Abstract
The circulating microRNAs (miRNAs) associated with type 2 diabetes (T2D) in elderly patients are still being defined. To identify novel miRNA biomarker candidates for monitoring responses to sitagliptin in such patients, we prospectively studied 40 T2D patients (age > 65) with HbA1c levels of 7.5–9.0% on metformin. After collection of baseline blood samples (t 0), the dipeptidyl peptidase-IV (DPP-IV) inhibitor (DPP-IVi) sitagliptin was added to the metformin regimen, and patients were followed for 15 months. Patients with HbA1c < 7.5% or HbA1c reduction > 0.5% after 3 and 15 months of therapy were classified as “responders” (group R, n = 34); all others were classified as “nonresponders” (group NR, n = 6). Circulating miRNA profiling was performed on plasma collected in each group before and after 15 months of therapy (t 0 and t 15). Intra- and intergroup comparison of miRNA profiles pinpointed three miRNAs that correlated with responses to sitagliptin: miR-378, which is a candidate biomarker of resistance to this DPP-IVi, and miR-126-3p and miR-223, which are associated with positive responses to the drug. The translational implications are as immediate as evident, with the possibility to develop noninvasive diagnostic tools to predict drug response and development of chronic complications.
1. Introduction
With >400 million patients worldwide, type 2 diabetes (T2D) is among the most frequently diagnosed metabolic disorders. T2D is a multifactorial disease: genetic, lifestyle, and environmental factors combine to render target tissues insensitive to insulin, resulting in increasingly high blood levels of glucose. The disease is associated with serious and frequently disabling long-term complications, including cardiovascular disease, renal failure, neuropathy, and blindness, and it is therefore one of the leading causes of the global increase in morbidity and mortality [1–3]. Outcomes could be improved by earlier diagnosis, while the disease is still in the initial phase, and more rational use of currently available therapies (i.e., targeting drugs to the patients most likely to benefit from them). For this reason, there is an urgent need for new biomarkers with potential applications in the prevention and early diagnosis of T2D and for predicting its response to therapy, especially for fragile elderly patients [1, 2, 4].
Recently, microRNAs (miRNAs)—short (21-22 nucleotides), single-stranded, noncoding RNAs—have been detected in human plasma and other biological fluids, and in some cases, their expression profiles prove to be disease-specific [5–9]. Compared with many more conventional biomarkers, miRNAs offer several advantages, such as high stability, even under drastic conditions. In addition, miRNAs have been identified as major regulators of pancreatic β-cell mass and function, that is, the two key factors in the pathogenesis of T2D [9–11]. For these reasons, they are considered a promising source of biomarkers for diagnosing and staging T2D as well as for predicting their response to therapy [1, 12].
Glucagon-like peptide-1 (GLP-1) is an incretin hormone that stimulates insulin release by β-cells in a glucose-dependent manner and, at the same time, reduces glucagon secretion by alpha-cells. GLP-1-based therapeutic strategies have an increasing role in T2D treatment and consist of GLP-1 agonists and dipeptidyl peptidase-IV (DPP-IV) inhibitors (gliptins) [13]. In detail, gliptins are able to rapidly deactivate GLP-1, resulting in the decrease in insulin production, and are a recent addition to the class of oral glucose-lowering drugs used to treat T2D. Several components of this class, including vildagliptin, sitagliptin, saxagliptin, alogliptin, and linagliptin, have already been approved for this indication by the US Food and Drug Administration or by the European Medicines Agency; others are awaiting for approval or still in development. Gliptins can be used as single-agent therapy or combined with other antidiabetic agents (e.g., metformin) when the latter fail to produce or maintain adequate glycemic control [14, 15]. DPP-IVi would indeed induce insulin secretion in a glucose-dependent manner, with minimal risk of hypoglycemia; accordingly, DPP-IVi can produce a significant reduction in HbA1c. Moreover, they are usually well tolerated, with no weight gain or gastrointestinal side effects [15–18]. Interestingly, they also appear to offer added benefits consisting of the epigenetically mediated restoration of normal gene activity in dysfunctional pancreatic islets [19]. A recent study in diabetic CD1 mice also indicates that gliptin therapy can ameliorate T2D-related kidney fibrosis [20], an effect that was mediated by the drug's induction of the expression of miR-29.
These findings prompted us to investigate the circulating miRNA profile of elderly patients with poorly controlled T2D and to identify the changes it undergoes during DPP-IVi therapy with sitagliptin. The plasma levels of three miRNAs (miR-1208, miR-550a-3p, and miR-30c-5p) displayed directionally similar trends in responders and nonresponders during the 15 months of sitagliptin treatment. These miRNAs thus appear to be modulated by sitagliptin, but not correlated with metabolic response. In contrast, three other miRNAs emerged as promising candidates for use as positive (miR-126-3p and miR-223) and negative (miR-378) biomarkers of responses to sitagliptin therapy in this elderly patient population.
2. Materials and Methods
2.1. Patients
The Ethics Committee of the Tor Vergata University of Rome Medical Center approved this study protocol, and written informed consent was obtained from all patients involved in the study. Patients (males and females) were eligible for enrollment in the study if they met all the following criteria: (1) age > 65 years, (2) a ≥1-year history of T2D, and (3) poor glycemic control (HbA1c levels ranging from 7.5% to 9.0%) on current treatment with maximum-dose metformin. The following exclusion criteria were applied at baseline and during follow-up: insulin therapy, major organ failure (e.g., congestive heart failure and respiratory and/or hepatic insufficiency), positive history for atrial fibrillation or a coronary or cerebrovascular event during the previous 6 months, known neoplastic disease, and/or acute infections.
Blood samples for HbA1c measurement and circulating miRNA profiling (details below) were drawn upon enrollment (t 0), and the patients were then started on sitagliptin (100 mg once daily) as an adjunct to their metformin therapy. Glycemic control was assessed 3 and 15 months later (t 3 and t 15, resp.) (as shown in Figure 1). Patients were classified as “responders” (R) if they exhibited good glycemic control at both time points, reflected by an HbA1c level of <7.5% or an HbA1c reduction of >0.5% relative to the level recorded at t 0. Patients who failed to meet these criteria were classified as “nonresponders” (NR). On the basis of these findings, plasma samples were pooled into the following five groups: (1) baseline samples from patients that emerged as responders (R-t 0 pool), (2) baseline samples from nonresponders (NR-t 0 pool), (3) t 3 samples from responders (R-t 3 pool), (4) t 15 samples from responders (R-t 15 pool), and (5) t 15 samples from nonresponders (NR-t 15 pool) (Figure 2). Characteristics of the patients enrolled in the study are reported in Table 1.
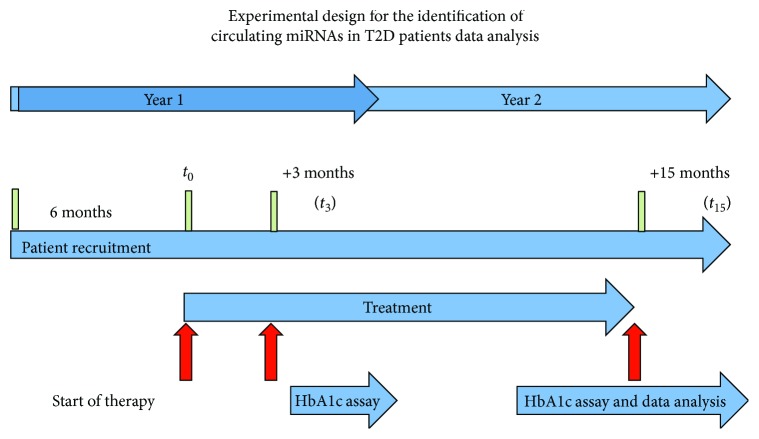
Figure 1.
Experimental design for the identification of circulating miRNAs in T2D patients. The duration of the study was 2 years. The blue horizontal arrow indicates the duration of the different phases of the project. The red vertical arrows indicate the main study periods. In year 1, patients were enrolled and started on metformin + sitagliptin (see Materials and Methods for details). After 3 and 15 months of treatment (t 3 and t 15), HbA1c values were reassessed and patients were classified as nonresponders (NR) or responders (R). Plasma miRNA levels at baseline (t 0) and t 15 from groups R and NR were compared as indicated. Comparison of plasma pools: (1) NR-t 0 versus R-t 0, (2) R-t 15 versus R-t 0, (3) NR-t 15 versus NR-t 0, and (4) NR-t 15 versus R-t 15.
Figure 2.
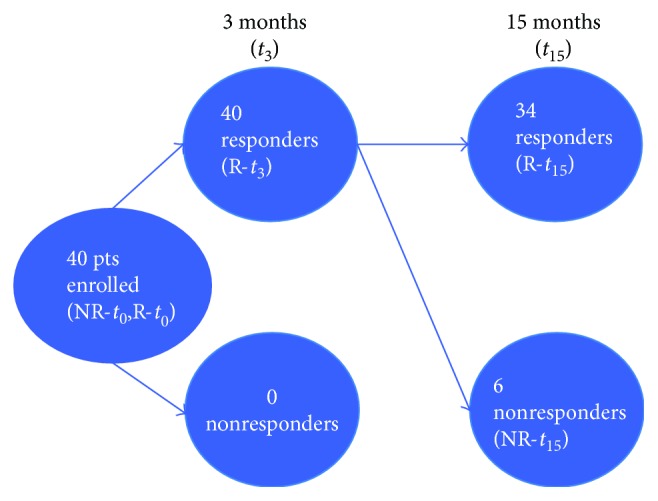
Glycemic control statuses of the patients at baseline and 3 and 15 months after initiation of metformin + sitagliptin. All 40 patients in poor metabolic control were enrolled. HbA1c levels were evaluated after 3 and 15 months from the addition of sitagliptin. On the basis of HbA1c values, patients were divided into responders and nonresponders. All patients showed an initial metabolic response to therapy (t 3), whereas after 15 months (t 15), 34/40 were responders. Based on this information, patients were divided into five groups: (1) t 0 responder samples (R-t 0), (2) t 0 nonresponder samples (NR-t 0), (3) t 3 responder samples (R-t 3), (4) t 15 responder samples (R-t 15), and (5) t 15 nonresponder samples (NR-t 15). miRNA profiling was performed at baseline and after 15 months of sitagliptin addition.
Table 1.
Characteristics of R and NR patients enrolled in the study.
| Parameters | Responders (R) | Nonresponders (NR) | ||||
|---|---|---|---|---|---|---|
| Age (y) | 66.62 ± 2.31 | 67.89 ± 2.24 | ||||
| Gender, males/females (n) | 17/17 | 4/2 | ||||
| Time since T2D diagnosis (y) | 11 ± 2 | 10 ± 3 | ||||
| Time points (in months) | t 0 | t 3 | t 15 | t 0 | t 3 | t 15 |
| Body weight (kg) | 72.30 ± 4.92 | 70.36 ± 4.89 | 70.43 ± 5.23 | 79.10 ± 12.19 | 78.57 ± 13.25 | 79.33 ± 12.99 |
| HbA1c (%) | 7.75 ± 0.38 | 6.38 ± 0.18 | 7.10 ± 0.28 | 7.59 ± 0.16 | 6.73 ± 0.39 | 7.81 ± 0.51 |
Data are means ± SD unless indicated otherwise.
2.2. Isolation and Profiling of Circulating miRNAs
For miRNA studies, 5 mL of blood was collected from each patient in EDTA-treated tubes. Within 30 minutes of collection, the samples were centrifuged for 10 minutes at 2000 ×g at room temperature (RT), and the plasma thus obtained was divided into 250 μL aliquots and stored at −80°C. To eliminate the risk of bias related to hemolysis [21], all plasma samples were visually assessed and those that were hemolyzed, icteric, or lipemic were excluded from the analysis. We also evaluated the expression of miRNAs susceptible to hemolysis, such as miR-324-3p, miR-454, and miR-652 [21]. Indeed, these miRNAs were not detected in our samples, confirming that none of the samples utilized in the study were hemolyzed.
Plasma samples from all patients in a given group (see above) were thawed on ice and pooled. A miRNA ABC purification kit (Applied Biosystems, Thermo Scientific) was used, according to the manufacturer's instructions. Briefly, 50 μL of each plasma pool was mixed with 100 μL of lysis buffer and centrifuged briefly before the addition of 100 nM of ath-miR-159a (used as a positive external control). Samples were then mixed with freshly prepared magnetic beads (80 × 106) and incubated in a magnetic rack (40 min at 30°C). Bead-hybridized miRNAs were then washed to remove any contaminants. Elution buffer (50 μL) was added, and the sample was incubated for 3 minutes in a ThermoMixer (1200 rpm, 70°C) and placed for 1 minute in a magnetic rack to clear solutions. The supernatants were then transferred into clean tubes and placed on ice.
miRNAs were reverse-transcribed using specific primers according to Applied Biosystems protocols. The cDNAs were preamplified using reagents from Applied Biosystems (Thermo Scientific), and the products were subjected to miRNA expression profiling. The latter was performed by RT-qPCR with Taqman Low-Density Array microfluidic cards (Human miR v3.0, Applied Biosystems), as previously described [22].
2.3. miRNA Expression Analysis
Statistical analysis was performed with StatMiner™ software, v. 5.0 (Integromics™) [23]. miRNA expression levels in plasma pools were subjected to global expression normalization, and relative levels were calculated with the comparative threshold cycle (Ct) method. miRNAs with Ct values > 33 were excluded. Differential expression between groups was assessed with the limma test and considered statistically significant when P values were <0.05. Heat maps were generated in the R environment (http://www.r-project.org/), using differentially expressed miRNAs as input. The Bray-Curtis and average linkage methods were used to cluster samples (hclust) and generate heat maps (heatmap.2).
3. Results and Discussion
All 40 enrolled patients completed 15 months of treatment with metformin + sitagliptin without occurrence of adverse events. After three months of the study treatment, all patients met the predefined criteria for good metabolic control. In contrast, at t 15, only 34/40 (85%) were still in good metabolic control; the remaining six (15%) had HbA1c levels that exceeded 7.5% (n = 6). On the basis of these findings, patients were divided into responders (group R), which included the 34 patients in good glycemic control at t 3 and t 15, and nonresponders (group NR, 6/40), whose initial response to the addition of sitagliptin at t 3 was not maintained at t 15.
3.1. Circulating miRNA Profiles of Groups R and NR
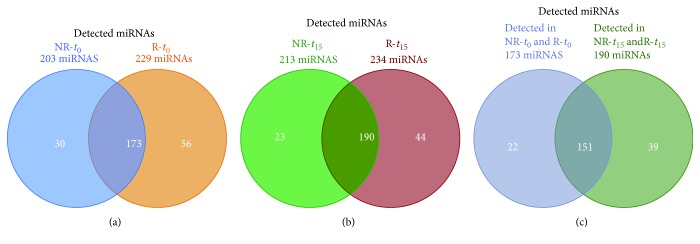
We then compared groups R and NR in terms of miRNA expression profiles in their t 0 plasma sample pools. Of the 754 miRNAs analyzed, 203 (27%) were detected in group NR and 229 (30%) in group R (Supplementary Tables 1 and 2, resp.). The reliability of our findings is supported by the fact that 173 miRNAs found in both groups of plasma samples (Figure 3(a)) included several miRNAs previously known to be associated with T2D; specifically, 36% of those were found in the BioM2MetDisease database (http://www.bio-bigdata.com/BioM2MetDisease/browse) and identified in a recent systematic review by He et al. [24] (Table 2). Similar findings emerged when we compared circulating miRNA profiles after 15 months of combined metformin + sitagliptin therapy. Indeed, the number of detectable miRNAs in the NR group (213/754, 28%) was comparable to that in the R group (234/754, 31%) (Supplementary Tables 3 and 4). In addition, the subset of miRNAs found in both t 15 pools (Figure 3(b)) comprised many of those with known links to T2D (39% in BioM2MetDisease and He et al. [24]) (Table 3). Finally, 151 miRNAs were detected in the plasma pools for both groups, before (t 0) and after the addition of sitagliptin (t 15) (Figure 3(c), Supplementary Table 6). Notably, miRNAs described to be linked to T2D in previous studies were detected also in our cohort of elderly T2D patients, further strengthening their potential role as T2D biomarkers.
Figure 3.
Venn diagram of circulating miRNAs detected in the elderly T2D cohort and in the R and NR subcohorts. Patients were classified as responders (R) or nonresponders (NR) based on their glycemic control status after 15 months of treatment with metformin + sitagliptin (t 15). Venn diagrams show the number of miRNAs detected in NR and R plasma samples collected (a) before the start of combined therapy (baseline, t 0) and (b) at t 15 and (c) at both t 0 and t 15.
Table 2.
miRNAs with known links to T2D found in baseline (t 0) plasma pools from groups NR and R.
| T2D-linked microRNAs at t 0 | |
|---|---|
| hsa-let-7d | |
| hsa-miR-103 | |
| hsa-miR-126 | |
| hsa-miR-130b | |
| hsa-miR-142-3p | |
| hsa-miR-144 | |
| hsa-miR-145 | |
| hsa-miR-146a | |
| hsa-miR-155 | |
| hsa-miR-17-5p | |
| hsa-miR-186 | |
| hsa-miR-191 | |
| hsa-miR-192 | |
| hsa-miR-195 | |
| hsa-miR-197 | |
| hsa-miR-20b | |
| hsa-miR-21 | |
| hsa-miR-222 | |
| hsa-miR-223 | |
| hsa-miR-23a | |
| hsa-miR-26a | |
| hsa-miR-27a | |
| hsa-miR-29a | |
| hsa-miR-30d | |
| hsa-miR-30e | |
| hsa-miR-342 | |
| hsa-miR-34a | |
| hsa-miR-375 | |
| hsa-miR-378 | |
| hsa-miR-423-5p | |
| hsa-miR-451a | |
| hsa-miR-483-3p | |
| hsa-miR-486 | |
| hsa-miR-571 | |
| hsa-miR-572 | |
| hsa-miR-593 | |
| hsa-miR-661 | |
| hsa-miR-770-5p | |
| hsa-miR-9 | |
| hsa-miR-92a |
Table 3.
miRNAs with known links to T2D found in t 15 plasma pools from groups NR and R.
| T2D-linked miRNAs at t 15 | |
|---|---|
| hsa-let-7d | |
| hsa-miR-103 | |
| hsa-miR-106b | |
| hsa-miR-122 | |
| hsa-miR-126 | |
| hsa-miR-130b | |
| hsa-miR-132 | |
| hsa-miR-140 | |
| hsa-miR-140-3p | |
| hsa-miR-142-3p | |
| hsa-miR-144 | |
| hsa-miR-145 | |
| hsa-miR-146a | |
| hsa-miR-155 | |
| hsa-miR-17-5p | |
| hsa-miR-181a | |
| hsa-miR-186 | |
| hsa-miR-18a | |
| hsa-miR-191 | |
| hsa-miR-192 | |
| hsa-miR-195 | |
| hsa-miR-197 | |
| hsa-miR-20b | |
| hsa-miR-21 | |
| hsa-miR-221 | |
| hsa-miR-222 | |
| hsa-miR-223 | |
| hsa-miR-23a | |
| hsa-miR-24 | |
| hsa-miR-26a | |
| hsa-miR-27a | |
| hsa-miR-28-3p | |
| hsa-miR-30d | |
| hsa-miR-30e | |
| hsa-miR-320 | |
| hsa-miR-342 | |
| hsa-miR-34a | |
| hsa-miR-375 | |
| hsa-miR-378 | |
| hsa-miR-423-5p | |
| hsa-miR-451a | |
| hsa-miR-483-3p | |
| hsa-miR-486 | |
| hsa-miR-571 | |
| hsa-miR-572 | |
| hsa-miR-593 | |
| hsa-miR-661 | |
| hsa-miR-770-5p | |
| hsa-miR-92a | |
| hsa-miR-96 |
3.2. Circulating miRNAs in Elderly T2D Patients: Differential Expression between Responders and Nonresponders at Baseline
Our next goal was to identify circulating miRNAs that might be correlated with a positive metabolic response to therapy. To this end, we first compared the plasma levels of the 173 miRNAs found in the t 0 plasma pools from groups R and NR. This analysis revealed 20 miRNAs that were differentially expressed in the two groups (Table 4, Figure 4(a)) and might thus be potentially useful for predicting the response to therapy. As noted in Table 4, plasma levels of eight of these 20 miRNAs are known to be potentially influenced by blood cell contamination [21]. However, this factor is unlikely to have played a role in this analysis, since occurrence of hemolysis was excluded in all plasma samples analyzed.
Table 4.
miRNAs that were differentially expressed in t 0 plasma pools from groups NR and R.
| Regulation | miRNA | Linear fold change | P value |
|---|---|---|---|
| Upregulated in NR-t 0 | hsa-miR-1208 | 2.05 | 1.84E − 02 |
| hsa-miR-1225-3p | 2.27 | 1.20E − 02 | |
| hsa-miR-1252-5p | 901.88 | 4.52E − 02 | |
| hsa-miR-338-5p | 7.56 | 4.22E − 02 | |
| hsa-miR-375 | 68.83 | 4.35E − 02 | |
| hsa-miR-378∗ | 15.26 | 3.81E − 02 | |
| hsa-miR-571 | 2.39 | 1.32E − 02 | |
| hsa-miR-595 | 540.84 | 4.66E − 02 | |
| hsa-miR-601∗ | 14179.56 | 4.97E − 02 | |
| hsa-miR-885-5p | 4.58 | 3.00E − 02 | |
|
| |||
| Downregulated in NR-t 0 | hsa-let-7b-5p∗ | 0.18 | 1.53E − 03 |
| hsa-let-7d-5p | 0.19 | 1.11E − 02 | |
| hsa-miR-1247-5p | 0.20 | 9.91E − 03 | |
| hsa-miR-16-5p∗ | 0.21 | 1.51E − 03 | |
| hsa-miR-223-5p | 0.32 | 1.83E − 02 | |
| hsa-miR-23a-5p∗ | 0.20 | 8.79E − 03 | |
| hsa-miR-30b-5p∗ | 0.36 | 2.74E − 02 | |
| hsa-miR-320a | 0.23 | 5.22E − 03 | |
| hsa-miR-451a∗ | 0.31 | 3.88E − 03 | |
| hsa-miR-93-5p∗ | 0.09 | 2.03E − 02 | |
∗Hemolysis-susceptible miRNAs as reported in Kirschner et al. [21].
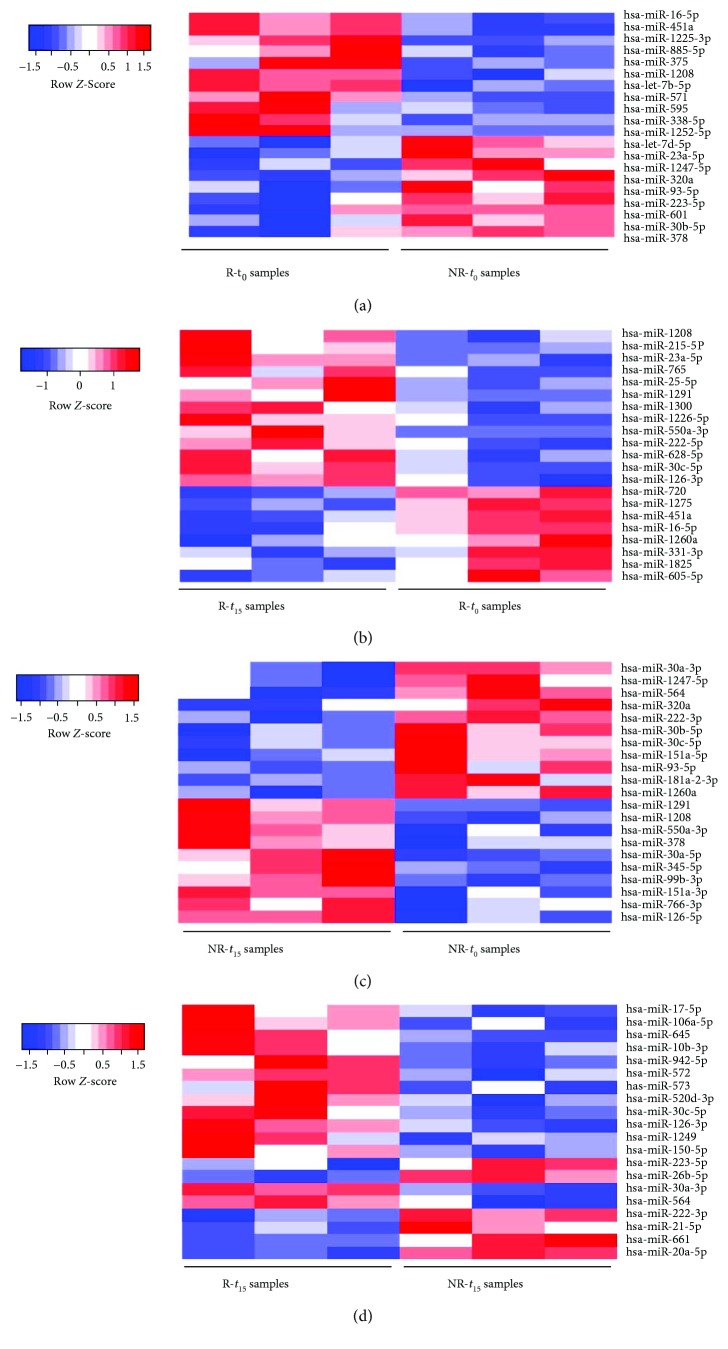
Figure 4.
Heat maps showing circulating microRNAs that were differentially expressed in plasma samples from (a) R and NR patients at baseline (t 0), prior to the addition of sitagliptin to the maximum-dose metformin regimen; (b) R at t 0 and after 15 months of sitagliptin (t 15); (c) NR patients at t 15 and t 0; and (d) R and NR patients at t 15.
Expression of 10 miRNAs was significantly lower in plasma from NR-t0 versus R-t0 (Table 4, Figure 4(a)); these included three miRNAs of particular interest: let-7d, miR-223, and miR-23a, whose expression was reported to be downregulated in T2D patients when compared to healthy individuals [6, 12, 25, 26]. Reduced expression of let-7d, miR-223, and miR-23a in NR patients suggests that these miRNAs may represent potential biomarkers of response to sitagliptin therapy.
Other miRNAs resulted to be significantly more expressed in NR-t 0 plasma samples. These included miR-375, whose high expression has been associated with β-cell dysfunction [27, 28]; miR-571, previously reported to be hyperexpressed in plasma samples from T2D patients compared to healthy controls [29]; and miR-378, which was associated with obesity-related insulin resistance [30, 31].
Increasing evidence has shown the involvement of miRNAs in the development of the endocrine pancreas, as well as in the regulation of insulin secretion, insulin signaling, and insulin gene transcription. Indeed, in Dicer-1 conditional knockout mice, loss of miRNAs in β-cells causes major defects in glucose homeostasis and in insulin secretion, with a marked reduction in insulin content. Moreover, studies in rodent models of T2D have revealed changes in miRNA expression in β-cells [32].
In this context, our data provide evidence that some miRNAs previously reported as regulators of glucose homeostasis in critical tissues (e.g., endocrine pancreas) may also represent circulating biomarkers for disease staging and/or for predicting response to glucose-lowering therapy.
3.3. Circulating miRNAs in the Elderly T2D Patients Who Responded to Sitagliptin Treatment
To identify miRNAs potentially modulated by sitagliptin therapy, we compared miRNA expression levels at t 0 and at t 15 in plasma samples from the R group of patients. Twenty-one miRNAs were differentially expressed between the two plasma pools (Table 5, Figure 4(b)). Of note, expression levels of miR-222, previously reported to be hyperexpressed in plasma samples from T2D patients versus healthy controls [33], were reduced at t 15 versus t 0.
Table 5.
miRNAs that were differentially expressed in t 0 and t 15 plasma pools from group R.
| Regulation | miRNA | Linear fold change | P value |
|---|---|---|---|
| Upregulated in R-t 15 | hsa-miR-126-3p | 3.30 | 2.05E − 02 |
| hsa-miR-30c-5p | 3.44 | 4.71E − 03 | |
| hsa-miR-331-3p∗ | 163.08 | 3.12E − 02 | |
|
| |||
| Downregulated in R-t 15 | hsa-miR-1208 | 0.53 | 3.55E − 02 |
| hsa-miR-1226-5p | 0.42 | 2.78E − 02 | |
| hsa-miR-1260a | 0.35 | 4.07E − 02 | |
| hsa-miR-1275 | 0.24 | 1.93E − 03 | |
| hsa-miR-1291 | 0.15 | 3.98E − 02 | |
| hsa-miR-1300 | 0.29 | 2.08E − 02 | |
| hsa-miR-16-5p∗ | 0.27 | 2.23E − 02 | |
| hsa-miR-1825 | 0.50 | 4.13E − 02 | |
| hsa-miR-215-5p | 0.05 | 3.76E − 02 | |
| hsa-miR-222-5p | 0.26 | 3.66E − 02 | |
| hsa-miR-23a-5p∗ | 0.18 | 4.28E − 03 | |
| hsa-miR-25-5p∗ | 0.03 | 3.26E − 02 | |
| hsa-miR-451a∗ | 0.35 | 9.49E − 03 | |
| hsa-miR-550a-3p | 0.34 | 1.27E − 02 | |
| hsa-miR-605-5p | 0.19 | 1.34E − 02 | |
| hsa-miR-628-5p | 0.00 | 3.07E − 02 | |
| hsa-miR-720 | 0.41 | 5.80E − 03 | |
| hsa-miR-765 | 0.01 | 4.16E − 02 | |
∗Hemolysis-susceptible miRNAs as reported in Kirschner et al. [21].
Conversely, three miRNAs were found significantly upregulated in the R-t 15 plasma pool (Table 5). These included miR-126-3p, which was reported to be decreased in T2D patients (with or without complications) versus healthy controls [34] and in T2D patients with major cardiovascular events [35]. Moreover, this miRNA has been proposed as a biomarker for the detection of prediabetes and diabetes. miR-30c, another miRNA significantly upregulated in the R-t 15 plasma pool, was recently shown to exert protective effects on diabetic nephropathy [36] and cardiomyopathy [37]. Interestingly, gliptins inhibit the degradation of several peptides and chemokines and reduce tissue inflammation by suppressing macrophage activation and M2 macrophage response. These findings suggest that glucagon-like peptide-1- (GLP-1-) based treatments provide additional benefits beyond glycemic control, including vascular protection and improved bone health [38].
3.4. Circulating miRNAs in Elderly T2D Patients Who Did Not Respond to Sitagliptin Treatment
We then compared miRNA expression levels at t 0 and at t 15 in plasma samples from the NR group of patients. As shown in Table 6 and in Figure 4(c), 21 miRNAs were differentially expressed between the two time points, with 5/21 (miR-1208, miR-550a-3p, miR-30c-5p, miR-1260a, and miR-1291) showing similar posttreatment changes to group R (Tables 4 and 5). Consequently, this observation suggests the exclusion of these five miRNAs as possible biomarkers of response to sitagliptin therapy. Nevertheless, the protective effects of miR-30c-5p on diabetic nephropathy and cardiomyopathy [36, 37] underline the need for further investigation on the functional role of miR-30c in elderly T2D patients.
Table 6.
miRNAs that were differentially expressed in t 0 and t 15 plasma pools from group NR.
| Regulation | miRNA | Linear fold change | P value |
|---|---|---|---|
| Upregulated in NR-t 15 | hsa-miR-1247-5p | 4.57 | 2.50E − 02 |
| hsa-miR-126-5p | 1.62 | 4.72E − 02 | |
| hsa-miR-151a-3p | 2.50 | 1.87E − 02 | |
| hsa-miR-151a-5p | 3.73 | 9.29E − 03 | |
| hsa-miR-181a-2-3p | 134.20 | 1.75E − 02 | |
| hsa-miR-222-3p | 1.85 | 1.94E − 02 | |
| hsa-miR-30a-3p∗ | 1.70 | 4.95E − 02 | |
| hsa-miR-30a-5p | 1.59 | 4.80E − 02 | |
| hsa-miR-30b-5p | 2.18 | 2.63E − 02 | |
| hsa-miR-30c-5p | 2.08 | 3.87E − 02 | |
| hsa-miR-320a | 2.95 | 3.40E − 02 | |
| hsa-miR-378∗ | 2.05 | 4.62E − 02 | |
| hsa-miR-564 | 3.39 | 1.41E − 02 | |
| hsa-miR-766-3p | 3.46 | 4.62E − 02 | |
| hsa-miR-93-5p∗ | 6.81 | 2.63E − 02 | |
|
| |||
| Downregulated in NR-t 15 | hsa-miR-1208 | 0.23 | 3.70E − 03 |
| hsa-miR-1260a | 0.37 | 8.75E − 03 | |
| hsa-miR-1291 | 0.28 | 1.11E − 02 | |
| hsa-miR-345-5p | 0.15 | 9.84E − 03 | |
| hsa-miR-550a-3p | 0.48 | 4.08E − 02 | |
| hsa-miR-99b-3p∗ | 0.29 | 7.73E − 03 | |
∗Hemolysis-susceptible miRNAs as reported in Kirschner et al. [21].
A comparison between the differentially expressed miRNAs of NR versus R at baseline (Table 4) and NR at t 15 versus baseline (Table 6) allowed us to observe that one of the miRNAs that were more abundant in NR plasma than in R plasma at baseline, miR-378, increased during the treatment period, reaching levels in the NR-t 15 plasma pool that were significantly higher than those in the NR-t 0 pool. This result allowed us to propose miR-378 as a negative biomarker candidate for response to sitagliptin therapy.
In this context, our results could open new perspectives providing the basis for further investigations of the reported dysregulated miRNAs as markers of response to gliptins.
3.5. Circulating miRNAs in Elderly T2D Patients after 15 Months of Sitagliptin Treatment
To identify miRNAs that might be affected by sitagliptin treatment, we compared circulating miRNA levels in plasma samples at t 15 from the two subgroups. Twenty miRNAs were differentially expressed in plasma from NR versus R patients (Table 7, Figure 4(d)). Ten of the 20 miRNAs were significantly more expressed in the plasma samples from the NR group. Among the upregulated miRNAs, we observed an increased expression of miR-661 and miR-572 in NR-t 15, in accordance with previous studies performed in T2D patients [26, 39].
Table 7.
miRNAs that were differentially expressed in the t 15 plasma pools from groups NR and R.
| Regulation | miRNA | Linear fold change | P value |
|---|---|---|---|
| Upregulated in NR-t 15 | hsa-miR-10b-3p | 2.49 | 3.39E − 02 |
| hsa-miR-1249 | 4.17 | 4.27E − 02 | |
| hsa-miR-30a-3p∗ | 3.84 | 1.62E − 03 | |
| hsa-miR-520d-3p | 6.78 | 1.79E − 02 | |
| hsa-miR-564 | 2.86 | 2.19E − 02 | |
| hsa-miR-572 | 2.15 | 1.81E − 02 | |
| hsa-miR-573 | 7.99 | 4.10E − 02 | |
| hsa-miR-645 | 2.85 | 1.90E − 02 | |
| hsa-miR-661 | 6.57 | 3.68E − 03 | |
| hsa-miR-942-5p | 2.63 | 1.70E − 02 | |
|
| |||
| Downregulated in NR-t 15 | hsa-miR-106a-5p∗ | 0.41 | 2.68E − 02 |
| hsa-miR-126-3p | 0.42 | 1.13E − 02 | |
| hsa-miR-150-5p | 0.49 | 4.21E − 02 | |
| hsa-miR-17-5p∗ | 0.43 | 3.28E − 02 | |
| hsa-miR-20a-5p∗ | 0.18 | 3.61E − 04 | |
| hsa-miR-21-5p∗ | 0.17 | 1.59E − 02 | |
| hsa-miR-222-3p | 0.35 | 5.14E − 03 | |
| hsa-miR-223-5p | 0.54 | 4.95E − 02 | |
| hsa-miR-26b-5p∗ | 0.41 | 4.04E − 03 | |
| hsa-miR-30c-5p | 0.45 | 2.62E − 02 | |
∗Hemolysis-susceptible miRNAs as reported in Kirschner et al. [21].
miRNAs that were hypoexpressed in the NR-t 15 pool included two of particular interest: miR-126-3p and miR-223. In the previous analysis of group R, miR-126-3p levels at t 15 were higher than those in the t 0 plasma pool, suggesting that this miRNA is a good candidate biomarker of successful metabolic response to therapy. Additionally, our previous comparison of t 0 plasma pools from the two groups revealed substantially lower levels of miR-223 in NR. This miRNA has already been associated with pancreatic islet β-cell function and glycemic control, and its expression is reportedly higher in individuals with pre-T2D and normal controls than in T2D patients [40, 41]. In light of these results, miR-223 appears to be a possible positive biomarker for monitoring patients' responsiveness to sitagliptin therapy.
Finally, we checked for a possible correlation between differentially expressed miRNAs, miR-126-3p, miR-223, and miR-378, and the patients' clinical features. We did not observe any statistically significant correlation between miR-126-3p, miR-223, and miR-378 and age, time from T2D diagnosis, body weight, and HbA1c levels.
An increasing body of evidence has linked diabetes to cardiovascular disease, renal failure, neuropathy, and osteoporosis, especially in elderly individuals, with a consequent increase in mortality, morbidity, and socioeconomic costs. Reliable biomarkers to predict drug response in these patients are urgently needed but still lacking. Glucose-lowering drugs, such as DPP-IVi, are known to differentially impact metabolic control and disease-related complications.
Our study is the first to provide a description of the circulating miRNAs that can be used as novel biomarkers for monitoring response to therapy in elderly T2D patients.
4. Conclusion
The results we obtained suggest that miR-378, miR-126-3p, and miR-223 represent candidate plasma biomarkers for disease staging and for predicting response to therapy in T2D elderly patients. High circulating levels of miR-378 appear to be a negative predictor of response to sitagliptin in elderly T2D patients. Indeed, miR-378 was more expressed in the NR-t 0 plasma pool than in the R-t 0 pool. In addition, its levels in the NR-t 15 plasma were even higher than those found in the NR-t 0 pool, and this result highlights its possible role as a biomarker of resistance to sitagliptin. In contrast, miR-126-3p and miR-223 seem to be markers of response to the drug. miR-126-3p levels have been reported to be lower in T2D patients than in healthy individuals [34]. Consistently, this miRNA was not differentially expressed in the plasma of R and NR patients at baseline. After 15 months of sitagliptin therapy, however, plasma levels in responders were significantly higher than those found in the NR group, suggesting that the addition of the DPP-IVi may have restored miR-126-3p levels to the range found in healthy subjects. As for miR-223, its expression at baseline was already significantly higher in the R group, and this difference persisted after 15 months of sitagliptin addition. This behavior is consistent with its potential role as a positive predictor of response to the drug.
Further work is needed to validate the role of these miRNAs as biomarkers in T2D patients, since this was a discovery study and the number of NR patients was limited. Interestingly, circulating miRNAs may reflect phenomena occurring at the level of those organs involved in T2D pathophysiology (e.g., endocrine pancreas, liver, and adipose tissue). Therefore, our data provide a snapshot of the circulating miRNAs that deserve further studies.
The translational relevance of our findings is immediate; in fact, data regarding miRNA profiles can produce results of potential impact not only predicting drug response of T2D elderly patients but also helping in selecting patients that could be suitable candidates to this therapy.
Acknowledgments
The work was partially supported by Fondazione Roma and University La Sapienza Research Grants. The authors would like to thank M. Everett Kent for editing revision.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this article.
Authors' Contributions
Giuseppina Catanzaro, Zein Mersini Besharat, Agnese Po, and Elisabetta Ferretti contributed equally to this work.
Supplementary Materials
Supplementary Tables 1–5 list the detected microRNAs in the plasma samples of interest (NR-t 0, R-t 0, NR-t 15, and R-t 15).
References
- 1.Guay C., Regazzi R. Circulating microRNAs as novel biomarkers for diabetes mellitus. Nature Reviews Endocrinology. 2013;9(9):513–521. doi: 10.1038/nrendo.2013.86. [DOI] [PubMed] [Google Scholar]
- 2.Pescador N., Pérez-Barba M., Ibarra J. M., Corbatón A., Martínez-Larrad M. T., Serrano-Ríos M. Serum circulating microRNA profiling for identification of potential type 2 diabetes and obesity biomarkers. PLoS One. 2013;8(10, article e77251) doi: 10.1371/journal.pone.0077251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kubota T., Kubota N., Kadowaki T. Imbalanced insulin actions in obesity and type 2 diabetes: key mouse models of insulin signaling pathway. Cell Metabolism. 2017;25(4):797–810. doi: 10.1016/j.cmet.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Zhu H., Leung S. W. Identification of microRNA biomarkers in type 2 diabetes: a meta-analysis of controlled profiling studies. Diabetologia. 2015;58(5):900–911. doi: 10.1007/s00125-015-3510-2. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell P. S., Parkin R. K., Kroh E. M., et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zampetaki A., Kiechl S., Drozdov I., et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circulation Research. 2010;107(6):810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 7.De Smaele E., Ferretti E., Gulino A. MicroRNAs as biomarkers for CNS cancer and other disorders. Brain Research. 2010;1338:100–111. doi: 10.1016/j.brainres.2010.03.103. [DOI] [PubMed] [Google Scholar]
- 8.Ciccacci C., Morganti R., di Fusco D., et al. Common polymorphisms in MIR146a, MIR128a and MIR27a genes contribute to neuropathy susceptibility in type 2 diabetes. Acta Diabetologica. 2014;51(4):663–671. doi: 10.1007/s00592-014-0582-2. [DOI] [PubMed] [Google Scholar]
- 9.Barbagallo D., Piro S., Condorelli A. G., et al. miR-296-3p, miR-298-5p and their downstream networks are causally involved in the higher resistance of mammalian pancreatic α cells to cytokine-induced apoptosis as compared to β cells. BMC Genomics. 2013;14(1):p. 62. doi: 10.1186/1471-2164-14-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guay C., Regazzi R. New emerging tasks for microRNAs in the control of β-cell activities. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2016;1861(12):2121–2129. doi: 10.1016/j.bbalip.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Sebastiani G., Po A., Miele E., et al. MicroRNA-124a is hyperexpressed in type 2 diabetic human pancreatic islets and negatively regulates insulin secretion. Acta Diabetologica. 2015;52(3):523–530. doi: 10.1007/s00592-014-0675-y. [DOI] [PubMed] [Google Scholar]
- 12.Yang Z., Chen H., Si H., et al. Serum miR-23a, a potential biomarker for diagnosis of pre-diabetes and type 2 diabetes. Acta Diabetologica. 2014;51(5):823–831. doi: 10.1007/s00592-014-0617-8. [DOI] [PubMed] [Google Scholar]
- 13.Butler P. C., Dry S., Elashoff R. GLP-1–based therapy for diabetes: what you do not know can hurt you. Diabetes Care. 2010;33(2):453–455. doi: 10.2337/dc09-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang F., He Y., Zhang R., Zeng Q., Zhao X. Combination therapy of metformin plus dipeptidyl peptidase-4 inhibitor versus metformin plus sulfonylurea and their association with a decreased risk of cardiovascular disease in type 2 diabetes mellitus patients. Medicine. 2017;96(36, article e7638) doi: 10.1097/MD.0000000000007638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Souza Cazarim M., da Cruz-Cazarim E. L. C., de Oliveira Baldoni A., et al. Cost-effectiveness analysis of different dipeptidyl-peptidase 4 inhibitor drugs for treatment of type 2 diabetes mellitus. Diabetes & Metabolic Syndrome: Clinical Research & Reviews. 2017;11(Supplement 2):S859–S865. doi: 10.1016/j.dsx.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 16.Mitrakou A. Pathogenesis of hyperglycaemia in type 2 diabetes. Diabetes, Obesity and Metabolism. 2002;4(4):249–254. doi: 10.1046/j.1463-1326.2002.00204.x. [DOI] [PubMed] [Google Scholar]
- 17.Kim Y.-G., Yoon D., Park S., et al. Dipeptidyl peptidase-4 inhibitors and risk of heart failure in patients with type 2 diabetes mellitus: a population-based cohort study. Circulation: Heart Failure. 2017;10(9, article e003957) doi: 10.1161/CIRCHEARTFAILURE.117.003957. [DOI] [PubMed] [Google Scholar]
- 18.Monami M., Ahrén B., Dicembrini I., Mannucci E. Dipeptidyl peptidase-4 inhibitors and cardiovascular risk: a meta-analysis of randomized clinical trials. Diabetes, Obesity and Metabolism. 2013;15(2):112–120. doi: 10.1111/dom.12000. [DOI] [PubMed] [Google Scholar]
- 19.Bansal V. S., Raja C. P., Venkataraman K., Vijayalakshmi M. Genes involved in pancreatic islet cell rejuvenation. The Indian Journal of Medical Research. 2013;137(4):695–703. [PMC free article] [PubMed] [Google Scholar]
- 20.Kanasaki K., Shi S., Kanasaki M., et al. Linagliptin-mediated DPP-4 inhibition ameliorates kidney fibrosis in streptozotocin-induced diabetic mice by inhibiting endothelial-to-mesenchymal transition in a therapeutic regimen. Diabetes. 2014;63(6):2120–2131. doi: 10.2337/db13-1029. [DOI] [PubMed] [Google Scholar]
- 21.Kirschner M. B., Edelman J. J. B., Kao S. C. H., Vallely M. P., van Zandwijk N., Reid G. The impact of hemolysis on cell-free microRNA biomarkers. Frontiers in Genetics. 2013;4:p. 94. doi: 10.3389/fgene.2013.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Catanzaro G., Besharat Z., Garg N., et al. MicroRNAs-proteomic networks characterizing human medulloblastoma-SLCs. Stem Cells International. 2016;2016:10. doi: 10.1155/2016/2683042.2683042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giannini G., Capalbo C., Ottini L., et al. Clinical classification of BRCA1 DNA missense variants: H1686Q is a novel pathogenic mutation occurring in the ontogenetically invariant THV motif of the N-terminal BRCT domain. Journal of Clinical Oncology. 2008;26(25):4212–4214. doi: 10.1200/JCO.2008.18.2089. [DOI] [PubMed] [Google Scholar]
- 24.He Y., Ding Y., Liang B., et al. A systematic study of dysregulated microRNA in type 2 diabetes mellitus. International Journal of Molecular Sciences. 2017;18(3):p. 456. doi: 10.3390/ijms18030456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu H., Shyh-Chang N., Segrè A. V., et al. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147(1):81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bijkerk R., Duijs J. M. G. J., Khairoun M., et al. Circulating microRNAs associate with diabetic nephropathy and systemic microvascular damage and normalize after simultaneous pancreas–kidney transplantation. American Journal of Transplantation. 2015;15(4):1081–1090. doi: 10.1111/ajt.13072. [DOI] [PubMed] [Google Scholar]
- 27.Zhao H., Guan J., Lee H. M., et al. Up-regulated pancreatic tissue microRNA-375 associates with human type 2 diabetes through β-cell deficit and islet amyloid deposition. Pancreas. 2010;39(6):843–846. doi: 10.1097/MPA.0b013e3181d12613. [DOI] [PubMed] [Google Scholar]
- 28.Higuchi C., Nakatsuka A., Eguchi J., et al. Identification of circulating miR-101, miR-375 and miR-802 as biomarkers for type 2 diabetes. Metabolism. 2015;64(4):489–497. doi: 10.1016/j.metabol.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Wang C., Wan S., Yang T., et al. Increased serum microRNAs are closely associated with the presence of microvascular complications in type 2 diabetes mellitus. Scientific Reports. 2016;6(1, article 20032) doi: 10.1038/srep20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu L.-l., Shi C.-m., Xu G.-f., et al. TNF-α, IL-6, and leptin increase the expression of miR-378, an adipogenesis-related microRNA in human adipocytes. Cell Biochemistry and Biophysics. 2014;70(2):771–776. doi: 10.1007/s12013-014-9980-x. [DOI] [PubMed] [Google Scholar]
- 31.Zanutto S., Pizzamiglio S., Ghilotti M., et al. Circulating miR-378 in plasma: a reliable, haemolysis-independent biomarker for colorectal cancer. British Journal of Cancer. 2014;110(4):1001–1007. doi: 10.1038/bjc.2013.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rottiers V., Näär A. M. MicroRNAs in metabolism and metabolic disorders. Nature Reviews Molecular Cell Biology. 2012;13(4):239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ortega F. J., Mercader J. M., Moreno-Navarrete J. M., et al. Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes Care. 2014;37(5):1375–1383. doi: 10.2337/dc13-1847. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y., Gao G., Yang C., et al. The role of circulating microRNA-126 (miR-126): a novel biomarker for screening prediabetes and newly diagnosed type 2 diabetes mellitus. International Journal of Molecular Sciences. 2014;15(6):10567–10577. doi: 10.3390/ijms150610567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olivieri F., Spazzafumo L., Bonafè M., et al. miR-21-5p and miR-126a-3p levels in plasma and circulating angiogenic cells: relationship with type 2 diabetes complications. Oncotarget. 2015;6(34):35372–35382. doi: 10.18632/oncotarget.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y., Yin Z., Li H., et al. miR-30c protects diabetic nephropathy by suppressing epithelial-to-mesenchymal transition in db/db mice. Aging Cell. 2017;16(2):387–400. doi: 10.1111/acel.12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raut S. K., Singh G. B., Rastogi B., et al. miR-30c and miR-181a synergistically modulate p53–p21 pathway in diabetes induced cardiac hypertrophy. Molecular and Cellular Biochemistry. 2016;417(1-2):191–203. doi: 10.1007/s11010-016-2729-7. [DOI] [PubMed] [Google Scholar]
- 38.de Graaf C., Donnelly D., Wootten D., et al. Glucagon-like peptide-1 and its class B G protein–coupled receptors: a long march to therapeutic successes. Pharmacological Reviews. 2016;68(4):954–1013. doi: 10.1124/pr.115.011395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding L., Ai D., Wu R., et al. Identification of the differential expression of serum microRNA in type 2 diabetes. Bioscience, Biotechnology, and Biochemistry. 2015;80(3):461–465. doi: 10.1080/09168451.2015.1107460. [DOI] [PubMed] [Google Scholar]
- 40.Nunez Lopez Y. O., Garufi G., Seyhan A. A. Altered levels of circulating cytokines and microRNAs in lean and obese individuals with prediabetes and type 2 diabetes. Molecular BioSystems. 2017;13(1):106–121. doi: 10.1039/C6MB00596A. [DOI] [PubMed] [Google Scholar]
- 41.Deiuliis J. A. MicroRNAs as regulators of metabolic disease: pathophysiologic significance and emerging role as biomarkers and therapeutics. International Journal of Obesity. 2016;40(1):88–101. doi: 10.1038/ijo.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables 1–5 list the detected microRNAs in the plasma samples of interest (NR-t 0, R-t 0, NR-t 15, and R-t 15).