Abstract
Screening of a wheat (Triticum aestivum) cDNA library for starch-branching enzyme I (SBEI) genes combined with 5′-rapid amplification of cDNA ends resulted in isolation of a 4,563-bp composite cDNA, Sbe1c. Based on sequence alignment to characterized SBEI cDNA clones isolated from plants, the SBEIc predicted from the cDNA sequence was produced with a transit peptide directing the polypeptide into plastids. Furthermore, the predicted mature form of SBEIc was much larger (152 kD) than previously characterized plant SBEI (80–100 kD) and contained a partial duplication of SBEI sequences. The first SBEI domain showed high amino acid similarity to a 74-kD wheat SBEI-like protein that is inactive as a branching enzyme when expressed in Escherichia coli. The second SBEI domain on SBEIc was identical in sequence to a functional 87-kD SBEI produced in the wheat endosperm. Immunoblot analysis of proteins produced in developing wheat kernels demonstrated that the 152-kD SBEIc was, in contrast to the 87- to 88-kD SBEI, preferentially associated with the starch granules. Proteins similar in size and recognized by wheat SBEI antibodies were also present in Triticum monococcum, Triticum tauschii, and Triticum turgidum subsp. durum.
Starch branching enzyme (SBE; 1,4-α-d-glucan-6-α-[1,4-α-glucan]-transferase; EC 2.4.1.18) is a key enzyme in the starch biosynthetic pathway. The enzyme acts on Glc polymers and catalyzes excision and transfer of glucan chains to the same or other glucan molecules. Translocated chains are attached to the polymer through α-1,6-glucosidic bonds to form branches on the α-1,4-linked Glc backbone. It is generally believed that SBE acts in concert with starch synthases and debranching enzymes to synthesize amylopectin polymers, which together with amylose molecules form the highly organized starch granules (Ball et al., 1996).
All of the reported SBE from plants to date can be divided into two classes, SBEI and SBEII, based on their amino acid sequences (Burton et al., 1995). Most of the characterized plant SBE are in the 80- to 100-kD molecular mass range and, like all of the enzymes of the α-amylase family, carry a catalytic (βα)8 barrel domain (Svensson, 1994). Several experiments performed with purified SBEI and SBEII suggest that the two SBE classes differ in their substrate specificity and kinetic properties. The biochemical data support that SBEI (denoted SBEII in pea) favors transfer of long glucan chains and prefers amylose over amylopectin as a substrate, whereas SBEII (denoted SBEI in pea) produces shorter branches and acts primarily on amylopectin (Smith, 1988; Guan and Preiss, 1993; Takeda et al., 1993a). The involvement of SBEII in the formation of amylopectin polymers is supported by analyses of starches produced in pea embryos and rice and maize endosperm with reduced SBEII activities. These studies have correlated the mutations with increased length of amylopectin chains and an apparent increase in amylose content (Bhattacharyya et al., 1990; Mizuno et al., 1993; Takeda et al., 1993b; Tomlinson et al., 1997). Similar results have also been obtained from studies of transgenic potato with reduced SBEII activity (Jobling et al., 1999). Transgenic potatoes with antisense inhibition of SBEI activity do not show any significant effect on amylose content or chain length distribution of amylopectin, although some alterations to starch properties can be demonstrated (Flipse et al., 1996; Safford et al., 1998). Thus, the two different SBE classes appear, at least in potato tubers, to play different roles in the formation of branched glucan polymers.
Characterization of SBEI genes in wheat (Triticum aestivum) has so far resulted in isolation of one cDNA clone from a wheat endosperm library (Repellin et al., 1997) and two different genomic clones (Båga et al., 1999; Rahman et al., 1999). Both of the cDNA and the genomic clones were predicted to encode precursor forms of SBEI, which are likely processed to 87-kD mature enzymes upon import into plastids. However, hexaploid wheat may contain up to seven additional SBEI, or SBEI-like, genes as suggested by DNA gel-blot analysis (Rahman et al., 1997). Several of the wheat SBEI genes appear to be clustered on chromosome 7, possibly including a gene predicted to encode a 74-kD SBEI-like protein, wSBEI-D2 (Rahman et al., 1997). In contrast to cDNA encoding the 87-kD SBEI, expression of wSBEI-D2 cDNA in a branching enzyme (BE)-deficient Escherichia coli strain does not result in production of BE activity. Thus, all of the SBEI-like genes in wheat may not encode active BE.
Several of the starch biosynthetic enzymes exist as soluble and starch granule-bound forms (Preiss, 1990). In wheat, both SBEI and SBEII are present in the soluble endosperm where several isoforms of SBEI can be identified by native PAGE (Morell et al., 1997; Nagamine et al., 1997). Polypeptides bound and extracted from wheat starch granules can be distinguished on SDS-PAGE gels as at least seven distinct protein bands, ranging in molecular mass from 60 to 149 kD. Only a 92-kD band has been associated with BE activity (Denyer et al., 1995) and shown to contain a protein related to maize SBEIIb (Rahman et al., 1995; Takaoka et al., 1997). The approximately 60, 80, 100, 108, and 115 protein bands have all been identified as various forms of starch synthases (Echt and Schwartz, 1981; Denyer et al., 1995; Yamamori and Endo, 1996).
In this paper, we reveal the identity of the 149-kD protein band reported to be present in wheat starch granules (Schofield and Greenwell, 1987), and show that it represents a previously uncharacterized form of SBEI in plants. The 152-kD isoform of SBEI predicted from the isolated cDNA was found to share features with both the 74-kD wSBEI-D2 and the 87-kD SBEI.
RESULTS
Isolation of a Partial SBEI cDNA Clone
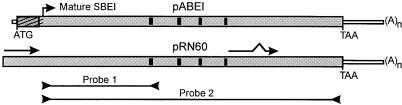
A wheat cDNA library was screened for Sbe1 clones using probe 1 (Fig. 1). The probe fragment was obtained from a reverse transcription (RT)-PCR reaction containing wheat kernel RNA and PCR primers based on previously characterized Sbe1 clones from wheat (Repellin et al., 1997; Båga et al., 1999). DNA sequence analysis of one of the isolated clones, pRN60 (Fig. 1), revealed a 2,962-bp insert that was 162 bp longer than a previously characterized full-length SBEI cDNA, pABEI (Repellin et al., 1997), isolated from the same library. The two cDNA clones matched almost perfectly from the 3′ end to 346 nucleotides from the 5′ end of the pRN60 cDNA (99.8% nucleotide identity and 100% encoded amino acid identity) at which point the two sequences diverged. In contrast to the pABEI cDNA, the 346-bp 5′ sequence of pRN60 cDNA did not seem to encode a transit peptide but instead matched sequences located further downstream on the same cDNA. The unusual 5′ sequence carried by pRN60 lacked stop codons in frame with the downstream SBEI coding region, which suggested that the isolated cDNA could be translated from the first base, and therefore, might not represent a full-length transcript.
Figure 1.
Schematic alignment of pABEI and pRN60 cDNA. Hatched area of pABEI coding region (gray box) represents sequence encoding a putative transit peptide and horizontal arrows on the pRN60 cDNA show location of imperfect direct repeats. The four black areas within the coding region represent sequences encoding the highly conserved regions of enzymes belonging to the α-amylase family (Svensson, 1994). DNA fragments used as probes in DNA and RNA hybridizations are indicated below.
RNA-Blot Analysis of Wheat Endosperm Reveals Two Sbe1 Transcripts
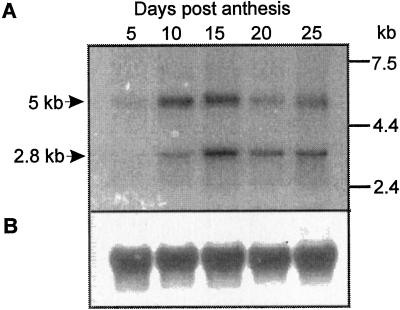
The existence of Sbe1 transcripts that were longer than those encoding the 87- to 88-kD SBEI isoforms was confirmed by an RNA gel-blot analysis. This analysis of wheat kernel RNA extracted at various time points during kernel development showed that a transcript of approximately 5 kb, in addition to the expected 2.8 Sbe1 mRNA, was recognized by the Sbe1-specific probe (Fig. 2A). The signals from both the 5- and 2.8-kb transcripts were very weak in samples of 5-d-old kernels, in which the endosperm is very immature, but were clearly seen in samples prepared from 10- to 25-d-old kernels. In kernels younger than 10 DPA, the 5-kb hybridization signals appeared stronger as compared with the signal from the 2.8-kb transcript.
Figure 2.
RNA-gel analysis of Sbe1 expression during wheat kernel development. A, Analysis of total RNA (20 μg) prepared from developing kernels harvested at different DPA. The blot was hybridized with probe 2 (Fig. 1) and estimated sizes of hybridizing RNA species are shown to the left. Migration of RNA size markers is indicated to the right. B, Same blot as above hybridized with a 25S rRNA DNA probe.
Isolation of Full-Length cDNA Corresponding to 4.6-kb Sbe1 Transcript
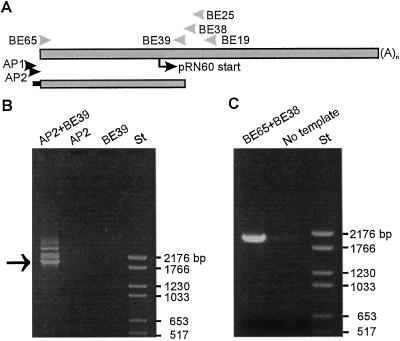
With the hypothesis that the pRN60 cDNA was a partial product of the approximately 5-kb Sbe1 transcript, we decided to isolate the 5′ end of this mRNA species using a 5′-RACE procedure. Gel analysis of products obtained from the final PCR reaction revealed one major fragment of 1.9 kb and three minor fragments (Fig. 3B, lane AP2+BE39). No products were obtained from control reactions using only one primer (Fig. 3B; lanes AP2 and BE39). The different PCR products were analyzed by DNA sequencing, which showed that only the 1.9-kb fragment carried Sbe1-like sequences. One of the 1.9-kb 5′-RACE products was found to correspond 100% to the 272-bp region overlapping the 5′ end of pRN60, and the composite cDNA sequence obtained with this product and the pRN60 cDNA gave a 4,563-bp-long sequence. This assembled sequence was denoted Sbe1c to distinguish it from our previously characterized wheat Sbe1 clones, Sbe1a (Båga et al., 1999) and Sbe1b (Repellin et al., 1997).
Figure 3.
Isolation of cDNA corresponding to 5′ end of 4.6-kb Sbe1c transcript. A, Schematic illustration of the 4.6-kb Sbe1c transcript and product obtained from 5′-RACE analysis. Start of pRN60 sequence and location of PCR primers used in the 5′-RACE and RT-PCR reactions are indicated. B, Gel analysis of 5′-RACE products obtained in reactions with primers indicated and poly(A+) RNA prepared from 12-d-old wheat kernels. Arrow indicates migration of product carrying the 5′ end of the 4.6-kb Sbe1c cDNA. Migration of standard DNA fragments are indicated to the right. C, Gel analysis of RT-PCR products obtained from reactions with PCR primers BE65 and BE38.
The 5′-RACE analysis suggested that several variants of the 4.6-kb Sbe1c transcript were produced in the wheat endosperm. This observation was further confirmed by RT-PCR analysis using the BE65/BE38 primer pair (Fig. 3A) and endosperm RNA. The 2.0-kb RT-PCR products generated from three independent RT-PCR experiments (Fig. 3C, lane BE65+ BE38) were found to be of at least three different variants that differed slightly in deduced amino acid sequence. One of the sequence variants matched exactly to the corresponding sequence on Sbe1c and thus independently confirmed the 2.0-kb 5′ sequence of Sbe1c.
The 4,563-bp SBEI cDNA Encodes a Protein with Two SBEI-Like Domains
DNA sequence analysis of the 4,563-bp Sbe1c cDNA (Fig. 4) revealed an open reading frame of 1,425 codons that was initiated from the 5′ end of the assembled sequence and terminated at nucleotides 4,278 to 4,280. The TAA stop codon was followed by a possible polyadenylation signal sequence, AAT- AAA, located 19-bp upstream of the polyadenylation tail. Initiation of translation was assigned for the first ATG codon (nucleotides 63–65), allowing translation of 1,405 codons of the open reading frame. Sequence analysis of the proposed amino-terminal region of SBEIc revealed a 50% sequence identity to transit peptides predicted from wheat Sbe1a and Sbe1b. Thus, SBEIc appeared, like the 87-kD SBEI, to be imported into plastids. Cleavage of the transit peptide was proposed to occur between amino acids Ala-67 and Ala-68 of the deduced SBEIc sequence (Ile–Ala–Ala↓Ala), as this site showed high resemblance to the consensus sequence for transit peptide cleavage sites, Val/Ile–X–Ala/Cys↓Ala (Gavel and von Heijne, 1990). Processing of the SBEIc precursor would leave a 1,338-amino-acid-long mature protein with a calculated molecular mass of 152 kD.
Figure 4.
Nucleotide sequence and deduced amino acid sequence of the 4.6-kb SBEIc transcript produced in the wheat endosperm. Possible polyadenylation sequence is underlined and proposed transit peptide cleavage site is indicated by a vertical arrow. Shadowed regions represent conserved sequences in enzymes belonging to the α-amylase family (Svensson, 1994). Start of pRN60 sequence and location of PCR primers used in the study are indicated.
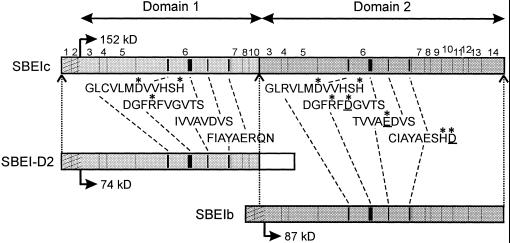
Analysis of the deduced mature SBEIc sequence disclosed the presence of two SBEI-like sequences, domains 1 and 2, encompassing amino acids 1 to 561 and 570 to 1,338, respectively, on the mature SBEIc (Fig. 5). As already mentioned, the sequence of the second domain was identical to that of the mature protein encoded by the pABEI cDNA. The main difference between the first domain and the second domain was the lack of a 21- and a 163-amino-acid-long sequence on domain 1. These two sequences corresponded to exon 9 and exons 11 to 14, respectively, on wheat genomic DNA coding for the 87-kD SBEI (Fig. 5). Further analysis of SBEIc showed that the first domain including the transit peptide was very similar to the first 629 amino acids (92% identical residues) of a 686-amino-acid-long SBEI-like protein, wSBEI-D2, presumed to be produced in the wheat endosperm (Rahman et al., 1997). The proposed translational start codons coincided for wSBEI- D2 and SBEIc cDNA, but no sequence corresponding to the 57-amino-acid long-carboxy-terminal residues of wSBEI-D2 was present on SBEIc.
Figure 5.
Schematic illustration of SBEIc precursor encoded by 4.6-kb Sbe1c transcript. DNA sequences corresponding to exons 1 to 14 on wheat genomic Sbe1 (Båga et al., 1999; Rahman et al., 1999) are indicated. Hatched area indicates location of predicted transit peptide and domains 1 and 2 encompass SBEI-like sequences. The location of the four highly conserved regions on (βα)8 barrels of amylolytic enzymes (Svensson, 1994) are indicated by black boxes, and their sequences are shown below. Highly conserved residues are indicated by asterisks and catalytic residues present only on domain 2 are underlined. SBEIc is aligned with the SBEI-like protein deduced from the wSBEI-D2 cDNA (Rahman et al., 1999) and the wheat 87-kD SBEIb (Repellin et al., 1997).
The first domain of SBEIc and the corresponding sequence on wSBEI-D2 differed from other characterized SBEI from plants at the four highly conserved regions on enzymes belonging to the α-amylase family, which include plant SBE (Svensson, 1994). It was especially notable that the Asp residues on regions 2 and 4 and the Glu residue on region 3, all proposed to be directly involved in hydrolysis of α-1,4 glucan bonds (Svensson, 1994), were replaced by nonequivalent residues (Fig. 5).
Expression of Sbe1c Complements a BE Mutation in E. coli
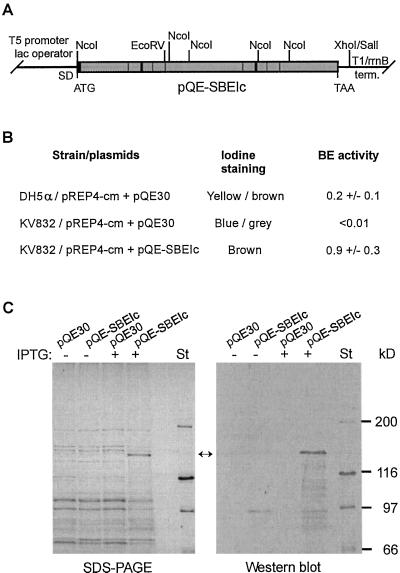
To examine if the isolated cDNA encoded an active enzyme, a prokaryotic expression vector, pQE-SBEIc, encoding a His-tagged mature SBEIc (amino acids 1–1,338) was constructed (Fig. 6A) and tested for activity in a E. coli BE-deficient mutant, KV832 (Kiel et al., 1987). Since high-level expression of the His-tagged SBEIc was found to severely affect cell growth, a construct expressing the Lac repressor (pREP4-cm) was also introduced into the cells to control transcription from the strong T5 promoter. SDS-PAGE and immunoblot analysis of extracts prepared from the transformed KV832 cells confirmed that a polypeptide of expected molecular mass (154 kD) was produced at a very low level in noninduced cells but was clearly seen in cells induced with isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h (Fig. 6C, lane 4). The BE-mutant carrying pREP4-cm and cloning vector pQE30 showed a blue/gray color upon iodine staining, indicating low or no branching of the glucan polymers (Fig. 6B). Expression of pQE-SBEIc in KV832 cells harboring pREP4-cm resulted in a brown color upon iodine staining, showing that the BE mutant had regained the ability to branch glucan molecules. The BE-positive strain, DH5α, transformed with pREP4-cm and pQE30A gave a yellow/brown color upon treatment with iodine, as expected from a strain able to produce glycogen-like polymers. The slight differences in iodine staining patterns of cells producing plant and bacterial BE has been suggested to reflect differences in enzyme specificity (Kossmann et al., 1991). Production of BE activity from cells expressing Sbe1c was confirmed by the phosphorylase a assay (Hawker et al., 1974), which revealed a >90-fold higher level of BE activity in soluble cell extracts of noninduced KV832 cells harboring pQE-SBEIc, as compared with KV832 cells lacking this construct (Fig. 6B). The BE-positive strain, DH5α, produced a 4.5-times lower level of BE activity than the complemented KV832 cells. The BE activity in induced cells expressing Sbe1c was not assessed, since most of the produced SBEIc in these cells was deposited into inclusion bodies (data not shown).
Figure 6.
Expression analysis of Sbe1c in E. coli. A, Schematic illustration of the expression vector pQE-SBEIc carrying sequences encoding mature SBEIc with His tag (black box) added at the amino-terminal end. B, Analysis of BE activity by iodine staining and phosphorylase a stimulation assay. BE activities were determined from the BE-positive strain DH5α and the BE-deficient strain KV832, transformed with plasmids indicated. Construct pREP4-cm expresses the Lac repressor and pQE30 is a cloning vector used for construction of pQE-SBEIc. The BE activity values and ses determined by the phosphorylase a stimulation assay (Hawker et al., 1974) are expressed as μmol Glc-1-P incorporated mg protein−1 min−1 and were determined from three separate experiments. C, SDS-PAGE and immunoblot analysis of recombinant wheat SBEIc produced in E. coli. Total cell extracts of noninduced and IPTG-induced cultures of the BE-deficient strain, KV832, harboring pREP4-cm and plasmid indicated were analyzed. The immunoblot analysis was done with antibodies prepared against wheat 87-kD SBEI. Migration of marker proteins revealed by amido black staining is shown to the right.
The 152-kD SBEI Is Associated with Starch Granules of the Wheat Endosperm
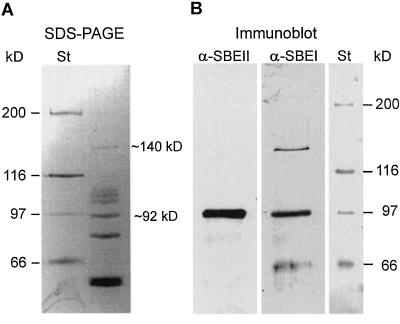
To test if the granule-bound protein of approximately 149 kD reported by Schofield and Greenwell (1987) could correspond to SBEIc, we analyzed starch granule extracts by SDS-PAGE and immunoblotting. Silver-staining of extracted and gel-separated proteins from granules of mature hexaploid wheat kernels resolved seven clearly visible protein bands, of which one band migrated as a 140-kD protein in our gel system (Fig. 7A).
Figure 7.
Immunoblot analysis of starch granule-bound proteins. A, Analysis of starch granule-bound proteins by SDS-PAGE and silver staining. Migration of marker proteins (St) is shown to the left. B, Immunoblot analysis of starch granule-bound proteins using antibodies prepared against wheat 87-kD SBEI and SBEII. Migration of marker proteins revealed by amido black staining is shown to the right.
An immunoblot analysis of the gel-separated proteins using polyclonal antiserum prepared against the wheat 87-kD SBEIb confirmed that the 140-kD protein band was related to SBEI (Fig. 7B, lane α-SBEI). The immunoblot analysis also revealed an interaction with the 92-kD protein band and several 62- to 67-kD protein bands of unknown identities. Since the 140-kD granule-bound protein corresponded reasonably well in mass to SBEIc and no SBEI corresponding in mass with SBEIc was found by immunoblot analysis of the soluble endosperm (data not shown), we reasoned that Sbe1c encoded a granule-bound form of SBEI. This prediction was later confirmed by N-terminal sequencing of the 140-kD protein band obtained from the wheat cv CDC Teal (Peng et al., 2000). Further analysis of the granule-bound proteins using polyclonal antibodies prepared against a 87-kD wheat SBEII revealed only an interaction with the 92-kD protein band (Fig. 7B, lane α-SBEII), as previously reported by Rahman et al. (1995). Thus, isoforms analogous to SBEIc and bound to starch granules did not seem to exist for SBEII in wheat.
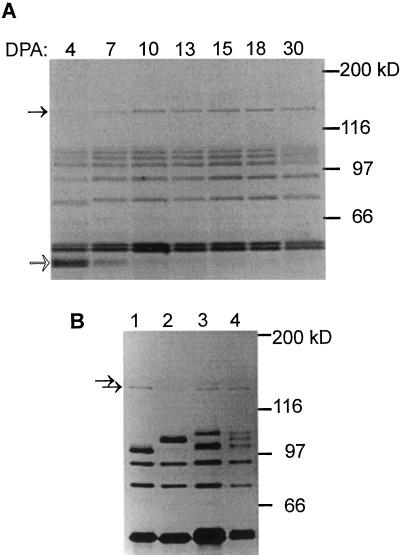
A gel analysis of granule-bound proteins extracted from developing kernels at different stages after anthesis showed no presence of SBEIc in starch prepared from kernels that were less than 5 d old. These young kernel samples contained a substantial amount of pericarp starch, as indicated by the presence of the 59-kD GBSSII (Nakamura et al., 1998; Fig. 8A). SBEIc appeared in total kernel starch between 5 and 7 DPA, and its abundance was relatively constant from there on. Thus, the accumulation of the large isoform of SBEI in the kernel starch coincided relatively well with the accumulation of the 4.6-kb SBEI transcript during kernel maturation (Fig. 2).
Figure 8.
SDS-PAGE analysis of starch granule proteins produced in wheat endosperm. A, Analysis of granule-bound proteins produced in developing endosperm of the hexaploid wheat cv Fielder. Solid arrow indicates migration of SBEIc isoforms and open arrow shows migration of the 59-kD GBSSII present in pericarp starch (Nakamura et al., 1998). B, SDS-PAGE analysis of granule-bound proteins extracted from mature kernels of T. monococcum Tm 23 (lane 1), T. tauschii (accession no. PI 511–380) (lane 2), T. turgidum subsp. durum cv Kyle (lane 3), and T. aestivum cv Fielder (lane 4). Arrows indicate proteins recognized by SBEI antibodies and with similar migration as SBEIc.
One or two proteins corresponding closely in migration with SBEIc were also found associated with starch granules of Triticum monococcum, Triticum tauschii, and Triticum turgidum subsp. Durum (Fig. 8b), and immunoblot analysis confirmed that these proteins were recognized by SBEI antibodies (data not shown). Thus, we concluded that SBEIc isoforms must be encoded by all three of the genomes of hexaploid wheat.
DISCUSSION
In this report, we show that a 140-kD protein band revealed by SDS-PAGE analysis of wheat starch granules represents a novel 152-kD isoform of SBEI in plants. SBEIc encoded by the isolated cDNA differed from previously characterized SBEI isoforms by its high molecular mass and by the presence of two domains of SBEI-like sequences.
The BE activity measured for SBEIc produced in E. coli was likely derived from the second domain, since the first domain lacked the catalytic residues required for BE activity. Nor have we been able to demonstrate any BE activity by expressing the first domain alone in E. coli (data not shown), whereas the second domain is an active enzyme (Repellin et al., 1997). Although the function of the first domain remains unknown, the fact that a closely related sequence exists on another putative wheat endosperm protein, wSBEI-D2, also of unknown function and inactive as BE in E. coli (Rahman et al., 1997), suggests that these SBEI-like polypeptides or protein domains may have a biological role. One possibility is that domain 1 of SBEIc, despite its apparent lack of catalytic BE residues, is able to interact with glucan polymers or other granule-bound proteins. This interaction with components of the starch biosynthetic pathway could differ from that of the 87- to 88-kD SBEI. Some support for this hypothesis is that the 152-kD SBEI is only found associated with the starch granules, whereas the 87- to 88-kD SBEI, which are almost identical to domain 2 on SBEIc, are preferentially located in the soluble fraction of the wheat endosperm (Morell et al., 1997). If domain 1 has any influence of the BE specificity exerted by domain 2, it could affect the structure and properties of synthesized amylopectin.
Domain 1 differs from domain 2 by the lack of a 21-amino-acid-long peptide and a 163-residue-long (approximately 17 kD) C-terminal sequence (Fig. 5). The 21-amino acid segment is part of a loop structure located between the eighth β-strand and the eighth 32 α-helix of the barrel. This loop differs in length between SBEI and SBEII isoforms (Burton et al., 1995) and has been suggested to determine spacing between the branches (Jespersen et al., 1993). The lack of the 163-amino acid segment may not impair the ability of domain 1 to bind glucan polymers, since removal of a similar approximately 20-kD C-terminal peptide from a 103-kD potato SBEI did not alter its activity (Khoshnoodi et al., 1996). However, how the missing segments on domain 1 may affect the interactions between domain 1 and glucan polymers or starch biosynthetic enzymes cannot be predicted until further investigations have been done.
Although it remains to be shown that SBEIc is a bifunctional protein, it would be of interest to determine how this type of protein has evolved. Inspection of the Sbe1c sequence would suggest that the sequence of the first domain derived from a wSBEI-D2-like gene was fused to Sbe1 gene at the junction of exons 2 and 3 (Fig. 5). It is also possible that Sbe1c is a product of a trans-splicing event between a wSBEI-D2-like mRNA and a 2.8-kb Sbe1 transcript. The development of this type of hybrid gene or transcript may have been facilitated by the close proximity of the wSBEI-D2-like and SBEI genes on chromosome 7 (Rahman et al., 1997).
Previous studies of SBE in wheat endosperm have localized SBEI isoforms only to the soluble phase of the granule (Morell et al., 1997). However, SBEII isoforms are found in both the soluble phase and in the starch granules. Of the two types of SBEII in wheat, SBEIIa and SBEIIb, only the latter appears to be granule-bound (Takaoka et al., 1997). Our study showed that the 152-kD SBEIc represents a granule-bound form of SBEI. This could also be true for a protein comigrating with SBEII on SDS-PAGE gels, but we cannot at this point exclude the possibility that it is a breakdown product of the 152-kD SBEIc. Genes encoding SBEI and SBEII isoforms in pea, maize, and wheat endosperm are differentially expressed with SBEII transcripts appearing earlier than SBEI transcripts (Burton et al., 1995; Gao et al., 1996; R.B. Nair, unpublished data). A similar difference in expression patterns for the granule-bound 152-kD SBEI and the soluble 87- to 88-kD SBEI was observed in our study (Fig. 2). The appearance of the 5-kb Sbe1 earlier than the 2.8-kb Sbe1 mRNA may be of significance for starch granule formation during endosperm development in wheat.
The analysis of SBEI transcripts produced in the developing wheat endosperm of the cv Fielder suggested that there are at least three different forms of SBEIc transcripts produced. These variants would encode proteins of very similar molecular masses (<1-kD difference), and thus, cannot be distinguished as separate bands on one-dimensional SDS-PAGE gels. Our analysis of starch granules of Triticum sp. suggested that variants of SBEIc also exist in both the diploid (T. monococcum, T. tauschii) and tetraploid (T. turgidum subsp. durum) wheat (Fig. 8B). For the tetraploid wheat cv Kyle, two separate protein bands were distinguished, and apparently, the difference between the SBEIc isoforms in this cultivar are more distinguishable on SDS-PAGE gels than those of the hexaploid wheat cv Fielder. A protein corresponding in migration with SBEIc is also present in starch granules of Triticale and rye (Schofield and Greenwell, 1987). The starch granules produced in Triticale, rye, and wheat show a bimodal granule size distribution and in that aspect differ from most other plants (Ellis et al., 1998). This apparent association between the presence of SBEIc isoforms and production of two size classes of starch granules is studied in an accompanied paper by Peng et al. (2000).
MATERIALS AND METHODS
Plant Materials
Wheat (Triticum aestivum L. cv Fielder) was grown in a greenhouse under optimal conditions as previously described (Nair et al., 1997). Developing kernels were harvested at various stages after anthesis, frozen in liquid nitrogen, and stored at −70°C until needed.
Screening of a Wheat cDNA Library
Approximately 200,000 plaques of a cDNA library, constructed from wheat poly(A+) RNA isolated from 12-d-old wheat kernels (Nair et al., 1997), were screened for Sbe1 clones by plaque hybridization (Sambrook et al., 1989). Probe 1 used in the library screening consisted of an 828-bp RT-PCR product, obtained from a reaction using 12-d-old wheat kernel RNA and the Sbe1-specific primers BE11 and BE12 (Figs. 1 and 4). Ten of the positive clones were plaque-purified, and their inserts were excised in vivo from the Uni-ZAP XR vector (Stratagene, La Jolla, CA). The clone with the longest insert was denoted pRN60 and chosen for further characterization.
DNA Sequence Analysis
Templates for sequencing were prepared by subcloning DNA fragments into the pBluescript II SK+ vector (Stratagene). DNA sequencing reactions were performed by the dye terminator cycle sequencing technique and analyzed on an automated DNA sequencer (Applied Biosystems, Foster City, CA). All of the reported sequences were determined on both of the strands and from overlapping templates. Nucleotide sequences were assembled and analyzed using the Lasergene software (DNASTAR Inc., Madison, WI). Pair-wise alignments of DNA and protein sequences were calculated by the Clustal method using a K-tuple value of 1, gap penalty value of 3, and window size of 5.
Isolation of RNA and RNA Gel-Blot Analysis
Total RNA was isolated from 12-d-old wheat kernels using a hot-phenol method as described (Båga et al., 1995). RNA gel-blot analysis was performed with 20 μg of total RNA fractionated on a 1% (w/v) agarose-2.2 m formaldehyde gel, transferred to a nylon membrane (Hybond N+, Amersham, Buckinghamshire, UK), hybridized with probe 2 (nucleotides 1,993–4,209 of Sbe1c; Fig. 1), and washed as described (Nair et al., 1997). To assure that about the same amount of RNA was loaded onto each lane, the hybridized blot was stripped and rehybridized with a 25S ribosomal DNA probe as described (Nair et al., 1997). Probes were radiolabeled using the Rediprime random primer labeling kit (Amersham).
5′-RACE
5′-RACE was performed with poly(A+) RNA extracted from 12-d-old wheat endosperm following the protocol supplied with the Marathon cDNA Amplification Kit (CLONTECH Laboratories, Palo Alto, CA). The first strand synthesis was primed with the Sbe1-specific BE19 primer (Figs. 3 and 4). After synthesis of the second strand, the double-stranded cDNA was ligated to the Marathon cDNA Adaptor (CLONTECH Laboratories), followed by a first-round PCR amplification performed with the adaptor primer AP1 (5′-CCATCCTAATACGACTCACTATAGGGC-3′; CLONTECH Laboratories) and the Sbe1-specific primer BE25 (Figs. 3 and 4). The reaction was initiated by a denaturation step at 94°C for 3 min followed, by 25 cycles of 94°C for 30 s, 62°C for 20 s, and 68°C for 3 min and a final 10-min extension at 68°C. Products derived from the 4.8-kb Sbe1 transcripts were separated from shorter products derived from the 2.8-kb Sbe1 mRNA by agarose gel electrophoresis. Products of 1.9 to 2.7 kb were gel-purified and used as a template in a nested amplification using nested adaptor primer AP2 (5′-ACTCACTATAGGGCTCGAGCGGC-3′; CLONTECH Laboratories) and the gene-specific primer BE39 (Figs. 3 and 4). The amplification conditions were 94°C for 3 min, 30 cycles of 94°C for 30 s, 65°C for 20 s, and 68°C for 3 min, followed by a final extension at 68°C for 10 min. Amplified fragments were separated by agarose gel electrophoresis, isolated, cloned, and analyzed by DNA sequencing.
RT-PCR
First strand cDNA, used as a template in the RT-PCR reactions, was synthesized from 1.0 μg of total RNA isolated from 12-d-old wheat endosperm. The RNA was primed with oligo(dT)12–18 and reverse-transcribed in a total volume of 20 μL using Superscript II (Life Technologies/Gibco-BRL, Cleveland). PCR reactions (25 μL) were performed with a 0.5-μL aliquot of the first-strand cDNA using the Long Expand Template PCR System (Roche Diagnostics Gmbh, Mannheim, Germany) and the primer pair BE65/BE38 (Figs. 3 and 4). Reactions were initiated by a denaturation step at 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 65°C for 20 s, 68°C for 2 min 30 s, and a final 10-min extension at 68°C. Amplified fragments were fractionated by agarose gel electrophoresis, isolated, cloned, and analyzed by DNA sequencing.
Construction of Expression Vectors
Assembly of the pQE-SBEIc plasmid (Fig. 6A) was initiated by PCR amplification (30 cycles of 94°C, 65°C for 20 s, 68°C for 2 min 30 s) of Sbe1c nucleotides 265 to 1,879, using the BE63/BE39 primer pair (Fig. 4). This reaction introduced a NcoI recognition site at the start of the sequence encoding the mature SBEIc. Thereafter, the NcoI-NcoI fragment carrying Sbe1c nucleotides 265 to 1,732 was isolated from the amplified product, filled-in, and inserted into a filled-in BamHI site of the His-tag expression vector pQE30 (Qiagen USA, Valencia, CA). Construction of pQE-SBEIc was completed by insertion of a 2.2-kb EcoRV-XhoI fragment (Sbe1c nucleotides 1,623–4,563 with XhoI site added at the end) into the EcoRV and SalI sites.
Construct pREP4-cm, encoding the Lac repressor, was derived from pREP4 (Qiagen USA) by replacing the NPTII gene carried on a ClaI-SmaI fragment with the chloroamphenicol resistance gene isolated as a PvuII-BstBI fragment from the pACYC184 vector.
Construction of pKKABEI, encoding the mature 87-kD wheat SBEI, was initiated by inserting nucleotides 221 to 923 (NcoI-KpnI fragment) of pABEI cDNA (Repellin et al., 1997) into NcoI-KpnI sites of the bacterial expression vector pKK388–1 (CLONTECH Laboratories). Then nucleotides 923 to 2,729, isolated as a KpnI fragment, were introduced to give pKKABEI. The SBEII expression vector, pQRN33, encoding the mature wheat SBEII was obtained by two cloning steps. First the pRN33 (Nair et al., 1997) nucleotides 317 to 1,442 carried by a HaeIII fragment were inserted into a filled-in BamHI site of the His-tag expression vector pQE31 (Qiagen USA). The resulting construct was restricted with KpnI and SmaI, followed by introduction of nucleotides 1,245 to 2,632 located on a KpnI-PvuII fragment, to give pQRN33.
Analysis of BE Activity Produced in Escherichia coli
The BE-deficient E. coli strain KV832 (Kiel et al., 1987) carrying pREP4-cm was transformed with pQE-SBEIc or the cloning vector pQE30. Plasmids pREP4-cm and pQE30 were also introduced into the BE-positive E. coli strain DH5α. The bacterial cultures were grown at 37°C in liquid YT medium (Sambrook et al., 1989) containing 1.0% (w/v) Glc, 100 μg/mL carbenicillin, and 25 μg/mL chloramphenicol, to an A600 = 0.6, and induced for 2 h by addition of IPTG to 1 mm final concentration. Production of SBEIc was verified by SDS-PAGE-gel analysis of cell lysates prepared from noninduced and induced cultures.
Visualization of BE activity in bacterial cells grown on solid media was done by iodine staining of colonies as described (Kossmann et al., 1991). The BE activity levels in cells from noninduced cultures was determined by the phosphorylase a stimulation assay (Hawker et al., 1974) performed at 30°C for 30 min using 2 and 5 μg of soluble protein extract. The cell extracts were prepared from cells of 1 mL of culture that were lysed by sonication in 0.25 mL of extraction buffer (50 mm Tris [tris(hydroxymethyl)aminomethane]-HCl, pH 7.5, 2 mm EDTA, 5 mm dithiothreitol, and 1 mm phenylmethylsulfonyl fluoride) and centrifuged at 15,000g for 20 min. Determination of protein concentration in the soluble extracts was done using the dye-binding assay (Bio-Rad Laboratories).
Isolation of Starch Granule Proteins and SDS-PAGE Analysis
Starch granules were extracted from mature and developing wheat kernels according to procedure described by Zhao and Sharp (1996) with the exception of the steeping step, which was only done with mature seeds. Extracted starch (10 mg) was resuspended in 150 μL of sample buffer (62.5 mm Tris-HCl, pH 8.0, 10% [w/v] SDS, 10% [v/v] glycerol, 5% [v/v] β-mercaptoethanol, and 0.005% [w/v] bromphenol blue), boiled for 7 min, cooled on ice for 5 min, and centrifuged at 15,000g for 20 min. Extracted proteins (40 μL) were loaded on a 10% SDS-PAGE gel (30:0.135 [w/w] acrylamide:bis-acrylamide) and subjected to electrophoresis. Separated proteins were visualized by silver staining.
Large Scale Production of Wheat SBE in E. coli
A culture of KV832 cells transformed with pKKABEI was grown at 37°C in Luria-Bertani medium containing 100 μg mL−1 ampicillin. At A600nm = 0.6, IPTG was added to a final concentration of 0.5 mm, and the culture was grown at 25°C for 14 h. Cells were harvested by centrifugation and SBEI was purified according to Guan et al. (1994). The final protein extract was loaded onto a 10% (w/v) preparative SDS-PAGE gel, and the 87-kD SBEI band was isolated by electroelution (model 422 Electro-eluter, Bio-Rad Laboratories). The protein eluate was concentrated using a Centriplus 30 column (Amicon, Beverly, MA) before immunization.
The SBEII expression vector, pQRN33, was introduced into the E. coli strain, M15, carrying pREP4 and grown at 22°C in medium containing 25 g/L tryptone, 15 g/L yeast extract, 5 g/L NaCl, 1% (v/v) Glc, 100 μg/mL ampicillin, and 25 μg/mL kanamycin. Cells were grown to A600nm = 0.7, IPTG was added to give a 1-mm final concentration, and the cells were grown for an additional 14 h. Harvested cells were lysed under denaturing conditions and the His-tagged SBEI was purified using the QIAexpress purification system (Qiagen USA). The guanidine hydrochloride denaturation buffer, column washing buffers, and elution buffer were all supplemented with 10 mm β-mercaptoethanol and 0.25% (v/v) Tween 20. The homogeneity of the column fractions used for immunization was verified by SDS-PAGE.
Preparation of SBEI and SBEII Antibodies
Approximately 100 μg of purified SBEI, or 250 μg of His-tagged SBEII in 500 μL of phosphate-buffered saline, was emulsified with an equal volume of Freund's complete adjuvant (DIFCO Laboratories, Detroit) and injected intradermally into cereal-starved rabbits. The injection was repeated twice at 2-week intervals using approximately 50 μg of antigen and an equal volume of Freund's incomplete adjuvant (DIFCO Laboratories). The antiserum was collected 2 weeks after the final injection.
Immunoblotting
Starch granule proteins separated by SDS-PAGE were transferred by vertical electroblotting (Sambrook et al., 1989) onto Immobilon nitrocellulose membranes (Millipore, Bedford, MA) at 1.4 V/cm for 2.5 h using buffer 3 described by Bolt and Mahoney (1997). The filters were blocked for 2 h in blocking buffer (5% [w/v] non-fat dry milk, 0.1% [v/v] Tween 20 in phosphate-buffered saline [Sambrook et al., 1989]) and subsequently incubated for 1 h with primary antibodies in blocking buffer (1:1,000 dilution). Blots were washed for 1 h in blocking buffer, followed by incubation with alkaline phosphatase-conjugated goat anti-rabbit antibodies (Stratagene) in blocking buffer (1:5,000 dilution). Thereafter, the membranes were washed with blocking buffer for 1 h and with 50 mm Tris-HCl, pH 7.5, 150 mm NaCl for 45 min. Immunoreactive bands were revealed by chemical staining with 5-bromo-4-chloro-3-indolyl phosphate and 4-nitroblue tetrazolium (Stratagene) following the supplier's instructions.
ACKNOWLEDGMENTS
We thank Drs. Patrick Covello and Pierre Fobert for their comments on the manuscript. Dr. B. Gill (Kansas State University) is acknowledged for providing T. tauschii seeds.
Footnotes
This work was supported by the National Research Council of Canada (NRCC no. 43786).
LITERATURE CITED
- Båga M, Chibbar RN, Kartha KK. Molecular cloning and expression analysis of peroxidase genes from wheat. Plant Mol Biol. 1995;29:647–662. doi: 10.1007/BF00041156. [DOI] [PubMed] [Google Scholar]
- Båga M, Glaze S, Mallard CS, Chibbar RN. A starch branching enzyme gene in wheat produces alternatively spliced transcripts. Plant Mol Biol. 1999;40:1019–1030. doi: 10.1023/a:1006286807176. [DOI] [PubMed] [Google Scholar]
- Ball S, Guan H-P, James M, Myers A, Keeling P, Mouille G, Buléon A, Colonna P, Preiss J. From glycogen to amylopectin: a model for the biogenesis of the plant starch granule. Cell. 1996;86:349–352. doi: 10.1016/s0092-8674(00)80107-5. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya MK, Smith AM, Ellis THN, Hedley C, Martin C. The wrinkled-seed character of pea described by Mendel is caused by a transposon-like insertion in a gene encoding starch-branching enzyme. Cell. 1990;60:115–122. doi: 10.1016/0092-8674(90)90721-p. [DOI] [PubMed] [Google Scholar]
- Bolt MW, Mahoney PA. High-efficiency blotting of proteins of diverse sizes following sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Anal Biochem. 1997;247:185–192. doi: 10.1006/abio.1997.2061. [DOI] [PubMed] [Google Scholar]
- Burton RA, Bewley JD, Smith AM, Bhattacharyya MK, Tatge H, Ring S, Bull V, Hamilton WDO, Martin C. Starch branching enzymes belonging to distinct enzyme families are differentially expressed during pea embryo development. Plant J. 1995;7:3–15. doi: 10.1046/j.1365-313x.1995.07010003.x. [DOI] [PubMed] [Google Scholar]
- Denyer K, Hylton CM, Jenner CF, Smith AM. Identification of multiple isoforms of soluble and granule-bound starch synthase in developing wheat endosperm. Planta. 1995;196:256–265. [Google Scholar]
- Echt CS, Schwartz D. Evidence for the inclusion of controlling elements within the structural gene at the waxylocus in maize. Genetics. 1981;99:275–284. doi: 10.1093/genetics/99.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RP, Cochrane MP, Dale MFB, Duffus CM, Lynn A, Morrison IM, Prentice RDM, Swanston JS, Tiller SA. Starch production and industrial use. J Sci Food Agric. 1998;77:289–311. [Google Scholar]
- Flipse E, Suurs L, Keetels CJAM, Kossmann J, Jacobsen E, Visser RGF. Introduction of sense and antisense cDNA for branching enzyme in the amylose-free potato mutant leads to physico-chemical changes in the starch. Planta. 1996;198:340–347. [Google Scholar]
- Gao M, Fisher DK, Kim K-N, Shannon JC, Guiltinan MJ. Evolutionary conservation and expression patterns of maize starch branching enzyme I and IIb genes suggests isoform specialization. Plant Mol Biol. 1996;30:1223–1232. doi: 10.1007/BF00019554. [DOI] [PubMed] [Google Scholar]
- Gavel Y, von Heijne G. A conserved cleavage-site motif in chloroplast transit peptides. FEBS Lett. 1990;261:455–458. doi: 10.1016/0014-5793(90)80614-o. [DOI] [PubMed] [Google Scholar]
- Guan HP, Baba T, Preiss J. Expression of branching enzyme I of maize endosperm in Escherichia coli. Plant Physiol. 1994;104:1449–1453. doi: 10.1104/pp.104.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan HP, Preiss J. Differentiation of the properties of the branching isozymes from maize (Zea mays) Plant Physiol. 1993;102:1269–1273. doi: 10.1104/pp.102.4.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawker JS, Ozbun JL, Ozaki H, Greenberg E, Preiss J. Interaction of spinach leaf adenosine diphosphate glucose α-1,4-glucan α-4-glucosyl transferase and α-1,4-glucan, α-1,4-glucan-6-glycosyl transferase in synthesis of branched α-glucan. Arch Biochem Biophys. 1974;160:530–551. doi: 10.1016/0003-9861(74)90430-5. [DOI] [PubMed] [Google Scholar]
- Jespersen HM, MacGregor EA, Henrissat B, Sierks MR, Svensson B. Starch- and glycogen-debranching and branching enzymes: prediction of structural features of the catalytic (βα)8-barrel domain and evolutionary relationship to other amylolytic enzymes. J Protein Chem. 1993;12:791–805. doi: 10.1007/BF01024938. [DOI] [PubMed] [Google Scholar]
- Jobling SA, Schwall GP, Westcott RJ, Sidebottom CM, Debet M, Gidley MJ, Jeffcoat R, Safford R. A minor form of starch branching enzyme in potato (Solanum tuberosum L.) tubers has a major effect on starch structure: cloning and characterization of multiple forms of SBEA. Plant J. 1999;18:163–171. doi: 10.1046/j.1365-313x.1999.00441.x. [DOI] [PubMed] [Google Scholar]
- Khoshnoodi J, Blennow A, Ek B, Rask L, Larsson H. The multiple forms of starch-branching enzyme I in Solanum tuberosum. Eur J Biochem. 1996;242:148–155. doi: 10.1111/j.1432-1033.1996.0148r.x. [DOI] [PubMed] [Google Scholar]
- Kiel JAKW, Vossen JPMJ, Venema G. A general method for the construction of Escherichia colimutants by homologous recombination and plasmid segregation. Mol Gen Genet. 1987;207:294–301. doi: 10.1007/BF00331592. [DOI] [PubMed] [Google Scholar]
- Kossmann J, Visser RGF, Müller-Röber B, Willmitzer L, Sonnewald U. Cloning and expression analysis of a potato cDNA that encodes branching enzyme: evidence for co-expression of starch biosynthetic genes. Mol Gen Genet. 1991;230:39–44. doi: 10.1007/BF00290648. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Kawasaki T, Shimada H, Satoh H, Kobayashi E, Okumura S, Arai Y, Baba T. Alteration of the structural properties of starch components by the lack of an isoform of starch branching enzyme in rice seeds. J Biol Chem. 1993;268:19084–19091. [PubMed] [Google Scholar]
- Morell MK, Blennow A, Kosar-Hashemi B, Samuel MS. Differential expression and properties of starch branching enzyme isoforms in developing wheat endosperm. Plant Physiol. 1997;113:201–208. doi: 10.1104/pp.113.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine T, Yoshida H, Komae K. Varietal differences and chromosome locations of multiple isoforms of starch branching enzyme in wheat endosperm. Phytochemistry. 1997;46:23–26. [Google Scholar]
- Nair RB, Båga M, Scoles GJ, Kartha KK, Chibbar RN. Isolation, characterization and expression analysis of a starch branching enzyme II cDNA from wheat. Plant Sci. 1997;122:153–163. [Google Scholar]
- Nakamura T, Vrinten P, Hayakawa K, Ikeda J. Characterization of a granule-bound starch synthase isoform found in the pericarp of wheat. Plant Physiol. 1998;118:451–459. doi: 10.1104/pp.118.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Gao M, Båga M, Hucl P, Chibbar RN. Starch-branching enzymes preferentially associated with A-type starch granules in wheat endosperm. Plant Physiol. 2000;124:265–272. doi: 10.1104/pp.124.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss J. Biology and molecular biology of starch synthesis and its regulation. In: Miflin BJ, editor. Oxford Surveys of Plant Molecular and Cell Biology. Vol. 7. Oxford: Oxford University Press; 1990. pp. 181–254. [Google Scholar]
- Rahman S, Abrahams S, Abbott D, Mukai Y, Samuel M, Morell M, Appels R. A complex arrangement of genes at a starch branching enzyme I locus in the D-genome donor of wheat. Genome. 1997;40:465–474. doi: 10.1139/g97-062. [DOI] [PubMed] [Google Scholar]
- Rahman S, Kosar-Hashemi B, Samuel MS, Hill A, Abbott DC, Skeritt JH, Preiss J, Appels R, Morell MK. The major proteins of wheat endosperm starch granules. Aust J Plant Physiol. 1995;22:793–803. [Google Scholar]
- Rahman S, Li Z, Abrahams S, Abbott D, Appels R, Morell MK. Characterization of a gene encoding wheat endosperm starch branching enzyme-I. Theor Appl Genet. 1999;98:156–163. [Google Scholar]
- Repellin A, Nair RB, Båga M, Chibbar RN. Isolation of a starch branching enzyme I cDNA from a wheat endosperm library (accession no. Y12320) (PGR 97-094) Plant Physiol. 1997;114:1135. [Google Scholar]
- Safford R, Jobling SA, Sidebottom CM, Westcott RJ, Cooke D, Tober KJ, Strongitharm BH, Russell AL, Gidley MJ. Consequences of antisense RNA inhibition of starch branching enzyme activity on properties of potato starch. Carbohydr Polym. 1998;35:155–168. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schofield JD, Greenwell P. Wheat starch granule proteins and their technological significance. In: Morton ID, editor. Cereals in a European Context. Chichester, UK: Ellis Horwood; 1987. pp. 407–420. [Google Scholar]
- Smith AM. Major differences in isoforms of starch-branching enzyme between developing embryos of round- and wrinkled-seeded peas (Pisum sativumL.) Planta. 1988;175:270–279. doi: 10.1007/BF00392437. [DOI] [PubMed] [Google Scholar]
- Svensson B. Protein engineering in the α-amylase family: catalytic mechanism, substrate specificity, and stability. Plant Mol Biol. 1994;25:141–157. doi: 10.1007/BF00023233. [DOI] [PubMed] [Google Scholar]
- Takaoka M, Watanabe S, Sassa H, Yamamori M, Nakamura T, Sasakuma T, Hirano H. Structural characterization of high molecular weight starch granule-bound proteins in wheat (Triticum aestivumL.) J Agric Food Chem. 1997;45:2929–2934. [Google Scholar]
- Takeda Y, Guan H-P, Preiss J. Branching of amylose by the branching isoenzymes of maize endosperm. Carbohydr Res. 1993a;240:253–263. [Google Scholar]
- Takeda C, Takeda Y, Hizukuri S. Structure of the amylopectin fraction of amylomaize. Carbohydr Res. 1993b;246:273–281. [Google Scholar]
- Tomlinson KL, Lloyd JR, Smith AM. Importance of isoforms of starch-branching enzyme in determining the structure of starch in pea leaves. Plant J. 1997;11:31–43. [Google Scholar]
- Yamamori M, Endo TR. Variation of starch granule proteins and chromosome mapping of their coding genes in common wheat. Theor Appl Genet. 1996;93:275–281. doi: 10.1007/BF00225757. [DOI] [PubMed] [Google Scholar]
- Zhao XC, Sharp PJ. An improved 1-D SDS-PAGE method for the identification of three bread wheat ‘waxy’ proteins. J Cereal Sci. 1996;23:191–193. [Google Scholar]