ABSTRACT
Apramycin, an aminocyclitol aminoglycoside, was rapidly bactericidal against Acinetobacter baumannii. In a neutropenic murine thigh infection model, treatment-associated A. baumannii CFU reductions of >4 log10 per thigh were observed for all exposures for which area under the curve (AUC)/MIC ratio was >50 and maximum concentration of drug in serum (Cmax)/MIC was ≈10 or higher. Based on these findings, we suggest that apramycin deserves further preclinical exploration as a repurposed therapeutic for multidrug-resistant Gram-negative pathogens, including A. baumannii.
KEYWORDS: antimicrobial, apramycin, maximum tolerated dose, mouse thigh model, pharmacodynamics, pharmacokinetics, resistance, time-kill
TEXT
There is a pressing need for new antimicrobials that target multidrug-resistant Gram-negative pathogens, including Acinetobacter baumannii (1). Apramycin is an aminocyclitol aminoglycoside used in veterinary medicine. It differs from 16S rRNA decoding A-site aminoglycosides approved for use in human therapy (e.g., gentamicin, tobramycin, amikacin) in several respects. First, at a molecular level, apramycin is believed to have only a minor effect on amino acid coding fidelity (2), yet it still demonstrates bactericidal activity for Escherichia coli (3). Second, apramycin appears to be neither ototoxic nor nephrotoxic (3–5), potentially based in part on greater selectivity for bacterial over mitochondrial ribosomes (3). Third, apramycin has a broad activity spectrum against multidrug-resistant human clinical isolates of A. baumannii, Pseudomonas aeruginosa, and carbapenem-resistant Enterobacteriaceae (6, 7). For multidrug- and extensively drug-resistant A. baumannii in particular, the apramycin MIC50/MIC90 (8/32 μg ml−1) was notably lower than that for gentamicin, tobramycin, and amikacin (≥64/>256 μg ml−1). Remarkably, only 2% of apramycin MICs for this highly resistant A. baumannii strain set were above the epidemiological cutoff value of 64 μg ml−1 (6).
Interestingly, apramycin, in contrast to other aminoglycosides, including plazomicin, retains activity in the presence of armA and rmtA-H 16S rRNA methylases, which are widely found in strains expressing NDM-1 (8, 9) and OXA-48 (10–15) carbapenemases and in some aminoglycoside-resistant A. baumannii strains (10, 16, 17). Only the npmA ribosomal methylase, through modification of a distinct nucleotide in the 16S RNA decoding A-site, undermines apramycin activity. However, at present, there is only one report of npmA in a clinical isolate (18, 19).
Despite these intriguing attributes, there is a paucity of toxicological, pharmacokinetic, and pharmacodynamic data for apramycin in the peer-reviewed literature. Therefore, we further characterized activity of apramycin against A. baumannii in in vitro time-kill assays and in the neutropenic mouse thigh infection model.
To evaluate in vitro bactericidal activity of apramycin, we selected three strains of A. baumannii that had representative apramycin MICs of 2, 16, and 64 μg ml−1, within the previously determined epidemiological cutoff value of 64 μg ml−1 (6), and were virulent in neutropenic CD-1 mice (see Table S1 in the supplemental material). Time-kill studies were performed according to CLSI guidelines (20), with CFU quantified using the drop-plate method (21). In time-kill analyses, apramycin demonstrated rapid bactericidal activity (99.9% killing) within 1 to 2 h of antibiotic exposure at 1× to 4× the broth microdilution MIC (Fig. 1).
FIG 1.
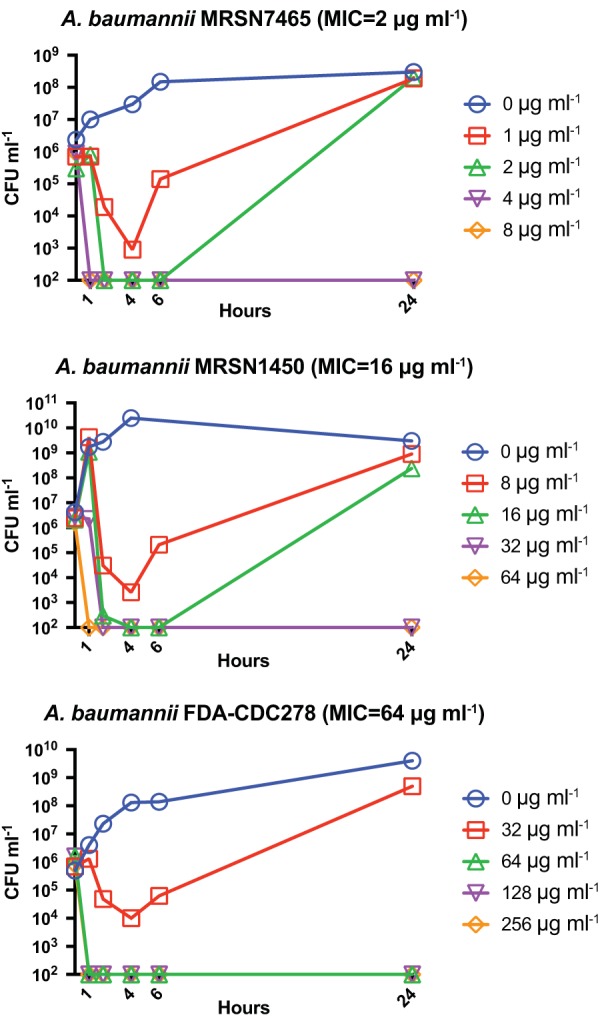
Time-kill studies. Apramycin demonstrates rapid bactericidal activity against representative A. baumannii strains. Data points plotted at 102 CFU correspond to the detection limit of the assay. Results shown are representative of two independent experiments.
To identify the single maximum tolerated dose (MTD) of apramycin, CD-1 mice (Charles River Laboratories, Inc., Kingston, NY), weighing 25 to 30 g, were injected intraperitoneally (i.p.) with ascending doses of apramycin. Over the next 72 h, no signs of distress were observed with doses up to 1,500 mg kg−1. Two of three mice from the 3,000-mg kg−1 group died ∼24 h postinjection. Thus, the single-dose MTD was 1,500 mg kg−1.
Apramycin was then given daily at 500 mg kg−1 i.p. for 14 consecutive days. Treated animals showed no signs of distress or change in body weight in comparison to controls during the experiment (see Fig. S1A in the supplemental material). On day 15, the mice were euthanized. Terminal measurements of serum creatinine (Fig. S1B) and organ histology, i.e., kidney (Fig. S1C) and liver (data not shown), were unremarkable. The multidose MTD was therefore ≥500 mg kg−1.
Pharmacokinetic and treatment studies were performed using CD-1 mice, rendered neutropenic with cyclophosphamide and mildly renal deficient with uranyl nitrate to more closely simulate human excretion kinetics (22). For pharmacokinetic studies, animals were injected subcutaneously (s.c.) with 20, 80, and 500 mg kg−1 apramycin (n = 3 per dose). Plasma apramycin concentrations were measured as described previously and are detailed in the supplemental material (23). Apramycin demonstrated first-order elimination kinetics (data not shown). Maximum concentrations of drugs in serum (Cmax) were 29 (±16), 141 (±19), and 2,100 μg ml−1 (±1,200); and area under the curve (AUC) values determined by the linear trapezoidal method were 138 (±97), 991 (±486), and 11,500 (±9,400) μg h ml−1, respectively.
For mouse thigh infection studies, mice were inoculated with 106 CFU of A. baumannii strains MSRN7465 and MSRN 1450 or 107 CFU of A. baumannii strain FDA-CDC278 and subsequently treated with apramycin 2 h postinfection with single doses of 20, 80, or 500 mg kg−1 s.c. Tissue was harvested 24 h after infection, ground, and serially diluted for CFU determination. Notably, apramycin showed a dramatic treatment effect against all three strains (Fig. 2). There was at least a 4-log10 reduction in CFU for all dosing in which AUC/MIC ratio was >50 and Cmax/MIC was ∼10 or more (24, 25).
FIG 2.
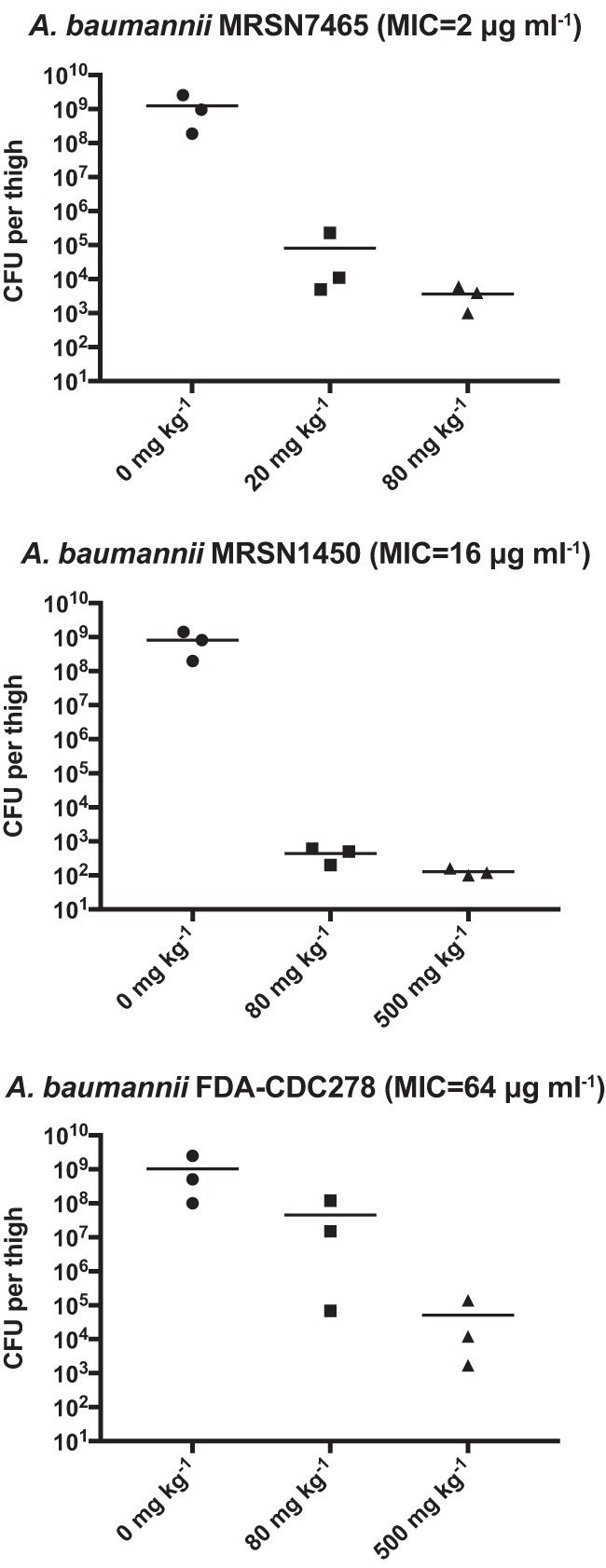
Apramycin demonstrates substantial treatment effect in the murine thigh model. Mice were infected with A. baumannii strains and dosed with apramycin 2 h later. CFU were enumerated 24 h postinfection. There were no CFU recovered from a single mouse infected with strain MRSN 7465 treated with 80 mg kg−1 apramycin and a single mouse infected with strain MRSN 1450 and treated with 500 mg kg−1 apramycin. Data points are plotted at the 103 CFU and 102 CFU assay limit of detection in these respective experiments.
Previously, therapeutic effects of apramycin against single strains of Staphylococcus aureus and Mycobacterium tuberculosis in murine infection models were described (26). In these studies, the apramycin MIC for S. aureus was 4 to 8 μg ml−1, and the therapeutic effect increased in an immunocompromised murine septicemia model in a stepwise fashion when dosed at 16, 32, or 80 mg kg−1. The M. tuberculosis MIC was not noted; however, a significant reduction in lung CFU occurred after dosing at 200 mg kg−1 for 9 days. Here, we provide evidence for an in vivo activity spectrum that also includes A. baumannii.
Several limitations of the study should be noted. First, absence of pathologies in MTD studies with relatively high systemic exposure provides some support for low toxicity. However, mice are insensitive to nephrotoxic effects of aminoglycosides (27–29). Therefore, our findings do not rule out the potential for kidney toxicity, an area that deserves further investigation in more relevant models (30). Furthermore, large doses were needed to obtain a 4-log10 reduction for strains with high MIC values. It is unclear how dosing would scale in future potential human use and ultimately what fraction of strains may prove treatable. Despite the preliminary nature of our findings, we believe, based on in vitro and in vivo data, that apramycin deserves further consideration as a repurposed therapeutic and as a starting point for future medicinal chemistry efforts targeting MDR Gram-negative pathogens such as A. baumannii.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Chief Academic Officer's Pilot Grant from Beth Israel Deaconess Medical Center to A.H.B., G.M.E., C.M., and J.E.K.
A.D.K. was supported by the Long Term Health Education and Training program from the U.S. Army as an American Society for Microbiology Committee on Postgraduate Educational Programs Fellow at Beth Israel Deaconess Medical Center. K.P.S. was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number F32 AI124590.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, United States Army, or Department of Defense.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02585-17.
REFERENCES
- 1.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 2.Perzynski S, Cannon M, Cundliffe E, Chahwala SB, Davies J. 1979. Effects of apramycin, a novel aminoglycoside antibiotic on bacterial protein synthesis. Eur J Biochem 99:623–628. doi: 10.1111/j.1432-1033.1979.tb13295.x. [DOI] [PubMed] [Google Scholar]
- 3.Matt T, Ng CL, Lang K, Sha SH, Akbergenov R, Shcherbakov D, Meyer M, Duscha S, Xie J, Dubbaka SR, Perez-Fernandez D, Vasella A, Ramakrishnan V, Schacht J, Bottger EC. 2012. Dissociation of antibacterial activity and aminoglycoside ototoxicity in the 4-monosubstituted 2-deoxystreptamine apramycin. Proc Natl Acad Sci U S A 109:10984–10989. doi: 10.1073/pnas.1204073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore RD, Smith CR, Lipsky JJ, Mellits ED, Lietman PS. 1984. Risk factors for nephrotoxicity in patients treated with aminoglycosides. Ann Intern Med 100:352–357. doi: 10.7326/0003-4819-100-3-352. [DOI] [PubMed] [Google Scholar]
- 5.Humes HD. 1988. Aminoglycoside nephrotoxicity. Kidney Int 33:900–911. doi: 10.1038/ki.1988.83. [DOI] [PubMed] [Google Scholar]
- 6.Kang AD, Smith KP, Eliopoulos GM, Berg AH, McCoy C, Kirby JE. 2017. In vitro apramycin activity against multidrug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa. Diagn Microbiol Infect Dis 88:188–191. doi: 10.1016/j.diagmicrobio.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Smith KP, Kirby JE. 2016. Evaluation of apramycin activity against carbapenem-resistant and -susceptible strains of Enterobacteriaceae. Diagn Microbiol Infect Dis 86:439–441. doi: 10.1016/j.diagmicrobio.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Livermore DM, Mushtaq S, Warner M, Zhang JC, Maharjan S, Doumith M, Woodford N. 2011. Activity of aminoglycosides, including ACHN-490, against carbapenem-resistant Enterobacteriaceae isolates. J Antimicrob Chemother 66:48–53. doi: 10.1093/jac/dkq408. [DOI] [PubMed] [Google Scholar]
- 9.Hu Y, Liu L, Zhang X, Feng Y, Zong Z. 2017. In vitro activity of neomycin, streptomycin, paromomycin and apramycin against carbapenem-resistant Enterobacteriaceae clinical strains. Front Microbiol 8:2275. doi: 10.3389/fmicb.2017.02275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z, Ling B, Zhou L. 2015. Prevalence of 16S rRNA methylase, modifying enzyme, and extended-spectrum beta-lactamase genes among Acinetobacter baumannii isolates. J Chemother 27:207–212. doi: 10.1179/1973947814Y.0000000190. [DOI] [PubMed] [Google Scholar]
- 11.Tada T, Tsuchiya M, Shimada K, Nga TTT, Thu LTA, Phu TT, Ohmagari N, Kirikae T. 2017. Dissemination of carbapenem-resistant Klebsiella pneumoniae clinical isolates with various combinations of carbapenemases (KPC-2, NDM-1, NDM-4, and OXA-48) and 16S rRNA methylases (RmtB and RmtC) in Vietnam. BMC Infect Dis 17:467. doi: 10.1186/s12879-017-2570-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poirel L, Schrenzel J, Cherkaoui A, Bernabeu S, Renzi G, Nordmann P. 2011. Molecular analysis of NDM-1-producing enterobacterial isolates from Geneva, Switzerland. J Antimicrob Chemother 66:1730–1733. doi: 10.1093/jac/dkr174. [DOI] [PubMed] [Google Scholar]
- 13.Seiffert SN, Marschall J, Perreten V, Carattoli A, Furrer H, Endimiani A. 2014. Emergence of Klebsiella pneumoniae co-producing NDM-1, OXA-48, CTX-M-15, CMY-16, QnrA and ArmA in Switzerland. Int J Antimicrob Agents 44:260–262. doi: 10.1016/j.ijantimicag.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Galani I, Anagnostoulis G, Chatzikonstantinou M, Petrikkos G, Souli M. 2016. Emergence of Klebsiella pneumoniae co-producing OXA-48, CTX-M-15, and ArmA in Greece. Clin Microbiol Infect 22:898–899. doi: 10.1016/j.cmi.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Dortet L, Poirel L, Al Yaqoubi F, Nordmann P. 2012. NDM-1, OXA-48 and OXA-181 carbapenemase-producing Enterobacteriaceae in Sultanate of Oman. Clin Microbiol Infect 18:E144–E148. doi: 10.1111/j.1469-0691.2012.03796.x. [DOI] [PubMed] [Google Scholar]
- 16.Landman D, Kelly P, Bäcker M, Babu E, Shah N, Bratu S, Quale J. 2011. Antimicrobial activity of a novel aminoglycoside, ACHN-490, against Acinetobacter baumannii and Pseudomonas aeruginosa from New York City. J Antimicrob Chemother 66:332–334. doi: 10.1093/jac/dkq459. [DOI] [PubMed] [Google Scholar]
- 17.Yu Y-s, Zhou H, Yang Q, Chen Y-g, Li L-j. 2007. Widespread occurrence of aminoglycoside resistance due to ArmA methylase in imipenem-resistant Acinetobacter baumannii isolates in China. J Antimicrob Chemother 60:454–455. doi: 10.1093/jac/dkm208. [DOI] [PubMed] [Google Scholar]
- 18.Wachino J, Shibayama K, Kurokawa H, Kimura K, Yamane K, Suzuki S, Shibata N, Ike Y, Arakawa Y. 2007. Novel plasmid-mediated 16S rRNA m1A1408 methyltransferase, NpmA, found in a clinically isolated Escherichia coli strain resistant to structurally diverse aminoglycosides. Antimicrob Agents Chemother 51:4401–4409. doi: 10.1128/AAC.00926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lioy VS, Goussard S, Guerineau V, Yoon E-J, Courvalin P, Galimand M, Grillot-Courvalin C. 2014. Aminoglycoside resistance 16S rRNA methyltransferases block endogenous methylation, affect translation efficiency and fitness of the host. RNA 20:382–391. doi: 10.1261/rna.042572.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clinical and Laboratory Standards Institute. 1999. Methods for determining the bactericidal activity of antimicrobial agents; approved guideline. CLSI document M-26A. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 21.Herigstad B, Hamilton M, Heersink J. 2001. How to optimize the drop plate method for enumerating bacteria. J Microbiol Methods 44:121–129. doi: 10.1016/S0167-7012(00)00241-4. [DOI] [PubMed] [Google Scholar]
- 22.Craig WA, Redington J, Ebert SC. 1991. Pharmacodynamics of amikacin in vitro and in mouse thigh and lung infections. J Antimicrob Chemother 27(Suppl C):29–40. doi: 10.1093/jac/27.suppl_C.29. [DOI] [PubMed] [Google Scholar]
- 23.Bernardi PM, Barreto F, Dalla Costa T. 2017. Application of a LC-MS/MS method for evaluating lung penetration of tobramycin in rats by microdialysis. J Pharm Biomed Anal 134:340–345. doi: 10.1016/j.jpba.2016.10.023. [DOI] [PubMed] [Google Scholar]
- 24.Drusano GL, Ambrose PG, Bhavnani SM, Bertino JS, Nafziger AN, Louie A. 2007. Back to the future: using aminoglycosides again and how to dose them optimally. Clin Infect Dis 45:753–760. doi: 10.1086/520991. [DOI] [PubMed] [Google Scholar]
- 25.Moore RD, Smith CR, Lietman PS. 1984. The association of aminoglycoside plasma levels with mortality in patients with gram-negative bacteremia. J Infect Dis 149:443–448. doi: 10.1093/infdis/149.3.443. [DOI] [PubMed] [Google Scholar]
- 26.Meyer M, Freihofer P, Scherman M, Teague J, Lenaerts A, Bottger EC. 2014. In vivo efficacy of apramycin in murine infection models. Antimicrob Agents Chemother 58:6938–6941. doi: 10.1128/AAC.03239-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki S, Takamura S, Yoshida J, Shinzawa Y, Niwa O, Tamatani R. 1995. Comparison of gentamicin nephrotoxicity between rats and mice. Comp Biochem Physiol C Pharmacol Toxicol Endocrinol 112:15–28. doi: 10.1016/0742-8413(95)00075-5. [DOI] [PubMed] [Google Scholar]
- 28.Schmitz C, Hilpert J, Jacobsen C, Boensch C, Christensen EI, Luft FC, Willnow TE. 2002. Megalin deficiency offers protection from renal aminoglycoside accumulation. J Biol Chem 277:618–622. doi: 10.1074/jbc.M109959200. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki S, Hatashima S, Shinzawa Y, Niwa O, Tamatani R. 1994. Toxicity of neomycin on enzyme activities of kidney and duodenal mucosa in vivo: organ specificity and species difference between rats and mice. Comp Biochem Physiol C Pharmacol Toxicol Endocrinol 109:77–92. doi: 10.1016/0742-8413(94)00037-B. [DOI] [PubMed] [Google Scholar]
- 30.Rodríguez Salgueiro S, González Núñez L. 2016. Animal models mimicking aminoglycoside-induced renal damage. J Nephropharmacol 5:1–3. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


