ABSTRACT
Hypermutable Pseudomonas aeruginosa strains are prevalent in patients with cystic fibrosis and rapidly become resistant to antibiotic monotherapies. Combination dosage regimens have not been optimized against such strains using mechanism-based modeling (MBM) and the hollow-fiber infection model (HFIM). The PAO1 wild-type strain and its isogenic hypermutable PAOΔmutS strain (MICmeropenem of 1.0 mg/liter and MICtobramycin of 0.5 mg/liter for both) were assessed using 96-h static-concentration time-kill studies (SCTK) and 10-day HFIM studies (inoculum, ∼108.4 CFU/ml). MBM of SCTK data were performed to predict expected HFIM outcomes. Regimens studied in the HFIM were meropenem at 1 g every 8 h (0.5-h infusion), meropenem at 3 g/day with continuous infusion, tobramycin at 10 mg/kg of body weight every 24 h (1-h infusion), and both combinations. Meropenem regimens delivered the same total daily dose. Time courses of total and less susceptible populations and MICs were determined. For the PAOΔmutS strain in the HFIM, all monotherapies resulted in rapid regrowth to >108.7 CFU/ml with near-complete replacement by less susceptible bacteria by day 3. Meropenem every 8 h with tobramycin caused >7-log10 bacterial killing followed by regrowth to >6 log10 CFU/ml by day 5 and high-level resistance (MICmeropenem, 32 mg/liter; MICtobramycin, 8 mg/liter). Continuous infusion of meropenem with tobramycin achieved >8-log10 bacterial killing without regrowth. For PAO1, meropenem monotherapies suppressed bacterial growth to <4 log10 over 7 to 9 days, with both combination regimens achieving near eradication. An MBM-optimized meropenem plus tobramycin regimen achieved synergistic killing and resistance suppression against a difficult-to-treat hypermutable P. aeruginosa strain. For the combination to be maximally effective, it was critical to achieve the optimal shape of the concentration-time profile for meropenem.
KEYWORDS: dynamic infection model, dosage regimen, combination therapy, Pseudomonas aeruginosa
INTRODUCTION
Pseudomonas aeruginosa has an extraordinary capacity to develop resistance to antipseudomonal agents (1). Treatment failure due to emergence and amplification of antibiotic-resistant mutants is a frequent outcome of P. aeruginosa infections (2–4) and is particularly important in the management of chronic infections requiring prolonged treatment (5). As antimicrobial resistance has increased worldwide, hypermutation has become increasingly recognized as a major problem for antimicrobial therapy (6, 7). Hypermutable strains (i.e., strains with up to 1,000-fold increased mutation rates caused by defects in DNA repair or error avoidance systems, commonly due to mutations in the mutS gene) of P. aeruginosa develop resistance even more rapidly than nonhypermutable strains (8–12). Hypermutable P. aeruginosa strains are prevalent in patients with cystic fibrosis (CF) and have been linked to increased multidrug resistance (MDR) and reduced lung function (8, 11–16).
Hypermutable P. aeruginosa strains can become resistant to all available antibiotics administered as monotherapy (15, 17). Given the serious clinical consequences arising from infections caused by hypermutable strains and their frequent development of MDR, there is an urgent need to optimize antibiotic therapy to improve efficacy, including suppression of the emergence of resistance (18, 19). However, exacerbations of chronic P. aeruginosa infections are typically treated with either nonoptimized monotherapy or empirically chosen nonoptimized combinations (20), risking the emergence of MDR hypermutable strains. We therefore aimed to (i) characterize the effect of different concentrations of meropenem and tobramycin, in monotherapy and combination, on bacterial killing and resistance emergence in hypermutable and nonhypermutable P. aeruginosa, (ii) develop mechanism-based models (MBM) that support the selection of an optimized combination dosage regimen that maximizes bacterial killing and minimizes the emergence of resistance, and (iii) evaluate optimized and standard combination regimens in the hollow-fiber infection model (HFIM).
(Part of this work has been presented as a poster at the 55th Interscience Conference on Antimicrobial Agents and Chemotherapy, 18 to 21 September 2015, San Diego, CA. This work was also subject to oral presentations at the 27th European Congress of Clinical Microbiology and Infectious Diseases, 22 to 25 April 2017, Vienna, Austria, and the Australian Society of Clinical and Experimental Pharmacologists and Toxicologists Annual Meeting, 29 November to 2 December 2015, Hobart, Australia.)
RESULTS
Static-concentration time-kill studies (SCTK).
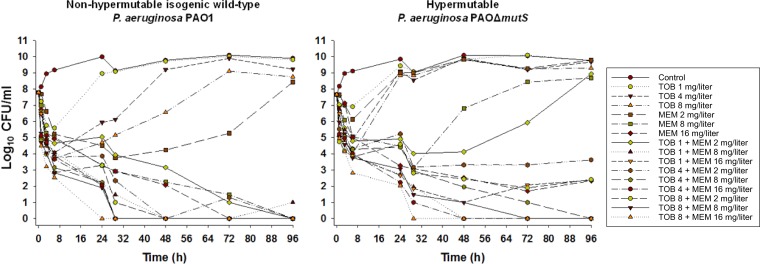
Total viable counts are presented in Fig. 1, and mutation frequencies (see Table S1 the supplemental material), log10 changes in viable counts (Table S2), and less-susceptible subpopulations (Fig. S1 and S2) are also presented.
FIG 1.
Bacterial counts with various concentrations of tobramycin and meropenem alone and in combination from the static time-kill model over 96 h. MEM, meropenem; TOB, tobramycin. For each strain, MICMEM is 1 mg/liter and MICTOB is 0.5 mg/liter.
PAO1 strain.
All tobramycin concentrations produced rapid (within 7 h) initial killing of strain PAO1 of ∼2 to 4 log10 CFU/ml, followed by regrowth close to growth control values, with the emergence of a high proportion of resistant bacteria. Meropenem at 2 mg/liter resulted in ∼4-log10 killing over the first 29 h, followed by regrowth due almost entirely to less susceptible bacteria; at 8 and 16 mg/liter, bacterial killing continued, with no colonies detected at 96 and 72 h, respectively. The combination of the two lowest concentrations of each antibiotic produced ∼3-log10 CFU/ml initial killing and a slow decline in bacterial numbers thereafter, with no viable bacteria detected at 96 h. All other combinations produced rapid initial killing of ∼4-log10 CFU/ml with synergy occurring at various times thereafter and no viable colonies from 24 to 48 h onwards.
PAOΔmutS strain.
Tobramycin at 1 mg/liter produced little PAOΔmutS strain bacterial killing without increases in resistant subpopulations. A 4 and 8 mg/liter initial ∼3- to 4-log10 killing was followed by rapid regrowth with emergence of a high proportion of resistant bacteria. Meropenem at 2 mg/liter produced ∼2-log10 killing at 6 h, whereas at 8 mg/liter ∼5-log10 killing occurred over 29 h. In both cases rapid regrowth followed, with virtually the entire population growing on the 2.5-mg/liter meropenem-containing agar plates. Meropenem at 16 mg/liter produced sustained killing over 96 h but not eradication, with less susceptible subpopulations emerging. Combinations containing tobramycin (all concentrations) plus meropenem at 2 mg/liter produced rapid ∼3 to 4 log10 CFU/ml killing over 6 h, followed by gradual regrowth (tobramycin at 1 mg/liter plus meropenem at 2 mg/liter) or suppression of regrowth (tobramycin at 4 or 8 mg/liter plus meropenem at 2 mg/liter) with synergy. In all combinations with 2 mg/liter meropenem, tobramycin-resistant subpopulations emerged by 48 h and subsequently increased, whereas subpopulations less susceptible to meropenem were detected after 24 h only with the two lowest tobramycin concentrations in combination. All other combinations, except 1 mg/liter tobramycin plus 16 mg/liter meropenem, produced synergistic killing from ∼24 to 48 h onwards and eventual eradication (no viable bacteria detected).
Mechanism-based mathematical modeling.
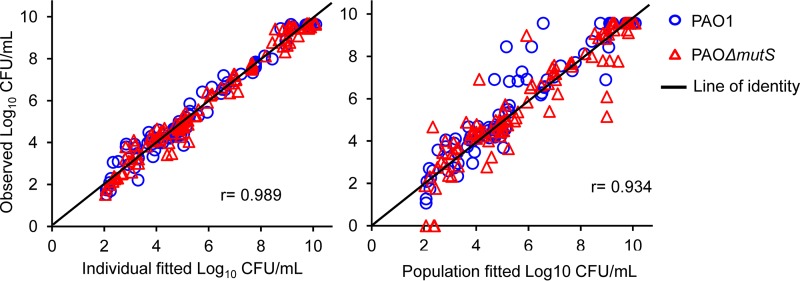
The SCTK data for both strains were comodeled and well described by the developed MBM, including subpopulation and mechanistic synergy (Fig. 2 and 3). A model with a single maximum killing rate constant (Kmax; i.e., using the same Kmax for each subpopulation and strain) per antibiotic was clearly inferior based on the significantly poorer (38 points; P < 0.0001) objective function (−1× log likelihood in S-ADAPT) and population fit plots. Thus, different estimates for Kmax were required. The model yielded unbiased and precise curve fits for the total bacterial counts of both strains. The coefficient of correlation for the observed versus individual fitted viable counts was 0.989. The parameter estimates are reported in Table S3. The extended MBM adequately captured the growth and killing of less susceptible bacteria (Fig. S3).
FIG 2.
(A) Fit plots for PAO1. (B) Fit plots for PAOΔmutS strain.
FIG 3.
Observed versus individual and population fitted viable counts for meropenem and tobramycin alone and in combinations against PAO1 and PAOΔmutS strains.
The MBM predicted failure of all monotherapies against both strains in the HFIM, except for suppression of regrowth of PAO1 over 10 days by meropenem at 3 g/day as a continuous infusion (Table 1). Against PAO1, both combination regimens were predicted to suppress regrowth over 10 days. In contrast, the MBM predicted suppression of regrowth of the PAOΔmutS strain would be achieved only by the optimized combination regimen with continuous infusion of meropenem (Table 1).
TABLE 1.
Meropenem and tobramycin dosage regimens, model predictions, and HFIM outcomes against PAO1 and PAOΔmutS strainsa
| Treatmentb | MEM fT>MIC (%) for PAO1 and PAOΔmutS (MIC, 1 mg/liter) |
TOB fAUC/MIC for PAO1 and PAOΔmutS (MIC, 0.5 mg/liter) | Predictions and outcomes for: |
||||
|---|---|---|---|---|---|---|---|
| PAO1 |
PAOΔmutS |
||||||
| fT>1× MIC | fT>5× MIC | Predicted from MBM | Observed in the HFIM | Predicted from MBM | Observed in the HFIM | ||
| MEM, 1 g, every 8 h | 61 | 36 | ∼5.2 log10 killing at 71 h, slow regrowth from 95 h, counts within ∼1 log10 of control values from 239 h | ∼5.2 log10 killing at 71 h, regrowth from 168 h, counts within ∼1 log10 of control values from 239 h | ∼3 log10 killing at 23 h (nadir), regrowth from 26 h, counts within ∼1 log10 of control values from 47 h | ∼1 log10 killing at 1.5 to 23 h, regrowth from 26 h, counts within ∼1 log10 of control values from 47 h | |
| MEM, 3 g/day, CI | 100 | 100 | ∼6.5 log10 killing at 71 h, suppressed regrowth over 239 h | ∼5.5 log10 killing at 71 h, limited regrowth at 239 h only | ∼3.7 log10 killing at 26 h (nadir), regrowth from 47 h, counts within ∼1 log10 of control from 71 h | ∼2 log10 killing at 23 h (nadir), regrowth from 26 h, counts within ∼1 log10 of control values from 71 h | |
| TOB, 10 mg/kg, every 24 h | 217 | ∼3.5 log10 killing, regrowth from 23 h, counts at control values from 47 h | ∼7.2 log10 killing, regrowth from 47 h, counts within ∼1.5 log10 of control from 71 h | ∼3.0 log10 killing at 5 h (nadir), regrowth from 23 h, counts at control values from 47 h | ∼4.5 log10 killing at 5 h (nadir), regrowth from 23 h, counts at control from 71 h | ||
| MEM, 1 g, every 8 h, plus TOB | 61 | 36 | 217 | ∼6.7 log10 killing (faster than MEM alone) at 47 h, counts of <1 log10 from 53 h until 239 h | ∼6.9 log10 killing (faster than MEM alone) at 47 h, counts of <1 log10 from 53 h until 239 h | ∼5.8 log10 killing at 53 h (nadir), ∼5 log10 killing at 71 h, regrowth above 4 log10 from 95 h, ∼7 log10 reached at 143 h | ∼5.7 log10 killing at 53 h, ∼7.5 log10 killing at 71 h (nadir), regrowth above 4 log10 from 95 h, ∼7 log10 reached at 119 h |
| MEM, 3 g/day, CI, plus TOB | 100 | 100 | 217 | ∼8.5 log10 killing (faster than MEM alone) from 47 h, counts of ∼1.0 log10 or below until 239 h | ∼8.5 log10 killing (faster than MEM alone) from 26 h, counts of ∼1.0 log10 or below until 239 h | ∼8.0 log10 killing at 71 h, counts of <1.0 log10 after that until 239 h | ∼8.0 log10 killing at 71 h, counts of ∼1.0 log10 or below from 119 h until 239 h |
MEM, meropenem; TOB, tobramycin; MBM, mechanism-based model; HFIM, hollow-fiber infection model; CI, continuous infusion; fT>1× MIC, the cumulative percentage of a 24-h period that unbound concentrations exceed 1× MIC; fT>5× MIC, the cumulative percentage of a 24-h period that unbound concentrations exceed 5× MIC; fAUC/MIC, ratio of the area under the unbound concentration-time curve to MIC. For less susceptible populations of PAO1, the extended MBM predicted the following: for 1 g MEM every 8 h, almost complete replacement of the total by populations less susceptible to MEM on day 10; for 3 g/day MEM CI, no quantifiable populations less susceptible to MEM from 23 h onwards; for 1 g MEM every 8 h plus TOB and 3 g/day MEM CI plus TOB, no quantifiable populations less susceptible to MEM and TOB from 23 h onwards; for 10 mg/kg TOB every 24 h, populations less susceptible to TOB within ∼1 log10 CFU/ml of the total population from 47 h onwards. For less susceptible populations of the PAOΔmutS strain, the extended MBM predicted the following: for 1 g MEM every 8 h and 3 g/day MEM CI, almost complete replacement of the total by populations less susceptible to MEM from 47 h onwards; for 10 mg/kg TOB every 24 h, populations less susceptible to TOB within ∼1 log10 CFU/ml of the total population from 47 h onwards; for 1 g MEM every 8 h plus TOB, populations less susceptible to MEM and TOB within ∼1 log10 CFU/ml of the total population from 71 h onwards; for 3 g/day MEM CI plus TOB, no quantifiable populations less susceptible to MEM and TOB from 23 h onwards.
Simulated half-lives: meropenem, 0.8 h; tobramycin, 2.5 h. No loading dose was administered for intermittent dosing. The meropenem CI (3 g/day) was started at the average steady-state concentration of ∼8 mg/liter.
Hollow-fiber in vitro infection model.
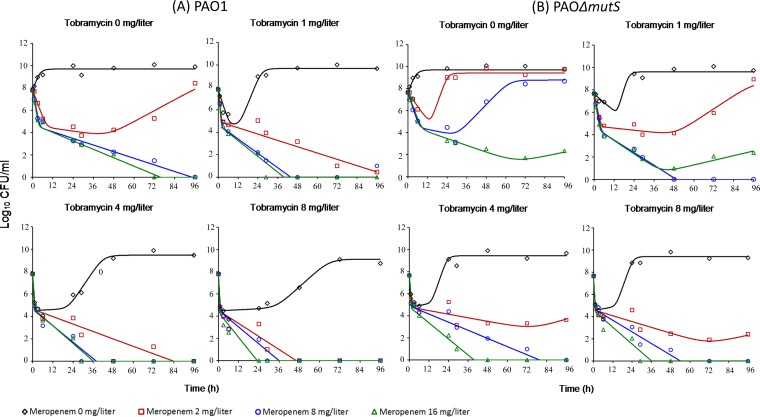
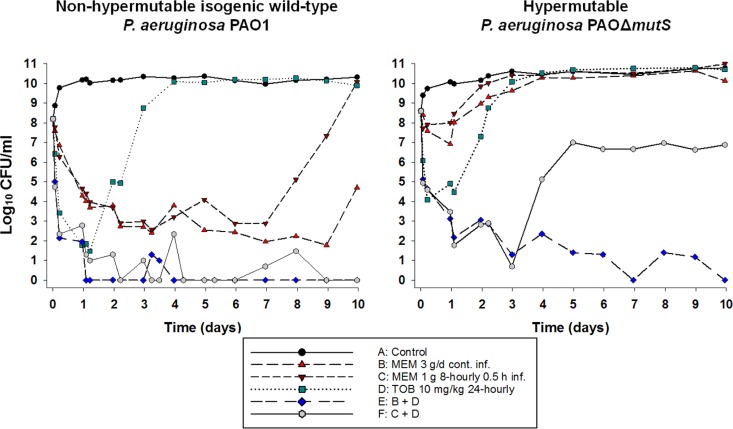
For each of the simulated profiles, the observed meropenem and tobramycin concentrations were, on average, within 20% of the targeted concentrations. Changes in viable counts, total and less susceptible populations, and MICs are shown in Fig. 4 and 5 and Table S6. Changes in mutation frequencies are shown in Tables S4 and S5.
FIG 4.
Bacterial counts following meropenem (3 g/day, continuous infusion, or 1 g via 30-min intravenous infusion every 8 h) and tobramycin (10 mg/kg every 24 h as a 1-h infusion) in monotherapies and combination studied in the HFIM. MEM, meropenem; TOB, tobramycin. For each strain, MICMEM is 1 mg/liter and MICTOB is 0.5 mg/liter.
FIG 5.
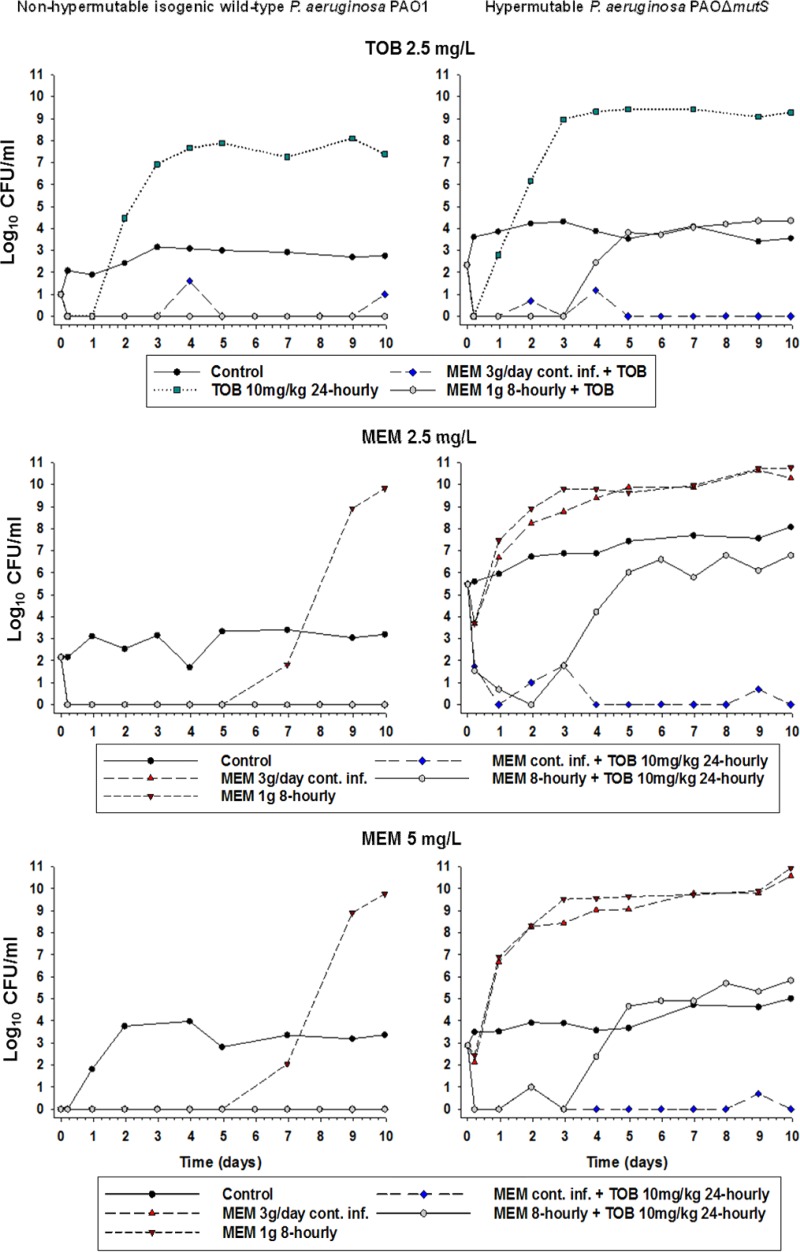
Bacterial counts on agar plates containing 2.5 mg/liter tobramycin, 2.5 mg/liter meropenem, and 5 mg/liter meropenem from the HFIM over 10 days. MEM, meropenem; TOB, tobramycin. For each strain, MICMEM is 1 mg/liter and MICTOB is 0.5 mg/liter.
PAO1.
The growth control grew rapidly to ∼10.5 log10 CFU/ml, with less susceptible populations for both meropenem (2.5 and 5 mg/liter on agar) and tobramycin (2.5 mg/liter on agar), plateauing at ∼3 to 4 log10 CFU/ml. Tobramycin monotherapy produced killing of ∼7 log10 over the first day, followed by rapid regrowth to control values. Regrowth was accompanied by an ∼5-log10 increase in less susceptible bacteria compared to the control. Both the intermittent and continuous meropenem regimens produced nearly identical killing of ∼5-log10 over the first 7 days, with few or no less susceptible bacteria detected. After that, rapid regrowth to control levels occurred with intermittent meropenem due almost entirely to an increase in less susceptible bacteria. Growth with the continuous meropenem regimen remained low (∼2.0 to 2.5 log10 CFU/ml) until day 9 before increasing to ∼5 log10 at day 10; no less susceptible bacteria were detected at any time. Both combination regimens suppressed regrowth over 10 days, with few if any colonies detected after 24 h.
PAOΔmutS strain.
For the PAOΔmutS strain, the growth control grew rapidly to ∼10.5 log10 CFU/ml, with less susceptible subpopulations plateauing at ∼7 to 8 log10 CFU/ml and ∼4 to 5 log10 CFU/ml on agar containing 2.5 mg/liter and 5 mg/liter meropenem, respectively. Less susceptible populations plateaued at ∼4 log10 CFU/ml on the tobramycin-containing agar. With monotherapy, initial killing of ∼4 log10 with tobramycin and <∼1.5 log10 with both meropenem regimens was followed in all cases by rapid regrowth due almost entirely to less susceptible bacteria. Both combinations produced nearly identical killing of ∼7.5 log10 over the first 3 days. With the standard combination (containing 1 g meropenem given every 8 h), rapid regrowth of ∼6 log10 due almost entirely to less susceptible subpopulations subsequently occurred, with growth plateauing at ∼7 log10. With the optimized combination, bacterial killing continued such that no viable bacteria were detected at 7 and 10 days.
DISCUSSION
The PAO1 wild-type reference strain and its isogenic hypermutable PAOΔmutS strain were used in this study. The PAOΔmutS strain differs from PAO1 only by the absence of mutS, which encodes a component of the mismatch repair system that detects and repairs replication errors. The strains shared the same MICs for each antibiotic. Deletion of mutS is one of the most frequent mutations in clinical hypermutable P. aeruginosa isolates, and it represents a nearly worst-case scenario, as it has a large impact on increasing mutation rate (12, 21, 22). The increased resistance of the PAOΔmutS strain is due to the ascent to dominance of resistant mutant subpopulations, but increases in minimum bactericidal concentrations observed with single agents (meropenem, imipenem, and ceftazidime) can be minimized using combinations of two bactericidal antipseudomonal agents (10). Current guidelines endorse the use of combination antipseudomonal therapy for the treatment of acute exacerbations in patients with CF (23), with antipseudomonal β-lactams and aminoglycosides most commonly used (24–26). However, combination dosage regimens have never been optimized. Antipseudomonal β-lactams are commonly administered via intermittent infusion (every 6 to 8 h over 5 to 30 min) (25).
Based on synergistic bacterial killing of the PAOΔmutS strain by clinically relevant meropenem and tobramycin concentrations in SCTK, we evaluated this combination in standard and optimized dosage regimens (the latter selected using MBM, which incorporated the SCTK data) in the HFIM. Short-term infusions remain the standard method of administration for β-lactams, including meropenem, in a large part of the world, as demonstrated by two recent studies that included up to 53 countries (27, 28). However, prolonged infusions are used in other countries. We performed simulations with our MBM for 0.5-h, 3-h, and continuous-infusion meropenem dosing in monotherapy and combination with tobramycin. Our MBM predicted that only the optimized combination regimen (containing meropenem at 3 g/day with continuous infusion) would suppress bacterial regrowth and minimize resistance emergence over 10 days for both strains. Regrowth and resistance emergence of the PAOΔmutS strain was predicted to occur with both the standard combination (containing 1 g meropenem every 8 h as 0.5-h infusions) and the combination including meropenem as prolonged infusions (1 g every 8 h as 3-h infusions; data not shown). Therefore, the combinations containing standard (short-term) and continuous-infusion meropenem were evaluated in the HFIM. Both of these combination regimens produced nearly identical killing of PAOΔmutS organisms across the first 3 days in the HFIM; extensive regrowth of less susceptible bacteria ensued with the standard combination, whereas killing continued across 10 days with the optimized combination regimen, similar to the MBM predictions.
Our results are instructive in a number of ways. First, they highlight the utility of MBM to select optimized regimens that maximize bacterial killing and minimize resistance emergence against hypermutable P. aeruginosa, an especially important finding given these strains can rapidly develop MDR. Second, they show the potentially different pharmacodynamic responses to therapy of hypermutable and nonhypermutable strains, with bacterial killing and emergence of resistance (for both mono- and combination therapy) differing substantially between otherwise identical strains. Third, as will be discussed below, they show the importance of shape in relation to concentration-time profiles.
PK/PD approaches to optimize the administration of β-lactams have typically sought to maximize the fT>MIC, i.e., the fraction of the dosing interval for which the unbound concentration remains above 1× MIC of the infecting pathogen (29, 30). For carbapenems such as meropenem, in vitro, ex vivo, and animal in vivo studies, usually of relatively short duration (≤24 h), have shown fT>MIC values of 20% and 40% to be necessary for bacteriostasis and near-maximal bacterial killing, respectively (31–33). In the present HFIM studies, meropenem monotherapy with both intermittent (fT>MIC of 61%) and continuous (fT>MIC of 100%) infusions did suppress regrowth over at least 7 days against the wild-type strain. However, even the continuous infusion, where concentrations remained at ∼8× MIC at all times, was completely ineffective against the hypermutable strain. This comprehensive failure of both intermittent and continuous infusion as monotherapy against the PAOΔmutS strain, due to the rapid ascent to dominance of mutants less susceptible to meropenem (Fig. 5), strongly argues against the use of monotherapy, especially against hypermutable strains.
While our data suggest that combination therapy is required for treatment of P. aeruginosa infections that are chronic or known to involve hypermutable strains, the design of such regimens must consider ways to minimize the emergence of resistance (10). With maximal bactericidal activity of the β-lactams occurring at 4× to 5× MIC (34, 35) and regrowth often observed as soon as concentrations fall below the MIC (36, 37), pharmacodynamic targets such as 100% fT>4–5×MIC and unbound minimum concentration (fCmin)/MIC have been investigated in relation to suppression of emergence of resistance (38–42). We observed that the standard meropenem regimen, in combination with tobramycin, eliminated the wild-type strain but not the hypermutable strain, which demonstrated significant regrowth comprising large numbers of bacteria less susceptible to both antibiotics. However, greatly enhanced bacterial killing and resistance suppression were achieved with the optimized combination. For the standard intermittent meropenem regimen in the HFIM, concentrations dropped below the MIC ∼4.2 h after administration, with essentially no drug remaining at the end of each dosing interval (Cmin of 0.07 mg/liter for meropenem every 8 h). However, with the optimized combination regimen, meropenem concentrations greatly exceeded the MIC (∼8× MIC) at all times, even when tobramycin concentrations had declined. For meropenem, an fCmin/MIC of ≥2 to 6 and 100% fT>5×MIC have been associated with suppression of resistance emergence (38–41), with both targets substantially exceeded with the continuous infusion regimen. Thus, despite each meropenem regimen having the same daily dose, the shape of the optimized meropenem concentration-time profile maximized fT>5×MIC and fCmin/MIC compared to the standard regimen, such that in combination with tobramycin, emergence of resistance was minimized. Achieving an optimal shape of the concentration-time profile for meropenem was therefore critical for the combination to be maximally effective.
Previous work has shown that combining antibiotics that require separate and independent mutations for resistance development may help minimize selection of mutants resistant to multiple drugs (10). Resistance to meropenem occurs primarily via reduced expression of the gene for the outer membrane porin OprD (which allows carbapenems to enter the cell) and enzymatic inactivation via carbapenemases (43). The most important resistance mechanisms of P. aeruginosa against aminoglycosides are increased expression of MexXY-OprM (particularly common in strains from patients with CF) (44), modification of the binding site, and enzymatic inactivation (43). The different resistance mechanisms of each antibiotic in the combination, together with the maintenance of higher meropenem concentrations, may have contributed to the substantially enhanced effectiveness of the optimized combination regimen against the hypermutable strain.
We have examined standard and optimized regimens against a hypermutable and a nonhypermutable P. aeruginosa strain. The focus on P. aeruginosa may be regarded as a limitation of the study, as the airways of patients with CF are often colonized with multiple bacterial species simultaneously (45). Nevertheless, P. aeruginosa is the predominating species in most adults with CF and remains the most common pathogen associated with morbidity and mortality among patients with CF (46–48). Future studies may be directed at evaluating the optimized combination regimen against other P. aeruginosa isolates or other species, alone or in combination. The HFIM utilized lacks an immune system, in particular granulocytes, which work in combination with antibiotic treatment to control bacterial infections. Therefore, future animal studies may be warranted to assess immune system effects on residual populations following initial bacterial killing by the antibiotics. However, given the ethical limitations on study duration inherent in animal studies, suppression of the emergence of resistance, a key component of the present study, is best examined in the HFIM where longer study durations can be employed.
In summary, standard regimens of meropenem and tobramycin, both as monotherapy and in combination, produced substantially less bacterial killing and suppression of emergence of resistance against a hypermutable P. aeruginosa strain than its isogenic wild-type reference strain. MBM was applied effectively to SCTK data to select an optimized dosage regimen. Against the hypermutable strain the optimized combination regimen, when subjected to human pharmacokinetics in the HFIM, prevented the regrowth and emergence of resistance observed with the standard combination regimen. Achieving an optimal shape of the concentration-time profile for meropenem was critical for the combination to be maximally effective against both strains. The optimized combination dosage regimen is expected to be highly promising for evaluation in future clinical studies.
MATERIALS AND METHODS
Antibiotics, bacterial strains, media, and susceptibility testing.
Stock solutions of meropenem (Hospira, Melbourne, Australia) and tobramycin (AK Scientific, Union City, MD, USA) were prepared as previously described (49). Viable counting was performed on cation-adjusted Mueller-Hinton agar (CAMHA; containing 25 mg/liter Ca2+ and 12.5 mg/liter Mg2+; Media Preparation Unit, University of Melbourne, Melbourne, Australia). Drug-containing agar plates were prepared by adding appropriate volumes of stock solution to CAMHA (BD, Sparks, MD, USA). All studies used CAMHB (BD, Sparks, MD, USA) containing 25 mg/liter Ca2+ and 12.5 mg/liter Mg2+.
The P. aeruginosa wild-type reference strain PAO1 and its PAOΔmutS isogenic hypermutable strain (constructed from PAO1 by Mena et al. [50] via mutS deletion) were used. The meropenem and tobramycin MICs prior to drug exposure were determined in duplicate on separate days (51). For both strains the meropenem and tobramycin MICs were 1 mg/liter and 0.5 mg/liter, respectively. Susceptibility and resistance were defined as MICs of ≤2 and >8 mg/liter for meropenem and ≤4 and >4 mg/liter for tobramycin according to EUCAST guidelines (52).
SCTK.
To characterize the effect of different meropenem and tobramycin concentrations in monotherapy and in combination on PAO1 and PAOΔmutS strains, static-concentration time-kill experiments (SCTK) were performed (53, 54). Bacteria were grown on CAMHA at 36°C for ∼20 h, followed by overnight incubation in a shaking water bath at 36°C in sterile CAMHB. The optical density of the bacterial suspension was measured using a spectrophotometer, and the suspension was appropriately diluted to achieve the targeted initial inoculum of ∼107.8 CFU/ml in 15 ml. Antibiotic stock solutions were added at the initiation of the experiments (0 h) to achieve concentrations of 2, 8, and 16 mg/liter for meropenem and 1, 4, and 8 mg/liter for tobramycin, i.e., concentrations within the range of those observed in patients (55, 56). Bacterial suspensions were centrifuged (10 min, 3,220 × g, 36°C) and resuspended in prewarmed, antibiotic-containing sterile CAMHB (100% of the initial antibiotic concentration) every 24 h (53). Meropenem was additionally supplemented at 30% of the initial concentration at 6, 30, 54, and 78 h to compensate for its known thermal degradation (57).
Samples for total viable counting were collected at 0, 1, 3, 6, 24, 29, 48, 72, and 96 h and for quantification of resistant bacteria at 0, 24, 48, and 96 h. To minimize antibiotic carryover, samples were twice centrifuged at 4,000 × g for 5 min and resuspended in sterile saline (36°C) (42). For total viable counting, 100 μl of undiluted or appropriately diluted sample was manually plated onto CAMHA. Given the expected low number of resistant bacteria in samples from some treatments, 200 μl of sample was plated onto CAMHA containing 2.5 mg/liter meropenem or 5 mg/liter tobramycin to increase sensitivity. Antibiotic-free and meropenem-containing plates were incubated at 36°C for 48 h and tobramycin-containing plates for 72 h, followed by manual counting of colonies. The limit of counting was 1.0 log10 CFU/ml for antibiotic-free (i.e., one colony per plate) and 0.7 log10 CFU/ml for antibiotic-containing plates. The log10(mutation frequency), i.e., log10(MF), was determined by the following equation: log10(MF) = log10(CFU/ml on antibiotic-containing agar) − log10(CFU/ml on antibiotic-free agar).
Microbiological responses were quantified using the log10 change method, which compares the change in log10(CFU/ml) from 0 h (CFU0) to time t (1, 3, 6, 24, 29, 48, 72 and 96 h; CFUt): log10 change = log10(CFUt) − log10(CFU0). Synergy was defined as a reduction of ≥2 log10 CFU/ml for the combination compared to the most active monotherapy component at the specified time (58).
MBM.
Simultaneous mechanism-based modeling (MBM) of SCTK data from both strains was performed using importance sampling (pmethod of 4) in S-ADAPT (v1.57) with S-ADAPT-TRAN (59–61). The S-ADAPT objective function value (−1× log likelihood), standard diagnostic plots, coefficients of correlation, biological plausibility of parameter estimates, and visual predictive checks were used for model evaluation. A life cycle growth model described bacterial growth and replication (62, 63). The final models included three subpopulations with different susceptibilities to meropenem and tobramycin and direct bacterial killing by both antibiotics. Subpopulation synergy and mechanistic synergy were incorporated to describe the effect of the combinations (54). Mechanistic synergy was described as tobramycin enhancing the target site penetration of meropenem via disrupting the bacterial outer membrane (53, 64, 65). The differential equations are available in the supplemental material. The models were then used in in silico simulations to predict the expected bacterial outcomes for clinically relevant monotherapy and combination regimens and the pharmacokinetics as observed in CF patients. Berkeley Madonna (v8.3.18) was used for all in silico simulations. We extended the MBM by simultaneously fitting the time courses of the total bacterial population and the populations less susceptible to meropenem or tobramycin (quantified on plates containing 2.5 mg/liter meropenem and 5 mg/liter tobramycin) for both strains, as previously described (49, 64).
Hollow-fiber in vitro infection model.
The free (non-protein-bound) meropenem and tobramycin concentration-time profiles were simulated based on pharmacokinetics reported from patients with CF (55, 56). The standard regimens were 1 g meropenem every 8 h as a 30-min infusion and 10 mg/kg tobramycin every 24 h as a 1-h infusion (Table 1); these doses correspond to those used clinically in CF patients (66). An optimized meropenem regimen of 3 g/day as continuous infusion, chosen based on MBM predictions, was also evaluated in monotherapy and in combination with tobramycin (Table 1). Based on in silico simulations, the meropenem and tobramycin concentration-time profiles, in monotherapy and combination therapy, were reproduced experimentally in the HFIM for each bacterial strain (simulated clearances and half-lives were the following: meropenem, 15.9 liters/h and 0.8 h; tobramycin, 4.9 liters/h and 2.5 h). The continuous infusion of meropenem was started at the steady-state concentration of ∼8 mg/liter. A growth control was included for each strain.
HFIM studies were conducted over 10 days as previously described (42, 67, 68), utilizing cellulosic cartridges (C3008-1; FiberCell Systems Inc., Frederick, MD, USA) in a humidified incubator at 36°C. For each strain, one colony was grown overnight in CAMHB at 36°C to prepare the bacterial stock solution. The optical density of the overnight culture was measured spectrophotometrically. A bacterial suspension was prepared to achieve the targeted initial inoculum of ∼108.4 CFU/ml, and 17 ml was injected into each HFIM cartridge. Samples for viable counting (1.0 to 1.5 ml) were collected at 0, 1.5, 5, 23, 26, 29, 47, 53, 71, 95, 119, 143, 167, 191, 215, and 239 h. Less susceptible bacteria were quantified on CAMHA containing meropenem at 2.5 and 5 mg/liter (2.5× and 5× MIC) and tobramycin at 2.5 mg/liter (5× MIC). The total and less susceptible bacteria and log10(MF) were quantified per the SCTK methods described above. MICs were determined at 0, 95, 215, and/or 239 h by taking a subset of at least three colonies from antibiotic-free and antibiotic-containing plates.
Meropenem and tobramycin assays for pharmacokinetics.
Samples (1.0 ml) were collected in duplicate from the outflow of the HFIM central reservoir and immediately stored at −80°C until assayed. Meropenem and tobramycin in CAMHB were measured using validated liquid chromatography-tandem mass spectrometry (LC-MS/MS) assays. An Agilent 1200 high-performance liquid chromatograph (HPLC) coupled with an Agilent 6430 triple-quadrupole mass spectrometer equipped with a turbo ion electrospray ionization (ESI) source (Agilent Technologies, Santa Clara, CA) was used.
For measurement of tobramycin and meropenem, 50 μl of internal standard solution (10 μg/ml metformin in acetonitrile) was added to 50 μl of sample and vortex mixed for 30 s. Fifty microliters of acetonitrile and 100 μl of 0.05% trichloroacetic acid then were added, followed by vortex mixing for 1 min before centrifugation for 10 min at 15,000 × g. One hundred microliters of the supernatant was transferred to a polypropylene HPLC vial, diluted with 100 μl of distilled water, and vortex mixed for 30 s, and then 5 μl was injected onto a Synergi Polar-RP column (150 by 2.0 mm, 4.0 μm; Phenomenex, Torrance, CA, USA) using a binary gradient mobile phase composed of 0.25% formic acid (A) and acetonitrile (B), programmed as A:B at 80:20 (0 to 1 and 3.6 to 10 min) and A:B at 10:90 (1.1 to 3.5 min). The flow rate of the mobile phase was 0.2 ml/min (0 to 2.5 and 4.9 to 10.0 min) and 0.5 ml/min (2.6 to 4.8 min). The column oven temperature was 30°C, and total run time was 10 min. The ESI source was operated in positive mode, and the ionization gas temperature was set at 350°C. Mass transitions of the precursor/product ion pairs were monitored at m/z of 468.3→163.1, 384.2→141.0, and 130.1→71.0 for tobramycin, meropenem, and metformin, respectively. Mass spectrometric data were processed by the MassHunter Quantitative Analysis software (Agilent Technologies, Santa Clara, CA, USA). For both antibiotics the lower limit of quantification was 50 ng/ml and the correlation coefficient was >0.99. The interday precision was 2.3 to 6.9% for meropenem and 2.5 to 7.7% for tobramycin; interday accuracy was 95.9 to 100% for meropenem and 94.3 to 103.5% for tobramycin.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Australian National Health and Medical Research Council (NHMRC) project grant APP1101553 (to C.B.L., J.D.B., J.B.B., A.O., and R.L.N.), the William Buckland Foundation (project reference CT 22943 to C.B.L.), and in part by NHMRC APP1045105 (to J.B.B., A.O., C.B.L., J.D.B., and R.L.N.). C.B.L. is the recipient of an Australian National Health and Medical Research Council Career Development Fellowship (APP1062509), and A.Y.P. is the recipient of an NHMRC Practitioner Fellowship (APP1117940).
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02055-17.
REFERENCES
- 1.Livermore DM. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin Infect Dis 34:634–640. doi: 10.1086/338782. [DOI] [PubMed] [Google Scholar]
- 2.Carmeli Y, Troillet N, Eliopoulos GM, Samore MH. 1999. Emergence of antibiotic-resistant Pseudomonas aeruginosa: comparison of risks associated with different antipseudomonal agents. Antimicrob Agents Chemother 43:1379–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fish DN, Piscitelli SC, Danziger LH. 1995. Development of resistance during antimicrobial therapy: a review of antibiotic classes and patient characteristics in 173 studies. Pharmacotherapy 15:279–291. [PubMed] [Google Scholar]
- 4.Juan C, Gutierrez O, Oliver A, Ayestaran JI, Borrell N, Perez JL. 2005. Contribution of clonal dissemination and selection of mutants during therapy to Pseudomonas aeruginosa antimicrobial resistance in an intensive care unit setting. Clin Microbiol Infect 11:887–892. doi: 10.1111/j.1469-0691.2005.01251.x. [DOI] [PubMed] [Google Scholar]
- 5.Gibson RL, Burns JL, Ramsey BW. 2003. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am J Respir Crit Care Med 168:918–951. doi: 10.1164/rccm.200304-505SO. [DOI] [PubMed] [Google Scholar]
- 6.Blazquez J. 2003. Hypermutation as a factor contributing to the acquisition of antimicrobial resistance. Clin Infect Dis 37:1201–1209. doi: 10.1086/378810. [DOI] [PubMed] [Google Scholar]
- 7.Chopra I, O'Neill AJ, Miller K. 2003. The role of mutators in the emergence of antibiotic-resistant bacteria. Drug Resist Updat 6:137–145. doi: 10.1016/S1368-7646(03)00041-4. [DOI] [PubMed] [Google Scholar]
- 8.Oliver A, Canton R, Campo P, Baquero F, Blazquez J. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1254. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 9.Miller JH. 1996. Spontaneous mutators in bacteria: insights into pathways of mutagenesis and repair. Annu Rev Microbiol 50:625–643. doi: 10.1146/annurev.micro.50.1.625. [DOI] [PubMed] [Google Scholar]
- 10.Oliver A, Levin BR, Juan C, Baquero F, Blazquez J. 2004. Hypermutation and the preexistence of antibiotic-resistant Pseudomonas aeruginosa mutants: implications for susceptibility testing and treatment of chronic infections. Antimicrob Agents Chemother 48:4226–4233. doi: 10.1128/AAC.48.11.4226-4233.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferroni A, Guillemot D, Moumile K, Bernede C, Le Bourgeois M, Waernessyckle S, Descamps P, Sermet-Gaudelus I, Lenoir G, Berche P, Taddei F. 2009. Effect of mutator P. aeruginosa on antibiotic resistance acquisition and respiratory function in cystic fibrosis. Pediatr Pulmonol 44:820–825. doi: 10.1002/ppul.21076. [DOI] [PubMed] [Google Scholar]
- 12.Macia MD, Blanquer D, Togores B, Sauleda J, Perez JL, Oliver A. 2005. Hypermutation is a key factor in development of multiple-antimicrobial resistance in Pseudomonas aeruginosa strains causing chronic lung infections. Antimicrob Agents Chemother 49:3382–3386. doi: 10.1128/AAC.49.8.3382-3386.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waine DJ, Honeybourne D, Smith EG, Whitehouse JL, Dowson CG. 2008. Association between hypermutator phenotype, clinical variables, mucoid phenotype, and antimicrobial resistance in Pseudomonas aeruginosa. J Clin Microbiol 46:3491–3493. doi: 10.1128/JCM.00357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henrichfreise B, Wiegand I, Pfister W, Wiedemann B. 2007. Resistance mechanisms of multiresistant Pseudomonas aeruginosa strains from Germany and correlation with hypermutation. Antimicrob Agents Chemother 51:4062–4070. doi: 10.1128/AAC.00148-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliver A. 2010. Mutators in cystic fibrosis chronic lung infection: prevalence, mechanisms, and consequences for antimicrobial therapy. Int J Med Microbiol 300:563–572. doi: 10.1016/j.ijmm.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 16.Hogardt M, Hoboth C, Schmoldt S, Henke C, Bader L, Heesemann J. 2007. Stage-specific adaptation of hypermutable Pseudomonas aeruginosa isolates during chronic pulmonary infection in patients with cystic fibrosis. J Infect Dis 195:70–80. doi: 10.1086/509821. [DOI] [PubMed] [Google Scholar]
- 17.Canton R, Cobos N, de Gracia J, Baquero F, Honorato J, Gartner S, Alvarez A, Salcedo A, Oliver A, Garcia-Quetglas E. 2005. Antimicrobial therapy for pulmonary pathogenic colonisation and infection by Pseudomonas aeruginosa in cystic fibrosis patients. Clin Microbiol Infect 11:690–703. doi: 10.1111/j.1469-0691.2005.01217.x. [DOI] [PubMed] [Google Scholar]
- 18.Bals R, Hubert D, Tummler B. 2011. Antibiotic treatment of CF lung disease: from bench to bedside. J Cyst Fibros 10(Suppl 2):S146–S151. doi: 10.1016/S1569-1993(11)60019-2. [DOI] [PubMed] [Google Scholar]
- 19.Prescott WA Jr, Gentile AE, Nagel JL, Pettit RS. 2011. Continuous-infusion antipseudomonal beta-lactam therapy in patients with cystic fibrosis. P T 36:723–763. [PMC free article] [PubMed] [Google Scholar]
- 20.Guillot E, Sermet I, Ferroni A, Chhun S, Pons G, Zahar JR, Jullien V. 2010. Suboptimal ciprofloxacin dosing as a potential cause of decreased Pseudomonas aeruginosa susceptibility in children with cystic fibrosis. Pharmacotherapy 30:1252–1258. doi: 10.1592/phco.30.12.1252. [DOI] [PubMed] [Google Scholar]
- 21.Denamur E, Matic I. 2006. Evolution of mutation rates in bacteria. Mol Microbiol 60:820–827. doi: 10.1111/j.1365-2958.2006.05150.x. [DOI] [PubMed] [Google Scholar]
- 22.Jolivet-Gougeon A, Kovacs B, Le Gall-David S, Le Bars H, Bousarghin L, Bonnaure-Mallet M, Lobel B, Guille F, Soussy CJ, Tenke P. 2011. Bacterial hypermutation: clinical implications. J Med Microbiol 60:563–573. doi: 10.1099/jmm.0.024083-0. [DOI] [PubMed] [Google Scholar]
- 23.Flume PA, Mogayzel PJ Jr, Robinson KA, Goss CH, Rosenblatt RL, Kuhn RJ, Marshall BC. 2009. Cystic fibrosis pulmonary guidelines: treatment of pulmonary exacerbations. Am J Respir Crit Care Med 180:802–808. doi: 10.1164/rccm.200812-1845PP. [DOI] [PubMed] [Google Scholar]
- 24.Van Meter DJ, Corriveau M, Ahern JW, Lahiri T. 2009. A survey of once-daily dosage tobramycin therapy in patients with cystic fibrosis. Pediatr Pulmonol 44:325–329. doi: 10.1002/ppul.20985. [DOI] [PubMed] [Google Scholar]
- 25.Zobell JT, Young DC, Waters CD, Ampofo K, Cash J, Marshall BC, Olson J, Chatfield BA. 2011. A survey of the utilization of anti-pseudomonal beta-lactam therapy in cystic fibrosis patients. Pediatr Pulmonol 46:987–990. doi: 10.1002/ppul.21467. [DOI] [PubMed] [Google Scholar]
- 26.Prescott WA., Jr 2011. National survey of extended-interval aminoglycoside dosing in pediatric cystic fibrosis pulmonary exacerbations. J Pediatr Pharmacol Ther 16:262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tabah A, De Waele J, Lipman J, Zahar JR, Cotta MO, Barton G, Timsit JF, Roberts JA. 2015. The ADMIN-ICU survey: a survey on antimicrobial dosing and monitoring in ICUs. J Antimicrob Chemother 70:2671–2677. doi: 10.1093/jac/dkv165. [DOI] [PubMed] [Google Scholar]
- 28.Abdul-Aziz MH, Lipman J, Akova M, Bassetti M, De Waele JJ, Dimopoulos G, Dulhunty J, Kaukonen KM, Koulenti D, Martin C, Montravers P, Rello J, Rhodes A, Starr T, Wallis SC, Roberts JA. 2016. Is prolonged infusion of piperacillin/tazobactam and meropenem in critically ill patients associated with improved pharmacokinetic/pharmacodynamic and patient outcomes? An observation from the defining antibiotic levels in intensive care unit patients (DALI) cohort. J Antimicrob Chemother 71:196–207. [DOI] [PubMed] [Google Scholar]
- 29.Leggett JE, Fantin B, Ebert S, Totsuka K, Vogelman B, Calame W, Mattie H, Craig WA. 1989. Comparative antibiotic dose-effect relations at several dosing intervals in murine pneumonitis and thigh-infection models. J Infect Dis 159:281–292. doi: 10.1093/infdis/159.2.281. [DOI] [PubMed] [Google Scholar]
- 30.Tozuka Z, Murakawa T. 2007. β-Lactam pharmacodynamics, p 129–146. In Nightingale CH, Ambrose PG, Drusano G, Murakami T (ed), Antimicrobial pharmacodynamics in theory and practice, 2nd ed Informa Healthcare, New York, NY. [Google Scholar]
- 31.Craig WA. 1997. The pharmacology of meropenem, a new carbapenem antibiotic. Clin Infect Dis 24(Suppl 2):S266–S275. doi: 10.1093/clinids/24.Supplement_2.S266. [DOI] [PubMed] [Google Scholar]
- 32.Roberts JA, Kirkpatrick CM, Roberts MS, Robertson TA, Dalley AJ, Lipman J. 2009. Meropenem dosing in critically ill patients with sepsis and without renal dysfunction: intermittent bolus versus continuous administration? Monte Carlo dosing simulations and subcutaneous tissue distribution. J Antimicrob Chemother 64:142–150. [DOI] [PubMed] [Google Scholar]
- 33.Drusano GL. 2004. Antimicrobial pharmacodynamics: critical interactions of “bug and drug.” Nat Rev Microbiol 2:289–300. [DOI] [PubMed] [Google Scholar]
- 34.Mouton JW, den Hollander JG. 1994. Killing of Pseudomonas aeruginosa during continuous and intermittent infusion of ceftazidime in an in vitro pharmacokinetic model. Antimicrob Agents Chemother 38:931–936. doi: 10.1128/AAC.38.5.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mouton JW, Vinks AA. 2007. Continuous infusion of beta-lactams. Curr Opin Crit Care 13:598–606. doi: 10.1097/MCC.0b013e3282e2a98f. [DOI] [PubMed] [Google Scholar]
- 36.Craig W. 1984. Pharmacokinetic and experimental data on beta-lactam antibiotics in the treatment of patients. Eur J Clin Microbiol 3:575–578. doi: 10.1007/BF02013628. [DOI] [PubMed] [Google Scholar]
- 37.Mouton JW, Vinks AA, Punt NC. 1997. Pharmacokinetic-pharmacodynamic modeling of activity of ceftazidime during continuous and intermittent infusion. Antimicrob Agents Chemother 41:733–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tam VH, Schilling AN, Neshat S, Poole K, Melnick DA, Coyle EA. 2005. Optimization of meropenem minimum concentration/MIC ratio to suppress in vitro resistance of Pseudomonas aeruginosa. Antimicrob Agents Chemother 49:4920–4927. doi: 10.1128/AAC.49.12.4920-4927.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tam VH, Schilling AN, Poole K, Nikolaou M. 2007. Mathematical modelling response of Pseudomonas aeruginosa to meropenem. J Antimicrob Chemother 60:1302–1309. doi: 10.1093/jac/dkm370. [DOI] [PubMed] [Google Scholar]
- 40.Louie A, Grasso C, Bahniuk N, Van Scoy B, Brown DL, Kulawy R, Drusano GL. 2010. The combination of meropenem and levofloxacin is synergistic with respect to both Pseudomonas aeruginosa kill rate and resistance suppression. Antimicrob Agents Chemother 54:2646–2654. doi: 10.1128/AAC.00065-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li C, Du X, Kuti JL, Nicolau DP. 2007. Clinical pharmacodynamics of meropenem in patients with lower respiratory tract infections. Antimicrob Agents Chemother 51:1725–1730. doi: 10.1128/AAC.00294-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bergen PJ, Bulitta JB, Kirkpatrick CM, Rogers KE, McGregor MJ, Wallis SC, Paterson DL, Lipman J, Roberts JA, Landersdorfer CB. 2016. Effect of different renal function on antibacterial effects of piperacillin against Pseudomonas aeruginosa evaluated via the hollow-fibre infection model and mechanism-based modelling. J Antimicrob Chemother 71:2509–2520. doi: 10.1093/jac/dkw153. [DOI] [PubMed] [Google Scholar]
- 43.Poole K. 2011. Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2:65. doi: 10.3389/fmicb.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poole K. 2005. Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 49:479–487. doi: 10.1128/AAC.49.2.479-487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Filkins LM, O'Toole GA. 2015. Cystic fibrosis lung infections: polymicrobial, complex, and hard to treat. PLoS Pathog 11:e1005258. doi: 10.1371/journal.ppat.1005258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhagirath AY, Li Y, Somayajula D, Dadashi M, Badr S, Duan K. 2016. Cystic fibrosis lung environment and Pseudomonas aeruginosa infection. BMC Pulm Med 16:174. doi: 10.1186/s12890-016-0339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lyczak JB, Cannon CL, Pier GB. 2002. Lung infections associated with cystic fibrosis. Clin Microbiol Rev 15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Hoiby N, Molin S. 2012. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol 10:841–851. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- 49.Rees VE, Bulitta JB, Oliver A, Tsuji BT, Rayner CR, Nation RL, Landersdorfer CB. 2016. Resistance suppression by high-intensity, short-duration aminoglycoside exposure against hypermutable and non-hypermutable Pseudomonas aeruginosa. J Antimicrob Chemother 71:3157–3167. doi: 10.1093/jac/dkw297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mena A, Smith EE, Burns JL, Speert DP, Moskowitz SM, Perez JL, Oliver A. 2008. Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. J Bacteriol 190:7910–7917. doi: 10.1128/JB.01147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clinical and Laboratory Standards Institute. 2017. Performance standards for antimicrobial susceptibility testing: 25th informational supplement (M100-S27). Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 52.EUCAST. 2017. Breakpoint tables for interpretation of MICs and zone diameters, version 6.0, 1 January 2016. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_7.1_Breakpoint_Tables.pdf.
- 53.Yadav R, Landersdorfer CB, Nation RL, Boyce JD, Bulitta JB. 2015. Novel approach to optimize synergistic carbapenem-aminoglycoside combinations against carbapenem-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 59:2286–2298. doi: 10.1128/AAC.04379-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Landersdorfer CB, Ly NS, Xu H, Tsuji BT, Bulitta JB. 2013. Quantifying subpopulation synergy for antibiotic combinations via mechanism-based modeling and a sequential dosing design. Antimicrob Agents Chemother 57:2343–2351. doi: 10.1128/AAC.00092-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bui KQ, Ambrose PG, Nicolau DP, Lapin CD, Nightingale CH, Quintiliani R. 2001. Pharmacokinetics of high-dose meropenem in adult cystic fibrosis patients. Chemotherapy 47:153–156. doi: 10.1159/000063216. [DOI] [PubMed] [Google Scholar]
- 56.Hennig S, Standing JF, Staatz CE, Thomson AH. 2013. Population pharmacokinetics of tobramycin in patients with and without cystic fibrosis. Clin Pharmacokinet 52:289–301. doi: 10.1007/s40262-013-0036-y. [DOI] [PubMed] [Google Scholar]
- 57.Viaene E, Chanteux H, Servais H, Mingeot-Leclercq MP, Tulkens PM. 2002. Comparative stability studies of antipseudomonal beta-lactams for potential administration through portable elastomeric pumps (home therapy for cystic fibrosis patients) and motor-operated syringes (intensive care units). Antimicrob Agents Chemother 46:2327–2332. doi: 10.1128/AAC.46.8.2327-2332.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pillai SK, Moellering RC, Eliopoulos GM. 2005. Antimicrobial combinations. In Lorian V. (ed), Antibiotics in laboratory medicine, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 59.Bauer RJ, Guzy S, Ng C. 2007. A survey of population analysis methods and software for complex pharmacokinetic and pharmacodynamic models with examples. AAPS J 9:E60–E83. doi: 10.1208/aapsj0901007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bulitta JB, Landersdorfer CB. 2011. Performance and robustness of the Monte Carlo importance sampling algorithm using parallelized S-ADAPT for basic and complex mechanistic models. AAPS J 13:212–226. doi: 10.1208/s12248-011-9258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bulitta JB, Bingolbali A, Shin BS, Landersdorfer CB. 2011. Development of a new pre- and post-processing tool (SADAPT-TRAN) for nonlinear mixed-effects modeling in S-ADAPT. AAPS J 13:201–211. doi: 10.1208/s12248-011-9257-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bulitta JB, Ly NS, Yang JC, Forrest A, Jusko WJ, Tsuji BT. 2009. Development and qualification of a pharmacodynamic model for the pronounced inoculum effect of ceftazidime against Pseudomonas aeruginosa. Antimicrob Agents Chemother 53:46–56. doi: 10.1128/AAC.00489-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bergen PJ, Bulitta JB, Kirkpatrick CMJ, Rogers KE, McGregor MJ, Wallis SC, Paterson DL, Nation RL, Lipman J, Roberts JA, Landersdorfer CB. 2017. Substantial impact of altered pharmacokinetics in critically ill patients on the antibacterial effects of meropenem evaluated via the dynamic hollow-fiber infection model. Antimicrob Agents Chemother 61:e02642-16. doi: 10.1128/AAC.02642-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yadav R, Bulitta JB, Nation RL, Landersdorfer CB. 2017. Optimization of synergistic combination regimens against carbapenem- and aminoglycoside-resistant clinical Pseudomonas aeruginosa isolates via mechanism-based pharmacokinetic/pharmacodynamic modeling. Antimicrob Agents Chemother 61:e01011-16. doi: 10.1128/AAC.01011-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bulitta JB, Ly NS, Landersdorfer CB, Wanigaratne NA, Velkov T, Yadav R, Oliver A, Martin L, Shin BS, Forrest A, Tsuji BT. 2015. Two mechanisms of killing of Pseudomonas aeruginosa by tobramycin assessed at multiple inocula via mechanism-based modeling. Antimicrob Agents Chemother 59:2315–2327. doi: 10.1128/AAC.04099-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rao GG, Ly NS, Bulitta JB, Soon RL, San Roman MD, Holden PN, Landersdorfer CB, Nation RL, Li J, Forrest A, Tsuji BT. 2016. Polymyxin B in combination with doripenem against heteroresistant Acinetobacter baumannii: pharmacodynamics of new dosing strategies. J Antimicrob Chemother 71:3148–3156. doi: 10.1093/jac/dkw293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cadwell JJS. 2012. The hollow fibre infection model for antimicrobial pharmacodynamics and pharmacokinetics. Adv Pharmacoepiderm Drug Safety S1:007. doi: 10.4172/2167-1052.S1-007. [DOI] [Google Scholar]
- 68.Louie A, Castanheira M, Liu W, Grasso C, Jones RN, Williams G, Critchley I, Thye D, Brown D, Vanscoy B, Kulawy R, Drusano GL. 2012. Pharmacodynamics of beta-lactamase inhibition by NXL104 in combination with ceftaroline: examining organisms with multiple types of beta-lactamases. Antimicrob Agents Chemother 56:258–270. doi: 10.1128/AAC.05005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.