Abstract
Background
Dysbiosis of the oral microbiome can lead to local oral disease and potentially to cancers of the head, neck, and digestive tract. However, little is known regarding exogenous factors contributing to such microbial imbalance.
Results
We examined the impact of alcohol consumption on the oral microbiome in a cross-sectional study of 1044 US adults. Bacterial 16S rRNA genes from oral wash samples were amplified, sequenced, and assigned to bacterial taxa. We tested the association of alcohol drinking level (non-drinker, moderate drinker, or heavy drinker) and type (liquor, beer, or wine) with overall microbial composition and individual taxon abundance. The diversity of oral microbiota and overall bacterial profiles differed between heavy drinkers and non-drinkers (α-diversity richness p = 0.0059 and β-diversity unweighted UniFrac p = 0.0036), and abundance of commensal order Lactobacillales tends to be decreased with higher alcohol consumption (fold changes = 0.89 and 0.94 for heavy and moderate drinkers, p trend = 0.005 [q = 0.064]). Additionally, certain genera were enriched in subjects with higher alcohol consumption, including Actinomyces, Leptotrichia, Cardiobacterium, and Neisseria; some of these genera contain oral pathogens, while Neisseria can synthesize the human carcinogen acetaldehyde from ethanol. Wine drinkers may differ from non-drinkers in microbial diversity and profiles (α-diversity richness p = 0.048 and β-diversity unweighted UniFrac p = 0.059) after controlling for drinking amount, while liquor and beer drinkers did not. All significant differences between drinkers and non-drinkers remained after exclusion of current smokers.
Conclusions
Our results, from a large human study of alcohol consumption and the oral microbiome, indicate that alcohol consumption, and heavy drinking in particular, may influence the oral microbiome composition. These findings may have implications for better understanding the potential role that oral bacteria play in alcohol-related diseases.
Electronic supplementary material
The online version of this article (10.1186/s40168-018-0448-x) contains supplementary material, which is available to authorized users.
Keywords: Oral microbiome, 16s rRNA genes, Alcohol consumption, Large population-based study
Background
More than 700 different bacterial species and a range of other microorganisms (archaea, fungi, and viruses) colonize the human oral cavity, known collectively as the oral microbiome [1, 2]. Oral microbiota play important roles in human health, including in immune response, carcinogen metabolism, and nutrient digestion [3–5]. Evidence indicates that oral microbiota dysbiosis is related to local oral diseases, such as periodontitis and dental caries [6] and potentially to systemic diseases, including gastrointestinal cancers [7, 8] and cardiovascular disease [9]. However, little is known regarding exogenous exposures that cause dysbiosis of the oral microbiota.
We hypothesized that alcohol drinking habits are associated with changes in the oral microbial community. Alcohol intake may impact the oral microbiota in several ways: by direct cytotoxic effects on bacteria [10], by disturbing saliva-bacterium interactions [11–14], and by providing ethanol as a substrate for bacterial metabolism [4]. Several previous studies, in both animals and humans, have also observed effects of alcohol consumption on oral bacteria. Animal studies showed that diet containing 20% ethanol increases colonization by Streptococcus mutans [15], a caries-related bacterium, and dramatically decreases the number of detectable bacterial species in the oral biofilms of rats [16]. Similarly, the association of excessive drinking (> 3 drinks per day) with poor oral health was observed in a population study [17]. In small-scale human studies, drinking at least one glass of red wine per day was associated with reduced species richness and reduction of certain anaerobic bacteria in sub- and supra-gingival plaque [18], while excessive co-use of tobacco and alcohol was associated with reduced species richness and decreased abundance of Neisseria, Aggregatibacter, and Fusobacteria, in oral mucosa biofilms [19]. Aside from direct effects, alcohol may indirectly impact the oral microbiota through disturbing the host defense system [20–23], subsequently resulting in host-mediated periodontitis [24, 25]. Large population-based studies have consistently demonstrated that alcohol consumption is associated with increased risk of periodontal disease in a dose-dependent fashion [24, 26]. Evidence shows that the oral microbiome is closely tied to oral health status [27, 28]. Despite this evidence suggesting an impact of alcohol on the oral microbiome, no study has comprehensively investigated the relationship of the oral microbiome to alcohol drinking habits in terms of drinking amount and types of alcoholic beverages consumed.
We tested the relationship of level and types of alcoholic beverages, with the oral microbiome in 1044 individuals from two large US national cohorts. The oral microbiome was characterized by bacterial 16S rRNA gene sequencing. Comparisons of overall community composition and taxon abundance were conducted across groups with respect to drinking status.
Methods
Study population
Participants were drawn from the American Cancer Society (ACS) Cancer Prevention Study II (CPS II) Nutrition cohort [29] and the National Cancer Institute (NCI) Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO) cohort [30]. As previously described [31], subjects included in the present analyses were originally selected from the CPS II and PLCO cohorts as cases or controls for collaborative nested case-control studies of the oral microbiome in relation to head and neck cancer and pancreatic cancer. Oral wash samples were collected by mail from 70,004 CPS II Nutrition cohort participants between 2000 and 2002 and in the PLCO control arm (n = 77,445) at recruitment from 1993 to 2001. After excluding participants without information on alcohol consumption status, 458 participants from CPS II (n = 169 from the head and neck study and n = 289 from the pancreas study) and 586 participants from PLCO (n = 231 from the head and neck study and n = 355 from the pancreas study) were included in the current study. All participants provided informed consent and all protocols were approved by the New York University School of Medicine Institutional Review Board.
Alcohol consumption and covariate assessment
Comprehensive demographic and lifestyle information was collected by baseline and follow-up questionnaires in the PLCO and CPS II Nutrition cohorts. Detailed information on alcohol consumption over the past year, including frequency of consumption, serving size, and type of alcoholic beverages consumed (wine, beer, and liquor), was ascertained via questionnaires that were most close in time to oral wash sample collection for both cohorts. The 137-item Food Frequency Questionnaire (FFQ) and Diet History Questionnaire (DHQ) were used in PLCO and CPS II Nutrition cohorts, respectively [32, 33]. According to the drinking level definition of the Dietary Guidelines for Americans 2010 [34], we defined moderate drinkers as > 0 but ≤ 1 drinks per day, on average, for women, and > 0 but ≤ 2 drinks per day, on average, for men. Women and men who had greater than one or two drinks per day, respectively, were considered heavy drinkers. This definition considers the gender differences in blood concentration of ethanol after drinking [35], which is equal to salivary ethanol for up to 5 h after consumption [36]. Drinkers exclusively consuming wine, beer, or liquor were defined as wine drinkers, beer drinkers, and liquor drinkers, respectively.
Oral microbiota characterization using 16S rRNA gene amplification and sequencing
Participants in both cohorts were asked to swish vigorously with 10 mL Scope mouthwash (P&G) for 30 s and then to expectorate into a specimen tube. Samples were shipped to each cohort’s biorepository, pelleted, and stored at − 80 °C until use [30, 37]. Bacterial genomic DNA was extracted from the samples using the MoBio PowerSoil DNA Isolation Kit (Carlsbad, CA), with the bead-beating method in the MoBio Powerlyzer instrument. As reported previously [38], 16S rRNA gene sequencing on the extracted DNA was performed. 16S rRNA amplicon libraries were generated using primers incorporating FLX Titanium adapters and a sample barcode sequence, allowing unidirectional sequencing covering variable regions V3 to V4 (Primers: 347F- 5′GGAGGCAGCAGTAAGGAAT-3′ and 803R- 5′CTACCGGGGTATCTAATCC-3′). Five nanograms of genomic DNA was used as the template in 25 uL PCR reaction buffer for 16S rRNA amplicon preparation. Cycling conditions were one cycle of 94 °C for 3 min, followed by 25 cycles of 94 °C for 15 s, 52 °C for 45 s, and 72 °C for 1 min followed by a final extension of 72 °C for 8 min. The generated amplicons were then purified using Agencourt AMPure XP kit (Beckman Coulter, CA). Purified amplicons were quantified by fluorometry using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, CA). Equimolar amounts (107 molecules/uL) of purified amplicons were pooled for sequencing. Pyrosequencing (Roche 454 GS FLX Titanium) was carried out according to the manufacturer’s instructions [39].
Upstream sequence analysis of microbiome data
16S rRNA gene amplicon sequences were processed and analyzed using the QIIME pipeline [40]. Multiplexed libraries were deconvoluted based on the barcodes assigned to each sample. Poor-quality sequences were excluded using the default parameters of the QIIME script split_libraries.py (minimum average quality score = 25, minimum/maximum sequence length = 200/1000 base pairs, no ambiguous base calls, and no mismatches allowed in the primer sequence). From 1044 pre-diagnostic oral wash samples, we obtained 11,395,395 high-quality 16S rRNA gene sequence reads (mean 10,915 [SD = 3049] sequences per sample), with similar number of reads in all cohorts [31]. Filtered sequence reads were clustered into operational taxonomic units (OTUs) and subsequently assigned to taxa by using the Human Oral Microbiome Database (HOMD) pre-defined taxonomy map of reference sequences with ≥ 97% identity [41]. Summary of sequence reads per sample which were assigned to HOMD reference is shown in Additional file 1: Table S1.
Quality control
Blinded positive quality control (QC) specimens were used across all sequencing batches. We previously reported good agreement of microbiome parameters in replicates from these QC subjects [31]. Negative control samples (with Scope mouthwash only) were used to detect possible bacteria in Scope and reagent, as well as environment contamination in all sequencing batches. No DNA was detected from the negative control samples. We further conducted an independent experiment which compared the oral microbiota in paired mouthwash samples and immediately frozen whole saliva samples from 10 healthy subjects. The results showed that oral microbial profiles in mouthwash samples were similar to the profiles in saliva samples (Additional file 1: Figure S1). Thus, the mouthwash samples are suitable to test the hypothesis in this study.
Statistical analysis
The effect of drinking level was assessed by comparing subjects in each drinking category (moderate, heavy) to non-drinkers. For trend tests, drinking level was treated as a continuous variable by assigning the numbers 0, 1, and 2 to non-, moderate, and heavy drinkers, respectively. The effect of drinking type (wine, beer, or liquor) was examined by modeling the three dichotomous variables of any consumption of wine, beer or liquor, while additionally adjusting for multiple drinking types using cross-product terms (e.g., formula to test effect of wine drinking: ~any_wine + any_beer + any_liquor + any_wine*any_beer + any_wine*any_liquor). To control for the effects of potential confounders, all models were adjusted for age, sex, race, BMI category (normal weight, overweight, obese), smoking status (never, former, current), education level (no college, some college, college graduate), and study (CPSII-a, CPSII-b, PLCO-a, PLCO-b); drinking type models were additionally adjusted for drinking amount (grams of ethanol per day).
We assessed α-diversity (within-subject diversity) using numbers of observed species (richness) and the inverse Simpson’s Index (evenness). These α-diversity indices were calculated in 500 iterations of rarefied OTU tables with the minimum sequencing depth of 1325 among all study subjects. The average over the iterations was taken for each participant. Linear regression with covariate adjustment was used to examine the difference of α-diversity indices among drinking groups. β-diversity (between-subject diversity) was assessed using unweighted and weighted UniFrac distance matrices accounting for both presence or absence of observed organisms and their abundance, respectively [42]. We performed Permutational Multivariate Analysis of Variance (PERMANOVA; adonis function, vegan package, R) [43] and partial constrained analysis of principal coordinates (partial CAP; “capscale” function, vegan package, R) [44] to examine statistically and visually whether bacterial community profiles differed by drinking level or type. Unlike the commonly used unconstrained principal coordinate analysis (PCoA) representation of UniFrac distances, partial CAP displays the variation attributable to alcohol consumption with covariate adjustment. We also conducted pairwise comparisons among drinking groups for each of the first three coordinates in unconstrained PCoA, using the Kruskal-Wallis post-hoc test (Dunn’s test). The community type analysis of the oral wash samples was performed with Dirichlet multinomial mixture (DMM) model using counts of sequencing reads at the genus level [45].
Operational taxonomic units (OTUs) were classified into 13 phyla, 23 classes, 37 orders, 76 families, 227 genera, and 341 species, according to their alignment with the HOMD reference database. We used the “DESeq” function within the DESeq2 package [46] in R to test for differentially abundant taxa by drinking level and types of alcoholic beverages, at each taxonomic level. This function models raw counts using a negative binomial distribution and adjusts internally for “size factors” which normalize for differences in sequencing depth between samples. We analyzed taxa within the major oral phyla (carried by more than 90% of study subjects). Additionally, we filtered to include only taxa with greater than 2 sequences in at least 100 participants using R command in DESeq “rowSums(counts(OTU.deseq) > 2) ≥ 100” to remove low-count taxa for the phylum through species level analysis. This resulted in inclusion of 5 phyla, 9 classes, 15 orders, 30 families, 96 genera, and 95 species in the analysis. DESeq2 default outlier replacement and filtering of count outliers were turned off. Taxa models with a maximum Cook’s distance > 10 were removed prior to p value adjustment for the false discovery rate (FDR). To account for the multiple comparisons at each taxonomic level, we considered an FDR-adjusted p value (q value) less than 0.10 as significant. Pearson’s linear correlation was used to explore the correlation among the log-transformed DESeq2-normalized taxa abundance. All statistical tests were two-sided, and all statistical analyses were carried out using R version 3.2.1.
To examine if smoking, future cancer case/control status, or oral health status (periodontal disease and caries) has confounding effects on the observed alcohol-microbiome associations, we conducted several sensitivity analyses: (a) excluding current smokers, (b) adjusting for future case/control status in models, (c) excluding subjects who had detectable periodontal pathogens Porphyromonas gingivalis and/or Aggregatibacter actinomycetemcomitans in their oral samples (surrogate markers for periodontal disease [47]), and (d) stratified analysis by the median abundance of Streptococcus mutans (surrogate marker for dental caries [48]). The latter two analyses used surrogate markers of oral disease as we lacked information on oral disease in our study.
Results
Participant characteristics
Of the 1044 participants in this study, 25.9% (n = 270) were non-drinkers, 58.8% (n = 614) were moderate drinkers, and 15.3% (n = 160) were heavy drinkers. The study participants were predominantly above middle age (mean 67.7, range 55–87) and White (95%), with age and race distributions not differing significantly by drinking level. The alcohol drinking groups had higher percentages of men and smokers and tended to be leaner and more educated than the non-drinker group (Table 1). Among alcohol drinkers, 13.0% (n = 101) were wine-only drinkers, 5.0% (n = 39) were beer-only drinkers, and 3.4% (n = 26) were liquor-only drinkers (Table 2).
Table 1.
Demographic characteristics of the study participants
| Non-drinkers (n = 270) | Moderate drinkers* (n = 614) | Heavy drinkers* (n = 160) | p value† | ||||
|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | ||
| Age‡ | 68.3 ± 7.0 | 67.6 ± 7.4 | 66.8 ± 6.8 | 0.092 | |||
| Gender | |||||||
| Male | 150 | 55.6 | 412 | 67.1 | 103 | 64.4 | |
| Female | 120 | 44.4 | 202 | 32.9 | 57 | 35.6 | 0.0044 |
| Race | |||||||
| White | 255 | 94.4 | 581 | 94.6 | 157 | 98.1 | |
| Non-White | 15 | 5.6 | 33 | 5.4 | 3 | 1.9 | 0.16 |
| BMI, kg/m2 | |||||||
| < 25 | 87 | 32.2 | 226 | 36.8 | 70 | 43.7 | |
| 25–< 30 | 114 | 42.2 | 273 | 44.5 | 64 | 40.0 | |
| ≥ 30 | 69 | 25.6 | 115 | 18.7 | 26 | 16.3 | 0.039 |
| Education | |||||||
| High school or less | 96 | 33.6 | 188 | 30.6 | 39 | 24.4 | 0.014 |
| Some college | 93 | 34.4 | 178 | 29.0 | 54 | 33.8 | |
| College graduate or higher | 81 | 30.0 | 248 | 40.4 | 67 | 41.9 | |
| Smoking status | |||||||
| Never | 157 | 58.1 | 266 | 43.3 | 38 | 23.8 | |
| Former | 92 | 34.1 | 301 | 49.0 | 98 | 61.3 | |
| Current | 21 | 7.8 | 47 | 7.7 | 24 | 15.0 | < 0.0001 |
.According to the Dietary Guidelines for Americans, moderate drinking was defined as > 0 to 1 drink per day for women and > 0 to 2 drinks per day for men; more than 1 drink per day for women and 2 drinks per day for men was defined as heavy drinking
†p values were from chi-square test or ANOVA
‡Mean and standard deviation were calculated
Table 2.
Types of alcoholic beverages consumed by drinkers. Subjects who ever consumed alcohol were included in the table (n = 774)
| Number of drinkers in each drinking type | Mean of pure ethanol consumption | ||
|---|---|---|---|
| N | % | gram/day | |
| Any wine drinkers | 645 | 83.3 | 15.5 |
| Wine only† | 101 | 13.0 | 3.8 |
| Wine and other types | 544 | 70.3 | 17.7 |
| Any beer drinkers | 565 | 73.0 | 19.9 |
| Beer only† | 39 | 5.0 | 20.7 |
| Beer and other types | 526 | 68.0 | 19.8 |
| Any liquor drinkers | 545 | 70.4 | 19.0 |
| Liquor only† | 26 | 3.4 | 7.0 |
| Liquor and other types | 519 | 67.0 | 19.6 |
†Wine, beer, or liquor drinkers exclusively consumed each type of beverages respectively
Drinking level analysis
We first examined the overall microbial composition according to drinking level. In the α-diversity analysis, richness (Fig. 1a, c) increased in heavy drinkers (HD) and moderate drinkers (MD) with statistical significance (meanHD = 100.3, p from linear regression = 0.0059 and meanMD = 100.1, p = 0.0073) relative to non-drinkers (meannon-drinker = 94.6). These differences in richness remained significant after further adjustment for case/control status (p = 0.0051 for HD and 0.0072 for MD). Evenness tended to be higher in drinkers compared to non-drinkers (Fig. 1b, d); however, the differences were not statistically significant (p = 0.092 and 0.062 for HD and MD, respectively). When assessing β-diversity according to the unweighted UniFrac distance, partial CAP revealed that HD and MD separated from non-drinkers on the first CAP axis, based on position of the group centroids (Fig. 2a). Similarly, HD and MD differed from non-drinkers in the first principal coordinate in PCoA (p = 0.029 and 0.038 respectively) (Fig. 2c). In PERMANOVA analysis of the unweighted UniFrac distance, HD significantly differed from non-drinkers after controlling for covariates (p = 0.0036), while MD did not (p = 0.22). The difference between HD and non-drinkers in PERMANOVA analysis persisted after further adjustment for case/control status (p = 0.0050). When assessing β-diversity according to the weighted UniFrac distance, HD, MD, and non-drinkers clustered together in partial CAP plot (Fig. 2b) and showed no differences in the first three principal coordinates in PCoA, with the exception of the third coordinate comparing MD and non-drinkers (p = 0.018) (Fig. 2d). In PERMANOVA analysis of the weighted UniFrac distance, neither HD nor MD were found to be different from non-drinkers (p = 0.22 and 0.24 for HD and MD, respectively). The differences in the microbial community composition according to alcohol drinking were confirmed by the results of community type classification (Additional file 1: Figure S2).
Fig. 1.
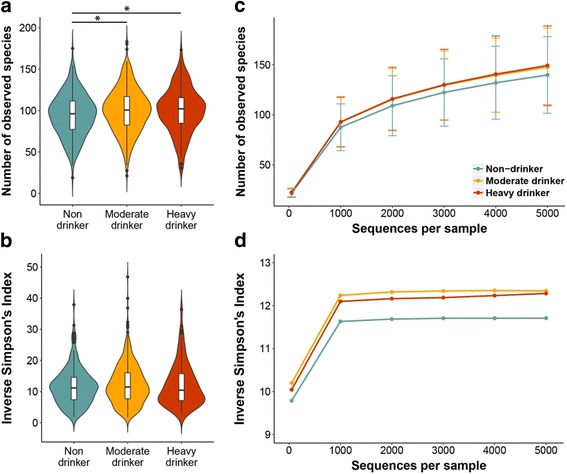
Richness and evenness of oral microbiome by alcohol drinking levels. a, b Violin plots of the number of observed species (richness) and inverse Simpson’s Index (evenness) in non-drinker, moderate drinker (MD) and heavy drinker (HD) groups. These indices were calculated for 500 iterations of rarefied OTU table with minimum sequencing depth of 1325 among all study subjects, and the average over the iterations was taken for each participant. Plotted are median, interquartile ranges, and the probability density of the indices at different values. Mean values of the richness in non-drinker, MD, and HD groups were 94.6, 100.1, and 100.3, respectively; mean values of the inverse Simpson’s Index in each group were 11.6, 12.3, and 12.1. One star (*) indicates p < 0.05 in linear regression model. c, d Rarefaction curves of number of observed species and inverse Simpson’s Index according to the number of reads per sample in non-drinkers, MD, and HD
Fig. 2.
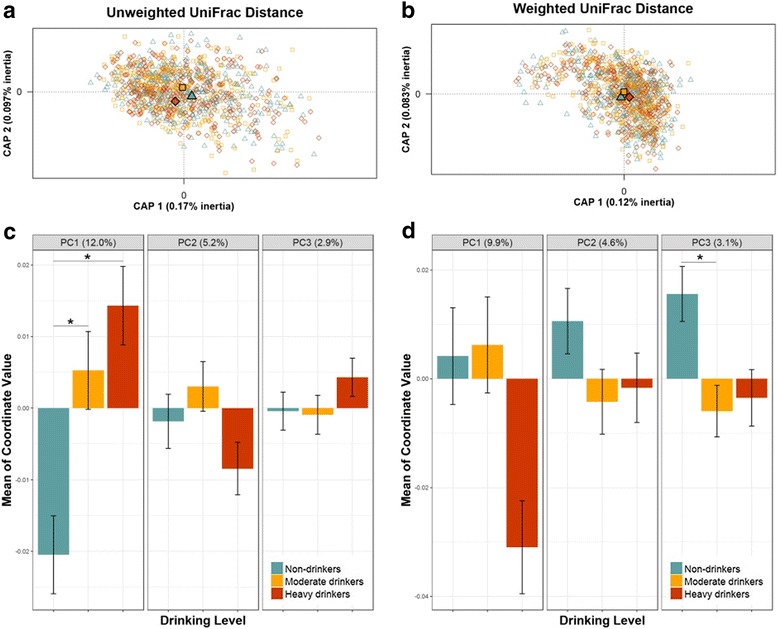
Partial constrained analysis of principal coordinates (CAP) and Principal Coordinate Analysis (PCoA) plots. a, b CAP plots using unweighted and weighted UniFrac phylogenetic distance matrices in all study participants. Drinking level was the constraining variable; age, race, gender, BMI, education, smoking status, and study were treated as partial variables. Filled shapes indicate centroids for each group. c, d Bar plots showing the means of the first, second, and third coordinates of PCoA for each drinking level using unweighted and weighted UniFrac phylogenetic distance matrices in all study participants. One star (*) indicates p < 0.05 in the Kruskal-Wallis post hoc test (Dunn’s test)
Associations between drinking level and the abundance of specific oral taxa were examined using negative binomial generalized linear models [46] (Table 3 and Fig. 3a). Abundance of class Bacilli in phylum Firmicutes was decreased with higher drinking levels (fold changes [FCs] = 0.92 and 0.94 for HD and MD, FDR-adjusted q trend = 0.050), as was its major order Lactobacillales (FCs = 0.89 and 0.94 for HD and MD, q = 0.064). Low-abundance genera Streptococcus and Lachnoanaerobaculum in Firmicutes were increased with higher drinking levels. Taxa in other phyla were also enriched with higher drinking level, including the genus Actinomyces (FCs = 1.41 and 1.15 for HD and MD, q = 0.041) and its constituent species A. graevenitzzi (FCs = 1.53 and 1.17 for HD and MD, q = 0.088). Leptotrichia (FCs = 1.61 and 1.24 for HD and MD, q = 0.0027) and low-abundance genera Cardiobacterium (FCs = 1.41 and 1.61 for HD and MD, q = 0.041) and Neisseria (FCs = 2.12 and 2.42 for HD and MD, q = 0.027) were increased in both heavy and moderate drinkers, although read counts tended to be low for these bacteria (Additional file 1: Figure S3). Additional taxa that were differentially abundant by drinking level are shown in Table 3 and Fig. 3a. The correlation matrix of the taxa associated with drinking level indicated that class Bacilli and order Lactobacillales, which were decreased with higher drinking level, generally had negative correlations with other taxa that were increased with higher drinking level (Fig. 3b). Results were not altered by further adjustment for case/control status.
Table 3.
Taxa related to alcohol drinking level in all study participants. The association between taxonomic abundance and alcohol drinking level was detected by DESeq function, adjusted for age, sex, race, BMI, education, smoking status, and study
| Taxon Class;Order;Family;Genus;Species | Mean counts† | Fold change (95% CI) | |||||
|---|---|---|---|---|---|---|---|
| Non-drinkers | Moderate drinkers | Heavy drinkers | Moderate vs. non-drinkers | Heavy vs. non-drinkers | p trend‡ | q trend§ | |
| Phylum Firmicutes | |||||||
| Bacilli | 5986.51 | 5578.79 | 5459.46 | 0.94 (0.89, 0.98) | 0.92 (0.86, 0.98) | 0.0055 | 0.050 |
| Bacilli;Lactobacillales | 5687.89 | 5340.36 | 5111.72 | 0.94 (0.88, 1.00) | 0.89 (0.82, 0.97) | 0.0050 | 0.064 |
| Bacilli;Lactobacillaes;Streptococcaceae;Streptococcus_gid_304 | 37.39 | 181.57 | 58.55 | 2.13 (1.66, 2.74) | 1.44 (1.03, 2.00) | < 0.001 | 0.015 |
| Bacilli;Lactobacillales;Carnobacteriaceae;Granulicatella;Adiacens_gid_296 | 133.08 | 110.70 | 105.93 | 0.84 (0.76, 0.93) | 0.80 (0.70, 0.92) | < 0.001 | 0.057 |
| Bacilli;Lactobacillales;Carnobacteriaceae;Granulicatella;Elegans_gid_337 | 15.86 | 23.52 | 22.88 | 1.54 (1.16, 2.04) | 1.74 (1.22, 2.48) | 0.0013 | 0.057 |
| Clostridia;Clostridiales;Lachnospiraceae[14];Lachnoanaerobaculum_gid_046 | 16.1 | 16.45 | 25.01 | 1.05 (0.86, 1.28) | 1.52 (1.16, 1.99) | 0.0063 | 0.042 |
| Phylum Bacteroidetes | |||||||
| Bacteroides;Bacteroidales;Bacteroidales[F-2];Bacteroidales[G-2]|| | 3.31 | 7.05 | 6.94 | 1.74 (1.30, 2.33) | 2.08 (1.43, 3.03) | < 0.0001 | 0.0027 |
| Bacteroides;Bacteroidales;Bacteroidales[F-2];Bacteroidales[G-2];taxon 274_gid_156 | 1.64 | 2.43 | 2.59 | 1.42 (1.09, 1.83) | 1.56 (1.13, 2.17) | 0.0043 | 0.094 |
| Bacteroides;Bacteroidales;Porphyromonadaceae;Tannerella_gid_164 | 2.28 | 3.00 | 3.61 | 1.28 (0.98, 1.67) | 1.60 (1.13, 2.27) | 0.0070 | 0.042 |
| Bacteroides;Bacteroidales;Prevotellaceae;Prevotella_gid_168 | 1.73 | 43.72 | 1.98 | 2.31 (1.64, 3.25) | 1.91 (1.24, 2.94) | < 0.001 | 0.0099 |
| Flavobacterial;Flavobacteriales;Flavobacteriaceae;Bergeyella | 3.62 | 4.62 | 5.29 | 1.29 (1.09, 1.54) | 1.46 (1.15, 1.84) | < 0.001 | 0.014 |
| Phylum Actinobacteria | |||||||
| Actinobacteria;Actinomycetale;Actinomycetaceae;Actinomyces | 103.79 | 123.63 | 131.63 | 1.15 (0.97, 1.37) | 1.41 (1.11, 1.78) | 0.0047 | 0.041 |
| Actinobacteria;Actinomycetale;Actinomycetaceae;Actinomyces;Graevenitzii_gid_484 | 37.25 | 47.59 | 55.77 | 1.17 (0.95, 1.45) | 1.53 (1.16, 2.02) | 0.0030 | 0.088 |
| Actinobacteria;Actinomycetales;Corynebacteriaceae;Corynebacterium¶ | 21.93 | 28.95 | 29.51 | 1.35 (1.12, 1.63) | 1.36 (1.06, 1.76) | 0.0064 | 0.042 |
| Phylum Fusobacteria | |||||||
| Fusobacteria;Fusobacteriales;Fusobacteriaceae;Fusobacterium_gid_114 | 184.52 | 369.96 | 229.27 | 1.48 (1.24, 1.78) | 1.22 (0.95, 1.56) | 0.018 | 0.090 |
| Fusobacteria;Fusobacteriales;Leptotrichiacefaae;Leptotrichia | 70.25 | 94.78 | 157.05 | 1.24 (1.05, 1.48) | 1.61 (1.27, 2.03) | < 0.0001 | 0.0027 |
| Fusobacteria;Fusobacteriales;Leptotrichiaceae;Leptotrichia_gid_126 | 7.97 | 10.31 | 12.10 | 1.30 (0.99, 1.72) | 1.58 (1.10, 2.27) | 0.010 | 0.055 |
| Phylum Proteobacteria | |||||||
| Grammaproteobacteria;Pasteurellales;Pasteurellaceae | 138.73 | 194.42 | 191.54 | 1.36 (1.12, 1.66) | 1.37 (1.07, 1.76) | 0.0064 | 0.052 |
| Gammaproteobacteria;Pasteurellales;Pasteurellaceae;Aggregatibacter_gid_286 | 3.21 | 10.94 | 5.63 | 2.83 (1.93, 4.15) | 1.81 (1.13, 2.89) | 0.0024 | 0.027 |
| Betaproteobacteria;Neisseriales;Neisseriaceae;Eikenella | 1.33 | 2.20 | 1.75 | 1.71 (1.33, 2.19) | 1.39 (1.00, 1.93) | 0.0099 | 0.055 |
| Betaproteobacteria;Neisseriales;Neisseriaceae;Kingella_gid_009 | 4.19 | 8.35 | 6.12 | 1.85 (1.38, 2.49) | 1.55 (1.06, 2.27) | 0.0057 | 0.042 |
| Betaproteobacteria;Neisseriales;Neisseriaceae;Neisseria_gid_382 | 1.90 | 5.20 | 5.55 | 2.42 (1.55, 3.75) | 2.12 (1.27, 3.54) | 0.0024 | 0.027 |
| Grammaproteobacteria;Cardiobacteriales;Cardiobacteriaceae;Cardiobacterium** | 5.41 | 7.47 | 7.04 | 1.60 (1.27, 2.01) | 1.41 (1.04, 1.92) | 0.0047 | 0.041 |
†Sequence read counts were normalized by dividing raw counts by DESeq size factors
‡Nominal p values from trend tests. In trend test, alcohol drinking was treated as a continuous variable by assigning the numbers 0, 1, and 2 to non-, moderate, and heavy drinkers, respectively. All taxa with an FDR-adjusted q < 0.10 are included in the table. See Additional file 1: Table S1 for mean counts and comparisons stratified by cohort
§FDR-adjusted p value. FDR adjustment was conducted at each taxonomic level (i.e., class, genus) separately
||Association was significant up to family Bacteroidales[F-2], because this genus is the single constituent member of its family
¶Association was significant up to family Corynebacteriaceae, because this genus is the single constituent member of its family
**Association was significant up to order Cardiobacteriales, because this genus is the single constituent member of its order
Fig. 3.
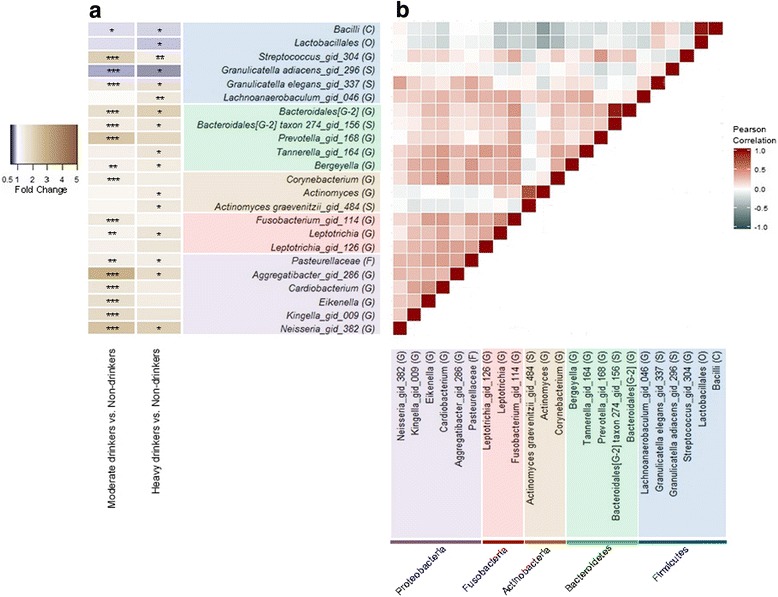
Heatmap of fold changes and the correlations of the taxa related to alcohol drinking level. a Fold change for moderate drinkers (N = 614) and heavy drinkers (N = 160) compared to non-drinkers (N = 270) was estimated by DESeq function, adjusting for age, sex, race, BMI, education, smoking status, and study. One star (*) indicates FDR-adjusted q < 0.10, two stars (**) indicate q < 0.05, and three stars (***) indicate q < 0.01, in the DESeq2 analysis. b Pearson’s linear correlation matrix of the selected taxa. For correlation analysis, counts were normalized for DESeq2 size factors and log2 transformed after adding a pseudocount of 1. Strong positive correlations are indicated by dark red and strong negative correlations by dark slate-gray. Color coding of taxa names represents the phylum to which each taxa belongs, as follows: Firmicutes (blue), Bacteroidetes (green), Actinobacteria (brown), Fusobacteria (red), and Proteobacteria (purple)
We examined the homogeneity of our results across two independent cohorts. The partial CAP plots indicated a similar trend of HD separated from non-drinkers on the first CAP axis in both cohorts (Additional file 1: Figure S4a–d), which was further supported by the PERMANOVA analysis showing that HD differed from non-drinkers according to the unweighted UniFrac distance in both CPS II and PLCO cohorts (p = 0.097 and p = 0.064 respectively). Most of the taxa associated with drinking level showed similar trends in both cohorts (Additional file 1: Figure S3 and Table S2), with the exception of family Bacteroidales[F-2], its constituent genus Bacteroidales[G-2], and genus Corynebacterium, which all were shown to increase significantly with drinking level in the PLCO, but not CPS II, cohorts.
In a sensitivity analysis to determine potential influences of smoking and oral disease on study results, the observed alcohol-microbiota associations (i.e., in α-diversity, β-diversity, and taxon abundance) were unchanged after excluding current smokers or subjects carrying periodontal pathogens (Additional file 1: Table S3). Additionally, similar trends for alcohol-microbiota associations were observed in those with low or high abundance of S. mutans, a marker of dental caries (Additional file 1: Table S3).
Drinking type analysis
We next investigated the overall microbial composition and taxon abundances for exclusive wine (n = 101), beer (n = 39), and liquor (n = 26) drinkers, relative to non-drinkers, after controlling for drinking amount. In the α-diversity analysis (Additional file 1: Figure S5), beer drinkers and liquor drinkers did not differ from non-drinkers in richness (p from linear regression = 0.87 and 0.91 for beer and liquor drinkers, respectively), while wine drinkers (mean = 100.9, p = 0.048) had increased richness compared to non-drinkers (mean = 94.6). Significance remained after further adjustment for case/control status (p = 0.048) and exclusion of current smokers (p = 0.053). Similarly, in covariate-adjusted PERMANOVA analysis of the unweighted UniFrac distance, wine drinkers differed from non-drinkers with marginal significance (p = 0.059), but liquor drinkers or beer drinkers did not (p = 0.73 and p = 0.59). Evenness (Additional file 1: Figure S5) and β-diversity estimated by weighted UniFrac distance did not reveal any differences by drinking type. Compared to non-drinkers, wine drinking was associated with decreased abundance of genus Peptococcus (FC = 0.45, q = 0.073); beer drinking was associated with decreased abundance of Porphyromonas (FC = 0.32, q = 0.071) and increased abundance of genus Parascardovia (FC = 5.87, q = 0.016); and liquor drinking was associated with decreased abundance of Lachnospiraceae[G-2] (FC = 0.39, q = 0.090) (Fig. 4 & Additional file 1: Table S4). Other taxa associated with wine, beer, or liquor drinking were in genera Corynebacterium, Prevotella, and Aggregatibacter and Eikenella, which had been identified as also associated with drinking level (Fig. 4 and Additional file 1: Table S4).
Fig. 4.
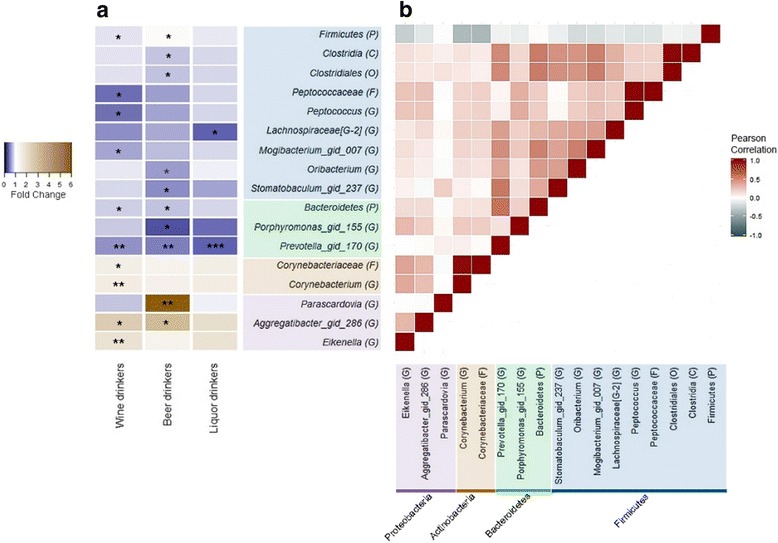
Heatmap of fold changes and the correlations of the taxa related to alcohol drinking type. a Fold change for exclusive wine drinkers (N = 101), beer drinkers (N = 39), and liquor drinkers (N = 26) compared to non-drinking group was estimated by DESeq function, adjusting for age, sex, race, BMI, education, smoking status, drinking amount, and study. One star (*) indicates FDR-adjusted q < 0.10, two stars (**) indicate q < 0.05, and three stars (***) indicates q < 0.01, in the DESeq2 analysis. b Pearson’s linear correlation matrix of the selected taxa. For correlation analysis, counts were normalized for DESeq2 size factors and log2 transformed after adding a pseudocount of 1. Strong positive correlations are indicated by dark red and strong negative correlations by dark slate-gray. Color coding of taxa names represents the phylum to which each taxa belongs, as follows: Firmicutes (blue), Bacteroidetes (green), Actinobacteria (brown), Fusobacteria (red), and Proteobacteria (purple)
Discussion
In this first large comprehensive human study, we observed that overall microbiome community composition in the oral cavity differed by drinking level, with consistent results across two independent cohorts and after exclusion of current smokers. Drinkers had decreased abundance of order Lactobacillales, the major order in the Firmicutes phylum. Other taxa, some of which are potentially pathogenic, were enriched with higher alcohol consumption. Additionally, wine drinkers may have a shifted microbiome community composition relative to non-drinkers. These results suggest that alcohol drinking and beverage type may influence the oral microbiota.
We studied the oral microbiome in mouthwash samples, which contain bacteria shed from adhering microbial communities on various oral sites, including tooth surfaces, gingival crevices, tongue dorsum, and buccal mucosa [49–51]. We found an increase in within-subject diversity (richness) and qualitative variation [42] in microbial community profiles (unweighted UniFrac) by drinking level. The observed increased diversity and altered profiles in drinkers may be due to direct effects of alcohol or may reflect poor oral health conditions in drinkers. Alcohol intake has been associated with risk of periodontal diseases and caries [17, 24, 26]. Additionally, poor oral health conditions, including higher plaque index, presence of decayed teeth, gingival bleeding, and deeper periodontal pockets, have been associated with higher phylogenetic diversity of salivary microbiota [27]. In a sensitivity analysis, we used periodontal pathogens (P. gingivalis and A. actinomycetemcomitans) and the abundance of S. mutans in saliva as surrogate markers for periodontitis and dental caries, respectively, and major findings remained unchanged. Thus, it is possible that alcohol consumption has a direct effect on oral bacteria composition independently of oral disease.
Our results indicate that alcohol consumption is associated with decreased abundance of Lactobacillales, a dominant order in class Bacilli and phylum Firmicutes. Bacteria in the order Lactobacillales (also named lactic acid bacteria or LAB), produce lactic acid as an end-product of carbohydrate fermentation [52] and are among the most common microbes employed as probiotics [53]. Firmicutes and Lactobacillales abundance have also been shown to decrease in the intestines of mice fed with ethanol chronically [54, 55], while increases were observed in potentially pathogenic bacteria, Proteobacteria and Actinobacteria, in response to ethanol feeding [54]. While the mechanism for ethanol-induced decreases in Lactobacillales are unclear, depletion of Lactobacillales may promote growth of alkaline-tolerant bacteria [54], which may explain the inverse correlations of Lactobacillales with the other taxa associated with alcohol drinking. In support of this, probiotic treatment of ethanol-fed mice with Lactobacillus rhamnosus GG decreased luminal pH and prevented ethanol-related pathogenic increases in the gut microbiome [54]. Evidence shows that Lactobacillales have a beneficial effect on oral health [56]. Some Lactobacillales can reduce the risk of caries development [57], possibly through competitive exclusion and displacement of pathogens, or by production of antibacterial compounds [58–60]. Additionally, oral Lactobacilli species can suppress the growth of periodontal pathogens [61], and oral intake of some Lactobacillales can promote reduction of gingival inflammation [62]. Thus, the alcohol-related depletion of beneficial commensal Lactobacillales may lead to further oral health inflammation-related disturbances.
We also identified some taxa enriched in drinkers. Ethanol may indirectly increase certain bacterial taxa by decreasing Lactobacillales thereby increasing pH, as mentioned above, or by inhibiting the antimicrobial properties of saliva and by disturbing the host-microbial balance. Both acute and chronic ethanol exposure can lead to functional changes in saliva, including decreased flow rate and impaired output of total protein, amylase, and electrolytes [12, 14]. Additionally, alcohol could impair neutrophil function (contributing to bacterial overgrowth and increased bacterial penetration) [20], reduce monocyte production of inflammatory cytokines (allowing for microbial proliferation) [21], and have adverse effects on teeth (stimulate bone resorption and suppress bone formation) [22] and the periodontium [23]. These detrimental effects of alcohol on host defense potentially lead to periodontitis [24]. Among the taxa enriched in drinkers, the genera Aggregatibacter, Actinomyces, Kingella, Leptotrichia, Cardiobacterium, Bacteroidales[G-2], and Prevotella may contain human pathogenic and cardiogenic pathogens. Aggregatibacter, a genus in the Pasteurellaceae family of the Proteobacteria phylum, is related to periodontal disease [63–66] and infective endocarditis [67, 68], as are species in Actinomyces, a genus from the Actinobacteria phylum [69, 70], and Kingella, a genus from the Proteobacteria phylum [68, 71, 72]. Leptotrichia is one of the two major genera in phylum Fusobacteria, which tends to cause disease in the presence of local or general pre-disposing factors and has been reported to be involved in various human infections [73]. The only two species in genus Cardiobacterium, C. hominis and C. valvarum, both can cause endocarditis [74–76]. The constituent species oral taxon 274 in Bacteroidales[G-2] was shown to be more prevalent in subgingival sites in periodontitis subjects than in healthy subjects [77]. Species of genus Prevotella are part of the indigenous microbiota of mucous membranes from oral cavity, while some black-pigmented Prevotella, for instance P. intermedia and P. melaninogenica, associate with periodontal disease [78]. Thus, the changes in the composition of oral microbiome related to alcohol consumption may potentially contribute to dental caries, periodontal diseases, and other health consequences.
In addition to these compositional changes related to alcohol use, alcohol consumption may result in the bacterial production from ethanol of acetaldehyde, a World Health Organization (WHO) group 1 human carcinogen [79]. Acetaldehyde is toxic, mutagenic, and carcinogenic in human cells and animal models [80–83]. Certain bacteria are involved in producing acetaldehyde from ethanol, while others can beneficially metabolize acetaldehyde to less toxic compounds. Neisseria is mainly a non-pathogenic genus in the oral cavity, but it has extremely high alcohol dehydrogenase (ADH) activity and produces significant amounts of acetaldehyde from ethanol [84]. The increased abundance of Neisseria with alcohol drinking that we have observed is consistent with the ADH activity of Neisseria and is supported by another human study [84]. Conversely, strains of gastrointestinal Lactobacillus, decreased in alcohol consumers in our study, show a high capacity to metabolize acetaldehyde to less toxic forms [85]. Thus, ethanol-related increase in abundance of Neisseria, which produces acetaldehyde, and decrease in abundance of LAB, involved in acetaldehyde degradation, suggest that oral bacteria may play a role in alcohol-related carcinogenesis through acetaldehyde.
Analyses by beverage type revealed that wine drinkers may have a higher richness and different microbial profiles from non-drinkers, with decreased abundance of phyla Bacteroidetes and Firmicutes, and family Peptostreptococcaceae. These findings are consistent with previous in vivo and in vitro studies investigating the antimicrobial properties of wine, showing that wine drinkers had decreased abundance of certain species in sub- and supra-gingival plaques, including species in Peptostreptococcaceae [18, 86]. The results on microbial richness in these studies are inconsistent with our findings, which may relate to different sample types used (subgingival plaque vs. mouthwash samples). Beer has a relatively low alcohol concentration but a complex chemical composition [87], containing a greater abundance of proteins, B type vitamins, fiber, minerals, antioxidants, and varied flavors than other alcoholic beverage types [88], which potentially may have differential effects on the growth of oral bacteria. Due to the high concentration of ethanol in liquor, the bacterial variations associated with liquor drinking may only more closely represent the effects of pure ethanol. Bacterial taxa similarly altered relative to non-drinkers in wine, beer, and liquor drinkers (e.g., Prevotella_gid_170) may simply reflect an effect of drinking alcohol vs. not drinking. Because our study had limited numbers of subjects who exclusively consumed beer, wine, or liquor, further study is required to disentangle the differential effect of each type of alcoholic beverage on oral microbial composition.
Our study has several strengths. First, the use of 16S rRNA gene sequencing for microbiome analysis allowed us to comprehensively study overall bacterial community composition and specific oral taxon abundances. Second, our very large sample size provided excellent statistical power to detect variation in the oral microbiome with alcohol drinking and allowed us to confirm our findings in multiple independent study groups. Finally, the detailed information on alcohol drinking amount and type allowed us to classify alcohol intake in detail, and the detailed demographic and lifestyle information allowed us to adjust for potential confounding factors. A limitation of our study is that it is observational, limiting our ability to establish a causal relationship. A trial where moderate or heavy drinkers were randomized to continue or stop drinking for a long enough time to influence the oral microbiome could provide more definitive information. Additionally, a few enriched bacteria in drinkers has overall low abundance. Further, the majority of this study population was White and above middle age, limiting the generalizability of our findings to other races and age groups. We lacked information on the oral health status of the study participants, though sensitivity analysis using bacterial markers as proxies of dental caries and periodontitis suggests that the observed alcohol-microbiota associations are independent of oral diseases. Lastly, the oral wash sample might not be representative of oral bacterial profiles in specific oral niches, such as the subgingival plaque, which is more directly involved in oral disease.
Conclusions
We found that alcohol consumption is related to overall oral microbiome community composition and to the abundance of specific oral taxa. Heavy drinking may influence bacterial composition, including potential depletion of beneficial commensal bacteria and increased colonization of potentially pathogenic bacteria. Such changes potentially contribute to alcohol-related diseases, including periodontal disease, head and neck cancer, and digestive tract cancers, but further research is needed to relate alcohol-related composition changes to disease phenotypes. The taxa we have identified can be further investigated to tease out their potential relationship with underlying oral health status and to elucidate their potential role in alcohol-related health consequences. Future studies should also investigate the impact of alcohol drinking on the metagenomic (functional) content of the oral microbiome. Improved understanding of the causes and health impacts of oral dysbiosis can lead to microbiome-targeted approaches for disease prevention.
Additional file
Tables S1-S4 and Figures S1-S5. Table S1. Number of sequence reads per sample assigned to reference taxonomy map. Table S2. Mean counts of taxa associated with drinking levels in all study participants and subjects in each cohort. Table S3. Associations between oral bacteria diversity and abundance of order Lactobacillales, genera Actinomyces and Neisseria with alcohol drinking levels in sensitivity analysis. Table S4. Mean counts and fold changes of taxa associated with type of alcoholic beverages in all study participants. Figure S1. Beta-diversity of oral bacterial communities in the paired mouthwash-saliva samples. Figure S2. Distribution of alcohol consumption in the enterotypes. Figure S3. Count boxplots of order Lactobacillales and selected genera that were associated with drinking level. Figure S4. Partial Constrained Analysis of Principal Coordinates plots. Figure S5. Richness and evenness of oral microbiome by alcohol drinking type. (DOCX 1148 kb)
Acknowledgements
We thank our colleague, Dr. Yilong Zhang, from the Division of Biostatistics, Department of Population Health, NYU School of Medicine, who provided insight and expertise that greatly assisted the analysis of data.
Funding
Research reported in this publication was supported in part by the US National Cancer Institute under award numbers R01CA159036, U01CA182370, R01CA164964, R03CA159414, P30CA016087, and R21CA183887, and by AACR/Pancreas Cancer Action Network Career Development Award.
Availability of data and materials
The data are available from the Sequence Read Archive (SRA) with accession number SRP133146 and SRP133149.
Disclaimer
The American Cancer Society (ACS) funds the creation, maintenance, and updating of the Cancer Prevention Study II cohort. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the US Department of Veterans Affairs, or the US Government.
Abbreviations
- ACS
American Cancer Society
- ADH
Alcohol dehydrogenase
- CAP
Constrained analysis of principal coordinates
- CI
Confidence interval
- CPS II
Cancer Prevention Study II
- FCs
Fold changes
- FDR
False discovery rate
- HD
Heavy drinkers
- HOMD
Human Oral Microbiome Database
- LAB
Lactic acid bacteria
- MD
Moderate drinkers
- NCI
National Cancer Institute
- OTUs
Operational taxonomic units
- PCoA
Principal coordinate analysis
- PERMANOVA
Permutational Multivariate Analysis of Variance
- PLCO
Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial
- WHO
World Health Organization
Authors’ contributions
JA had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. XF, RH, and JA conceived of the study concept and design. JW, EJ, SG, MP, LY, and ZP carried out the acquisition of data. XF, BP, EJ, SG, MP, AA, JW, LY, ZP, RH, and JA carried out the analysis and interpretation of data. XF, BP, RH, and JA participated in the drafting of the manuscript. XF, BP, AA, JW, EJ, SG, MP, LY, ZP, RH, and JA carried out the critical revision of the manuscript for important intellectual content. XF, BP, AA, RH, and JA participated in the statistical analysis. JA obtained funding. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The studies described in this manuscript were approved by the institutional review boards (IRB) of the NYU School of Medicine, and all participants provided written informed consent (IRB study number: S12-00721 and 10-01834).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s40168-018-0448-x) contains supplementary material, which is available to authorized users.
Contributor Information
Xiaozhou Fan, Email: Xiaozhou.Fan@nyumc.org.
Brandilyn A. Peters, Email: Brandilyn.Peters@nyumc.org
Eric J. Jacobs, Email: Eric.Jacobs@cancer.org
Susan M. Gapstur, Email: Susan.Gapstur@cancer.org
Mark P. Purdue, Email: purduem@mail.nih.gov
Neal D. Freedman, Email: freedmanne@mail.nih.gov
Alexander V. Alekseyenko, Email: alekseye@musc.edu
Jing Wu, Email: Jing.Wu@nyumc.org.
Liying Yang, Email: Liying.Yang@nyumc.org.
Zhiheng Pei, Email: Zhiheng.Pei@nyumc.org.
Richard B. Hayes, Email: Richard.B.Hayes@nyumc.org
Jiyoung Ahn, Phone: 212-263-3390, Email: Jiyoung.Ahn@nyumc.org.
References
- 1.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. The human oral microbiome. J Bacteriol. 2010;192(19):5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker JL, Bor B, Agnello M, Shi W, He X. Ecology of the oral microbiome: beyond bacteria. Trends Microbiol. 2017;25(5):362–374. doi: 10.1016/j.tim.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slocum C, Kramer C, Genco CA. Immune dysregulation mediated by the oral microbiome: potential link to chronic inflammation and atherosclerosis. J Intern Med. 2016;280(1):114–128. doi: 10.1111/joim.12476. [DOI] [PubMed] [Google Scholar]
- 4.Homann N, Tillonen J, Meurman JH, Rintamaki H, Lindqvist C, Rautio M, Jousimies-Somer H, Salaspuro M. Increased salivary acetaldehyde levels in heavy drinkers and smokers: a microbiological approach to oral cavity cancer. Carcinogenesis. 2000;21(4):663–668. doi: 10.1093/carcin/21.4.663. [DOI] [PubMed] [Google Scholar]
- 5.Moye ZD, Zeng L, Burne RA. Fueling the caries process: carbohydrate metabolism and gene regulation by Streptococcus mutans. J Oral Microbiol. 2014;6(1):24878. doi: 10.3402/jom.v6.24878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wade WG. The oral microbiome in health and disease. Pharmacol Res. 2013;69(1):137–143. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Ahn J, Chen CY, Hayes RB. Oral microbiome and oral and gastrointestinal cancer risk. Cancer Causes Control. 2012;23(3):399–404. doi: 10.1007/s10552-011-9892-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan X, Alekseyenko AV, Wu J, Peters BA, Jacobs EJ, Gapstur SM, Purdue MP, Abnet CC, Stolzenberg-Solomon R, Miller G, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut. 2018;67(1):120–127. doi: 10.1136/gutjnl-2016-312580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koren O, Spor A, Felin J, Fak F, Stombaugh J, Tremaroli V, Behre CJ, Knight R, Fagerberg B, Ley RE, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4592–4598. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ingram LO. Ethanol tolerance in bacteria. Crit Rev Biotechnol. 1990;9(4):305–319. doi: 10.3109/07388558909036741. [DOI] [PubMed] [Google Scholar]
- 11.Scannapieco FA. Saliva-bacterium interactions in oral microbial ecology. Crit Rev Oral Biol Med. 1994;5(3–4):203–248. doi: 10.1177/10454411940050030201. [DOI] [PubMed] [Google Scholar]
- 12.Enberg N, Alho H, Loimaranta V, Lenander-Lumikari M. Saliva flow rate, amylase activity, and protein and electrolyte concentrations in saliva after acute alcohol consumption. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2001;92(3):292–298. doi: 10.1067/moe.2001.116814. [DOI] [PubMed] [Google Scholar]
- 13.Tenovuo J. Antimicrobial function of human saliva—how important is it for oral health? Acta Odontol Scand. 1998;56(5):250–256. doi: 10.1080/000163598428400. [DOI] [PubMed] [Google Scholar]
- 14.Maier H, Born IA, Mall G. Effect of chronic ethanol and nicotine consumption on the function and morphology of the salivary glands. Klin Wochenschr. 1988;66(Suppl 11):140–150. [PubMed] [Google Scholar]
- 15.Kantorski KZ, de Souza DM, Yujra VQ, Junqueira JC, Jorge AO, da Rocha RF. Effect of an alcoholic diet on dental caries and on Streptococcus of the mutans group. Study in rats. Braz Oral Res. 2007;21(2):101–105. doi: 10.1590/S1806-83242007000200002. [DOI] [PubMed] [Google Scholar]
- 16.Jabbour Z, Nascimento C, Kotake BGS, El-Hakim M, Henderson JE, de Albuquerque Junior RF. Assessing the oral microbiota of healthy and alcohol-treated rats using whole-genome DNA probes from human bacteria. Arch Oral Biol. 2013;58(3):317–323. doi: 10.1016/j.archoralbio.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 17.Jansson L. Association between alcohol consumption and dental health. J Clin Periodontol. 2008;35(5):379–384. doi: 10.1111/j.1600-051X.2008.01210.x. [DOI] [PubMed] [Google Scholar]
- 18.Signoretto C, Bianchi F, Burlacchini G, Sivieri F, Spratt D, Canepari P. Drinking habits are associated with changes in the dental plaque microbial community. J Clin Microbiol. 2010;48(2):347–356. doi: 10.1128/JCM.00932-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas AM, Gleber-Netto FO, Fernandes GR, Amorim M, Barbosa LF, Francisco AL, de Andrade AG, Setubal JC, Kowalski LP, Nunes DN, et al. Alcohol and tobacco consumption affects bacterial richness in oral cavity mucosa biofilms. BMC Microbiol. 2014;14:250. doi: 10.1186/s12866-014-0250-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szabo G. Consequences of alcohol consumption on host defence. Alcohol Alcohol. 1999;34(6):830–841. doi: 10.1093/alcalc/34.6.830. [DOI] [PubMed] [Google Scholar]
- 21.Szabo G, Mandrekar P, Girouard L, Catalano D. Regulation of human monocyte functions by acute ethanol treatment: decreased tumor necrosis factor-alpha, interleukin-1 beta and elevated interleukin-10, and transforming growth factor-beta production. Alcohol Clin Exp Res. 1996;20(5):900–907. doi: 10.1111/j.1530-0277.1996.tb05269.x. [DOI] [PubMed] [Google Scholar]
- 22.Pepersack T, Fuss M, Otero J, Bergmann P, Valsamis J, Corvilain J. Longitudinal study of bone metabolism after ethanol withdrawal in alcoholic patients. J Bone Miner Res Off J Am Soc Bone Miner Res. 1992;7(4):383–387. doi: 10.1002/jbmr.5650070405. [DOI] [PubMed] [Google Scholar]
- 23.Ogden GR, Wight AJ, Rice P. Effect of alcohol on the oral mucosa assessed by quantitative cytomorphometry. J Oral Pathol Med. 1999;28(5):216–220. doi: 10.1111/j.1600-0714.1999.tb02027.x. [DOI] [PubMed] [Google Scholar]
- 24.Pitiphat W, Merchant AT, Rimm EB, Joshipura KJ. Alcohol consumption increases periodontitis risk. J Dent Res. 2003;82(7):509–513. doi: 10.1177/154405910308200704. [DOI] [PubMed] [Google Scholar]
- 25.Bartold PM, Van Dyke TE. Periodontitis: a host-mediated disruption of microbial homeostasis. Unlearning learned concepts. Periodontol. 2013;62(1):203–217. doi: 10.1111/j.1600-0757.2012.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tezal M, Grossi SG, Ho AW, Genco RJ. Alcohol consumption and periodontal disease. The third national health and nutrition examination survey. J Clin Periodontol. 2004;31(7):484–488. doi: 10.1111/j.1600-051X.2004.00503.x. [DOI] [PubMed] [Google Scholar]
- 27.Takeshita T, Kageyama S, Furuta M, Tsuboi H, Takeuchi K, Shibata Y, Shimazaki Y, Akifusa S, Ninomiya T, Kiyohara Y, et al. Bacterial diversity in saliva and oral health-related conditions: the Hisayama study. Sci Rep. 2016;6:22164. doi: 10.1038/srep22164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8(7):481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- 29.Calle EE, Rodriguez C, Jacobs EJ, Almon ML, Chao A, McCullough ML, Feigelson HS, Thun MJ. The American cancer society cancer prevention study II nutrition cohort: rationale, study design, and baseline characteristics. Cancer. 2002;94(9):2490–2501. doi: 10.1002/cncr.101970. [DOI] [PubMed] [Google Scholar]
- 30.Hayes RB, Reding D, Kopp W, Subar AF, Bhat N, Rothman N, Caporaso N, Ziegler RG, Johnson CC, Weissfeld JL, et al. Etiologic and early marker studies in the prostate, lung, colorectal and ovarian (PLCO) cancer screening trial. Control Clin Trials. 2000;21(6 Suppl):349S–355S. doi: 10.1016/S0197-2456(00)00101-X. [DOI] [PubMed] [Google Scholar]
- 31.Wu J, Peters BA, Dominianni C, Zhang Y, Pei Z, Yang L, Ma Y, Purdue MP, Jacobs EJ, Gapstur SM, et al. Cigarette smoking and the oral microbiome in a large study of American adults. ISME J. 2016;10(10):2435–2446. doi: 10.1038/ismej.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haftenberger M, Heuer T, Heidemann C, Kube F, Krems C, Mensink GB. Relative validation of a food frequency questionnaire for national health and nutrition monitoring. Nutr J. 2010;9:36. doi: 10.1186/1475-2891-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subar AF, Thompson FE, Kipnis V, Midthune D, Hurwitz P, McNutt S, McIntosh A, Rosenfeld S. Comparative validation of the Block, Willett, and National Cancer Institute food frequency questionnaires: the eating at America’s table study. Am J Epidemiol. 2001;154(12):1089–1099. doi: 10.1093/aje/154.12.1089. [DOI] [PubMed] [Google Scholar]
- 34.United States. Department of Health and Human Services., United States. Department of Agriculture., United States. Dietary Guidelines Advisory Committee . Dietary guidelines for Americans, 2010. 7. Washington, D.C: Goverment Printing Office; 2010. [Google Scholar]
- 35.Mumenthaler MS, Taylor JL, O'Hara R, Yesavage JA. Gender differences in moderate drinking effects. Alcohol Res Health. 1999;23(1):55–64. [PMC free article] [PubMed] [Google Scholar]
- 36.Jones AW. Distribution of ethanol between saliva and blood in man. Clin Exp Pharmacol Physiol. 1979;6(1):53–59. doi: 10.1111/j.1440-1681.1979.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 37.Feigelson HS, Rodriguez C, Welch R, Hutchinson A, Shao W, Jacobs K, Diver WR, Calle EE, Thun MJ, Hunter DJ, et al. Successful genome-wide scan in paired blood and buccal samples. Cancer Epidemiol Biomark Prev. 2007;16(5):1023–1025. doi: 10.1158/1055-9965.EPI-06-0859. [DOI] [PubMed] [Google Scholar]
- 38.Wu J, Lin I, Hayes RB, Ahn J. Comparison of DNA extraction methods for human oral microbiome research. Br J Med Med Res. 2014;4(10):1980–1991. doi: 10.9734/BJMMR/2014/5333. [DOI] [Google Scholar]
- 39.Nossa CW, Oberdorf WE, Yang LY, Aas JA, Paster BJ, DeSantis TZ, Brodie EL, Malamud D, Poles MA, Pei ZH. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World J Gastroenterol. 2010;16(33):4135–4144. doi: 10.3748/wjg.v16.i33.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database. 2010;2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73(5):1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26(1):32–46. [Google Scholar]
- 44.Anderson MJ, Willis TJ. Canonical analysis of principal coordinates: a useful method of constrained ordination for ecology. Ecology. 2003;84(2):511–525. doi: 10.1890/0012-9658(2003)084[0511:CAOPCA]2.0.CO;2. [DOI] [Google Scholar]
- 45.Holmes I, Harris K, Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS One. 2012;7(2):e30126. doi: 10.1371/journal.pone.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Newman MG. Carranza’s clinical periodontology. 11. Philadelphia: London: Saunders; 2012. [Google Scholar]
- 48.Loesche WJ. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50(4):353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou Y, Gao H, Mihindukulasuriya KA, La Rosa PS, Wylie KM, Vishnivetskaya T, Podar M, Warner B, Tarr PI, Nelson DE, et al. Biogeography of the ecosystems of the healthy human body. Genome Biol. 2013;14(1):R1. doi: 10.1186/gb-2013-14-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, Huttenhower C, Izard J. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13(6):R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol. 2003;30(7):644–654. doi: 10.1034/j.1600-051X.2003.00376.x. [DOI] [PubMed] [Google Scholar]
- 52.Wright AV, Lars A. Lactic acid bacteria. In: Lactic acid bacteria, microbiological and functional aspects. Boca Raton: CRC Press, Taylor & Francis Group; 2011. p. 1–16.
- 53.Ljungh A, Wadstrom T. Lactic acid bacteria as probiotics. Curr Issues Intest Microbiol. 2006;7(2):73–89. [PubMed] [Google Scholar]
- 54.Bull-Otterson L, Feng W, Kirpich I, Wang Y, Qin X, Liu Y, Gobejishvili L, Joshi-Barve S, Ayvaz T, Petrosino J, et al. Metagenomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of lactobacillus rhamnosus GG treatment. PLoS One. 2013;8(1):e53028. doi: 10.1371/journal.pone.0053028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yan AW, Fouts DE, Brandl J, Starkel P, Torralba M, Schott E, Tsukamoto H, Nelson KE, Brenner DA, Schnabl B. Enteric dysbiosis associated with a mouse model of alcoholic liver disease. Hepatology. 2011;53(1):96–105. doi: 10.1002/hep.24018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jukka HM, Iva S. Lactic acid bacteria. Boca Raton: CRC Press; 2011. Lactic acid bacteria in oral health; pp. 403–421. [Google Scholar]
- 57.Nase L, Hatakka K, Savilahti E, Saxelin M, Ponka A, Poussa T, Korpela R, Meurman JH. Effect of long-term consumption of a probiotic bacterium, Lactobacillus rhamnosus GG, in milk on dental caries and caries risk in children. Caries Res. 2001;35(6):412–420. doi: 10.1159/000047484. [DOI] [PubMed] [Google Scholar]
- 58.Simark-Mattsson C, Emilson CG, Hakansson EG, Jacobsson C, Roos K, Holm S. Lactobacillus-mediated interference of mutans streptococci in caries-free vs. caries-active subjects. Eur J Oral Sci. 2007;115(4):308–314. doi: 10.1111/j.1600-0722.2007.00458.x. [DOI] [PubMed] [Google Scholar]
- 59.Strahinic I, Busarcevic M, Pavlica D, Milasin J, Golic N, Topisirovic L. Molecular and biochemical characterizations of human oral lactobacilli as putative probiotic candidates. Oral Microbiol Immunol. 2007;22(2):111–117. doi: 10.1111/j.1399-302X.2007.00331.x. [DOI] [PubMed] [Google Scholar]
- 60.Lang C, Bottner M, Holz C, Veen M, Ryser M, Reindl A, Pompejus M, Tanzer JM. Specific Lactobacillus/Mutans Streptococcus co-aggregation. J Dent Res. 2010;89(2):175–179. doi: 10.1177/0022034509356246. [DOI] [PubMed] [Google Scholar]
- 61.Koll-Klais P, Mandar R, Leibur E, Marcotte H, Hammarstrom L, Mikelsaar M. Oral lactobacilli in chronic periodontitis and periodontal health: species composition and antimicrobial activity. Oral Microbiol Immunol. 2005;20(6):354–361. doi: 10.1111/j.1399-302X.2005.00239.x. [DOI] [PubMed] [Google Scholar]
- 62.Twetman S, Derawi B, Keller M, Ekstrand K, Yucel-Lindberg T, Stecksen-Blicks C. Short-term effect of chewing gums containing probiotic Lactobacillus reuteri on the levels of inflammatory mediators in gingival crevicular fluid. Acta Odontol Scand. 2009;67(1):19–24. doi: 10.1080/00016350802516170. [DOI] [PubMed] [Google Scholar]
- 63.Zambon JJ. Actinobacillus-actinomycetemcomitans in human periodontal-disease. J Clin Periodontol. 1985;12(1):1–20. doi: 10.1111/j.1600-051X.1985.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 64.Slots J, Reynolds HS, Genco RJ. Actinobacillus-actinomycetemcomitans in human periodontal-disease—a cross-sectional microbiological investigation. Infect Immun. 1980;29(3):1013–1020. doi: 10.1128/iai.29.3.1013-1020.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Slots J, Zambon JJ, Rosling BG, Reynolds HS, Christersson LA, Genco RJ. Actinobacillus-actinomycetemcomitans in human periodontal-disease—association, serology, leukotoxicity, and treatment. J Periodontal Res. 1982;17(5):447–448. doi: 10.1111/j.1600-0765.1982.tb02022.x. [DOI] [PubMed] [Google Scholar]
- 66.Zambon JJ, Christersson LA, Slots J. Actinobacillus-actinomycetemcomitans in human periodontal-disease-prevalence in patient groups and distribution of biotypes and serotypes within families. J Periodontol. 1983;54(12):707–711. doi: 10.1902/jop.1983.54.12.707. [DOI] [PubMed] [Google Scholar]
- 67.Norskov-Lauritsen N. Classification, identification, and clinical significance of haemophilus and aggregatibacter species with host specificity for humans. Clin Microbiol Rev. 2014;27(2):214–240. doi: 10.1128/CMR.00103-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Revest M, Egmann G, Cattoir V, Tattevin P. HACEK endocarditis: state-of-the-art. Expert Rev Anti-Infect Ther. 2016;14(5):523–30. [DOI] [PubMed]
- 69.Vielkind P, Jentsch H, Eschrich K, Rodloff AC, Stingu CS. Prevalence of Actinomyces spp. in patients with chronic periodontitis. Int J Med Microbiol. 2015;305(7):682–688. doi: 10.1016/j.ijmm.2015.08.018. [DOI] [PubMed] [Google Scholar]
- 70.Kottam A, Kaur R, Bhandare D, Zmily H, Bheemreddy S, Brar H, Herawi M, Afonso L. Actinomycotic endocarditis of the eustachian valve: a rare case and a review of the literature. Tex Heart Inst J. 2015;42(1):44–49. doi: 10.14503/THIJ-13-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yagupsky P, Porsch E, St Geme JW., 3rd Kingella kingae: an emerging pathogen in young children. Pediatrics. 2011;127(3):557–565. doi: 10.1542/peds.2010-1867. [DOI] [PubMed] [Google Scholar]
- 72.Chen C. Distribution of a newly described species, Kingella oralis, in the human oral cavity. Oral Microbiol Immunol. 1996;11(6):425–427. doi: 10.1111/j.1399-302X.1996.tb00206.x. [DOI] [PubMed] [Google Scholar]
- 73.Eribe ER, Olsen I. Leptotrichia species in human infections. Anaerobe. 2008;14(3):131–137. doi: 10.1016/j.anaerobe.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 74.Brouqui P, Raoult D. Endocarditis due to rare and fastidious bacteria. Clin Microbiol Rev. 2001;14(1):177–207. doi: 10.1128/CMR.14.1.177-207.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Berbari EF, Cockerill FR, Steckelberg JM. Infective endocarditis due to unusual or fastidious microorganisms. Mayo Clin Proc. 1997;72(6):532–542. doi: 10.4065/72.6.532. [DOI] [PubMed] [Google Scholar]
- 76.Bothelo E, Gouriet F, Fournier PE, Roux V, Habib G, Thuny F, Metras D, Raoult D, Casalta JP. Endocarditis caused by Cardiobacterium valvarum. J Clin Microbiol. 2006;44(2):657–658. doi: 10.1128/JCM.44.2.657-658.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li CL, Liang JP, Jiang YT. Association of uncultivated oral phylotypes AU126 and X112 with periodontitis. Oral Dis. 2006;12(4):371–374. doi: 10.1111/j.1601-0825.2005.01205.x. [DOI] [PubMed] [Google Scholar]
- 78.Mario JAC, Maria RLS, Elerson G-J. Prevotella. In: Molecular detection of human bacterial pathogens. Boca Raton: CRC Press, Taylor & Francis Group; 2011. p. 585–97.
- 79.World Health Organization Iarc monographs on the evaluation of carcinogenic risks to humans. Iarc Monog Eval Carc. 2010;95:9–38. [Google Scholar]
- 80.Higginson J, DeVita VT, Jr. IARC monographs on the evaluation of carcinogenic risk of chemicals to humans. Am Ind Hyg Assoc J 1980, 41(5):A26, A28, A30 passim. [PubMed]
- 81.Helander A, Lindahlkiessling K. Increased frequency of acetaldehyde-induced sister-chromatid exchanges in human-lymphocytes treated with an aldehyde dehydrogenase inhibitor. Mutat Res. 1991;264(3):103–107. doi: 10.1016/0165-7992(91)90124-M. [DOI] [PubMed] [Google Scholar]
- 82.Dellarco VL. A mutagenicity assessment of acetaldehyde. Mutat Res. 1988;195(1):1–20. doi: 10.1016/0165-1110(88)90013-9. [DOI] [PubMed] [Google Scholar]
- 83.Obe G, Jonas R, Schmidt S. Metabolism of ethanol in vitro produces a compound which induces sister-chromatid exchanges in human peripheral lymphocytes in vitro: acetaldehyde not ethanol is mutagenic. Mutat Res. 1986;174(1):47–51. doi: 10.1016/0165-7992(86)90075-8. [DOI] [PubMed] [Google Scholar]
- 84.Muto M, Hitomi Y, Ohtsu A, Shimada H, Kashiwase Y, Sasaki H, Yoshida S, Esumi H. Acetaldehyde production by non-pathogenic Neisseria in human oral microflora: implications for carcinogenesis in upper aerodigestive tract. Int J Cancer. 2000;88(3):342–350. doi: 10.1002/1097-0215(20001101)88:3<342::AID-IJC4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 85.Nosova T, Jousimies-Somer H, Jokelainen K, Heine R, Salaspuro M. Acetaldehyde production and metabolism by human indigenous and probiotic lactobacillus and Bifidobacterium strains. Alcohol Alcohol. 2000;35(6):561–568. doi: 10.1093/alcalc/35.6.561. [DOI] [PubMed] [Google Scholar]
- 86.Munoz-Gonzalez I, Thurnheer T, Bartolome B, Moreno-Arribas MV. Red wine and oenological extracts display antimicrobial effects in an oral bacteria biofilm model. J Agric Food Chem. 2014;62(20):4731–4737. doi: 10.1021/jf501768p. [DOI] [PubMed] [Google Scholar]
- 87.Gerhauser C. Beer constituents as potential cancer chemopreventive agents. Eur J Cancer. 2005;41(13):1941–1954. doi: 10.1016/j.ejca.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 88.Preedy VR. Beer in health and disease prevention. Cambridge: Academic Press; 2011.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1-S4 and Figures S1-S5. Table S1. Number of sequence reads per sample assigned to reference taxonomy map. Table S2. Mean counts of taxa associated with drinking levels in all study participants and subjects in each cohort. Table S3. Associations between oral bacteria diversity and abundance of order Lactobacillales, genera Actinomyces and Neisseria with alcohol drinking levels in sensitivity analysis. Table S4. Mean counts and fold changes of taxa associated with type of alcoholic beverages in all study participants. Figure S1. Beta-diversity of oral bacterial communities in the paired mouthwash-saliva samples. Figure S2. Distribution of alcohol consumption in the enterotypes. Figure S3. Count boxplots of order Lactobacillales and selected genera that were associated with drinking level. Figure S4. Partial Constrained Analysis of Principal Coordinates plots. Figure S5. Richness and evenness of oral microbiome by alcohol drinking type. (DOCX 1148 kb)
Data Availability Statement
The data are available from the Sequence Read Archive (SRA) with accession number SRP133146 and SRP133149.


