ABSTRACT
A novel 65.8-kb multidrug resistance transposon, designated Tn6450, was characterized in a Proteus mirabilis isolate from chicken in China. Tn6450 contains 18 different antimicrobial resistance genes, including cephalosporinase gene blaDHA-1 and fluoroquinolone resistance genes qnrA1 and aac(6′)-Ib-cr. It carries a class 1/2 hybrid integron composed of intI2 and a 3′ conserved segment of the class 1 integron. Tn6450 is derived from Tn7 via acquisition of new mobile elements and resistance genes.
KEYWORDS: Proteus mirabilis, Tn7, multidrug resistance, transposon
TEXT
Proteus mirabilis is a member of the family Enterobacteriaceae that is widely distributed in the natural environment and the intestinal tract of humans and animals. P. mirabilis is recognized as an opportunistic pathogen associated with urinary tract infection (1), nosocomial infection (2), and food poisoning (3). Transposons, known as “jumping genes,” are specific DNA segments inserting into one or more sites in one or more genomes (4). They usually carry transposase that catalyzes the transposition event. Transposons are important mobile genetic elements that mediate the horizontal transfer of antimicrobial resistance genes (5). In the present study, we report a novel transposon harboring 18 different resistance genes in P. mirabilis.
P. mirabilis strain SNYG17 was isolated from a cloacal swab of a healthy chicken in Sichuan province in the nationwide surveillance of animal-origin bacterial antimicrobial resistance of 2016. MICs to various antimicrobials were determined by broth microdilution according to CLSI guidelines (6–8). Strain SNYG17 was resistant to ampicillin (MIC, >256 μg/ml), amoxicillin-clavulanic acid (>256/128 μg/ml), cefoxitin (128 μg/ml), cefotaxime (16 μg/ml), ceftriaxone (8 μg/ml), kanamycin (128 μg/ml), gentamicin (>256 μg/ml), streptomycin (>256 μg/ml), trimethoprim-sulfamethoxazole (>16/304 μg/ml), and florfenicol (>256 μg/ml); intermediate to amikacin (32 μg/ml); and susceptible to imipenem (<0.125 mg/liter), aztreonam (0.25 mg/liter), nalidixic acid (16 mg/liter), ciprofloxacin (0.5 μg/ml), and norfloxacin (4 μg/ml). Salmonella genomic island 1 (SGI1) and Proteus genomic island 1 (PGI1) were important mobilizable elements conferring multidrug resistance (MDR) phenotypes in P. mirabilis (9, 10). The presence of MDR genomic islands SGI1/PGI1 and plasmids were determined by PCR amplification targeting SGI1/PGI1 integrase genes (11) and plasmid replicons (12), respectively. PCR showed negative results, suggesting that strain SNYG17 did not harbor SGI1/PGI1 genes or plasmids.
The whole genome of SNYG17 was sequenced using a PacBio RS II sequencing instrument with a 100-fold average read depth. The reads were assembled into one scaffold using the software SMRT portal v.3.2.0. Nineteen different antimicrobial resistance genes were identified by ResFinder 2.1 (http://www.genomicepidemiology.org/), including blaDHA-1 (cephalosporin resistance), qnrA1 (fluoroquinolone), aac(6′)-Ib-cr (fluoroquinolone and aminoglycosides), floR (chloramphenicol/florfenicol), mphE and msrE (macrolide), and lunF (lincosamide) genes. Sequence analysis showed that 19 resistance genes, except for tet(J), were carried by a novel MDR transposon that was designated Tn6450 (Fig. 1), according to the nomenclature of transposons (http://transposon.lstmed.ac.uk/) (4).
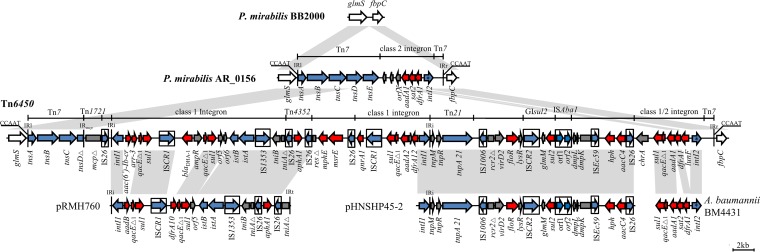
FIG 1.
Genetic structure of Tn6450. Structures are drawn to scale from GenBank accession numbers MF805806 (Tn6450), CP004022 (P. mirabilis BB2000), CP021852 (P. mirabilis AR_0156), AY123253 (pRMH760), KU341381 (pHNSHP45-2), and AJ289189 (A. baumannii BM4431). Arrows, genes and open reading frames; arrowheads, orientations of transcription. Shading, shared regions with >99.9% identity. Horizontal lines, different regions corresponding to Tn7, Tn1721, Tn21, and integrons. Antimicrobial resistance genes are red, and transposase or integrase genes are blue. The IS elements are indicated by boxes around the blue arrows. IRl and IRr, inverted repeats defining the left and right ends of class 1 integron, respectively; IRmcp, inverted repeat of Tn1721.
Tn6450 is 65,817 bp (corresponding to bases 2,006 to 67,822 in GenBank accession number MF805806) with an average GC content of 52.73% that differs from that of the rest of the P. mirabilis chromosome (GC content, 38.88%) (13). It has partial characteristics of the Tn7 transposon (Fig. 1), which is inserted downstream of the glmS gene, encoding glucosamine 6-phosphate synthase and surrounded by 5-bp direct repeats (CCAAT). Tn7 contains a transposition module formed by the tnsA, tnsB, tnsC, tnsD, and tnsE genes (14), in which tnsD and tnsE encode transposition target site selection proteins (15, 16). The truncation of tnsD and loss of tnsE might enhance the stability of Tn6450 in the P. mirabilis chromosome.
The tnsD in Tn6450 is interrupted by insertion of the inverted repeat (IRmcp) of Tn1721. The putative insertion site of Tn1721 is an AT-rich 5-bp target site (TTAGA) reported previously (17). On the right side of the IRmcp, the mcp gene of Tn1721 encoding a methyl-accepting chemotaxis protein is truncated at its 5′ end by insertion of IS26. Five copies of IS26 are present in Tn6450, which promotes the accumulation of several transposons and resistance genes (18). Tn6450 harbors two class 1 integrons, both of which lose the right-hand inverted repeat (IRt) resulting from the IS26-mediated rearrangements (Fig. 1). The left one is an In5-type class 1 integron containing aac(6′)-Ib-cr and arr-3 cassettes and two copies of the 3′ conserved segments (3′-CS) separated by ISCR1 and a region containing blaDHA-1 and ampR genes. The tniA gene is interrupted by Tn4352B containing kanamycin and neomycin resistance determinant aphA1, which is identical to In34 (pRMH760) (19). The other one is a Tn21 class 1 integron that contains dfrA12 and aadA2 cassettes and resistance gene qnrA1 linked to ISCR1. The 20,849-bp region (from 39,463 to 60,311 bp in MF805806), containing floR, sul2, hph, and aacC4 resistance genes, shows almost 100% identity to the corresponding region of seven IncHI2 plasmids, i.e., pHNSHP45-2 (KU341381), pWJ1 (KY924928), pHNYJC8 (KY019259), pHNLDF400 (KY019258), pXGE1mcr (KY990887), pHXY0908 (KM877269), and pSJ_255 (CP011062), with only 5 to 7 single-base changes, suggesting that this region in Tn6450 and IncHI2 plasmids might derive from the same source.
An interesting hybrid integron composed of a nonfunctional intI2 and 3′-CS of class 1 integron is found in Tn6450, carrying lunF, dfrA1, and aadA1 cassettes (Fig. 1). Another hybrid integron previously characterized in a clinical Acinetobacter baumannii strain harbored dfrA1, sat2, and aadA1 cassettes (20). The hybrid structure likely resulted from cointegrated formation catalyzed by the IntI1 integrase between a class 2 integron and a class 1 integron, with the aadA1 cassette at the left end of the 3′-CS.
The stability of Tn6450 was determined by continuous passage propagation lasting 20 days (40 passages) in the absence of antimicrobial pressure. Two hundred clones in the 41st passage were picked to detect the presence of Tn6450 by PCR and sequence with the primers listed in Table S1 in the supplemental material. The results of all PCRs were positive. The possible excision of Tn6450 from P. mirabilis chromosome was not detected in the 41st passage culture by use of a two-step PCR (Table S1), indicating that Tn6450 can be stable in P. mirabilis in the absence of antimicrobial pressure.
In conclusion, we characterized a novel MDR transposon, Tn6450, in P. mirabilis, which was derived from Tn7 via the acquisition of new mobile elements (class 1 and 2 integrons, transposons, and insertion sequence elements) and resistance genes, including the clinically important cephalosporinase gene blaDHA-1 and fluoroquinolone resistance genes qnrA1 and aac(6′)-Ib-cr. An interesting class 1/2 hybrid integron was found in Tn6450. More attention should be paid to the MDR transposon derived from Tn7, given that the Tn7 transposon, in which class 2 integrons have been embedded (21), has been found in various Gram-negative pathogenic bacteria (22).
Accession number(s).
The complete nucleotide sequence of Tn6450 characterized in this study was submitted to GenBank and assigned accession number MF805806.
Supplementary Material
ACKNOWLEDGMENTS
We thank Adam P. Roberts (University College London) for helpful suggestions regarding the nomenclature of Tn6450.
This work was supported by the earmarked fund for the Modern Agro-Industry Technology Research System (project no. CARS-40-K14), the general program of National Natural Science Foundation of China (grant no. 31572547), the Special Fund for Agro-Scientific Research in the Public Interest of China (grant no. 201303044), and the Scientific Research Foundation of Sichuan University (grant no. 2017SCU12006).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02192-17.
REFERENCES
- 1.Schaffer JN, Pearson MM. 2015. Proteus mirabilis and urinary tract infections. Microbiol Spectr 3. doi: 10.1128/microbiolspec.UTI-0017-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu YY, Cai JC, Zhang R, Zhou HW, Sun Q, Chen GX. 2012. Emergence of Proteus mirabilis Harboring blaKPC-2 and qnrD in a Chinese hospital. Antimicrob Agents Chemother 56:2278–2282. doi: 10.1128/AAC.05519-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, Zhang S, Yu J, Zhang H, Yuan Z, Sun Y, Zhang L, Zhu Y, Song H. 2010. An outbreak of Proteus mirabilis food poisoning associated with eating stewed pork balls in brown sauce, Beijing. Food Control 21:302–305. doi: 10.1016/j.foodcont.2009.06.009. [DOI] [Google Scholar]
- 4.Roberts AP, Chandler M, Courvalin P, Guedon G, Mullany P, Pembroke T, Rood JI, Smith CJ, Summers AO, Tsuda M, Berg DE. 2008. Revised nomenclature for transposable genetic elements. Plasmid 60:167–173. doi: 10.1016/j.plasmid.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillings MR, Paulsen IT, Tetu SG. 2017. Genomics and the evolution of antibiotic resistance. Ann NY Acad Sci 1388:92–107. doi: 10.1111/nyas.13268. [DOI] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. CLSI M100-S25. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals—4th ed CLSI VET01-A4. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. CLSI VET01-S2. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 9.Ahmed AM, Hussein AI, Shimamoto T. 2007. Proteus mirabilis clinical isolate harbouring a new variant of Salmonella genomic island 1 containing the multiple antibiotic resistance region. J Antimicrob Chemother 59:184–190. [DOI] [PubMed] [Google Scholar]
- 10.Siebor E, Neuwirth C. 2014. Proteus genomic island 1 (PGI1), a new resistance genomic island from two Proteus mirabilis French clinical isolates. J Antimicrob Chemother 69:3216–3220. doi: 10.1093/jac/dku314. [DOI] [PubMed] [Google Scholar]
- 11.Schultz E, Cloeckaert A, Doublet B, Madec JY, Haenni M. 2017. Detection of SGI1/PGI1 elements and resistance to extended-spectrum cephalosporins in Proteae of animal origin in France. Front Microbiol 8:32. doi: 10.3389/fmicb.2017.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 13.Pearson MM, Sebaihia M, Churcher C, Quail MA, Seshasayee AS, Luscombe NM, Abdellah Z, Arrosmith C, Atkin B, Chillingworth T, Hauser H, Jagels K, Moule S, Mungall K, Norbertczak H, Rabbinowitsch E, Walker D, Whithead S, Thomson NR, Rather PN, Parkhill J, Mobley HL. 2008. Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J Bacteriol 190:4027–4037. doi: 10.1128/JB.01981-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peters JE, Craig NL. 2001. Tn7: smarter than we thought. Nat Rev Mol Cell Biol 2:806–814. doi: 10.1038/35099006. [DOI] [PubMed] [Google Scholar]
- 15.Waddell CS, Craig NL. 1988. Tn7 transposition: two transposition pathways directed by five Tn7-encoded genes. Genes Dev 2:137–149. doi: 10.1101/gad.2.2.137. [DOI] [PubMed] [Google Scholar]
- 16.Peters JE. 2014. Tn7. Microbiol Spectr 2. doi: 10.1128/microbiolspec.MDNA3-0010-2014. [DOI] [PubMed] [Google Scholar]
- 17.Allmeier H, Cresnar B, Greck M, Schmitt R. 1992. Complete nucleotide sequence of Tn1721: gene organization and a novel gene product with features of a chemotaxis protein. Gene 111:11–20. doi: 10.1016/0378-1119(92)90597-I. [DOI] [PubMed] [Google Scholar]
- 18.Harmer CJ, Hall RM. 2016. IS 26-mediated formation of transposons carrying antibiotic resistance genes. mSphere 1:e00038–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Partridge SR, Hall RM. 2003. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrob Agents Chemother 47:342–349. doi: 10.1128/AAC.47.1.342-349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ploy MC, Denis F, Courvalin P, Lambert T. 2000. Molecular characterization of integrons in Acinetobacter baumannii: description of a hybrid class 2 integron. Antimicrob Agents Chemother 44:2684–2688. doi: 10.1128/AAC.44.10.2684-2688.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansson K, Sundstrom L, Pelletier A, Roy PH. 2002. IntI2 integron integrase in Tn7. J Bacteriol 184:1712–1721. doi: 10.1128/JB.184.6.1712-1721.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramirez MS, Pineiro S, Argentinian Integron Study G, Centron D. 2010. Novel insights about class 2 integrons from experimental and genomic epidemiology. Antimicrob Agents Chemother 54:699–706. doi: 10.1128/AAC.01392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.