russell BM, Edstein MD, Straimer J, Fidock DA, Diagana TT, Bifani P. 2017. The Plasmodium PI(4)K inhibitor KDU691 selectively inhibits dihydroartemisinin-pretreated Plasmodium falciparum ring-stage parasites. Sci Rep 7:2325. doi: 10.1038/s41598-017-02440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paquet T, Le Manach C, Cabrera DG, Younis Y, Henrich PP, Abraham TS, Lee MCS, Basak R, Ghidelli-Disse S, Lafuente-Monasterio MJ, Bantscheff M, Ruecker A, Blagborough AM, Zakutansky SE, Zeeman AM, White KL, Shackleford DM, Mannila J, Morizzi J, Scheurer C, Angulo-Barturen I, Martinez MS, Ferrer S, Sanz LM, Gamo FJ, Reader J, Botha M, Dechering KJ, Sauerwein RW, Tungtaeng A, Vanachayangkul P, Lim CS, Burrows J, Witty MJ, Marsh KC, Bodenreider C, Rochford R, Solapure SM, Jimenez-Diaz MB, Wittlin S, Charman SA, Donini C, Campo B, Birkholtz LM, Hanson KK, Drewes G, Kocken CHM, Delves MJ, Leroy D, Fidock DA, Waterson D, Street LJ, Chibale K. 2017. Antimalarial efficacy of MMV390048, an inhibitor of Plasmodium phosphatidylinositol 4-kinase. Sci Transl Med 9:eaad9735. doi: 10.1126/scitranslmed.aad9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hain AU, Bartee D, Sanders NG, Miller AS, Sullivan DJ, Levitskaya J, Meyers CF, Bosch J. 2014. Identification of an Atg8-Atg3 protein-protein interaction inhibitor from the Medicines for Malaria Venture Malaria Box active in blood and liver stage Plasmodium falciparum parasites. J Med Chem 57:4521–4531. doi: 10.1021/jm401675a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jain J, Jain SK, Walker LA, Tekwani BL. 2017. Inhibitors of ubiquitin E3 ligase as potential new antimalarial drug leads. BMC Pharmacol Toxicol 18:40. doi: 10.1186/s40360-017-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaMonte GM, Almaliti J, Bibo-Verdugo B, Keller L, Zou BY, Yang J, Antonova-Koch Y, Orjuela-Sanchez P, Boyle CA, Vigil E, Wang L, Goldgof GM, Gerwick L, O'Donoghue AJ, Winzeler EA, Gerwick WH, Ottilie S. 2017. Development of a potent inhibitor of the Plasmodium proteasome with reduced mammalian toxicity. J Med Chem 60:6721–6732. doi: 10.1021/acs.jmedchem.7b00671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, O'Donoghue AJ, van der Linden WA, Xie SC, Yoo E, Foe IT, Tilley L, Craik CS, da Fonseca PC, Bogyo M. 2016. Structure- and function-based design of Plasmodium-selective proteasome inhibitors. Nature 530:233–236. doi: 10.1038/nature16936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tschan S, Brouwer AJ, Werkhoven PR, Jonker AM, Wagner L, Knittel S, Aminake MN, Pradel G, Joanny F, Liskamp RM, Mordmuller B. 2013. Broad-spectrum antimalarial activity of peptido sulfonyl fluorides, a new class of proteasome inhibitors. Antimicrob Agents Chemother 57:3576–3584. doi: 10.1128/AAC.00742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bobenchik AM, Witola WH, Augagneur Y, Nic Lochlainn L, Garg A, Pachikara N, Choi JY, Zhao YO, Usmani-Brown S, Lee A, Adjalley SH, Samanta S, Fidock DA, Voelker DR, Fikrig E, Ben Mamoun C. 2013. Plasmodium falciparum phosphoethanolamine methyltransferase is essential for malaria transmission. Proc Natl Acad Sci U S A 110:18262–18267. doi: 10.1073/pnas.1313965110. [DOI] [PMC free article] [PubMed] [Google Scholar]