ABSTRACT
Cryptococcus neoformans and Cryptococcus gattii species complexes are the etiologic agents of cryptococcosis. We have deciphered the roles of three ABC transporters, Afr1, Afr2, and Mdr1, in the representative strains of the two species, C. neoformans H99 and C. gattii R265. Deletion of AFR1 in H99 and R265 drastically reduced the levels of resistance to three xenobiotics and three triazoles, suggesting that Afr1 is the major drug efflux pump in both strains. Fluconazole susceptibility was not affected when AFR2 or MDR1 was deleted in both strains. However, when these genes were deleted in combination with AFR1, a minor additive effect in susceptibility toward several drugs was observed. Deletion of all three genes in both strains caused further increases in susceptibility toward fluconazole and itraconazole, suggesting that Afr2 and Mdr1 augment Afr1 function in pumping these triazoles. Intracellular accumulation of Nile Red significantly increased in afr1Δ mutants of both strains, but rhodamine 6G accumulation increased only in the mdr1Δ mutant of H99. Thus, the three efflux pumps play different roles in the two strains when exposed to different azoles and xenobiotics. AFR1 and AFR2 expression was upregulated in H99 and R265 when treated with fluconazole. However, MDR1 expression was upregulated only in R265 under the same conditions. We screened a library of transcription factor mutants and identified several mutants that manifested either altered fluconazole sensitivity or an increase in the frequency of fluconazole heteroresistance. Gene expression analysis suggests that the three efflux pumps are regulated independently by different transcription factors in response to fluconazole exposure.
KEYWORDS: ABC transport, Afr1, Afr2, Mdr1, azoles, efflux pump
INTRODUCTION
Cryptococcosis is caused by two sibling species complexes, Cryptococcus neoformans and Cryptococcus gattii, which can be readily differentiated by their genetic and phenotypic characteristics (1, 2). Although the two species differ significantly in their pathobiology, the most common manifestation of the disease caused by both species globally is meningoencephalitis, which is treated with the same antifungal drugs. The most common treatment includes amphotericin B (AMB) with or without 5-fluorocytosine (5-FC) for induction therapy and fluconazole (FLC) for maintenance therapy (3). Amphotericin B is a fungicidal polyene that interacts with ergosterol, an essential membrane sterol of fungal cells (4, 5); 5-fluorocytosine is a pyrimidine analog which can inhibit RNA and DNA synthesis (6, 7); and FLC is a fungistatic triazole that inhibits lanosterol 14α-demethylase encoded by ERG11 in the ergosterol biosynthesis pathway (8). Recurrences of cryptococcosis during FLC maintenance therapy, however, have been increasingly reported (9–13).
The principal mechanisms responsible for the emergence of stable azole resistance in most fungal cells involve alterations in Erg11 or membrane proteins that efflux the drugs (7, 14, 15). In contrast, the majority of azole-resistant C. neoformans strains isolated from therapy failure cases have been unstable, and stable resistant strains due to gene mutations have been infrequently reported (16–18). The unstable resistance to azoles in isolates causing cryptococcosis has been described as heteroresistance, which represents a transient adaptation to the drug (16, 19). Analysis of the heteroresistant clones that emerged in vitro during growth in the presence of FLC as well as those that emerged in the mouse brain during FLC treatment indicated that they were not due to reversible mutations in ERG11 or efflux pump genes but were due to a duplication of chromosome 1 (Chr1). Chr1 harbors ERG11 as well as AFR1, which encodes the major azole transporter (16, 20, 21), and both genes play a crucial role in Chr1 duplication (16).
Several studies have revealed that strains of C. gattii generally show both a higher MICs toward various azoles (22–24) and higher levels of FLC heteroresistance than the C. neoformans species complex (25). Earlier studies have aimed to explain the reasons for higher azole MICs in C. gattii than in C. neoformans by comparing the levels of ERG11 mRNA between the two species (24). These studies found higher ERG11 mRNA levels in C. gattii than in C. neoformans strains regardless of their geographic origin. However, ERG11 mRNA levels did not correlate with FLC MICs within any group. Furthermore, the ERG11 gene sequence variation observed in the wild-type (WT) C. gattii strains also showed no correlation with FLC MICs (24), suggesting that neither ERG11 overexpression nor ERG11 sequence variation can explain the higher azole MICs in C. gattii. These data raise the question whether differences in the function of efflux pumps play a role in C. gattii's lower susceptibility toward azole drugs.
The role of Afr1, an ABC transporter, as a primary azole efflux pump has been well documented in both species complexes. AFR1 overexpression caused increased FLC MICs in C. neoformans (26), while deletion of the gene significantly lowered FLC MICs in vitro in both species (19, 27). Mice infected with a C. neoformans afr1 mutant responded to FLC treatment significantly better than those infected with the wild-type strain, although the virulence of the afr1Δ mutant remained the same as that of the wild type (26). Moreover, AFR1 and PDR11 (synonym of the AFR1 ortholog in C. gattii) caused higher azole MICs when expressed in Saccharomyces cerevisiae and lower intracellular FLC accumulation than controls (21, 28). Interestingly, the fluconazole-resistant AFR1-overexpressing mutant strain was less sensitive than the wild type and the afr1Δ strain to microglia-mediated anticryptococcal activity (29). This is likely due to reduced acidification and delayed maturation of phagosomes (29).
The protein encoded by AFR1 shares about 35% identity with those encoded by CgSNQ2 and ScSNQ2, the ABC transporters reported in Candida glabrata and S. cerevisiae, respectively (30). CgSnq2 and ScSnq2 are known to transport 4-nitroquinoline N-oxide and other chemicals in addition to azoles (28, 31–33). There are several other ABC transporters in C. neoformans and C. gattii, and two of these transporters, Afr2 and Mdr1, along with Afr1, expressed higher azole resistance and lowered intracellular azole accumulation when expressed in S. cerevisiae (21, 28). However, MDR1 or AFR2 overexpression has not been reported to cause azole resistance in C. neoformans (28) or in C. gattii, and deletion of MDR1 in C. gattii displayed FLC susceptibility similar to that of the wild type (27). These data suggest that unlike the AFR1 product, the roles of the other two ABC transporters, encoded by AFR2 and MDR1, as azole efflux pumps in Cryptococcus species are ambiguous at best.
It is important, therefore, to decipher the roles of the three ABC transporters, Afr1, Afr2, and Mdr1, as azole efflux pumps in both species of cryptococcosis agents. In the present study, we constructed deletion mutants of the three genes individually or in combinations for both species and compared the responses of the mutants toward treatment with FLC as well as several other xenobiotics. We also determined the functions of these efflux pumps by measuring the intracellular accumulation of two fluorescent dyes. Furthermore, we measured the effect of FLC treatment on the expression of each gene and identified the transcription factors (TFs) that are relevant for expression of the three efflux pumps.
RESULTS
Deletion of efflux pump genes affects drug resistance levels.
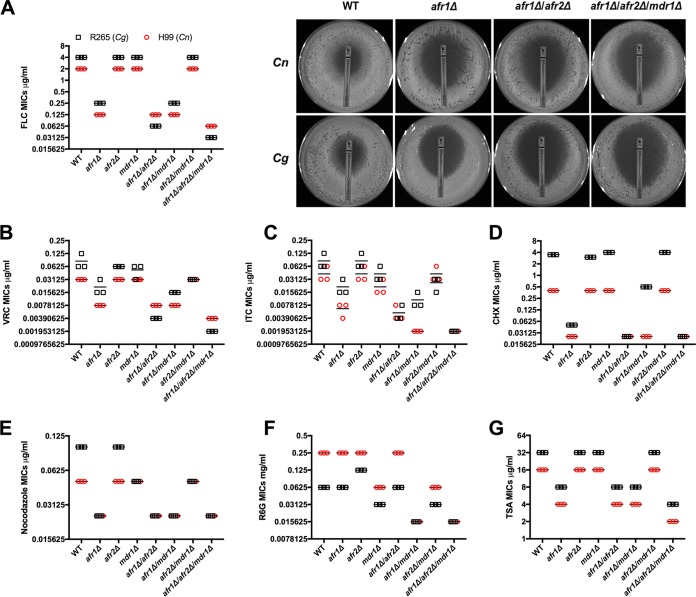
Since the substrate specificity of cryptococcal efflux pumps has been poorly characterized, we constructed deletion mutants of the three well-known efflux pump genes in Cryptococcus, AFR1, AFR2, and MDR1, either individually or in combinations in two representative strains of cryptococcosis agents, C. neoformans H99 and C. gattii R265. To assess their function and substrate specificity, we measured MICs of the wild-type strains and all the deletion strains to eight different selected compounds. They included five of the most commonly used antifungal antibiotics (FLC, itraconazole [ITC], voriconazole [VRC], 5-fluorocytosine, and amphotericin B), one eukaryote protein synthesis inhibitor (cycloheximide [CHX]), one antineoplastic agent (nocodazole, which exerts its effect in cells by interfering with the polymerization of microtubules), one less commonly known antifungal antibiotic (trichostatin A [TSA], which selectively inhibits the class I and II mammalian histone deacetylase families of enzymes), and one potent inhibitor of mitochondrial oxidative phosphorylation (rhodamine 6G [R6G]). We found that MICs of R265 were higher than those of H99 against most of the tested compounds, except for R6G (Fig. 1 and Table 1).
FIG 1.
Deletion of efflux pump genes affects drug resistance levels. The MICs of different compounds in each indicated strain were determined (red circles, H99 and its derivatives; black squares, R265 and its derivatives). The experiments were repeated three times, and the bars represent the average MICs. The details for each strain's MICs are listed in Table 1. The pictures on the right side of panel A are the fluconazole Etest results from a representative experiment after 3 days of incubation. FLC, fluconazole; VRC, voriconazole; ITC, itraconazole; CHX, cycloheximide; R6G, rhodamine 6G; TSA, trichostatin A.
TABLE 1.
MIC of antifungals and xenobiotics in C. neoformans and C. gattii mutant strains
| Genotype | MICs (μg/ml)a |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| FLCb | VRCb | ITCb | AMBc | 5-FCb | CHXc | Nocodazolec | R6Gc (mg/ml) | TSAc | |
| WT (C. neoformans H99) | 2 | 0.031 | 0.031–0.063 | 0.125 | 2 | 0.4 | 0.05 | 0.25 | 16 |
| Cnafr1Δ | 0.125 | 0.008 | 0.004–0.008 | 0.125 | 2–4 | 0.025 | 0.025 | 0.25 | 4 |
| Cnafr2Δ | 2 | 0.031 | 0.031–0.063 | 0.125 | 2 | 0.4 | 0.05 | 0.25 | 16 |
| Cnmdr1Δ | 2 | 0.031 | 0.016–0.031 | 0.125 | 2 | 0.4 | 0.05 | 0.063 | 16 |
| Cnafr1Δ Cnafr2Δ | 0.125 | 0.008 | 0.004–0.008 | 0.063 | 2 | 0.025 | 0.025 | 0.25 | 4 |
| Cnafr1Δ Cnmdr1Δ | 0.125 | 0.008 | 0.002 | 0.125 | 2 | 0.025 | 0.025 | 0.016 | 4 |
| Cnafr2Δ Cnmdr1Δ | 2 | 0.031 | 0.031–0.063 | 0.125 | 2 | 0.4 | 0.05 | 0.063 | 16 |
| Cnafr1Δ Cnafr2Δ Cnmdr1Δ | 0.063 | 0.004 | 0.002 | 0.125 | 2 | 0.025 | 0.025 | 0.016 | 2 |
| WT (C. gattii R265) | 4 | 0.063–0.125 | 0.063–0.125 | 0.5 | 4 | 3.5 | 0.1 | 0.063 | 32 |
| Cgafr1Δ | 0.25 | 0.016–0.031 | 0.016–0.031 | 0.5 | 4 | 0.05 | 0.025 | 0.063 | 8 |
| Cgafr2Δ | 4 | 0.063 | 0.063–0.125 | 0.5 | 4 | 3 | 0.1 | 0.125 | 32 |
| Cgmdr1Δ | 4 | 0.031–0.063 | 0.031–0.063 | 0.25 | 4 | 4 | 0.05 | 0.031 | 32 |
| Cgafr1Δ Cgafr2Δ | 0.063 | 0.004 | 0.004–0.008 | 0.25 | 2–4 | 0.025 | 0.025 | 0.063 | 8 |
| Cgafr1Δ Cgmdr1Δ | 0.25 | 0.016 | 0.008–0.016 | 0.25 | 4 | 0.5 | 0.025 | 0.016 | 8 |
| Cgafr2Δ Cgmdr1Δ | 4 | 0.031 | 0.016–0.031 | 0.25 | 4 | 4 | 0.05 | 0.031 | 32 |
| Cgafr1Δ Cgafr2Δ Cgmdr1Δ | 0.031 | 0.002 | 0.002 | 0.25 | 2 | 0.025 | 0.025 | 0.016 | 4 |
FLC, fluconazole; VRC, voriconazole; ITC, itraconazole; AMB, amphotericin B; 5-FC, 5-fluorocytosine; CHX, cycloheximide; R6G, rhodamine 6G; TSA, trichostatin A. All experiments were repeated three times.
MIC-2.
MIC-0.
Among the single-gene deletion mutants, afr1Δ mutants of both H99 and R265 resulted in drastically higher susceptibility to the tested compounds except for R6G, 5-fluorocytosine, and amphotericin B (Fig. 1 and Table 1), suggesting that Afr1 is the major efflux pump for the tested chemicals in both species. Deletion of AFR2 alone had no effect on the susceptibility to most of the tested compounds in both H99 and R265, except that the C. gattii afr2Δ (Cgafr2Δ) mutant displayed slightly lower MICs to cycloheximide and higher MICs to R6G than the wild-type R265. Single deletion of MDR1 in H99 caused no change in MICs to most of the drugs, except for itraconazole and R6G. Single deletion of MDR1 in R265 caused higher susceptibility to nocodazole and R6G but higher resistance to cycloheximide. Additionally, all the mutants tested had MICs for 5-fluorocytosine and amphotericin B that were similar to those of the wild-type strains, suggesting that AFR1, AFR2, and MDR1 are not important for pumping 5-fluorocytosine and amphotericin B.
Double deletion of AFR1 and AFR2 in H99 showed the same phenotype toward most of the tested drugs as the AFR1 single deletion mutant except for slightly lower MICs for amphotericin B (Fig. 1 and Table 1). In R265, the drug susceptibility phenotype of the Cgafr1Δ Cgafr2Δ mutant was similar to that of the Cgafr1Δ mutant except that the double deletion mutant had lower MICs for FLC, voriconazole, itraconazole, and cycloheximide than the Cgafr1Δ mutant. Double deletion of AFR1 and MDR1 did not cause further increases in susceptibility to FLC, voriconazole, nocodazole, trichostatin A, and amphotericin B in both genetic backgrounds. However, the afr1Δ mdr1Δ mutant exhibited further decreases in MICs to R6G compared to the AFR1 or MDR1 single-gene deletion mutants of both H99 and R265. In addition, only Cnafr1Δ Cnmdr1Δ mutant further decreased the MICs to itraconazole, and Cgafr1Δ Cgmdr1Δ did not. Interestingly, the MICs for cycloheximide in the Cgafr1Δ Cgmdr1Δ double deletion mutant were between those of the Cgafr1Δ and Cgmdr1Δ mutants. Double deletion of AFR2 and MDR1 in H99 and R265 caused MICs similar to those of their respective single mdr1Δ deletion mutants.
Deletion of all three efflux pump genes in H99 and R265 significantly increased susceptibility to FLC, voriconazole, and itraconazole, suggesting that the two efflux pumps Afr2 and Mdr1 augment the function of the major drug efflux pump, Afr1, in response to all three tested azoles. However, the triple deletion of the three pump genes did not further exacerbate the susceptibility to the other compounds tested. Additionally, we determined the posaconazole (PSC) susceptibility phenotype by using Etest strips and found that the posaconazole susceptibility patterns of all efflux pump mutants were similar to those for itraconazole in both species. However, afr1Δ mdr1Δ MICs were higher than those of afr1Δ afr2Δ mutant strains (MICs of 0.023 μg/ml for the afr1Δ mdr1Δ and 0.125 μg/ml for the afr1Δ afr2Δ mutants of H99 and MICs of 0.032 μg/ml for the afr1Δ mdr1Δ and 0.125 μg/ml for the afr1Δ afr2Δ mutants of R265). Taken together, these results indicate that AFR1, AFR2, and MDR1, either individually or in combination, contribute differently to the tolerance against different xenobiotics in the two species.
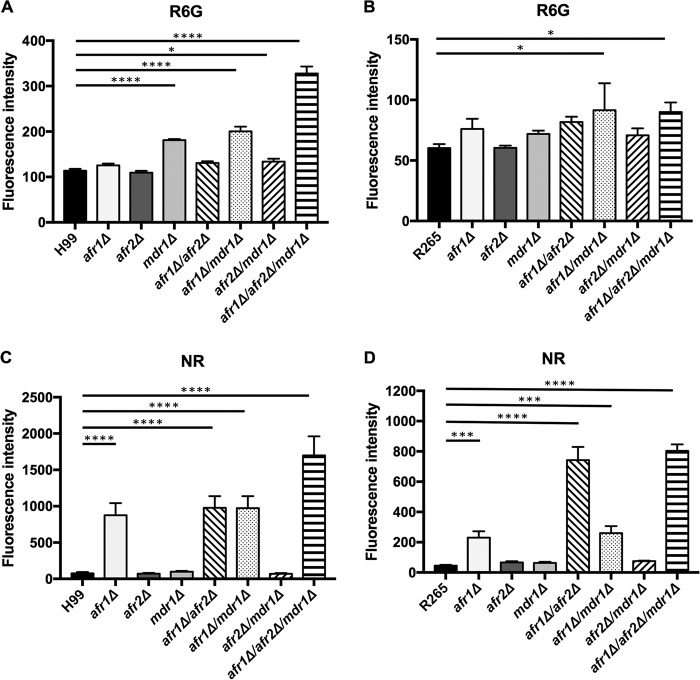
Deletion of efflux pump genes affects drug accumulation.
Rhodamine 6G and Nile Red are fluorescent dyes which have been used to determine the function of efflux pumps (34–38). It is possible that modification of the genomic constituents of the efflux pumps could alter the levels of accumulation of the fluorescent dyes and reflect the function of the efflux pumps. To determine the contributions of the three genes in pumping xenobiotics, we measured intracellular accumulation of R6G and Nile Red by flow cytometry. As shown in Fig. 2A, no difference in the intracellular R6G levels was observed between the wild-type H99 and the Cnafr1Δ or Cnafr2Δ mutant. However, all the mutants containing a CnMDR1 deletion accumulated more R6G than the wild type, and the triple deletion mutant accumulated the highest levels of R6G. The observed increases in R6G accumulation in all the mutants containing a CnMDR1 deletion corroborated the results of R6G susceptibility testing (Fig. 1F), which indicated the direct contribution of CnMDR1 as an efflux pump gene for R6G in C. neoformans.
FIG 2.
Deletion of efflux pump genes affects drug accumulation. (A) Wild-type and deletion strains were incubated with 10 μM R6G (A and B) or 7 μM Nile Red (C and D) in YEPD medium for 30 min at 30°C, and the cells were analyzed by flow cytometry. (A and C) H99; (B and D) R265. Values represent the means ± standard deviations from three independent experiments. *, P < 0.0442; ***, P < 0.0006; ****, P < 0.0001 (Holm-Sidak's multiple-comparison test).
In R265, deletion of each individual efflux pump gene showed no significant difference in R6G accumulation compared to that in the wild type, while a slight increase of R6G accumulation was seen in the Cgafr1Δ Cgmdr1Δ mutant and the triple deletion mutant (Fig. 2B). These results are in contrast to the aforementioned observations that the Cgmdr1Δ mutant had reduced R6G MICs and the Cgafr2Δ mutant had increased MICs for R6G (Fig. 1). Interestingly, although H99 accumulated more R6G than R265 (Fig. 2A and B), H99 was more resistant to R6G than R265 (Fig. 1F). Thus, there is no clear relationship between R6G accumulation levels and susceptibility to the dye in both species.
The patterns of Nile Red accumulation were very different than those of R6G accumulation in both species (Fig. 2). Among the single-gene deletion mutants, deletion of AFR1 in both H99 and R265 significantly increased the accumulation of Nile Red, indicating the importance of Afr1 as an efflux pump for this dye. Deletion of AFR2 or MDR1 in addition to AFR1 in H99 did not further increase the accumulation of Nile Red, but the Cgafr1Δ Cgafr2Δ mutant accumulated markedly increased levels of the dye compared to those in the R265 Cgafr1Δ mutant, suggesting that AFR2 also contributed to Nile Red pumping in R265 but not in H99. Interestingly, the triple deletion mutants of H99 displayed the highest levels of Nile Red accumulation, suggesting that the three efflux pump genes may function synergistically. In contrast, the accumulation of Nile Red was similar in the Cgafr1Δ Cgafr2Δ and Cgafr1Δ Cgafr2Δ Cgmdr1Δ mutants, suggesting that only AFR1 and AFR2 contributed to Nile Red efflux in R265.
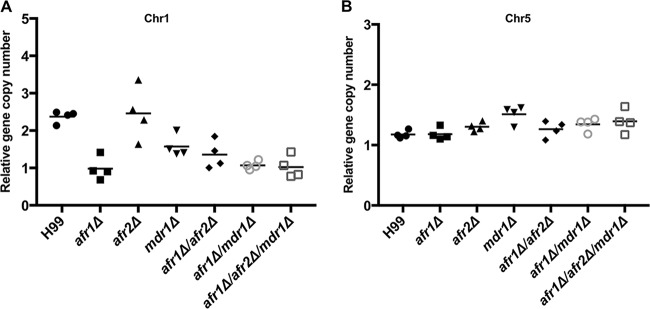
Deletion of efflux pump genes influences formation of chromosome disomy.
Recent studies have shown that H99 is innately heteroresistant to azoles (19). We found that all efflux pump deletion mutants of H99 remained heteroresistant to FLC, and the levels of heteroresistance to FLC in most of the deletion mutants were reduced compared to that in H99, except for the Cnafr2Δ mutant (Table 2). It has also been shown that deletion of CnAFR1, a gene located on chromosome 1 (Chr1) in H99, abrogated Chr1 duplication in FLC-resistant clones (16). To determine if deletion of AFR2 or MDR1 affects the status of aneuploidy in FLC-resistant clones in H99, we determined the chromosome copy number indirectly by using single-colony quantitative PCR (qPCR) to measure the copy numbers of genes located on specific chromosome (16). Since AFR1 and MDR1 are located on Chr1 and AFR2 is located on Chr5, we designed Chr1- and Chr5-specific probes to monitor the status of these two chromosomes. Because Chr3 disomy was rarely observed in all the FLC-resistant clones (16), a Chr3-specific probe was used as an internal control. Figure 3A shows that the relative copy number of Chr1 is close to 2 in H99 FLC-resistant clones and is close to 1 in all the FLC-resistant clones containing an AFR1 deletion, confirming the previous findings on the important role of AFR1 in the formation Chr1 disomy in the presence of FLC (16). Interestingly, the Chr1 copy number of FLC-resistant clones of the mdr1Δ mutant was 1.6. Since quantitative PCRs were carried out by using independently isolated single colonies, the copy number of 1.6 suggested that the population of the analyzed mdr1Δ colonies contained a mixture of cells with and without duplicated Chr1. Therefore, the presence or absence of MDR1 appears to influence the formation of Chr1 disomy under FLC stress. Although the levels of FLC resistance were further reduced in the Cnafr1Δ Cnafr2Δ mutant compared to Cnafr1Δ mutant, which suggested that AFR2 plays an additive role in FLC heteroresistance (Table 2), deletion of AFR2 did not alter the copy number of Chr1 or Chr5 in any genetic background (Fig. 3). These findings indicate that there is no relationship between the status of AFR2 and the formation of disomic chromosomes.
TABLE 2.
Levels of heteroresistance of C. neoformans strains
| Genotype | Heteroresistance (μg/ml) |
|---|---|
| WT (H99) | 32 |
| afr1Δ | 1 |
| afr2Δ | 32 |
| mdr1Δ | 24 |
| afr1Δ afr2Δ | 0.75 |
| afr1Δ mdr1Δ | 1 |
| afr1Δ afr2Δ mdr1Δ | 0.5 |
FIG 3.
Deletion of efflux pump genes influences the formation of disomic chromosomes. Quantitative PCR from single colonies was performed using FLC-resistant clones derived from the indicated strains. Probes specific for different chromosomes were chosen to assess the changes in genomic copy number. The PCR results with the Chr1- and Chr5-specific probes were compared to those with the Chr3-specific probe, which served as unduplicated internal control in the indicated strains. (A) Copy number of the gene on Chr1. (B) Copy number of the gene on Chr5. Values represent the means ± standard deviations for four independently isolated FLC-resistant clones of each mutant.
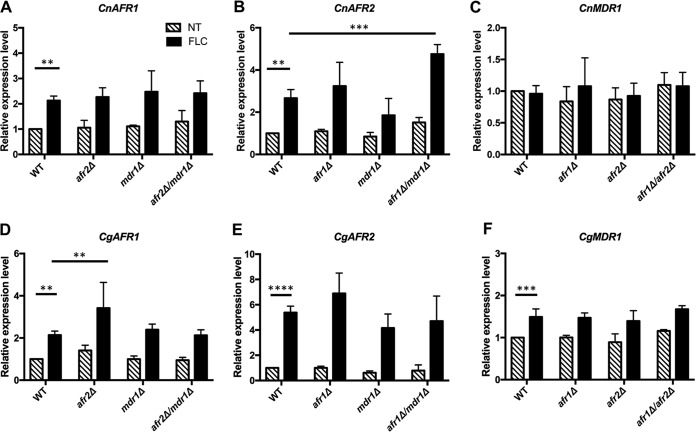
Expression and regulation of efflux genes.
To gain insight into how the expression of each efflux pump gene is regulated, we first investigated the influence of FLC treatment on the expression of each efflux pump gene in the wild type and mutants by quantitative reverse transcription-PCR (qRT-PCR). FLC treatment caused greater than a 2-fold increase in the transcription levels of both CnAFR1 and CnAFR2, but not CnMDR1, in H99 (Fig. 4A, B, and C). In contrast, all three efflux pump genes in R265 showed significantly increased transcription levels in response to FLC treatment (Fig. 4D, E, and F). Deletion of each efflux pump gene did not affect the expression of the other pump genes in H99 and R265 in the absence of FLC. Upon FLC treatment, however, the expression levels of CnAFR2 increased 5-fold in the Cnafr1Δ Cnmdr1Δ mutant compared to the wild type, but no such increase was observed in the single deletion mutant of CnAFR1 or CnMDR1. This suggests that H99 cells respond to the effect of CnAFR1 and CnMDR1 double deletion by elevating the expression of CnAFR2 in the presence of FLC (Fig. 4B). In contrast, the expression patterns of CnAFR1 or CnMDR1 were similar to those of the wild type under FLC treatment when other efflux pump genes were deleted (Fig. 4A and C). In R265, treatment with FLC significantly increased the expression of CgAFR1 in the Cgafr2Δ mutant but not in the Cgafr2Δ Cgmdr1Δ mutant compared to the wild type (Fig. 4D). The expression patterns of CgAFR1, CgAFR2, or CgMDR1 in other deletion mutants were similar to those in the wild type in the presence of FLC (Fig. 4D, E, and F). These results suggested that the deletion of AFR1 and AFR2 imposed small but different effects in H99 versus R265 on the expression of other efflux pump genes in the presence of FLC.
FIG 4.
Gene expression levels of AFR1, AFR2, and MDR1. Cells of the indicated strains from the exponential growth phase were either untreated (NT) or treated (FLC) with 32 μg/ml FLC for 2 h. RNA was isolated, and the expression levels of each gene were determined by qRT-PCR. The expression levels of each gene under each condition were normalized to that of the actin gene and compared to the expression levels of the wild type without FLC treatment. (A, B, and C) Expression levels of AFR1, AFR2, and MDR1, respectively, in H99. (D, E, and F) Expression levels of AFR1, AFR2, and MDR1, respectively, in R265. Values represent the means ± standard deviations for three biological replicates. **, P < 0.0091; ***, P < 0.0007; ****, P < 0.0001 (uncorrected Fisher's least significant difference [LSD]).
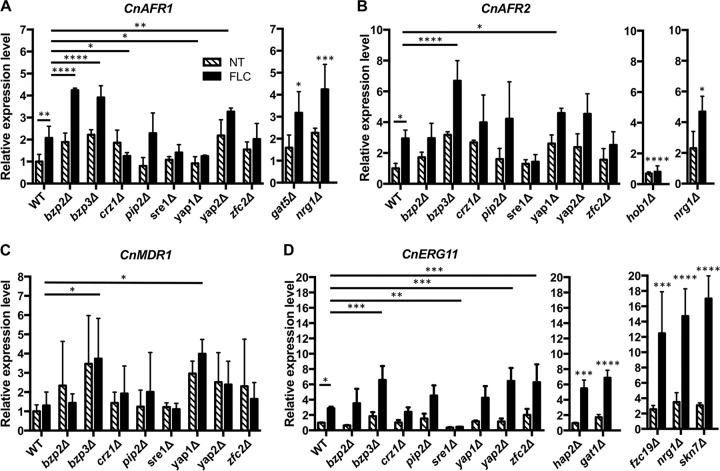
Our transcriptional analysis revealed that expression levels of CnAFR1 and CnAFR2 were upregulated in response to FLC treatment in H99. To identify the transcription factors (TFs) regulating the expression of the efflux pump genes, we screened the H99 TF deletion library generated by Jung et al. (39) and identified mutants exhibiting altered FLC sensitivity or increased FLC heteroresistance. Several mutants showing a clear phenotype in each category were identified. We examined the expression patterns of CnAFR1, CnAFR2, and CnMDR1 in these deletion mutants by qRT-PCR. Among the 8 TF mutants that displayed increased sensitivity to FLC, the aforementioned 2-fold increase in the expression of CnAFR1 upon FLC treatment was diminished in crz1Δ and yap1Δ mutants (Fig. 5A). Since CnAFR1 plays a clear role in FLC tolerance, the significant decreases in expression of CnAFR1 may be partially responsible for the increased susceptibility to FLC in crz1Δ and yap1Δ mutants. Under similar conditions, we did not observe any TF that influenced the expression patterns of CnAFR2 and CnMDR1 (Fig. 5B and C). Interestingly, the expression levels of CnAFR1 were significantly increased in bzp2Δ, bzp3Δ, and yap2Δ mutants compared to those in the wild type under both FLC-treated and untreated conditions. Similarly, the expression levels of CnAFR2 and CnMDR1 were significantly increased in bzp3Δ and yap1Δ mutants compared to those in the wild type under both FLC-treated and untreated conditions. Therefore, the bzp3Δ mutant showed increased expression of all three efflux pump genes. However, the relationship between such increases in gene expression and the FLC sensitivity phenotype in the bzp3Δ mutant is not clear. Since FLC targets lanosterol 14α-demethylase, Erg11, assessment of the ERG11 expression patterns among the TF mutants might shed light on the mechanism of FLC resistance. We measured the expression levels of CnERG11 in H99 and the FLC-sensitive TF mutants. FLC treatment upregulated CnERG11 expression 3-fold in the wild type (Fig. 5D). CnERG11 expression was significantly reduced in the sre1Δ mutant compared to wild type under the FLC treatment. Therefore, the hypersensitive phenotype of the sre1Δ mutant to FLC may be partially due to the significant reduction of CnERG11 expression. We also noted that expression levels of CnERG11 increased significantly in the bzp3Δ, yap2Δ, and zfc2Δ mutants compared to the wild type in the presence of FLC.
FIG 5.
Expression of efflux pump genes and ERG11 is independently regulated by different transcription factors. Exponential-growth-phase cells of transcription factor mutant strains were either untreated (NT) or treated (FLC) with 32 μg/ml FLC for 2 h. RNA was isolated, and the expression levels of each gene were determined by qRT-PCR. The expression levels of each gene were normalized to that of the actin gene and compared to the expression levels of the wild type without FLC treatment. (A) Expression levels of CnAFR1; (B) expression levels of CnAFR2; (C) expression levels of CnMDR1; (D) expression levels of CnERG11. Values represent the means ± standard deviations for three biological replicates. *, P < 0.0476; **, P < 0.0098; ***, P < 0.0006; ****, P < 0.0001 (uncorrected Fisher's LSD). The details for the expression levels for each gene in the FLC-resistant mutants and mutants with increased frequencies of FLC heteroresistance are shown in Fig. S1 in the supplemental material.
Among the 3 FLC-resistant TF mutants, the expression patterns of all the efflux pump genes were similar to that in the wild type under both FLC-treated and untreated conditions except that upregulation of CnAFR2 expression was not observed in FLC-treated hob1Δ cells (Fig. 5B [middle panel]; see Fig. S1A to C in the supplemental material). Under the same conditions, the expression levels of CnERG11 in the hap2Δ and gat1Δ mutants were significantly increased compared to that in the wild type (Fig. 5D [middle panel] and Fig. S1D). Furthermore, among the 4 TF mutants and showing increased frequency of heteroresistance to FLC, only the nrg1Δ mutant had significantly increased expression of CnAFR1, CnAFR2, and CnERG11 compared to that in the wild type under both FLC-treated and untreated conditions (Fig. 5 and S1). The expression of the four studied genes in other heteroresistant mutants was similar to that in the wild type under both FLC-treated and untreated conditions. Taken together, these results suggest that expression of AFR1, AFR2, MDR1, and ERG11 is independently regulated by different transcription factors.
DISCUSSION
The main objective of the present study was to determine the functions of three ABC transporters by gene deletion, singly or in combinations, in the two most studied strains of cryptococcosis, H99 and R265, as well as to compare the responses of the deletion mutants to various azoles since the two species have shown different levels of azole susceptibility (22, 24, 40). Furthermore, the substrate specificities of the three cryptococcal pumps associated with azole resistance were analyzed, since this has not been well studied in either species.
We examined the sensitivity levels of our strains to several triazoles and various nonazole xenobiotics and found that R265 is generally more resistant than H99 to most compounds besides the azoles tested. From the studies of deletion strains, we were able to decipher the roles of the three ABC transporter pumps toward these chemicals. Deletion of AFR1 in both H99 and R265 resulted in hypersensitivity to most tested drugs and xenobiotics, except for R6G, 5-fluorocytosin,e, and amphotericin B. Interestingly, the single deletion of MDR1 in H99 resulted in two different effects on azole resistance: higher susceptibility to itraconazole and posaconazole but no change in MICs to FLC and voriconazole. Structurally, itraconazole and posaconazole are similar and are distinct from FLC and voriconazole, which resemble each other. The structural differences in these two groups of azoles may affect the binding affinity to the Mdr1 pump. MDR1 deletion in H99 also resulted in higher susceptibility to R6G. Single deletion of MDR1 in R265 had no effect on susceptibility to triazoles but resulted in a higher susceptibility to nocodazole and R6G and caused higher resistance to cycloheximide. Single deletion of AFR2 caused higher resistance to R6G only in the R265 background. These results indicate that Afr1 is a crucial pump in both species that is required for azole efflux and is important for handling other xenobiotics, including cycloheximide, nocodazole, and trichostatin A. Moreover, Mdr1 and Afr2 play minor but different roles in drug tolerance in the two species. However, Afr1, Afr2, and Mdr1 appeared to play no clear role in susceptibility toward amphotericin B and 5-fluorocytosine.
Alterations in efflux pump function and drug accumulation levels are directly linked to mechanisms of drug resistance (41). We showed increased intracellular accumulation of R6G in all the mutants containing a CnMDR1 deletion, which also had markedly reduced tolerance toward R6G in H99. But the relationship between R6G accumulation and the tolerance toward the dye was not as clear in R265. We also showed that deletion of AFR1 did not significantly altered the R6G accumulation in both H99 and R265, suggesting that Afr1 is not important for R6G efflux. Interestingly, the protein sequence of Afr1 is closest to that of the ABC transporter Snq2 described in S. cerevisiae and Candida glabrata, which reportedly pump out R6G (31, 42), but the Afr1 sequence homology with the two Snq2 proteins is only about 35% (30). Therefore, the differences in substrate specificity between Afr1 and Snq2 are not unusual. However, our results on R6G accumulation contradict a previous report where a C. neoformans afr1Δ strain had higher accumulation of R6G (30). The discrepancy between the two studies is likely due to the difference in the genetic backgrounds of the deletion mutants. We deleted AFR1 in the FLC-susceptible wild-type strain, and the resulting deletion mutant accumulated amounts of R6G similar to those in wild type. In contrast, the AFR1 gene was deleted in the azole-resistant mutant strain in the previous work, and the levels of R6G accumulation in the afr1 deletion mutant were higher than those in the resistant parental strain but similar to those in the original FLC-susceptible strain (30). It is possible that wild-type strains may have other pumps which can compensate for the efflux function for R6G in an afr1Δ mutant. This explanation is supported by the finding that the wild type and the afr1Δ mutant had the same MICs for R6G in our studies.
The patterns of the Nile Red accumulation were clearly different from the patterns of R6G accumulation in the two species. Interestingly, the relative accumulation levels of Nile Red in each deletion mutant strain were in parallel with the relative levels of FLC MICs in both the H99 and R265 backgrounds, suggesting that Nile Red may be used as a reliable indicator for FLC susceptibility. Despite repeated attempts, we were unable to determine the MICs of Nile Red due to its poor solubility in water and precipitation in RPMI culture medium. Thus, we failed to establish the correlation between Nile Red accumulation and each strain's MICs for this dye. Although it is most likely that these pumps are involved in fluconazole efflux, we cannot rule out the possibility that these pumps may efflux some noxious material that accumulates in the presence of fluconazole, since we did not measure uptake or efflux of FLC in these strains. It is also possible that these pumps are involved in other functions yet to be identified. These two possibilities may account for the lack of correlation with the R6G dye measurements.
The most commonly observed mechanisms of FLC resistance in C. neoformans are overexpression of drug efflux pump genes and mutations in ERG11 (17, 43–45). In C. neoformans, upregulation of AFR1 contributes to FLC resistance (26). However, correlation between the expression of each efflux pump gene and FLC susceptibility has not yet been identified. We showed that the expression levels of two of three pumps, encoded by AFR1 and AFR2, increased upon FLC treatment in C. neoformans, while expression of all three efflux genes was higher in the presence of FLC in C. gattii. Since the triple deletion mutants of efflux pump genes in both species exhibited the lowest FLC MICs of all the deletion mutants, it is likely that the upregulation of AFR2 and MDR1 expression upon FLC treatment contributed to the FLC tolerance in both species. Through screening of a library of transcription factor mutants, we identified transcription factors regulating the expression of efflux pumps genes. CnAFR1 expression in response to FLC was significantly downregulated in deletion mutants of CRZ1 and YAP1. CRZ1 and YAP1 have been previously characterized as transcription factors functioning in response to environmental stress, including thermal, hypoxic and FLC, and oxidative stress (46–48). In addition, we found that the FLC-sensitive phenotype of crz1Δ and yap1Δ mutants was reverted to near-wild-type levels when CnAFR1 was overexpressed in crz1Δ and yap1Δ mutants (see Fig. S2 in the supplemental material). These results indicated that CnAFR1 was positively regulated by CRZ1 and YAP1.
Taken together, our findings suggest that AFR1 is the major pump for azole efflux and that the other two efflux pumps play additive roles in the management of FLC stress in both species of Cryptococcus. In addition, the expression of the three genes is regulated independently by stress response transcription factors. However, the detailed role of each pump in the FLC resistance mechanism remains unclear. Furthermore, the physiological significance of the efflux pump genes also remains to be determined.
MATERIALS AND METHODS
Strains, media, growth conditions, and chemicals.
All strains used in this study are listed in Table S1 in the supplemental material. C. neoformans and C. gattii strains were refreshed from −80°C by subculturing on solid yeast extract-peptone-dextrose (YEPD) (2% peptone, 2% dextrose, 1% yeast extract, and 2% agar) and maintained on YEPD or yeast nitrogen base (YNB) (6.7 g/liter) agar plates at 30°C before each experiment.
Fluconazole (FLC), voriconazole (VRC), itraconazole (ITC), amphotericin B (AMB), cycloheximide, nocodazole, rhodamine 6G (R6G), and trichostatin A (TSA) were prepared in dimethyl sulfoxide (DMSO) at 50 mg/ml, 25 mg/ml, 4 mg/ml, 40 mg/ml, 10 mg/ml, 5 mg/ml, 4.8 mg/ml, and 2 mg/ml, respectively. 5-Fluorocytosine (5-FC) was dissolved in sterile distilled water at 5 mg/ml. Stock solutions were kept at −20°C. RPMI 1640 was used as a medium to dilute drugs and inocula of strains in MIC tests.
Construction of deletion mutants and library screening.
Three ABC transporter-encoding genes, AFR1 (CNAG_00730 and CNBG_1200), AFR2 (CNAG_00869 and CNBG_6088), and MDR1 (CNAG_00796 and CNBG_1138) were disrupted in the H99 (VNI) or R265 (VGIIa) background by homologous recombination using a gene-specific deletion cassette amplified by overlapping PCR. The G418 (NEO), nourseothricin (NAT) and hygromycin (HYG) cassette was used as a selectable marker in biolistic transformation with a Bio-Rad model PDS-1000/He biolistic particle delivery system. All overlapping PCR primers used in deletion strain construction are listed in Table S2 in the supplemental material. Deletions in all mutants were confirmed by PCR and Southern blot analysis.
We screened the deletion library of H99 transcription factors generated by Jung et al. (39) by replica stamping of the individual clones on medium containing 28, 32, or 36 μg/ml FLC. The plates were incubated at 30°C and photographed after 2, 3, and 6 days of incubation. The clones that showed a decrease or increase in growth rate or an increase in the frequency of heteroresistance at each tested FLC concentration were identified.
Drug susceptibility assays.
The CLSI M27-A3 reference method was used to determine the MIC of each chemical (49). Briefly, chemicals were dispensed in 96-well microtiter plates with 2-fold serial dilutions of drug. The final drug concentration was prepared from stock solution in RPMI 1640 medium. The concentration ranges tested in this study were as follows: FLC, 64 μg/ml to 0.0000305 μg/ml; ITC, VRC, and 5-FC, 16 μg/ml to 0.0000075 μg/ml; AMB, 16 μg/ml to 0.0313 μg/ml; CHX, 2 μg/ml to 0.0125 μg/ml; nocodazole, 0.8 μg/ml to 0.0031 μg/ml; R6G, 1 mg/ml to 0.0019 mg/ml; and TSA, 32 μg/ml to 0.0625 μg/ml. We used 5 × 102 cells from overnight cultures in each test. Plates were sealed with an air-permeable membrane and incubated at 37°C for 72 h. MICs of AMB, CHX, nocodazole, R6G, and TSA were recorded as the lowest concentration required for 100% inhibition of growth (MIC-0). MICs of 5-FC and the azoles were defined as the lowest concentration required for 50% reduction in turbidity compared to the growth control well (MIC-2).
The MICs of FLC and posaconazole (PSC) were also determined on YNB agar plates with Epsilometer test strips (Etest strips; AB Biodisk). About 2 × 106 cells were plated on YNB before application of azole Etest strips. Plates were incubated at 30°C for 72 h before being photographed.
RNA extraction and qPCR.
For RNA extraction, cells were grown overnight in YEPD medium on a shaker at 30°C. Overnight cultures were diluted in 10 ml fresh YEPD medium to an optical density at 600 nm (OD600) of 0.2 and grown at 30°C to an OD600 of 0.5. The cultures were divided into two parts; one part was allowed to grow continuously under the same conditions, while the other was supplemented with 32 μg/ml of FLC and incubated for indicated time periods. Cells were washed with sterile H2O, and cell pellets were frozen, lyophilized, and then lysed by beating with glass beads. Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA), and the RNA pellet was resuspended in DNase- and RNase-free H2O and quantified with a NanoDrop 2000 UV-visible spectrophotometer. RNA was separated by 1% formaldehyde agarose gel electrophoresis to confirm the quantity and purity. Equal amounts (6 μg) of total RNA were treated with Turbo RNase-free DNase (Ambion) to remove the genomic DNA. cDNAs were synthesized using a high-capacity cDNA reverse transcription kit (Applied Biosystems). Quantitative RT-PCR was performed using Fast SYBR green qPCR master mix and an Applied Biosystems 7500 real-time PCR system. Each reaction was run in triplicate with three independent cultures, and a cDNA sample was analyzed in triplicate for each primer set. The PCR efficiency and threshold cycle (CT) determination were performed using an algorithm as described previously (50). Data were normalized to actin gene (ACTIN1) levels and expressed as the amount relative to RNA levels of H99 or R265.
To quantify the gene copy number on specific chromosomes, FLC-resistant colonies were isolated and qPCRs were performed as previously described (16). Briefly, cells from individual colonies were suspended in 50 μl of 10 mM EDTA buffer, boiled for 6 min, and then centrifuged. The supernatant was diluted 10-fold in Tris-EDTA (TE) buffer, and 5 μl of diluted DNA sample was added to 20 μl of the qPCR mixture (TaqMan universal PCR master mix; Applied Biosystems). The relative copy number of each gene was determined by comparing with the control gene CNAG_02959 on Chr3A. All primer and probe sequences are listed in Tables S2 and S3 in the supplemental material.
Flow cytometry analysis of the efflux of R6G and Nile Red.
Each strain was grown overnight in YEPD medium at 30°C with shaking. Cultures were diluted to an OD600 of 0.5 in YEPD. Cells were incubated in the presence of R6G (10 μM; 10 mM stock in DMSO) or Nile Red (7 μM; 3.5 mM stock in DMSO) for 30 min at 30°C with shaking. The reaction was stopped by cooling on ice, and then the mixture was diluted 40-fold in cold 1× phosphate-buffered saline (PBS). The accumulation of R6G and Nile Red was immediately assessed by flow cytometry. For each sample, 10,000 events were collected, and data were analyzed using FlowJo software.
Statistical analysis.
All experiments were performed in triplicate, and statistics and graphs were obtained using Graph Pad Prism 7.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Division of Intramural Research (DIR), NIAID, NIH.
We thank Y. S. Bahn for providing the deletion library of transcription factors and A. Varma for critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01751-17.
REFERENCES
- 1.Kwon-Chung KJ, Bennett JE, Wickes BL, Meyer W, Cuomo CA, Wollenburg KR, Bicanic TA, Castaneda E, Chang YC, Chen J, Cogliati M, Dromer F, Ellis D, Filler SG, Fisher MC, Harrison TS, Holland SM, Kohno S, Kronstad JW, Lazera M, Levitz SM, Lionakis MS, May RC, Ngamskulrongroj P, Pappas PG, Perfect JR, Rickerts V, Sorrell TC, Walsh TJ, Williamson PR, Xu J, Zelazny AM, Casadevall A. 2017. The case for adopting the “species complex” nomenclature for the etiologic agents of cryptococcosis. mSphere 2:e00357-16. doi: 10.1128/mSphere.00357-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kwon-Chung KJ, Saijo T. 2015. Is Cryptococcus gattii a primary pathogen? J Fungi (Basel) 1:154–167. doi: 10.3390/jof1020154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perfect JR, Dismukes WE, Dromer F, Goldman DL, Graybill JR, Hamill RJ, Harrison TS, Larsen RA, Lortholary O, Nguyen MH, Pappas PG, Powderly WG, Singh N, Sobel JD, Sorrell TC. 2010. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the infectious diseases society of america. Clin Infect Dis 50:291–322. doi: 10.1086/649858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Norman AW, Demel RA, de Kruyff B, Geurts van Kessel WS, van Deenen LL. 1972. Studies on the biological properties of polyene antibiotics: comparison of other polyenes with filipin in their ability to interact specifically with sterol. Biochim Biophys Acta 290:1–14. doi: 10.1016/0005-2736(72)90046-6. [DOI] [PubMed] [Google Scholar]
- 5.Gruda I, Gauthier E, Elberg S, Brajtburg J, Medoff G. 1988. Effects of the detergent sucrose monolaurate on binding of amphotericin B to sterols and its toxicity for cells. Biochem Biophys Res Commun 154:954–958. doi: 10.1016/0006-291X(88)90232-X. [DOI] [PubMed] [Google Scholar]
- 6.Akins RA. 2005. An update on antifungal targets and mechanisms of resistance in Candida albicans. Med Mycol 43:285–318. doi: 10.1080/13693780500138971. [DOI] [PubMed] [Google Scholar]
- 7.Sanglard D. 2002. Clinical relevance of mechanisms of antifungal drug resistance in yeasts. Enferm Infecc Microbiol Clin 20:462–469. (Quiz 20:470–479.) [DOI] [PubMed] [Google Scholar]
- 8.Zonios DI, Bennett JE. 2008. Update on azole antifungals. Semin Respir Crit Care Med 29:198–210. doi: 10.1055/s-2008-1063858. [DOI] [PubMed] [Google Scholar]
- 9.Paugam A, Dupouy-Camet J, Blanche P, Gangneux JP, Tourte-Schaefer C, Sicard D. 1994. Increased fluconazole resistance of Cryptococcus neoformans isolated from a patient with AIDS and recurrent meningitis. Clin Infect Dis 19:975–976. [DOI] [PubMed] [Google Scholar]
- 10.Schell RE, Tran NV, Bramhall JS. 1986. Amphotericin-B induced changes in renal membrane ion permeation—model of nephrotoxicity. J Dent Res 65:249–249. [DOI] [PubMed] [Google Scholar]
- 11.Mondon P, Petter R, Amalfitano G, Luzzati R, Concia E, Polacheck I, Kwon-Chung KJ. 1999. Heteroresistance to fluconazole and voriconazole in Cryptococcus neoformans. Antimicrob Agents Chemother 43:1856–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamazumi T, Pfaller MA, Messer SA, Houston AK, Boyken L, Hollis RJ, Furuta I, Jones RN. 2003. Characterization of heteroresistance to fluconazole among clinical isolates of Cryptococcus neoformans. J Clin Microbiol 41:267–272. doi: 10.1128/JCM.41.1.267-272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bicanic T, Harrison T, Niepieklo A, Dyakopu N, Meintjes G. 2006. Symptomatic relapse of HIV-associated cryptococcal meningitis after initial fluconazole monotherapy: the role of fluconazole resistance and immune reconstitution. Clin Infect Dis 43:1069–1073. doi: 10.1086/507895. [DOI] [PubMed] [Google Scholar]
- 14.White TC, Marr KA, Bowden RA. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev 11:382–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cowen LE, Sanglard D, Calabrese D, Sirjusingh C, Anderson JB, Kohn LM. 2000. Evolution of drug resistance in experimental populations of Candida albicans. J Bacteriol 182:1515–1522. doi: 10.1128/JB.182.6.1515-1522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sionov E, Lee H, Chang YC, Kwon-Chung KJ. 2010. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog 6:e1000848. doi: 10.1371/journal.ppat.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sionov E, Chang YC, Garraffo HM, Dolan MA, Ghannoum MA, Kwon-Chung KJ. 2012. Identification of a Cryptococcus neoformans cytochrome P450 lanosterol 14alpha-demethylase (Erg11) residue critical for differential susceptibility between fluconazole/voriconazole and itraconazole/posaconazole. Antimicrob Agents Chemother 56:1162–1169. doi: 10.1128/AAC.05502-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brandt ME, Pfaller MA, Hajjeh RA, Hamill RJ, Pappas PG, Reingold AL, Rimland D, Warnock DW, Cryptococcal Disease Active Surveillance Group. 2001. Trends in antifungal drug susceptibility of Cryptococcus neoformans isolates in the United States: 1992 to 1994 and 1996 to 1998. Antimicrob Agents Chemother 45:3065–3069. doi: 10.1128/AAC.45.11.3065-3069.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sionov E, Chang YC, Garraffo HM, Kwon-Chung KJ. 2009. Heteroresistance to fluconazole in Cryptococcus neoformans is intrinsic and associated with virulence. Antimicrob Agents Chemother 53:2804–2815. doi: 10.1128/AAC.00295-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Florio AR, Ferrari S, De Carolis E, Torelli R, Fadda G, Sanguinetti M, Sanglard D, Posteraro B. 2011. Genome-wide expression profiling of the response to short-term exposure to fluconazole in Cryptococcus neoformans serotype A. BMC Microbiol 11:97. doi: 10.1186/1471-2180-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basso LR Jr, Gast CE, Bruzual I, Wong B. 2015. Identification and properties of plasma membrane azole efflux pumps from the pathogenic fungi Cryptococcus gattii and Cryptococcus neoformans. J Antimicrob Chemother 70:1396–1407. doi: 10.1093/jac/dku554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris JR, Lockhart SR, Debess E, Marsden-Haug N, Goldoft M, Wohrle R, Lee S, Smelser C, Park B, Chiller T. 2011. Cryptococcus gattii in the United States: clinical aspects of infection with an emerging pathogen. Clin Infect Dis 53:1188–1195. doi: 10.1093/cid/cir723. [DOI] [PubMed] [Google Scholar]
- 23.Bernal-Martinez L, Gomez-Lopez A, Castelli MV, Mesa-Arango AC, Zaragoza O, Rodriguez-Tudela JL, Cuenca-Estrella M. 2010. Susceptibility profile of clinical isolates of non-Cryptococcus neoformans/non-Cryptococcus gattii Cryptococcus species and literature review. Med Mycol 48:90–96. doi: 10.3109/13693780902756073. [DOI] [PubMed] [Google Scholar]
- 24.Gast CE, Basso LR Jr, Bruzual I, Wong B. 2013. Azole resistance in Cryptococcus gattii from the Pacific Northwest: investigation of the role of ERG11. Antimicrob Agents Chemother 57:5478–5485. doi: 10.1128/AAC.02287-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varma A, Kwon-Chung KJ. 2010. Heteroresistance of Cryptococcus gattii to fluconazole. Antimicrob Agents Chemother 54:2303–2311. doi: 10.1128/AAC.00153-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanguinetti M, Posteraro B, La Sorda M, Torelli R, Fiori B, Santangelo R, Delogu G, Fadda G. 2006. Role of AFR1, an ABC transporter-encoding gene, in the in vivo response to fluconazole and virulence of Cryptococcus neoformans. Infect Immun 74:1352–1359. doi: 10.1128/IAI.74.2.1352-1359.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang ML, Uhrig J, Vu K, Singapuri A, Dennis M, Gelli A, Thompson GR III. 2015. Fluconazole susceptibility in Cryptococcus gattii is dependent on the ABC transporter Pdr11. Antimicrob Agents Chemother 60:1202–1207. doi: 10.1128/AAC.01777-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cannon RD, Lamping E, Holmes AR, Niimi K, Baret PV, Keniya MV, Tanabe K, Niimi M, Goffeau A, Monk BC. 2009. Efflux-mediated antifungal drug resistance. Clin Microbiol Rev 22:291–321. doi: 10.1128/CMR.00051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orsi CF, Colombari B, Ardizzoni A, Peppoloni S, Neglia R, Posteraro B, Morace G, Fadda G, Blasi E. 2009. The ABC transporter-encoding gene AFR1 affects the resistance of Cryptococcus neoformans to microglia-mediated antifungal activity by delaying phagosomal maturation. FEMS Yeast Res 9:301–310. doi: 10.1111/j.1567-1364.2008.00470.x. [DOI] [PubMed] [Google Scholar]
- 30.Posteraro B, Sanguinetti M, Sanglard D, La Sorda M, Boccia S, Romano L, Morace G, Fadda G. 2003. Identification and characterization of a Cryptococcus neoformans ATP binding cassette (ABC) transporter-encoding gene, CnAFR1, involved in the resistance to fluconazole. Mol Microbiol 47:357–371. doi: 10.1046/j.1365-2958.2003.03281.x. [DOI] [PubMed] [Google Scholar]
- 31.Torelli R, Posteraro B, Ferrari S, La Sorda M, Fadda G, Sanglard D, Sanguinetti M. 2008. The ATP-binding cassette transporter-encoding gene CgSNQ2 is contributing to the CgPDR1-dependent azole resistance of Candida glabrata. Mol Microbiol 68:186–201. doi: 10.1111/j.1365-2958.2008.06143.x. [DOI] [PubMed] [Google Scholar]
- 32.Servos J, Haase E, Brendel M. 1993. Gene SNQ2 of Saccharomyces cerevisiae, which confers resistance to 4-nitroquinoline-N-oxide and other chemicals, encodes a 169 kDa protein homologous to ATP-dependent permeases. Mol Gen Genet 236:214–218. doi: 10.1007/BF00277115. [DOI] [PubMed] [Google Scholar]
- 33.Cui Z, Hirata D, Miyakawa T. 1999. Functional analysis of the promoter of the yeast SNQ2 gene encoding a multidrug resistance transporter that confers the resistance to 4-nitroquinoline N-oxide. Biosci Biotechnol Biochem 63:162–167. doi: 10.1271/bbb.63.162. [DOI] [PubMed] [Google Scholar]
- 34.Holmes AR, Lin YH, Niimi K, Lamping E, Keniya M, Niimi M, Tanabe K, Monk BC, Cannon RD. 2008. ABC transporter Cdr1p contributes more than Cdr2p does to fluconazole efflux in fluconazole-resistant Candida albicans clinical isolates. Antimicrob Agents Chemother 52:3851–3862. doi: 10.1128/AAC.00463-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maesaki S, Marichal P, Vanden Bossche H, Sanglard D, Kohno S. 1999. Rhodamine 6G efflux for the detection of CDR1-overexpressing azole-resistant Candida albicans strains. J Antimicrob Chemother 44:27–31. doi: 10.1093/jac/44.1.27. [DOI] [PubMed] [Google Scholar]
- 36.Daban JR. 2001. Fluorescent labeling of proteins with Nile red and 2-methoxy-2,4-diphenyl-3(2H)-furanone: physicochemical basis and application to the rapid staining of sodium dodecyl sulfate polyacrylamide gels and Western blots. Electrophoresis 22:874–880. doi:. [DOI] [PubMed] [Google Scholar]
- 37.Verstrepen KJ, Van Laere SD, Vercammen J, Derdelinckx G, Dufour JP, Pretorius IS, Winderickx J, Thevelein JM, Delvaux FR. 2004. The Saccharomyces cerevisiae alcohol acetyl transferase Atf1p is localized in lipid particles. Yeast 21:367–377. doi: 10.1002/yea.1100. [DOI] [PubMed] [Google Scholar]
- 38.Ivnitski-Steele I, Holmes AR, Lamping E, Monk BC, Cannon RD, Sklar LA. 2009. Identification of Nile red as a fluorescent substrate of the Candida albicans ATP-binding cassette transporters Cdr1p and Cdr2p and the major facilitator superfamily transporter Mdr1p. Anal Biochem 394:87–91. doi: 10.1016/j.ab.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jung KW, Yang DH, Maeng S, Lee KT, So YS, Hong J, Choi J, Byun HJ, Kim H, Bang S, Song MH, Lee JW, Kim MS, Kim SY, Ji JH, Park G, Kwon H, Cha S, Meyers GL, Wang LL, Jang J, Janbon G, Adedoyin G, Kim T, Averette AK, Heitman J, Cheong E, Lee YH, Lee YW, Bahn YS. 2015. Systematic functional profiling of transcription factor networks in Cryptococcus neoformans. Nat Commun 6:6757. doi: 10.1038/ncomms7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gomez-Lopez A, Zaragoza O, Dos Anjos Martins M, Melhem MC, Rodriguez-Tudela JL, Cuenca-Estrella M. 2008. In vitro susceptibility of Cryptococcus gattii clinical isolates. Clin Microbiol Infect 14:727–730. doi: 10.1111/j.1469-0691.2008.02021.x. [DOI] [PubMed] [Google Scholar]
- 41.Shapiro RS, Robbins N, Cowen LE. 2011. Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol Mol Biol Rev 75:213–267. doi: 10.1128/MMBR.00045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsujimoto Y, Shimizu Y, Otake K, Nakamura T, Okada R, Miyazaki T, Watanabe K. 2015. Multidrug resistance transporters Snq2p and Pdr5p mediate caffeine efflux in Saccharomyces cerevisiae. Biosci Biotechnol Biochem 79:1103–1110. doi: 10.1080/09168451.2015.1010476. [DOI] [PubMed] [Google Scholar]
- 43.Espinel-Ingroff A, Aller AI, Canton E, Castanon-Olivares LR, Chowdhary A, Cordoba S, Cuenca-Estrella M, Fothergill A, Fuller J, Govender N, Hagen F, Illnait-Zaragozi MT, Johnson E, Kidd S, Lass-Florl C, Lockhart SR, Martins MA, Meis JF, Melhem MS, Ostrosky-Zeichner L, Pelaez T, Pfaller MA, Schell WA, St-Germain G, Trilles L, Turnidge J. 2012. Cryptococcus neoformans-Cryptococcus gattii species complex: an international study of wild-type susceptibility endpoint distributions and epidemiological cutoff values for fluconazole, itraconazole, posaconazole, and voriconazole. Antimicrob Agents Chemother 56:5898–5906. doi: 10.1128/AAC.01115-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodero L, Mellado E, Rodriguez AC, Salve A, Guelfand L, Cahn P, Cuenca-Estrella M, Davel G, Rodriguez-Tudela JL. 2003. G484S amino acid substitution in lanosterol 14-alpha demethylase (ERG11) is related to fluconazole resistance in a recurrent Cryptococcus neoformans clinical isolate. Antimicrob Agents Chemother 47:3653–3656. doi: 10.1128/AAC.47.11.3653-3656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamb DC, Corran A, Baldwin BC, Kwon-Chung J, Kelly SL. 1995. Resistant P45051A1 activity in azole antifungal tolerant Cryptococcus neoformans from AIDS patients. FEBS Lett 368:326–330. doi: 10.1016/0014-5793(95)00684-2. [DOI] [PubMed] [Google Scholar]
- 46.Lev S, Desmarini D, Chayakulkeeree M, Sorrell TC, Djordjevic JT. 2012. The Crz1/Sp1 transcription factor of Cryptococcus neoformans is activated by calcineurin and regulates cell wall integrity. PLoS One 7:e51403. doi: 10.1371/journal.pone.0051403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moranova Z, Virtudazo E, Hricova K, Ohkusu M, Kawamoto S, Husickova V, Raclavsky V. 2014. The CRZ1/SP1-like gene links survival under limited aeration, cell integrity and biofilm formation in the pathogenic yeast Cryptococcus neoformans. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 158:212–220. [DOI] [PubMed] [Google Scholar]
- 48.Paul S, Doering TL, Moye-Rowley WS. 2015. Cryptococcus neoformans Yap1 is required for normal fluconazole and oxidative stress resistance. Fungal Genet Biol 74:1–9. doi: 10.1016/j.fgb.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.CLSI. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts, 3rd ed Approved standard M27-A3. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 50.Zhao S, Fernald RD. 2005. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol 12:1047–1064. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.