ABSTRACT
Mycobacterium avium subsp. hominissuis mainly causes disseminated infection in immunocompromised hosts, such as individuals with human immunodeficiency virus (HIV) infection, and pulmonary infection in immunocompetent hosts. However, many aspects of the different types of M. avium subsp. hominissuis infection remain unclear. We examined the antibiotic susceptibilities and genotypes of M. avium subsp. hominissuis isolates from different hosts by performing drug susceptibility testing using eight antibiotics (clarithromycin, rifampin, ethambutol, streptomycin, kanamycin, amikacin, ethionamide, and levofloxacin) and variable-number tandem-repeat (VNTR) typing analysis for 46 isolates from the sputa of HIV-negative patients with pulmonary M. avium subsp. hominissuis disease without previous antibiotic treatment and 30 isolates from the blood of HIV-positive patients with disseminated M. avium subsp. hominissuis disease. Interestingly, isolates from pulmonary M. avium subsp. hominissuis disease patients were more resistant to seven of the eight drugs, with the exception being rifampin, than isolates from HIV-positive patients. Moreover, VNTR typing analysis showed that the strains examined in this study were roughly classified into three clusters, and the genetic distance from reference strain 104 for isolates from pulmonary M. avium subsp. hominissuis disease patients was statistically significantly different from that for isolates from HIV-positive patients (P = 0.0018), suggesting that M. avium subsp. hominissuis strains that cause pulmonary and disseminated disease have genetically distinct features. Significant differences in susceptibility to seven of the eight drugs, with the exception being ethambutol, were noted among the three clusters. Collectively, these results suggest that an association between the type of M. avium subsp. hominissuis infection, drug susceptibility, and the VNTR genotype and the properties of M. avium subsp. hominissuis strains associated with the development of pulmonary disease are involved in higher levels of antibiotic resistance.
KEYWORDS: Mycobacterium avium subsp. hominissuis, pulmonary disease, disseminated disease, antibiotic susceptibility, variable-number tandem repeats
INTRODUCTION
Nontuberculous mycobacteria (NTM) are ubiquitous in the environment, including natural water, soil, and household dust (1, 2), and can cause significant disease in humans and animals (3). NTM infection was thought to be caused by NTM residing in the environment (2, 4, 5). However, person-to-person transmission has recently been reported among cystic fibrosis patients infected with Mycobacterium abscessus (6). The incidence of NTM pulmonary infection is increasing annually in many countries, including the United States and Japan (7–10). In Japan, the causative NTM strain for pulmonary disease with the highest incidence is M. avium (approximately 60%), followed by M. intracellulare, M. kansasii, and M. abscessus, and the incidence per 100,000 population increased remarkably from 5.7 in 2007 to 14.7 in 2014 (9, 11).
M. avium is the most clinically significant NTM species in humans and animals and consists of four subspecies (M. avium subsp. avium, M. avium subsp. silvaticum, M. avium subsp. hominissuis, and M. avium subsp. paratuberculosis), each of which has specific pathogenic and host range characteristics (12–14). Among the M. avium subspecies, M. avium subsp. hominissuis has been isolated from patients with respiratory disease, human immunodeficiency virus (HIV) infection, and lymphadenitis (3), as well as from asymptomatic pigs with granulomatous lesions (15). M. avium subsp. hominissuis mainly causes disseminated infection via the gastrointestinal route in immunocompromised hosts or pigs and pulmonary infection via the respiratory route in immunocompetent hosts. Recent studies have reported the isolation of genetically different M. avium subsp. hominissuis strains from different hosts or different countries and regions (16, 17), showing the genetic diversity of M. avium subsp. hominissuis. We have previously reported genetic differences between strain TH135, isolated from a patient with pulmonary M. avium subsp. hominissuis disease, and strain 104, obtained from an HIV-positive patient, by comparing the genomes of the strains (18). Such genetic differences may affect not only the pathological manifestation of M. avium subsp. hominissuis infection but also produce various phenotypes of M. avium subsp. hominissuis, such as various antibiotic susceptibility phenotypes. Elucidation of the phenotypic differences would facilitate the understanding of M. avium subsp. hominissuis infections and provide a valuable insight into antibiotic treatments.
The guidelines for antibiotic treatment of pulmonary M. avium subsp. hominissuis disease recommend macrolide-based multidrug therapy, comprising macrolides, such as clarithromycin or azithromycin, in combination with rifampin and ethambutol. In addition, aminoglycosides, such as streptomycin or amikacin, are recommended for patients with severe disease (19). However, the therapeutic efficacy of the drug above certain levels is unknown (19–21). Moreover, the clinical course of patients with pulmonary M. avium subsp. hominissuis disease is diverse, and some patients remain stable without treatment, while the symptoms cause deterioration in others, despite long-term multidrug therapy, leading to severe lung damage (3, 22, 23). This is possibly the result of host factors as well as bacterial factors. In our previous study, comparative genome analysis revealed the presence of potential genetic determinants, including a pMAH135 plasmid (24), associated with the progression of pulmonary M. avium subsp. hominissuis disease (25). These results suggest the involvement of bacterial factors in the progression of pulmonary M. avium subsp. hominissuis disease.
This study sought to examine the features of antibiotic susceptibility and the genotype in M. avium subsp. hominissuis isolates from hosts with different types of M. avium subsp. hominissuis infection. Thus, we performed drug susceptibility testing and variable-number tandem-repeat (VNTR) typing analysis of 46 isolates from HIV-negative patients with pulmonary M. avium subsp. hominissuis disease without previous antibiotic treatment and 30 isolates from HIV-positive patients with disseminated M. avium subsp. hominissuis disease.
RESULTS
Drug susceptibility of M. avium subsp. hominissuis isolates from different origins.
We examined the characteristics of antibiotic susceptibility of M. avium subsp. hominissuis isolates from different hosts by measuring the MICs of eight drugs (clarithromycin, rifampin, ethambutol, streptomycin, kanamycin, amikacin, ethionamide, and levofloxacin) for 46 isolates from the sputa of HIV-negative patients who were diagnosed with pulmonary M. avium subsp. hominissuis disease but received no antibiotic treatment, as well as 30 isolates from the blood of HIV-positive patients with disseminated M. avium subsp. hominissuis disease, by the broth dilution method (see Tables S1 and S2 in the supplemental material). Interestingly, ethambutol and ethionamide resistance was observed in 84.8% (39/46) and 65.2% (30/46) of the isolates from pulmonary M. avium subsp. hominissuis disease patients, respectively, and 43.3% (13/30) and 33.3% (10/30) of the isolates from HIV-positive patients, respectively (Table 1). A significantly higher percentage of strains with resistance to both drugs was found among the isolates from patients with pulmonary M. avium subsp. hominissuis disease than among the isolates from HIV-positive patients. Resistance to streptomycin, kanamycin, and amikacin was observed in 30.4% (14/46), 32.6% (15/46), and 15.2% (7/46) of the isolates from patients with pulmonary M. avium subsp. hominissuis disease, respectively, and 16.7% (5/30), 13.3% (4/30), and 10.0% (3/30) of the isolates from HIV-positive patients, respectively; the former group of isolates showed stronger resistance to all three drugs. In contrast, clarithromycin resistance was observed in 10.0% (3/30) of the isolates from HIV-positive patients and in only 1 strain (2.2%) among the isolates from pulmonary M. avium subsp. hominissuis disease patients. Regarding rifampin and levofloxacin, almost all isolates in both groups showed susceptibility.
TABLE 1.
Comparison of drug resistance and susceptibility in isolates from different hosts
| Antimicrobial agent | No. (%) of isolatesa |
P valueb (pMAH vs HIV) | |||
|---|---|---|---|---|---|
| pMAH (n = 46) |
HIV (n = 30) |
||||
| R | S | R | S | ||
| Clarithromycin | 1 (2.2) | 45 (97.8) | 3 (10.0) | 27 (90.0) | 0.294 |
| Rifampin | 1 (2.2) | 45 (97.8) | 0 (0) | 30 (100) | 1 |
| Ethambutol | 39 (84.8) | 7 (15.2) | 13 (43.3) | 17 (56.7) | ≤0.001 |
| Streptomycin | 14 (30.4) | 32 (69.6) | 5 (16.7) | 25 (83.3) | 0.278 |
| Kanamycin | 15 (32.6) | 31 (67.4) | 4 (13.3) | 26 (86.7) | 0.065 |
| Amikacin | 7 (15.2) | 39 (84.8) | 3 (10.0) | 27 (90.0) | 0.731 |
| Ethionamide | 30 (65.2) | 16 (34.8) | 10 (33.3) | 20 (66.7) | 0.009 |
| Levofloxacin | 0 (0) | 46 (100) | 1 (3.3) | 29 (96.7) | 0.395 |
The breakpoints of the antimicrobial agents were determined according to the criteria described in the BrothMIC NTM system manual and Materials and Methods. pMAH, isolates from the sputa of patients with pulmonary M. avium subsp. hominissuis disease; HIV, isolates from the blood of HIV-positive patients with disseminated M. avium subsp. hominissuis disease; R, resistant; S, susceptible.
P values were calculated using Fisher's exact test.
Next, the Mann-Whitney U test was performed to analyze the difference in drug susceptibility among the isolates from the two different groups of hosts. Table 2 shows the mean log2 values ± standard deviations of the MIC of individual drugs for isolates from each group. There was a significant difference in the log2 MICs of seven of the eight drugs, with the exception being rifampin, among the isolates from pulmonary M. avium subsp. hominissuis disease patients and HIV-positive patients; the former group of isolates had higher MIC values of all seven drugs. These findings reveal clear differences in susceptibility between the isolates from both groups and an overall higher rate of antibiotic resistance among the isolates from patients with pulmonary M. avium subsp. hominissuis disease. This suggests an association between drug susceptibility and the type of M. avium subsp. hominissuis infection.
TABLE 2.
Mean log2 values of MICs of test drugs for isolates from different hosts
| Antimicrobial agent | Log2 MIC (mean ± SD) |
P valuea (pMAH vs HIV) | |
|---|---|---|---|
| pMAHb | HIVc | ||
| Clarithromycin | −0.83 ± 1.44 | −1.75 ± 2.74 | 0.004 |
| Rifampin | −2.22 ± 2.42 | −2.56 ± 1.97 | 0.710 |
| Ethambutol | 3.33 ± 0.97 | 2.37 ± 1.54 | ≤0.001 |
| Streptomycin | 1.93 ± 1.12 | 0.63 ± 1.83 | ≤0.001 |
| Kanamycin | 2.93 ± 1.14 | 1.27 ± 1.86 | ≤0.001 |
| Amikacin | 2.33 ± 1.10 | 1.33 ± 1.63 | 0.008 |
| Ethionamide | 2.76 ± 0.77 | 2.10 ± 0.99 | 0.003 |
| Levofloxacin | 0.63 ± 1.02 | −0.27 ± 1.88 | 0.037 |
P values for log2 MIC values between two different groups were calculated using the Mann–Whitney U test.
pMAH, isolates from the sputa of patients with pulmonary M. avium subsp. hominissuis disease.
HIV, isolates from the blood of HIV-positive patients with disseminated M. avium subsp. hominissuis disease.
VNTR genotypes of M. avium subsp. hominissuis isolates from different origins.
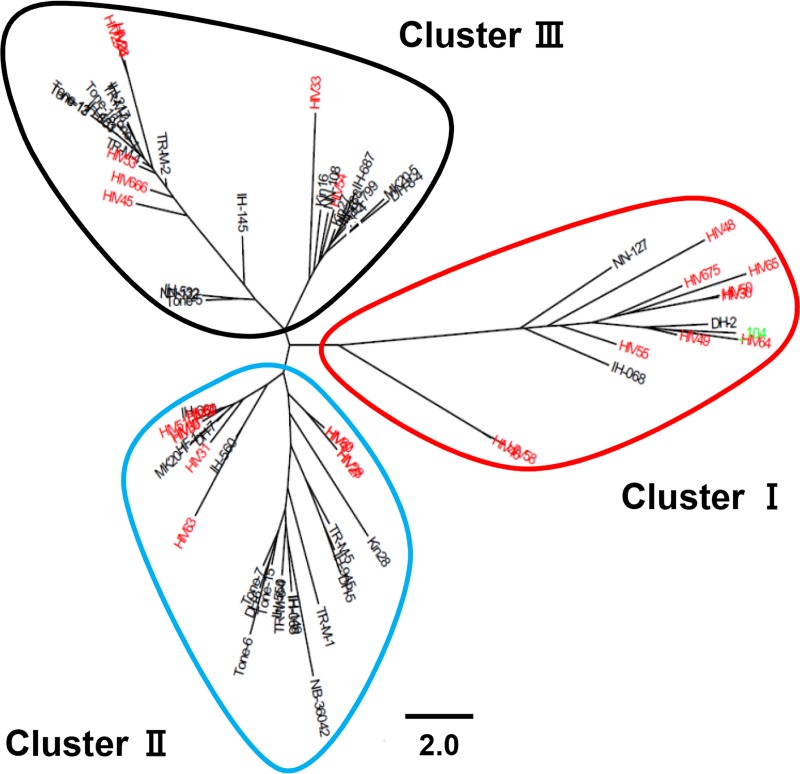
In a recent study conducting VNTR typing analysis using 13 M. avium tandem repeat (MATR) loci (MATR-VNTR typing analysis), Adachi et al. reported that M. avium subsp. hominissuis isolates from pulmonary M. avium subsp. hominissuis disease patients at the National Hospital Organization (NHO) Higashinagoya National Hospital in the Aichi Prefecture of Japan, HIV-positive patients, and pigs have different VNTR genotypes (26). The isolates from HIV-positive patients used in the present study were almost identical to the strains used by Adachi et al. (26). In this study, we used different strains isolated from pulmonary M. avium subsp. hominissuis disease patients without previous antibiotic treatment at 9 NHO hospitals throughout Japan and performed VNTR typing analysis using 15 MATR loci to examine the VNTR genotypes of the isolates from hosts with different types of M. avium subsp. hominissuis infection. As shown in Fig. 1, 76 strains examined in this study were roughly classified into three clusters: cluster I, cluster II, and cluster III. The proportion of isolates from pulmonary M. avium subsp. hominissuis disease patients and that of isolates from HIV-positive patients in each cluster were 6.5% (3/46) and 36.7% (11/30), respectively, for cluster I, 41.3% (19/46) and 36.7% (11/30), respectively, for cluster II, and 52.2% (24/46) and 26.7% (8/30), respectively, for cluster III (Table S3). The ratio of isolates from HIV-positive patients to those from pulmonary M. avium subsp. hominissuis disease patients was significantly higher in cluster I than in the other two clusters (P = 0.0017 by Fisher's exact test) (Table S3), indicating that a group of isolates from HIV-positive patients has a unique VNTR genotype. Furthermore, to compare the genetic distances of the isolates from pulmonary M. avium subsp. hominissuis disease patients and HIV-positive patients, we performed a multiple-comparison analysis using strain 104 as a clinically unbiased standard to estimate the Manhattan distance of the individual isolates of each cluster. The genetic distance from strain 104 for isolates from pulmonary M. avium subsp. hominissuis disease patients was statistically significantly different from that for isolates from HIV-positive patients (P = 0.0018) (Fig. S1). These results suggest that M. avium subsp. hominissuis strains that cause pulmonary and disseminated disease have genetically distinct features.
FIG 1.
Cluster analysis of M. avium subsp. hominissuis isolates from different hosts based on MATR-VNTR profiles. The M. avium subsp. hominissuis isolates comprised 46 strains (black) from pulmonary M. avium subsp. hominissuis disease patients and 30 strains (red) from HIV-positive patients, including strain 104 (green) as a reference. The phylogenetic distribution was created from distance matrix files by use of the Fitch-Margoliash algorithm according to the MATR-VNTR markers. The scale bar indicates the Manhattan distance. M. avium subsp. hominissuis isolates were classified into clusters I to III by MATR-VNTR typing analysis.
Next, we analyzed the association between the VNTR genotype and drug susceptibility in strains within each cluster by the Kruskal-Wallis test, revealing a significant difference in the log2 MICs of seven of the eight drugs, with the exception being ethambutol (Table 3). The Mann-Whitney U test was then used to further compare two clusters. The strains in cluster I were more susceptible to ethionamide and levofloxacin than the strains in cluster II and were more susceptible to clarithromycin, streptomycin, kanamycin, amikacin, ethionamide, and levofloxacin than the strains in cluster III. The strains in cluster II were more susceptible to clarithromycin, rifampin, streptomycin, and amikacin but more resistant to ethionamide than the strains in cluster III. A comparison of the presence of resistant and susceptible strains among the clusters revealed that cluster III had the highest percentage of strains resistant to ethambutol, streptomycin, kanamycin, and amikacin and cluster I had the lowest (Table S4). Furthermore, a significant difference in ethionamide resistance was found among the clusters, with the highest percentage of resistant strains being found in cluster II. Accordingly, intergroup comparisons revealed that strains in cluster I had the lowest MIC values and those in cluster III tended to have the highest values. These results are related to the proportions of isolates from pulmonary M. avium subsp. hominissuis disease patients and those of isolates from HIV-positive patients in each cluster. In good agreement with the findings presented above, the proportion of isolates from HIV-positive patients was the highest in cluster I and the lowest in cluster III (Table S3).
TABLE 3.
Mean log2 values of MICs of test drugs for isolates within the three VNTR clusters
| Antimicrobial agent | Log2 MIC (mean ± SD) |
P valuea |
|||||
|---|---|---|---|---|---|---|---|
| Clusterb I | Cluster II | Cluster III | Clusters I vs II vs III | Clusters I vs II | Clusters I vs III | Clusters II vs III | |
| Clarithromycin | −1.94 ± 1.95 | −1.44 ± 2.21 | −0.63 ± 1.94 | 0.011 | 0.398 | 0.010 | 0.020 |
| Rifampin | −2.95 ± 1.35 | −3.03 ± 1.73 | −1.46 ± 2.69 | 0.046 | 0.758 | 0.106 | 0.020 |
| Ethambutol | 2.57 ± 1.02 | 2.97 ± 1.59 | 3.09 ± 1.12 | 0.156 | 0.449 | 0.058 | 0.224 |
| Streptomycin | 0.64 ± 0.84 | 1.16 ± 1.71 | 2.0 ± 1.5 | 0.003 | 0.137 | ≤0.001 | 0.038 |
| Kanamycin | 1.36 ± 1.01 | 2.03 ± 1.94 | 2.91 ± 1.4 | 0.002 | 0.076 | ≤0.001 | 0.058 |
| Amikacin | 1.07 ± 0.92 | 1.73 ± 1.46 | 2.50 ± 1.32 | 0.001 | 0.093 | ≤0.001 | 0.028 |
| Ethionamide | 1.79 ± 0.80 | 3.0 ± 0.69 | 2.34 ± 0.9 | ≤0.001 | ≤0.001 | 0.044 | 0.004 |
| Levofloxacin | −0.64 ± 1.22 | 0.73 ± 1.46 | 0.25 ± 1.44 | 0.003 | 0.001 | 0.022 | 0.119 |
P values for the log2 MIC values between the indicated clusters were calculated using the Kruskal-Wallis test for the differences between three clusters and the Mann-Whitney U test for the differences between two clusters.
Each cluster was classified by phylogenetic analysis, as shown in Fig. 1.
Association between the presence of ISMav6 and drug susceptibility.
Based on a previous report on the association between the presence of ISMav6 and drug susceptibility (27), we investigated such possible associations for M. avium subsp. hominissuis isolates from different hosts in the present study. We previously reported the presence of ISMav6, which is a novel insertion sequence, with 60 point mutations compared with the nucleotide sequence of the original IS901 in Japanese human clinical M. avium subsp. hominissuis isolates (28). ISMav6 was present in 41.3% of the isolates from patients with pulmonary M. avium subsp. hominissuis disease and 40.0% of those from HIV-positive patients, with no significant intergroup difference. Comparisons of the isolates from the different hosts revealed that isolates harboring IsMav6 were significantly more resistant to clarithromycin, rifampin, streptomycin, kanamycin, and amikacin than isolates not harboring IsMav6 (Table 4), showing the association between the presence of ISMav6 and drug susceptibility. Intriguingly, a comparison of the presence of resistant and susceptible strains between the two groups revealed that higher percentages of strains possessing ISMav6 than isolates not possessing ISMav6 were resistant to clarithromycin, streptomycin, kanamycin, and amikacin (Table S5). No such trend could be found for rifampin or levofloxacin because there was only one strain resistant to these antibiotics. The result was consistent with that shown in Table 4, once these drugs were removed from the analysis. We then investigated the association between the ISMav6 and VNTR genotypes. The highest prevalence (22/31, 71%) of ISMav6 was observed among strains in cluster III, which had the highest MIC values toward individual drugs, whereas no strains harboring ISMav6 were observed among the isolates in cluster I, which overall had the lowest MIC values (Table S6). These findings suggest the involvement of ISMav6 in the association between the VNTR genotype and drug susceptibility.
TABLE 4.
Association between presence of ISMav6 and drug susceptibility
| Antimicrobial agent | Log2 MIC (mean ± SD) |
P valuea (ISMav6-positive vs -negative isolates) | |
|---|---|---|---|
| ISMav6-positive isolates | ISMav6-negative isolates | ||
| Clarithromycin | −0.46 ± 2.55 | −1.70 ± 1.53 | 0.009 |
| Rifampin | −1.31 ± 2.64 | −3.07 ± 1.59 | 0.003 |
| Ethambutol | 2.87 ± 1.36 | 3.0 ± 1.28 | 0.488 |
| Streptomycin | 1.74 ± 1.92 | 1.20 ± 1.25 | 0.034 |
| Kanamycin | 2.68 ± 1.85 | 2.0 ± 1.49 | 0.017 |
| Amikacin | 2.39 ± 1.65 | 1.62 ± 1.13 | 0.005 |
| Ethionamide | 2.45 ± 1.03 | 2.53 ± 0.84 | 0.698 |
| Levofloxacin | 0.32 ± 1.45 | 0.24 ± 1.51 | 0.888 |
P values were calculated using the Mann-Whitney U test.
DISCUSSION
In this study, we compared the MICs of eight drugs for 46 isolates from patients with pulmonary M. avium subsp. hominissuis disease without previous antibiotic treatment and 30 isolates from HIV-positive patients with disseminated M. avium subsp. hominissuis disease. Interestingly, isolates from pulmonary M. avium subsp. hominissuis disease patients were more resistant to all drugs except rifampin than isolates from HIV-positive patients. Thus, isolates from different hosts showed distinct differences in drug susceptibility, and isolates from pulmonary M. avium subsp. hominissuis disease patients had higher levels of drug resistance. Such differences in drug susceptibility are thought to be caused by differences in genetic characteristics. In our previous study, comparative genome analysis of strain TH135, isolated from a patient with pulmonary M. avium subsp. hominissuis disease, and strain 104, derived from an HIV-positive patient, showed that many strain-specific regions including virulence-associated genes were present in the genomes of both strains (18). These results suggest that M. avium subsp. hominissuis strains that cause pulmonary and disseminated disease possess genetically distinct features, and these differences possibly reflect different drug susceptibilities among the isolates from both groups. These findings suggest an association between drug susceptibility and the types of M. avium subsp. hominissuis infection, and the genetic characteristics of M. avium subsp. hominissuis strains associated with the development of pulmonary disease may be involved in higher levels of antibiotic resistance.
We performed MATR-VNTR typing analysis to compare the genotypes of M. avium subsp. hominissuis isolates from different hosts. MATR-VNTR typing analysis showed that the strains examined in this study were roughly classified into three clusters. Cluster I contained 3 isolates (DH-2, IH-068, and NN-127) from pulmonary M. avium subsp. hominissuis disease patients and 11 isolates from HIV-positive patients. The ratio of isolates from HIV-positive patients to those from pulmonary M. avium subsp. hominissuis disease patients was significantly higher in cluster I than in the other two clusters, indicating that a group of isolates from HIV-positive patients exhibited a unique VNTR genotype. We previously performed phylogenetic analysis based on single nucleotide variants using the genome information for isolates from pulmonary M. avium subsp. hominissuis disease patients, the same isolates used in the present study, and M. avium subsp. hominissuis strains isolated abroad, including the United States, Belgium, and Germany (25). Most isolates in Japan were grouped into clusters different from those containing strains isolated abroad. However, 3 strains, DH-2, IH-068, and NN-127, were grouped into the clusters containing strains isolated abroad, suggesting that the strains in cluster I are genetically similar to those isolated abroad. Previous studies reported the association between genotype and drug susceptibility in NTM (29, 30). In the present study, significant differences in the log2 MICs of seven of eight of the drugs tested, with the exception being ethambutol, were observed among the isolates in the three clusters grouped by VNTR typing of M. avium subsp. hominissuis isolates. Moreover, intergroup comparisons revealed that the strains in cluster I had the lowest MIC values and those in cluster III tended to have the highest values. This result correlated with the proportion of isolates from HIV-positive patients in each cluster.
Given that a report on the association between ISMav6 and drug susceptibility has been previously published (27), we investigated the association between the presence of IsMav6 and differences in drug susceptibilities among M. avium subsp. hominissuis isolates from different hosts. The presence of ISMav6 did not differ among the isolates, indicating that differences in drug susceptibility and the presence of ISMav6 are unrelated. However, isolates possessing IsMav6 were significantly more resistant to five of the drugs tested than those not possessing IsMav6, indicating an association between IsMav6 and drug susceptibility. Furthermore, isolates with ISMav6 had higher percentages of strains resistant to clarithromycin, streptomycin, kanamycin, and amikacin than isolates without ISMav6. Because resistance to macrolides and aminoglycosides is caused by mutations in the genes encoding 23S rRNA and 16S rRNA (31, 32), respectively, this result suggests a possible correlation between mutations of these genes and the presence of ISMav6. The proportion of isolates possessing IsMav6 relative to all M. avium subsp. hominissuis isolates is higher in Japan and South Korea than in the United States, with almost no report of IsMav6 being made for M. avium subsp. hominissuis isolates recovered in Germany or the Netherlands (17, 27, 33), suggesting that drug susceptibility varies among different geographical regions and that M. avium subsp. hominissuis isolates in Japan and South Korea are more resistant to antibiotics than M. avium subsp. hominissuis isolates in other countries with a low rate of possession of IsMav6. This also suggests that IsMav6 serves as a predictor of drug susceptibility, and to clarify this, further study is needed to compare the findings of drug susceptibility in the present study with those obtained from M. avium subsp. hominissuis strains isolated abroad.
This study has some limitations. Isolates from patients with pulmonary M. avium subsp. hominissuis disease were obtained before the administration of antibiotics. However, it is not known whether multidrug therapy with antibiotics was performed in HIV-positive patients, and if so, the effect of antibiotics on the drug susceptibility of the isolates from these hosts cannot be disregarded. However, in such cases, isolates would have higher MIC values than isolates from patients not treated with antibiotics. In other words, the difference in drug susceptibility between the two groups would be greater if the isolates were collected from HIV-positive patients not treated with multidrug therapy using antibiotics. Therefore, we think that this does not alter the conclusions of this study. Also, we did not investigate the mechanisms underlying the resistance of M. avium subsp. hominissuis isolates to individual antibiotics. Thus, this investigation can be regarded as a preliminary study.
In conclusion, we observed a difference in drug susceptibility and VNTR genotypes between M. avium subsp. hominissuis isolates from pulmonary M. avium subsp. hominissuis disease patients and those from HIV-positive patients. These differences indicate the association with types of M. avium subsp. hominissuis infection, and the genetic characteristics of M. avium subsp. hominissuis strains that are involved in the establishment of pulmonary disease in immunocompetent hosts or disseminated disease in immunocompromised hosts are thought to influence these differences. Additionally, the enhanced antibiotic resistance of the isolates from pulmonary M. avium subsp. hominissuis disease patients might potentially make treatment of pulmonary M. avium subsp. hominissuis disease patients difficult. Further study is needed to examine the genes involved in drug resistance by comparing the genome of each M. avium subsp. hominissuis isolate with the genomes of other isolates and to examine M. avium subsp. hominissuis strains isolated abroad to clarify whether the results obtained in the present study are specific to strains prevalent in Japan.
MATERIALS AND METHODS
Bacterial strains.
As reported previously (34), 46 M. avium subsp. hominissuis isolates from HIV-negative patients with pulmonary M. avium subsp. hominissuis disease were provided by nine National Hospital Organization (NHO) hospitals across Japan. These clinical isolates were obtained from the sputa of 46 patients who did not undergo treatment immediately after a diagnosis of pulmonary M. avium subsp. hominissuis disease (corresponding to the diagnostic criteria of the American Thoracic Society and the Infectious Diseases Society of America [19]) between July 2008 and September 2009. Also, 30 M. avium subsp. hominissuis isolates from the blood of HIV-positive patients, including strain 104, derived from an AIDS patient and used as a standard (35), with disseminated M. avium subsp. hominissuis disease were provided by the National Center for Global Health and Medicine, formerly the International Medical Center of Japan. However, it is not known whether antibiotic therapy was provided to the HIV-positive patients. Moreover, the age, sex, and location of the patients with pulmonary M. avium subsp. hominissuis disease were known, while those of the HIV-positive patients were not. Only one strain per patient was analyzed in this study.
Identification of subspecies of M. avium, growth condition, and DNA isolation.
The subspecies of the M. avium isolates was identified to be M. avium subsp. hominissuis by sequence analysis of the 3′ fragment of the hsp65 gene (36). The organism was grown in Middlebrook 7H9 liquid medium supplemented with 10% oleic acid-albumin-dextrose-catalase enrichment (Difco, Sparks, MD) at 37°C. DNA was extracted using InstaGene Matrix (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer's instructions.
Drug susceptibility testing.
The BrothMIC NTM system (Kyokuto Pharmaceutical Industrial Co., Ltd., Tokyo, Japan) was used to determine the susceptibility of the M. avium subsp. hominissuis strains to clarithromycin, rifampin, ethambutol, streptomycin, kanamycin, amikacin, ethionamide, and levofloxacin, according to the manufacturer's instructions for each antimicrobial agent; this procedure is in compliance with the standard protocol of the Clinical and Laboratory Standards Institute, formerly called the National Committee for Clinical Laboratory Standards (37, 38). Serial dilutions of each antimicrobial and Middlebrook 7H9 broth (Difco) were reconstituted by inoculation of 0.1 ml of a cell suspension prepared in distilled water by 1:100 dilution of a 0.5 McFarland suspension in air-dried microplates. After inoculation, the isolates were incubated at 37°C at pH 7.4 for clarithromycin and at pH 6.6 for the remaining antimicrobials. Growth endpoints were read visually after incubation for 7 days. The MIC breakpoints of the drugs indicating resistance were determined according to the criteria described in the BrothMIC NTM system manual, as follows: clarithromycin, ≥32 μg/ml; rifampin, ≥8 μg/ml; ethambutol, ≥8 μg/ml; streptomycin, ≥8 μg/ml; kanamycin, ≥16 μg/ml; amikacin, ≥16 μg/ml; ethionamide, ≥8 μg/ml; and levofloxacin, ≥8 μg/ml.
VNTR genotyping.
Variable-number tandem-repeat (VNTR) typing analysis using M. avium tandem repeats (MATR) was carried out using 15 VNTR loci (MATR-1 to MATR-16, except for MATR-10) and the corresponding primer sets, as described previously (39). After the number of base pairs in the target VNTR loci was estimated according to their relationship to molecular weight markers by agarose gel electrophoresis, the number of repetitions of various VNTR loci in each strain was determined and regarded as an allele profile. The amplification product of M. avium subsp. paratuberculosis ATCC 19698, for which the number of repetitions of each VNTR locus had been determined by sequence analysis, was used as a positive control. The Manhattan distance was determined on the basis of each obtained allele profile, and the genotypic diversity of the M. avium subsp. hominissuis isolates was analyzed with a Fitch-Margoliash algorithm using PHYLIP software (version 3.68). Branches were supported by the bootstrap value as 1,000 replicates of a randomly assembled data set. The phylogenetic distribution was generated according to the genotypic diversity of the isolates using FigTree software (version 1.3.1).
Detection of ISMav6.
The presence of the ISMav6 gene in M. avium subsp. hominissuis isolates was determined by using specific PCR primers, as described previously (28). The resulting PCR products were purified using a GenElute PCR DNA purification kit (Sigma-Aldrich, St. Louis, MO), and direct sequencing analysis was performed using the same primers used for PCR. The resulting nucleotide sequences were compared with the sequence data for ISMav6. The suitability of the present DNA samples for screening clinical isolates by PCR was determined by amplification of the hsp65 gene, the gene used to identify the subspecies of M. avium isolates.
Statistical analysis.
The Mann-Whitney U test and the Kruskal-Wallis test were used for comparison of the mean log2 values of the MICs of the test drugs for M. avium subsp. hominissuis strains derived from two different hosts and the three VNTR clusters, respectively. The genetic distances estimated from the Manhattan distance matrix data for the M. avium subsp. hominissuis isolates were analyzed using the Mann-Whitney U test. Fisher's exact test was used for categorical variables. All statistical analyses were performed using GraphPad Prism (version 5.0) software (GraphPad Software, San Diego, CA). P values of <0.05 were considered significant.
Ethics.
This study was approved by the Ethics Review Committee for Human Research of the NHO Higashinagoya National Hospital, and written informed consent was obtained from all patients with pulmonary M. avium subsp. hominissuis disease.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Teruo Kirikae, Satoru Fujiuchi, Katsuhiro Kuwabara, Yuka Sasaki, Emiko Toyota, Ryoji Maekura, Kazunari Tsuyuguchi, Masahiro Shirai, Takefumi Saitoh, Seiji Kawabata, Yoshiaki Tao, Shuichi Takigawa, and Mitsunori Sakatani for kindly providing us with clinical isolates and clinical information. We also thank Miki Takami and Mahoh Kondoh for technical assistance.
This work was supported by JSPS KAKENHI grant number 15K08049 (to K.U.).
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02035-17.
REFERENCES
- 1.Ichiyama S, Shimokata K, Tsukamura M. 1988. The isolation of Mycobacterium avium complex from soil, water, and dusts. Microbiol Immunol 32:733–739. doi: 10.1111/j.1348-0421.1988.tb01434.x. [DOI] [PubMed] [Google Scholar]
- 2.von Reyn CF, Maslow JN, Barber TW, Falkinham JO, Arbeit RD. 1994. Persistent colonization of potable water as a source of Mycobacterium avium infection in AIDS. Lancet 343:1137–1141. doi: 10.1016/S0140-6736(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 3.Stout JE, Hamilton CD. 2011. Mycobacterium avium complex disease, p 531–564. In Schlossberg D. (ed), Tuberculosis and nontuberculous mycobacterial infection, 6th ed ASM Press, Washington, DC. [Google Scholar]
- 4.Maekawa K, Ito Y, Hirai T, Kubo T, Imai S, Tatsumi S, Fujita K, Takakura S, Niimi A, Iinuma Y, Ichiyama S, Togashi K, Mishima M. 2011. Environmental risk factors for pulmonary Mycobacterium avium-intracellulare complex disease. Chest 140:723–729. doi: 10.1378/chest.10-2315. [DOI] [PubMed] [Google Scholar]
- 5.Nishiuchi Y, Maekura R, Kitada S, Tamaru A, Taguri T, Kira Y, Hiraga T, Hirotani A, Yoshimura K, Miki M, Ito M. 2007. The recovery of Mycobacterium avium-intracellulare complex (MAC) from the residential bathrooms of patients with pulmonary MAC. Clin Infect Dis 45:347–351. doi: 10.1086/519383. [DOI] [PubMed] [Google Scholar]
- 6.Bryant JM, Grogono DM, Greaves D, Foweraker J, Roddick I, Inns T, Reacher M, Haworth CS, Curran MD, Harris SR, Peacock SJ, Parkhill J, Floto RA. 2013. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet 381:1551–1560. doi: 10.1016/S0140-6736(13)60632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prevots DR, Shaw PA, Strickland D, Jackson LA, Raebel MA, Blosky MA, Montes de Oca R, Shea YR, Seitz AE, Holland SM, Olivier KN. 2010. Nontuberculous mycobacterial lung disease prevalence at four integrated health care delivery systems. Am J Respir Crit Care Med 182:970–976. doi: 10.1164/rccm.201002-0310OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashi M, Takayanagi N, Kanauchi T, Miyahara Y, Yanagisawa T, Sugita Y. 2012. Prognostic factors of 634 HIV-negative patients with Mycobacterium avium complex lung disease. Am J Respir Crit Care Med 185:575–583. doi: 10.1164/rccm.201107-1203OC. [DOI] [PubMed] [Google Scholar]
- 9.Namkoong H, Kurashima A, Morimoto K, Hoshino Y, Hasegawa N, Ato M, Mitarai S. 2016. Epidemiology of pulmonary nontuberculous mycobacterial disease, Japan. Emerg Infect Dis 22:1116–1117. doi: 10.3201/eid2206.151086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adjemian J, Olivier KN, Seitz AE, Holland SM, Prevots DR. 2012. Prevalence of nontuberculous mycobacterial lung disease in U.S. Medicare beneficiaries. Am J Respir Crit Care Med 185:881–886. doi: 10.1164/rccm.201111-2016OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kajiki A. 2011. Non-tuberculous mycobacteriosis. What has been coming out. Kekkaku 86:113–125. [PubMed] [Google Scholar]
- 12.Mijs W, de Haas P, Rossau R, Van der Laan T, Rigouts L, Portaels F, van Soolingen D. 2002. Molecular evidence to support a proposal to reserve the designation Mycobacterium avium subsp. avium for bird-type isolates and ‘M. avium subsp hominissuis’ for the human/porcine type of M. avium. Int J Syst Evol Microbiol 52(Pt 5):1505–1518. [DOI] [PubMed] [Google Scholar]
- 13.Thorel MF, Krichevsky M, Levy-Frebault VV. 1990. Numerical taxonomy of mycobactin-dependent mycobacteria, emended description of Mycobacterium avium, and description of Mycobacterium avium subsp. avium subsp. nov., Mycobacterium avium subsp. paratuberculosis subsp. nov., and Mycobacterium avium subsp. silvaticum subsp. nov. Int J Syst Bacteriol 40:254–260. doi: 10.1099/00207713-40-3-254. [DOI] [PubMed] [Google Scholar]
- 14.Cocito C, Gilot P, Coene M, de Kesel M, Poupart P, Vannuffel P. 1994. Paratuberculosis. Clin Microbiol Rev 7:328–345. doi: 10.1128/CMR.7.3.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hibiya K, Utsunomiya K, Yoshida T, Toma S, Higa F, Tateyama M, Fujita J. 2010. Pathogenesis of systemic Mycobacterium avium infection in pigs through histological analysis of hepatic lesions. Can J Vet Res 74:252–257. [PMC free article] [PubMed] [Google Scholar]
- 16.Iwamoto T, Nakajima C, Nishiuchi Y, Kato T, Yoshida S, Nakanishi N, Tamaru A, Tamura Y, Suzuki Y, Nasu M. 2012. Genetic diversity of Mycobacterium avium subsp. hominissuis strains isolated from humans, pigs, and human living environment. Infect Genet Evol 12:846–852. doi: 10.1016/j.meegid.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 17.Ichikawa K, van Ingen J, Koh WJ, Wagner D, Salfinger M, Inagaki T, Uchiya K, Nakagawa T, Ogawa K, Yamada K, Yagi T. 2015. Genetic diversity of clinical Mycobacterium avium subsp. hominissuis and Mycobacterium intracellulare isolates causing pulmonary diseases recovered from different geographical regions. Infect Genet Evol 36:250–255. doi: 10.1016/j.meegid.2015.09.029. [DOI] [PubMed] [Google Scholar]
- 18.Uchiya K, Takahashi H, Yagi T, Moriyama M, Inagaki T, Ichikawa K, Nakagawa T, Nikai T, Ogawa K. 2013. Comparative genome analysis of Mycobacterium avium revealed genetic diversity in strains that cause pulmonary and disseminated disease. PLoS One 8:e71831. doi: 10.1371/journal.pone.0071831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ Jr, Winthrop K. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 20.Glassroth J. 2008. Pulmonary disease due to nontuberculous mycobacteria. Chest 133:243–251. doi: 10.1378/chest.07-0358. [DOI] [PubMed] [Google Scholar]
- 21.Thomson RM, Yew WW. 2009. When and how to treat pulmonary non-tuberculous mycobacterial diseases. Respirology 14:12–26. doi: 10.1111/j.1440-1843.2008.01408.x. [DOI] [PubMed] [Google Scholar]
- 22.Weiss CH, Glassroth J. 2012. Pulmonary disease caused by nontuberculous mycobacteria. Expert Rev Respir Med 6:597–613. doi: 10.1586/ers.12.58. [DOI] [PubMed] [Google Scholar]
- 23.Griffith DE, Brown-Elliott BA, Langsjoen B, Zhang Y, Pan X, Girard W, Nelson K, Caccitolo J, Alvarez J, Shepherd S, Wilson R, Graviss EA, Wallace RJ Jr. 2006. Clinical and molecular analysis of macrolide resistance in Mycobacterium avium complex lung disease. Am J Respir Crit Care Med 174:928–934. doi: 10.1164/rccm.200603-450OC. [DOI] [PubMed] [Google Scholar]
- 24.Uchiya K, Takahashi H, Nakagawa T, Yagi T, Moriyama M, Inagaki T, Ichikawa K, Nikai T, Ogawa K. 2015. Characterization of a novel plasmid, pMAH135, from Mycobacterium avium subsp. hominissuis. PLoS One 10:e0117797. doi: 10.1371/journal.pone.0117797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uchiya K, Tomida S, Nakagawa T, Asahi S, Nikai T, Ogawa K. 2017. Comparative genome analyses of Mycobacterium avium reveal genomic features of its subspecies and strains that cause progression of pulmonary disease. Sci Rep 7:39750. doi: 10.1038/srep39750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adachi T, Ichikawa K, Inagaki T, Moriyama M, Nakagawa T, Ogawa K, Hasegawa Y, Yagi T. 2016. Molecular typing and genetic characterization of Mycobacterium avium subsp. hominissuis isolates from humans and swine in Japan. J Med Microbiol 65:1289–1295. doi: 10.1099/jmm.0.000351. [DOI] [PubMed] [Google Scholar]
- 27.Kim SY, Jeong BH, Park HY, Jeon K, Han SJ, Shin SJ, Koh WJ. 2016. Association of ISMav6 with the pattern of antibiotic resistance in Korean Mycobacterium avium clinical isolates but no relevance between their genotypes and clinical features. PLoS One 11:e0148917. doi: 10.1371/journal.pone.0148917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ichikawa K, Yagi T, Moriyama M, Inagaki T, Nakagawa T, Uchiya KI, Nikai T, Ogawa K. 2009. Characterization of Mycobacterium avium clinical isolates in Japan using subspecies-specific insertion sequences, and identification of a new insertion sequence, ISMav6. J Med Microbiol 58:945–950. doi: 10.1099/jmm.0.008623-0. [DOI] [PubMed] [Google Scholar]
- 29.Tatano Y, Sano C, Yasumoto K, Shimizu T, Sato K, Nishimori K, Matsumoto T, Yano S, Takeyama H, Tomioka H. 2012. Correlation between variable-number tandem-repeat-based genotypes and drug susceptibility in Mycobacterium avium isolates. Eur J Clin Microbiol Infect Dis 31:445–454. doi: 10.1007/s10096-011-1326-7. [DOI] [PubMed] [Google Scholar]
- 30.Wu TS, Leu HS, Chiu CH, Lee MH, Chiang PC, Wu TL, Chia JH, Su LH, Kuo AJ, Lai HC. 2009. Clinical manifestations, antibiotic susceptibility and molecular analysis of Mycobacterium kansasii isolates from a university hospital in Taiwan. J Antimicrob Chemother 64:511–514. doi: 10.1093/jac/dkp238. [DOI] [PubMed] [Google Scholar]
- 31.Meier A, Kirschner P, Springer B, Steingrube VA, Brown BA, Wallace RJ Jr, Bottger EC. 1994. Identification of mutations in 23S rRNA gene of clarithromycin-resistant Mycobacterium intracellulare. Antimicrob Agents Chemother 38:381–384. doi: 10.1128/AAC.38.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prammananan T, Sander P, Brown BA, Frischkorn K, Onyi GO, Zhang Y, Bottger EC, Wallace RJ Jr. 1998. A single 16S ribosomal RNA substitution is responsible for resistance to amikacin and other 2-deoxystreptamine aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J Infect Dis 177:1573–1581. doi: 10.1086/515328. [DOI] [PubMed] [Google Scholar]
- 33.Kolb J, Hillemann D, Mobius P, Reetz J, Lahiri A, Lewin A, Rusch-Gerdes S, Richter E. 2014. Genetic characterization of German Mycobacterium avium strains isolated from different hosts and specimens by multilocus sequence typing. Int J Med Microbiol 304:941–948. doi: 10.1016/j.ijmm.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Moriyama M, Ogawa K, Nakagawa T, Nikai T, Uchiya K. 2016. Association between a pMAH135 plasmid and the progression of pulmonary disease caused by Mycobacterium avium. Kekkaku 91:9–15. [PubMed] [Google Scholar]
- 35.Horan KL, Freeman R, Weigel K, Semret M, Pfaller S, Covert TC, van Soolingen D, Leao SC, Behr MA, Cangelosi GA. 2006. Isolation of the genome sequence strain Mycobacterium avium 104 from multiple patients over a 17-year period. J Clin Microbiol 44:783–789. doi: 10.1128/JCM.44.3.783-789.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turenne CY, Semret M, Cousins DV, Collins DM, Behr MA. 2006. Sequencing of hsp65 distinguishes among subsets of the Mycobacterium avium complex. J Clin Microbiol 44:433–440. doi: 10.1128/JCM.44.2.433-440.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawata N, Kawahara S, Tada A, Takigawa N, Shibayama T, Soda R, Takahashi K. 2006. Antimycobacterial susceptibility against nontuberculous mycobacteria using BrothMIC NTM. Kekkaku 81:329–335. [PubMed] [Google Scholar]
- 38.National Committee for Clinical Laboratory Standards. 2003. Susceptibility testing of mycobacteria, Nocardiae and other aerobic actinomycetes; approved standard. Document M24A. National Committee for Clinical Laboratory Standards, Wayne, PA. [PubMed] [Google Scholar]
- 39.Inagaki T, Nishimori K, Yagi T, Ichikawa K, Moriyama M, Nakagawa T, Shibayama T, Uchiya K, Nikai T, Ogawa K. 2009. Comparison of a variable-number tandem-repeat (VNTR) method for typing Mycobacterium avium with mycobacterial interspersed repetitive-unit-VNTR and IS1245 restriction fragment length polymorphism typing. J Clin Microbiol 47:2156–2164. doi: 10.1128/JCM.02373-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.