Abstract
Otoconia are minute bio-crystals composed of glycoproteins, proteoglycans and CaCO3, and are indispensable for sensory processing in the utricle and saccule. Otoconia abnormalities and degeneration can cause or facilitate crystal dislocation to the ampulla, leading to vertigo and imbalance in humans. In order to better understand the molecular mechanism controlling otoconia formation and maintenance, we have examined the spatial and temporal expression differences of otoconial genes in the mouse inner ear at developmental, mature and aging stages using whole transcriptome sequencing (RNA-Seq) and quantitative RT-PCR. We show that the expression levels of most otoconial genes are much higher in the utricle and saccule compared to other inner ear tissues before postnatal stages in C57Bl/6J mice, and the expression of a few of these genes is restricted to the embryonic utricle and saccule. After the early postnatal stages, expression of all otoconial genes in the utricle and saccule is drastically reduced, while a few genes gain expression dominance in the aging ampulla, indicating a potential for ectopic debris formation in the latter tissue at old ages. The data suggest that the expression of otoconial genes is tightly regulated spatially and temporally during developmental stages and can become unregulated at aging stages.
Keywords: otoconial genes, development, RNA-Seq, temporal, spatial, aging
Introduction
The utricle and saccule in the vertebrate inner ear are responsible for the sense of linear acceleration and gravity. The two vestibular end organs distinguish themselves from the other inner ear tissues, such as the semicircular canals, ampulla, and cochlea, by possessing tens of thousands of minute bio-crystals called otoconia. Head tilt and linear head motion cause displacement of the otoconial complex against the underlying sensory epithelia (macula), producing a shearing force which deflects the hair bundles and subsequently depolarizes the sensory hair cells. Hence, the size, shape and organization of the otoconia crystals affect the stimulus input to the afferent nerve and the precision of perception of motion (Anniko et al. 1988;Jones et al. 1999;Jones et al. 2004;Kozel et al. 1998;Simmler et al. 2000;Zhao et al. 2008). Furthermore, dislocation of otoconia into the ampulla generates false information of angular acceleration in response to head position changes with respect to gravity. An example for the latter is benign paroxysmal positional vertigo (BPPV), the most common cause of vertigo in humans (Salvinelli et al. 2004;Schuknecht 1962;Schuknecht 1969).
The above underscores the critical importance of studying the molecular mechanisms responsible for otoconia formation, anchoring and maintenance. To date, over 24 genes in mice have been identified to affect otoconia (Lundberg et al. 2015). About half of these genes encode matrix proteins, which function as otoconial constituent and anchoring proteins (Lundberg et al. 2006;Takemura et al. 1994;Thalmann et al. 2006;Verpy et al. 1999;Wang et al. 1998;Xu et al. 2010;Zhao et al. 2007). The other half are critical regulators of otoconia formation, mutations in which usually lead to absent otoconia. These regulators consist of ion channels and pumps [Atp2b2 (aka PMCA2), Slc26a4 (Pendrin), Slc30a4, TRPVs], reactive oxygen species (ROS)-producing enzymes and partners (Nox3 and partners Noxo1, Noxa1 and p22phox), and trafficking proteins (Otopetrin-1, Pallidin, Atg4 & 5) (Everett et al. 2001;Huang and Gitschier 1997;Hurle et al. 2003;Kiss et al. 2006;Kozel et al. 1998;Marino et al. 2010;Nakano et al. 2008;Paffenholz et al. 2004).
Most of the constituent proteins interact with each other to form an organic framework for the efficient deposition of mineral crystallites in an ordered manner (Deans et al. 2010;Tohse et al. 2008;Yang et al. 2011). The regulatory proteins are proposed to impact otoconia development by adjusting the endolymphatic ion concentrations to establish an appropriate micro-environment for crystal seeding and growth, or by modulating the secretion of otoconins (Dror et al. 2010;Kim et al. 2010;Nakano et al. 2008;Paffenholz et al. 2004;Sollner et al. 2004). The anchoring proteins comprise the honeycomb and fibrous meshwork layers (collectively called the otoconial membrane) in the utricle and saccule for the site-specific anchoring of otoconia (Xu et al. 2010;Simmler et al. 2000;Zwaenepoel et al. 2002;Legan and Richardson 1997;Legan and Richardson 1997).
The identification and functional studies of otoconial proteins in recent years have shed some light on the bio-mineralization processes. However, the whole picture regarding otoconia formation and maintenance is far from being complete. In order to better understand mechanisms underlying the temporal and spatial specificity of otoconial morphogenesis, we examined the expression changes of critical otoconial genes in different inner ear compartments at developmental, adult and aging stages in this study.
Results
Differential expression of otoconial genes in the murine inner ear
The onset of otoconia seeding can be tracked back to embryonic day 14.5 (E14.5) or 15.5, and the seeds continue to grow until at around postnatal day 7 (P7) (Anniko 1980;Takumida and Harada 1984). To identify genes possibly responsible for the spatial specific development of otoconia, we performed RNA-Seq to compare the expression levels of otoconia-related genes in the utricle and saccule, as compared to the cochlea, in C57BL/6J mice at the age of postnatal day 0 (P0). As shown in Table 1, most otoconial genes [except genes encoding Sparcl1 (Sc1), Sparc, Spp1, Lum, Car3, Trpv2 and the tectorins] are more highly expressed in the utricle + saccule than the cochlea. Otop1, Otol1 and Nox3 show the greatest difference that is not a result of alternative isoform expression, and Otop1 and Nox3 are essentially absent in the cochlea. The full names of the otoconial genes are listed in Supplementary Table 1, and the raw RNA-Seq data are presented in Supplementary Table 2.
Table 1.
Expression of otoconial genes by RNA-Seq analysis (as fragments per kilobase per million reads, or FPKM) of epithelial cells from the utricle and saccule (U+S), as compared to the cochlea (Co), in C57BL/6J mice at age P0.
| Product Type | Gene | U+S | Co | U+S/Co |
|---|---|---|---|---|
| Constituent proteins | Oc90 | 10405.5 | 7672.4 | 1.4 |
| Otol1 | 296.9 | 3.3 | 91.4 | |
| Sparc | 4059.5 | 8192.5 | 0.5 | |
| Sparcl1 | 665.7 | 1154.2 | 0.6 | |
| Spp1 (splice variant 1) | 392.0 | 815.7 | 0.5 | |
| Spp1 (splice variant 2) | 35.3 | 83.2 | 0.4 | |
| Dmp1 | 2.4 | 0.3 | 9.4 | |
| Lum | 105.1 | 1167.8 | 0.1 | |
| Fmod | 14.1 | 2.8 | 5.1 | |
| Omd | 3.7 | 1.0 | 3.8 | |
| Ahsg | 0.2 | 0.0 | N/A | |
|
| ||||
| Regulatory proteins | Otop1 | 20.4 | 0.1 | 183.5 |
| Nox3 | 25.0 | 0.5 | 48.8 | |
| Cyba | 230.2 | 114.6 | 2.0 | |
| Noxo1 | 78.4 | 14.4 | 5.4 | |
| Noxa1 (splice variant 1) | 1.5 | 0.0 | 3068.6 | |
| Noxa1 (splice variant 2) | 0.0 | 1.2 | 0.0 | |
| Slc26a4 | 16.8 | 1.5 | 10.9 | |
| Slc30a4 | 3.2 | 1.2 | 2.8 | |
| Pldn | 24.6 | 8.6 | 2.9 | |
| Atg4a | 7.7 | 4.9 | 1.6 | |
| Atg4b | 30.4 | 13.1 | 2.3 | |
| Atg4c | 13.4 | 3.9 | 3.4 | |
| Atg4d | 9.1 | 1.1 | 8.0 | |
| Atp2b2 | 6.6 | 1.5 | 4.3 | |
| Car2 | 172.9 | 142.2 | 1.2 | |
| Car3 | 1403.4 | 4716.3 | 0.3 | |
| Car7 | 73.6 | 1.3 | 55.0 | |
| Car12 | 91.0 | 11.7 | 7.8 | |
| Car13 | 24.3 | 8.1 | 3.0 | |
| Car14 | 87.7 | 77.4 | 1.1 | |
| Trpv2 | 0.9 | 3.3 | 0.3 | |
| Trpv4 | 24.1 | 6.3 | 3.8 | |
| Trpv6 | 2.0 | 0.1 | 33.2 | |
|
| ||||
| Anchoring proteins | Otog | 86.5 | 6.6 | 13.0 |
| Otogl | 10.8 | 4.7 | 2.3 | |
| Otoa | 229.1 | 98.1 | 2.3 | |
| Tecta | 103.3 | 265.5 | 0.4 | |
| Tectb | 620.8 | 728.7 | 0.9 | |
Note: Otol1, Lum, Fmod and Omd are also anchoring proteins, and Tecta is also an otoconial component protein. The full names of the genes are listed in Supplementary Table 1.
We also compared gene expression in the utricle and saccule in young (P0) versus old C57BL/6J mice (8 months old, as 8M in Table 2). RNA-Seq results revealed that the expression levels of most otoconia-related genes are greatly reduced in older mice. The raw RNA-Seq data are also presented in Supplementary Table 2.
Table 2.
Expression levels of otoconial genes by RNA-Seq analysis (as fragments per kilobase per million reads, or FPKM) of epithelial cells from the C57BL/6J utricle and saccule at age P0 and 8 months (8M).
| Product Type | Gene | P0 | 8M | 8M/P0 |
|---|---|---|---|---|
| Constituent proteins | Oc90 | 10405.5 | 13.4 | 0.0 |
| Otol1 | 296.9 | 63.6 | 0.2 | |
| Sparc | 4059.5 | 1451.5 | 0.4 | |
| Sparcl1 | 665.7 | 378.9 | 0.6 | |
| Spp1 (splice variant 1) | 392.0 | 5433.6 | 13.9 | |
| Spp1 (splice variant 2) | 35.3 | 234.2 | 6.6 | |
| Dmp1 | 2.4 | 6.3 | 2.6 | |
| Lum | 105.1 | 27.2 | 0.3 | |
| Fmod | 14.1 | 3.8 | 0.3 | |
| Omd | 3.7 | 20.8 | 5.6 | |
| Ahsg | 0.2 | 0.0 | 0.0 | |
|
| ||||
| Regulatory proteins | Otop1 | 20.4 | 2.2 | 0.1 |
| Nox3 | 25.0 | 33.2 | 1.3 | |
| Cyba | 230.2 | 418.6 | 1.8 | |
| Noxo1 | 78.4 | 40.0 | 0.5 | |
| Noxa1 (splice variant 1) | 1.5 | 0.0 | 0.0 | |
| Noxa1 (splice variant 2) | 0.0 | 0.1 | N/A | |
| Slc26a4 | 16.8 | 51.1 | 3.0 | |
| Slc30a4 | 3.2 | 2.7 | 0.9 | |
| Pldn | 24.6 | 11.6 | 0.5 | |
| Atg4a | 7.7 | 6.9 | 0.9 | |
| Atg4b | 30.4 | 18.4 | 0.6 | |
| Atg4c | 13.4 | 4.1 | 0.3 | |
| Atg4d | 9.1 | 6.0 | 0.7 | |
| Atp2b2 | 6.6 | 15.8 | 2.4 | |
| Car2 | 172.9 | 180.9 | 1.0 | |
| Car3 | 1403.4 | 1278.8 | 0.9 | |
| Car7 | 73.6 | 47.3 | 0.6 | |
| Car12 | 91.0 | 65.6 | 0.7 | |
| Car13 | 24.3 | 47.0 | 1.9 | |
| Car14 | 87.7 | 266.7 | 3.0 | |
| Trpv2 | 0.9 | 1.4 | 1.6 | |
| Trpv4 | 24.1 | 7.5 | 0.3 | |
| Trpv6 | 2.0 | 0.1 | 0.1 | |
|
| ||||
| Anchoring proteins | Otog | 86.5 | 67.4 | 0.8 |
| Otoa | 229.1 | 109.5 | 0.5 | |
| Otogl | 10.8 | 2.6 | 0.2 | |
| Tecta | 103.3 | 14.7 | 0.1 | |
| Tectb | 620.8 | 132.6 | 0.2 | |
Expression levels of critical otoconial genes were individually confirmed by quantitative RT-PCR (qRT-PCR) and are described in sections below. The murine inner ear cell types expressing some of the genes presented below at embryonic stages have been previously reported: Oc90 (Ignatova et al. 2004;Verpy et al. 1999;Wang et al. 1998), Otolin (Deans et al. 2010;Yang et al. 2011;Zhao et al. 2007), Sc1 (Xu et al. 2010), Otop1 (Hurle et al. 2003;Kim et al. 2010), and Nox3 (Banfi et al. 2004). Oc90 is expressed in all non-sensory epithelium, whereas the other genes are expressed in the sensory epithelium.
Temporal changes in Oc90 transcript versus protein
Quantitative RT-PCR was performed to confirm the RNA-Seq results, and to further examine the spatial and temporal expression changes of critical otoconial genes in different inner ear compartments including the utricle and saccule (U+S), ampulla, canal and cochlea. Epithelia of these tissues from mice of various ages at E17.5, P0, P7, 1.5 months and 18 months old (abbreviated as 1.5M and 18M, respectively, in the figures) were analyzed. In each inner ear compartment, as shown in Figure 1A-D, the expression level of the predominant otoconial protein Oc90 was the highest at E17.5 and drastically decreased thereafter.
Figure 1. Temporal changes in Oc90 transcript (A-D) and protein (E-H) in mouse inner ear tissues.
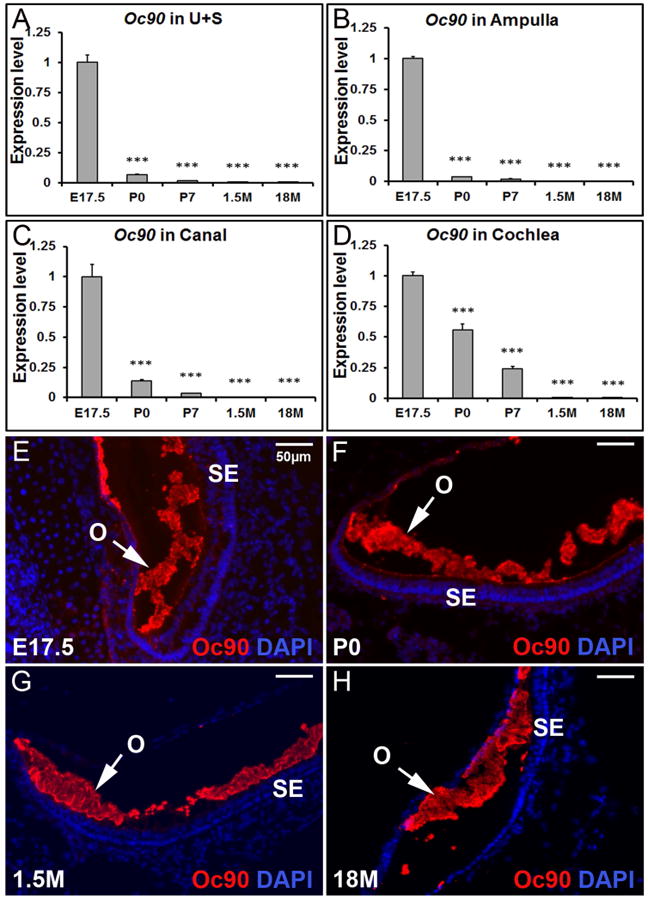
Based on qRT-PCR, Oc90 is expressed mostly before birth in the utricle and saccule (A), ampulla (B), canal (C) and cochlea (D). *** denote p<0.001 when compared with E17.5 (n=6 for each age group). U+S, utricle and saccule. Based on fluorescent immunostaining, Oc90 protein is abundant in the developing (E & F), mature (G) and aging (H) otoconia (utricle shown). O, otoconia; SE, sensory epithelia.
A comparison of the RNA-Seq data of 8 commonly used reference genes in the utricle and saccule at P0 vs. 8 months (Supplementary Table 3), and vs. the cochlea at P0 (Supplementary Table 2), showed that Actb is one of the best reference genes for the qRT-PCR analysis. The expression levels of Actb in the vestibular tissues (utricle, saccule and ampulla) are similar to each other, but are lower than that in the cochlea. Therefore, the expression levels of otoconial genes may be even lower in the cochlea than presented in the figures for qRT-PCR. Also, there is an increase in Actb levels in adult versus embryonic hair cells in both the vestibule and cochlea (Andrade 2015), but its levels are similar from birth to adult stages (Supplementary Table 3), so the actual reduction in Oc90 and other otoconial gene expression presented below in postnatal and adult stages, as compared to embryonic stages, is estimated to be even greater.
The distribution pattern of Oc90 protein in murine otoconia at different ages was examined by fluorescent immunostaining of frozen inner ear sections. Oc90 showed intense staining in both the developing and mature otoconia (Figure 1E-H) despite the very low level of Oc90 mRNA expression at adult stages.
Our previous work demonstrated that at embryonic stages, Oc90 mRNA levels in the utricle and saccule are significantly higher than the non-otolithic inner ear tissues (Yang et al. 2011). To test whether the same spatial pattern remains at later stages, we examined the expression levels in the postnatal, adult and aging inner ear tissues (Figure 2). Data presented in each panel of Figure 2 are not just re-analysis of Figure 1, but were independently obtained in separate qRT-PCR plates (n=3 × two replicate plates) to minimize experiment-introduced variations. At P0, the expression level of Oc90 in the utricle and saccule was twice as high as that in the ampulla, but only about 1.2 of that in the cochlea and about the same as that in the canals (Figure 2B). At P7, the spatial difference of Oc90 mRNA levels in these tissues was reversed (Figure 2C vs. 2A) and the order of low to high levels was now utricle/saccule, ampulla, canals and cochlea.
Figure 2. Spatial changes in Oc90 expression at different ages.
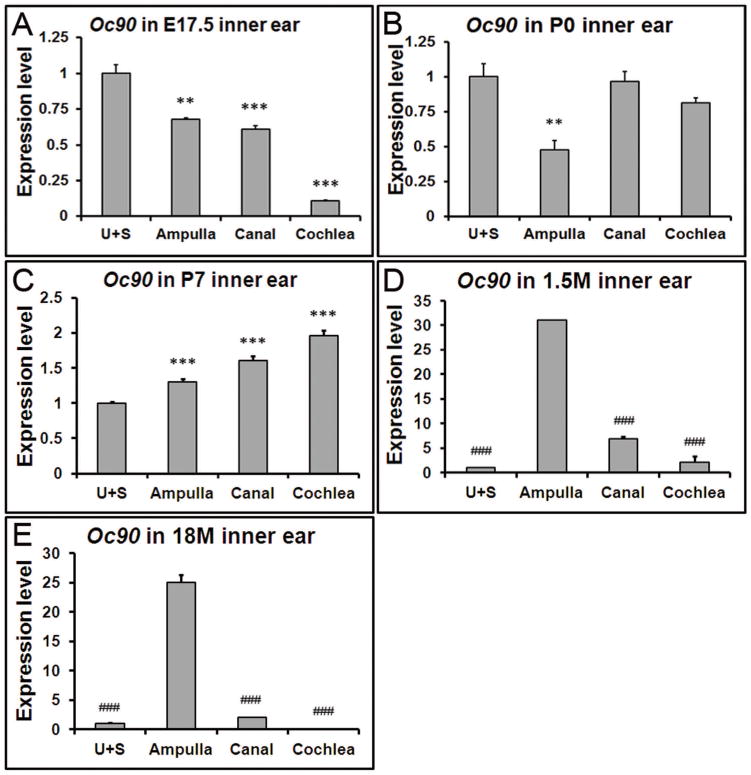
Data presented in each panel here are not just re-analysis of Figure 1, but were independently obtained in separate qRT-PCR plates for accurate comparison. (A) Oc90 shows the highest expression level in the utricle and saccule (U+S) at E17.5, but such a pattern is reversed at postnatal (C), adult (D) and aging stages (E). (D, E) In the 1.5 and 18 month-old inner ears, Oc90 shows the highest expression level in the ampulla. ** and *** denote p<0.01, <0.001 when compared with the U+S and ### indicates p<0.001 compared with the ampulla (n=6 for each group).
Predominant expression of Otolin in the embryonic utricle and saccule
Otolin, an inner ear-specific collagen, is a constituent of both otoconia and otoconial membrane (Deans et al. 2010;Xu et al. 2010;Zhao et al. 2007). Although it is not as abundant as Oc90 in the mouse otoconia crystal, studies performed on its homolog in zebrafish have demonstrated that this protein may be indispensable for crystal growth, maintenance and anchoring (Murayama et al. 2005).
Throughout life, Otolin showed the highest expression level in the utricle and saccule (Figure 3A-E), especially so at embryonic and early postnatal stages. There was a relative increase in Otolin transcript in the ampulla after developmental stages due to the reduction of the transcript in the utricle and saccule at each age. The expression of Otolin in the utricle and saccule was strikingly higher at E17.5 than neonatal and adult stages (Figure 3F, *** p<0.001). Interestingly, compared to earlier stages including neonatal, postnatal and adult stages, there's a notable amount of re-expression at 18 months old (Figure 3F, ˆˆˆ p<0.001).
Figure 3. The spatiotemporal expression pattern of Otolin transcript in the inner ear.
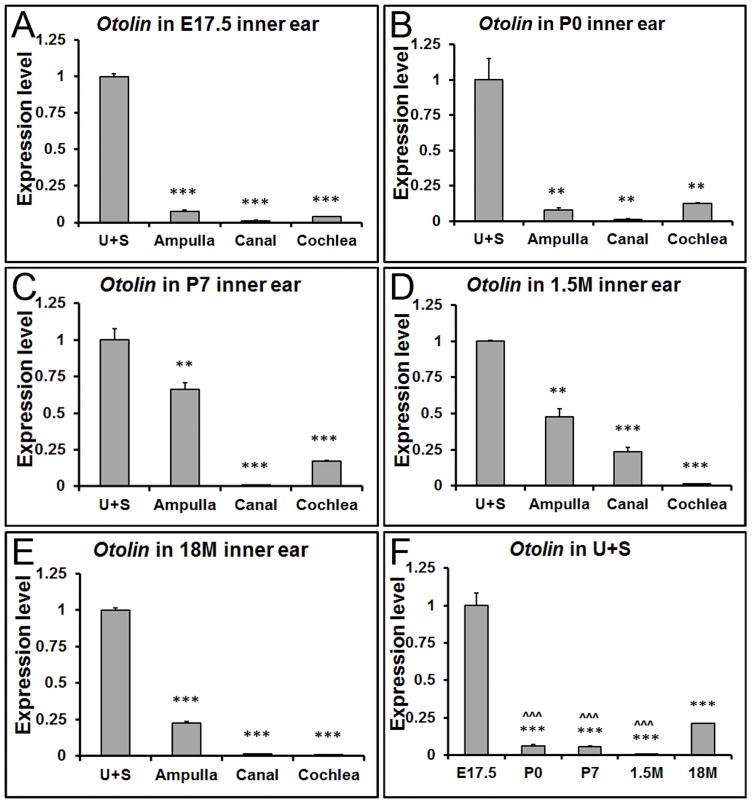
(A-E) qRT-PCR shows that Otolin has the highest expression level in the utricle and saccule at all ages, especially at developmental stages. (F) In the utricle and saccule, Otolin is mainly expressed at embryonic stages. **, *** denote p<0.01 and p<0.001 compared to the utricle and saccule (U+S), respectively (n=6 for each tissue and age group). ˆˆˆ indicates p < 0.001 when compared with 18M.
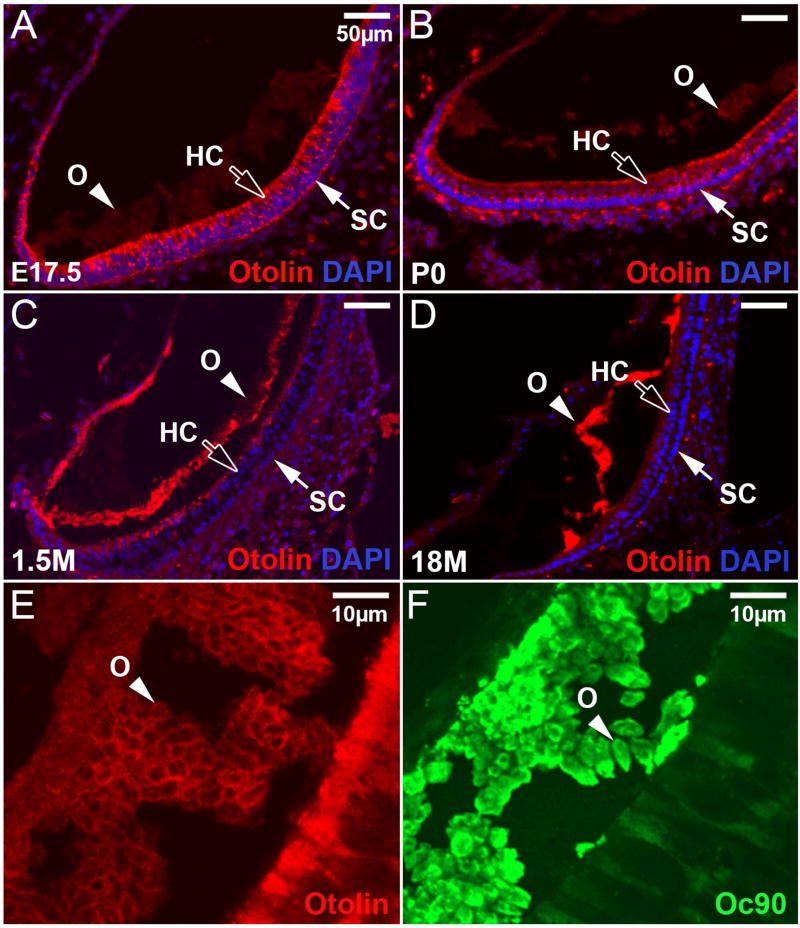
Distribution of Otolin in the inner ear was detected by immunohistochemistry using a polyclonal antibody against the N–terminal peptide (18 amino acids) of mouse Otolin. At E17.5, a time when otoconia seeding peaked, Otolin showed strong staining in hair cells, supporting cells and transitional cells of the utricle and saccule (Figure 4A, the utricle is shown), but the staining was faint in the lumen. By P0, more protein had been secreted into the lumen, and its presence in the immature otoconia was clearly discernible (Figure 4B). In the 1.5-month-old (1.5M) utricle and saccule, much Otolin had accumulated in the lumen, with a small amount still remaining in the apical side of the sensory epithelium (Figure 4C). In the aging mice, however, Otolin showed strong staining in the otoconial membrane, but was not detectable inside the epithelial cells (Figure 4D). Such a spatiotemporal change of Otolin distribution demonstrates that, although the expression of Otolin mRNA primarily occurred at the peak time of otoconia genesis, the accumulation of Otolin protein in otoconia appeared to last a much longer period of time.
Figure 4. Localization of Otolin protein in the utricle at different ages.
(A) Otolin is present in hair cells (HC), supporting cells (SC), and transitional epithelia at E17.5, but the signal was faint in otoconia at this age. (B) At P0, Otolin signal is slightly stronger in the otoconia (O), but less intense in the underlying epithelia when compared to the corresponding tissues at E17.5. (C) At 1.5 months old, Otolin accumulates further in otoconia and in the apical extracellular matrix of the sensory epithelium of the utricle. (D) In the aging utricle, Otolin is only detectable in otoconia and otoconial membrane, but not in the epithelia. (E) Otolin accumulates mostly in the matrix rim of otoconial crystallites at E17.5, whereas Oc90 signals (F) are more diffuse. Bars = 50μm in A-D and 20 μm in E, F.
A higher magnification view showed that most of the Otolin staining was in the matrix rim of each immature crystallite at E17.5 (Figure 4E), whereas Oc90 signals were more diffuse (Figure 4F). These crystallites fuse and give rise to the mature otoconia (Zhao et al. 2007). It is possible that Otolin in the rim may facilitate such fusion and expansion, or Otolin in the otoconial matrix may be oligomerized or bound to Oc90 and other molecules in a way that hindered antibody recognition. Previous studies have shown the presence of Otolin in the otoconial membrane (Zhao et al. 2007) and the fibrils interconnecting otoconia (Andrade LR et al. 2012). These data, together with the re-expression of Otolin in the aging utricle and saccule (Figure 3F), suggest the possibility that Otolin may be important for otoconia growth and maintenance in mice as well.
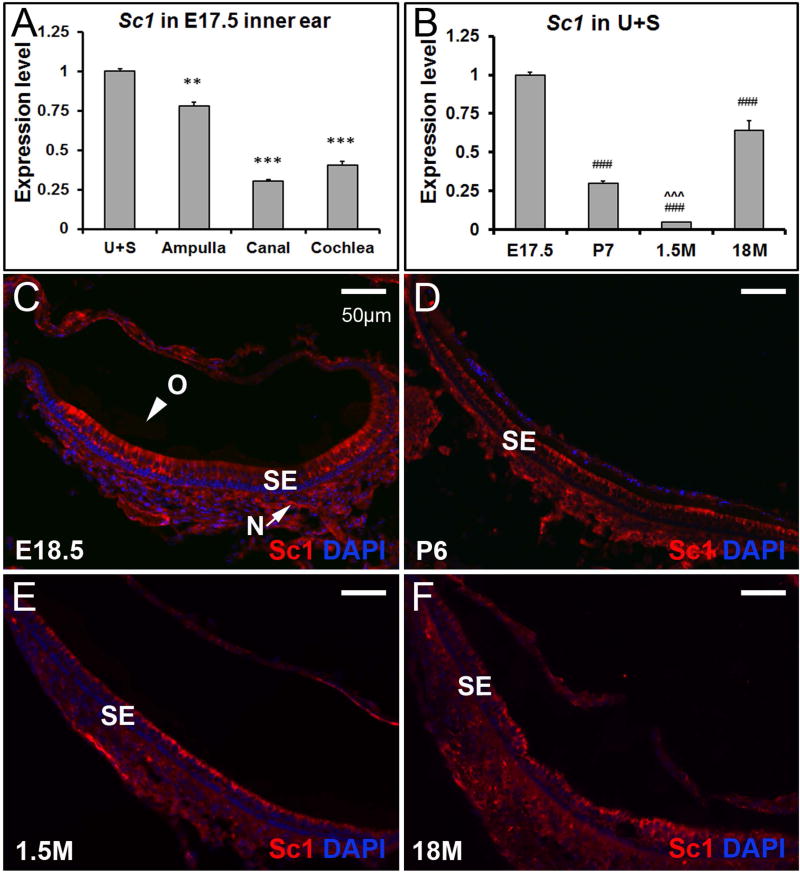
The expression of Sparc-like 1 (Sc1) in the inner ear at all stages examined
Sc1, a member of the Sparc family, is widely expressed in the brain and various types of neurons, and has recently been identified as a matrix constituent of the mouse otoconia (Thalmann et al. 2006;Xu et al. 2010). At embryonic stages, Sc1 was expressed in all of the inner ear tissues, but its mRNA level in the utricle and saccule was significantly higher than that in the ampulla (** p<0.01), canal (*** p<0.001) or cochlea (*** p<0.001) (Figure 5A). At later stages, the expression levels of Sc1 in some of the non-otolithic inner ear tissues, especially cochlea, were higher than those in the utricle and saccule (e.g. Table 1. Additional data are not shown). Sc1 mRNA expression was up-regulated in the utricle and saccule at 18 months of age compared to earlier stages, especially 1.5 months old (Figure 5B, ### p<0.001). This temporal expression change in the otolithic tissues is reminiscent of that for Otolin. Immunostaining showed faint signals of Sc1 in otoconia at all ages, which was visible under the microscope but not in the pictures in Figure 5. The staining was intense in the epithelial cells, more so at E18.5 and P6. Sc1 deposition in otoconia is drastically increased in Oc90 null mice (Xu et al. 2010). Here, the re-expression of these otoconial constituents may implicate their importance in repairing or sustaining otoconia in the aging mice.
Figure 5. The expression pattern of Sc1 in the inner ear.
(A) At E17.5, expression of Sc1 is significantly higher in the utricle and saccule by qRT-PCR, but the spatial specification is not as striking as that of Otolin. ** and *** denote p<0.01 and <0.001 when compared with the utricle and saccule (n=6 in each group). (B) Sc1 is re-expressed in the utricle and saccule at 18 months old by qRT-PCR. ### indicates p < 0.001 when compared with E17.5, and ˆˆˆ denote p < 0.001 when compared with 18M (n=6 for each age group). (C-F) Sc1 protein is hardly detectable in otoconia (O), but is present in the sensory epithelium (SE) and nerve fibers (N) (utricle shown).
Restriction of Otopetrin 1 (Otop1) expression to the developing utricle and saccule
In addition to the constituent proteins, some regulatory proteins are indispensible for otoconia formation. For example, Otop1 is essential for otoconia formation by modulating the intra- and extracellular calcium concentration (Hughes et al. 2007;Kim et al. 2010), or by regulating the secretion of otoconial proteins (Sollner et al. 2004). Our qRT-PCR results showed that the expression of Otop1 was restricted to the embryonic utricle and saccule (Figure 6A, 6B), with extremely low or no expression at other stages or in the other inner ear tissues, respectively (data not shown). These data may help explain why mutations in the gene selectively impair otoconia morphogenesis without affecting other endorgans such as the auditory system (Hurle et al. 2003). Otop1 protein has been detected in mostly supporting cells of the murine utricle and saccule (Kim et al. 2010), but in both supporting and hair cells in zebrafish (Sollner et al. 2004).
Figure 6. The expression of Otop1 is restricted to the embryonic utricle and saccule by qRT-PCR.
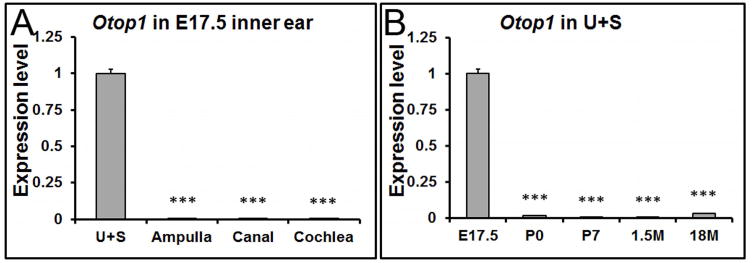
(A) At E17.5, the expression of Otop1 is confined to the utricle (U) and saccule (S). *** denote p<0.001 compared with the utricle and saccule (n=6 in each tissue group). (B) In the utricle and saccule (U+S), Otop1 shows the highest expression level at embryonic stages. ### denote p<0.001 compared with E17.5 (n=6 in each age group).
The unexpected expression pattern of NADPH oxidase 3 (Nox3)
Nox3, a plasma membrane-associated enzyme which catalyzes the production of reactive oxygen species (ROS) by using NADPH as the electron donor, is another essential regulatory protein of otoconia. Mice with mutations in the Nox3 gene and related family members, including Noxo1 and p22phox, show severe balance deficits because of the total absence of otoconia in the utricle and saccule (Kiss et al. 2006;Nakano et al. 2008;Paffenholz et al. 2004). Unlike the other otoconial genes, qRT-PCR results revealed a surprising expression pattern of Nox3 in the mouse inner ear: The highest mRNA level was in the semi-circular canals at all stages and in the aging ampulla, but not in the utricle and saccule (Figure 7 A-E). Furthermore, in the 18M inner ear, the expression level of Nox3 in the ampulla was significantly increased (Figure 7E, ### p<0.001). Its partners Noxo1 and p22phox also showed age-related increase of expression in the ampulla (data not shown). Fluorescent immunostaining showed that Nox3 protein was located at the apical membrane of hair cells in the utricle, saccule and crista (Figure 7F, utricle is shown and labeled as “U”), and in the epithelia cells of the semi-circular canal duct as well (Figure 7G).
Figure 7. Nox3 shows the highest expression level in the semi-circular canals and aging ampulla.
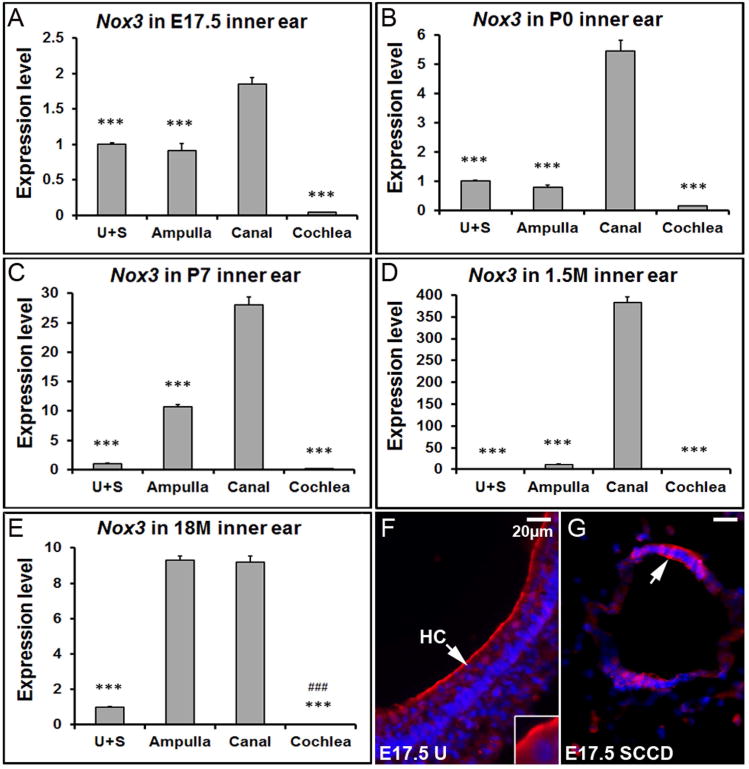
(A-E) qRT-PCR shows that Nox3 has a significantly higher expression level in the canals compared to other inner ear tissues at different stages. *** denote p<0.001 when compared with the canals (n=6 each tissue group). In the 18 month-old mice, the ampullar expression level of Nox3 is also much higher than the utricle/saccule, and cochlea. ### denote p<0.001 when compared with the ampulla (n=6 each tissue group). (F) The E17.5 utricle (U) was stained with a Nox3 antibody (red) and DAPI (blue). (G) Immunostaining of the semi-circular canal duct (SCCD) at E17.5 with Nox3 and DAPI. HC, hair cell; SCCD, semi-circular canal duct.
Discussion
The mechanisms leading to spatial specific formation and maintenance of otoconia are not yet understood. In the present study, we show that most of the otoconial genes except Nox3 present much higher expression levels in the utricle and saccule compared to other inner ear tissues, including the ampulla, canal, and cochlea at embryonic stages. The expression pattern of Otop1 and Otolin is especially restricted, with the former only in the embryonic utricle and saccule (Figures 6 and 3). The otoconial component/anchor protein α-Tectorin (Legan et al. 2000) is also only expressed at embryonic and early postnatal stages (Rau et al. 1999) in the mouse utricle and saccule, but the protein is more abundant in the cochlea. Another anchor protein, Otogelin, is continuously expressed in the developing and adult vestibule (Simmler et al. 2000;El-Amraoui et al. 2001), implicating a constant need for a high level of the protein to correctly anchor otoconia. Overall, the tight regulation of the spatiotemporal expression of otoconial genes, in addition to the higher endolymphatic Ca2+ concentration in the vestibule (∼280 μM in the embryonic vestibule) compared to the cochlea (∼25 μM) (Ferrary et al. 1988;Marcus and Wangemann 2009;Salt et al. 1989), are likely critical in ensuring otoconia crystallization to initiate only in the embryonic utricle and saccule and not in any other tissues or at any other time. Our data suggest that otoconia formation is regulated by a tissue- and age-specific program of gene expression.
When the utricular and saccular expression of otoconial genes is extremely low, otoconia maintenance and repair may become compromised, making the crystals more susceptible to adverse conditions and prone to degeneration. Degenerative otoconia have been observed in the aging inner ear in both animals (Jang YS et al. 2006;Lim 1984;Takumida and Zhang 1997) and humans (Ross et al. 1976;Anniko et al. 1984;Igarashi et al. 1993). Otoconial degeneration may make the crystals easier to dislocate. Indeed, BPPV occurs much more commonly in older people (Baloh et al. 1987;Ogun et al. 2014), and in people exposed to ototoxic drugs or with certain illnesses such as Meniere's disease (Calzada et al. 2012;Gross et al. 2000;Hughes CA and Proctor L 1997;Karlberg et al. 2000;Wu et al. 2007).
The consequences of adverse conditions such as aging may go beyond the passive degeneration and dislocation of otoconia. We detected up-regulation of the mRNA expression of multiple critical otoconial genes in non-otolithic inner ear tissues, especially the ampulla and canals, at aging stages compared to young adult stages. This poses the potential to cause de novo formation of otoconia-like particles in the aging ampulla when permissive conditions exist, which may be an additional event facilitating BPPV besides the well-known theory of otoconia fallen from the utricle into the ampulla.
In summary, we have obtained data on the spatiotemporal expression changes of critical otoconial genes in the mouse inner ear. The qRT-PCR findings are in agreement with that of RNA-Seq where applicable. While most of the data are limited to the genes currently known to be critical for otoconia formation, the information may help understand the restriction of otoconia formation to the embryonic utricle and saccule under normal conditions, and may serve as a foundation to design methods to stimulate otoconia neogenesis or stimulate anchor protein expression when needed. In addition to insufficient gene expression for otoconia maintenance and anchoring in the wanted sites, we show for the first time unexpected expression of these genes in non-otolithic tissues under aging. Hence, the findings may stimulate future studies to promote new thinking regarding BPPV etiology.
Methods
Mice
C57BL/6J mice were used to obtain the reported results. The mice were purchased from the Jackson Laboratory (Bar Harbor, Maine, USA) and maintained in the vivarium at Boys Town National Research Hospital (BTNRH). All animal procedures were approved by the Institutional Animal Care and Use Committee at BTNRH in accordance with federal and international guidelines.
Microdissection
Animals were anesthetized with ketamine (167mg/kg) and xylazine (4mg/kg) and then decapitated. Inner ears were dissected in phosphate-buffered saline (PBS) for the reported experiments.
For fluorescent immunostaining, the inner ears were fixed in 4% paraformaldehyde, dehydrated in 30% sucrose prepared in PBS, embedded in O.C.T. compound at below -20°C, and sectioned at 9 μm. Fixed inner ears from mice older than P4 were decalcified in 0.25 M EDTA (pH7.4) for 2 hr to overnight, depending on the age of animals.
For RNA-Seq and real-time quantitative RT-PCR (qRT-PCR), fresh epithelial tissues from the utricle, saccule, ampulla, semi-circular canal, and cochlea were dissected in PBS containing RNasin (Promega, Madison, WI), and were collected in Trizol Reagent (Invitrogen, Carlsbad, CA) for RNA preparation.
RNA preparation and next-generation sequencing (RNA-Seq)
Epithelial tissues of the inner ear were pooled for each endorgan per age group: P0 (n=8) and 8 months old (8M, n=7) C57BL/6J mice, kept in Trizol reagent and submitted on dry ice to Otogenetics Corporation (Norcross, GA, USA) for RNA extraction, RNA-Seq and analysis. Briefly, total RNA was extracted from the tissues using the E.Z.N.A. Total RNA Kit II (Omega Biotek, Norcross, GA, USA). Genomic DNA was removed using the RNase-Free DNase Set (Qiagen, Valencia, CA, USA). Concentrated ultrapure RNA was obtained by using RNA Clean and Concentrator columns (Zymo Research Corporation, Irvine, CA USA). The integrity and purity of total RNA were assessed using an Agilent Bioanalyzer 2100 and OD260/280.
Approximately 1-2 μg of cDNA was generated from 100ng of total RNA using Clontech SMARTer PCR cDNA kit (Clontech Laboratories, Inc., Mountain View, CA USA). The cDNA was fragmented using the Covaris AFA processing method (Covaris, Inc., Woburn, MA USA), profiled using an Agilent Bioanalyzer 2100, and subjected to Illumina library preparation using NEBNext reagents (New England Biolabs, Ipswich, MA USA). The quality, quantity and size distribution of the Illumina libraries were determined using an Agilent Bioanalyzer 2100. The libraries were then submitted for Illumina HiSeq2000 sequencing according to the standard operation. More than 20 (usually 25-37) million paired-end 100bp reads were generated per sample and checked for data quality using FASTQC (Babraham Institute, Cambridge, UK). Sequences were subjected to data analysis using the platform provided by DNAnexus (DNAnexus, Inc, Mountain View, CA USA). Sequences were mapped against reference genome mm9 with Star (2.4.0j), expression levels of genes and transcripts were obtained with Cufflinks (2.2.1), and comparison across groups was made with Cufflinks. cuffdiff (2.2.1).
Real-time quantitative RT-PCR (qRT-PCR)
Total RNA was prepared using the Trizol Reagent (Invitrogen), and first-strand cDNA was synthesized from total RNA with random hexamer primers using the SuperScript III First-Strand Synthesis System (Invitrogen). Probe & primer sets for qPCR were purchased from Applied Biosystems as TaqMan Gene Expression assays (Applied Biosystems, Foster City, CA), and probes were designed to minimize detection of genomic DNA (e.g. probes spanned 2 or more exons). Also, reactions without reverse-transcriptase were included. Amplification was done according to the manufacturer's protocol with minor modifications based on cDNA samples and primer/probe conditions. Amplification included TaqMan probe & primer set, TaqMan Universal PCR Master Mix and cDNA (∼50 ng). The standard mode of an ABI Prism 7900HT Sequence Detection System (Applied Biosystems) was used, and β-actin (Actb) was amplified in parallel as an endogenous control for the quantification of the relative gene expression between samples. The cycle number at which the reaction crosses a predetermined cycle threshold (CT) was identified for each gene, and the expression value of each target gene relative to Actb was determined using the equation 2-Δ ΔCT.
The probe sequences were:
Oc90 – TGAAATTTTTGACTGCCTAGGTTCC (ABI i.d. Mm01200961_m1),
Otol1 – AGAAAGGAGAGAAAGGACTAAAGGG (ABI i.d. Mm01222538_m1),
Sc1 – GCCACCTCTCCGCAGATCTAGCCAG (ABI i.d. Mm00447780_m1),
Otop1 – ACACTGGAAAGGTTTGGGGTGATCC (ABI i.d. Mm00554705_m1),
Nox3 – ATAGCTGTCAATTCAGTTATCCACA (ABI i.d. Mm01339126_m1),
Actb – TTACTGAGCTGCGTTTTACACCCTT (ABI i.d. Mm00607939_s1).
Immunohistochemistry
Frozen tissue sections were blocked in PBS containing 5% BSA + 0.25% Triton-X-100 at room temperature for 30 minutes, and incubated in 5% BSA + 0.25% Triton-X-100 at 4°C overnight with one of the following antibodies: rabbit-derived polyclonal anti-mouse Oc90 (1:500) (Zhao et al. 2007), rabbit-derived polyclonal anti-mouse Otolin (1:100) (Zhao et al. 2007), goat-derived polyclonal anti-mouse Sparc-like 1 (Sc1, 1:100) (R&D Systems, Minneapolis, MN), or rabbit-derived polyclonal anti-human/mouse Nox3 (1:100) (Abcam, Cambridge, MA). Specificity of these antibodies was confirmed by Western blotting as described in the quoted publication or on the stated supplier's websites.
After 3 washes in PBS, Alexa-488 or 568 (Molecular Probes, Carlsbad, CA) conjugated secondary antibodies were added at a dilution of 1:600 and incubated at room temperature for 1 hour in the dark. Pre-immune (Oc90 and Otolin) or non-immune (Sc1 and Nox3) sera instead of primary antibodies were used in some sections as negative controls. Slides were mounted in Fluoromount-G and pictures were taken using a Zeiss Axio Observer Z1 inverted microscope.
To minimize measurement variation, all tissue sections under comparison were processed strictly under the same conditions (e.g., identical immunostaining procedures, identical microscope scanning parameters, and the same number of fluorescent exposures). Cross sections that covered both the striola and peri-macular region were analyzed to take into consideration possible intensity differences caused by the position and orientation of cells and sections. Three or more animals were examined for each age and tissue type.
Data Analysis
Reported data represent the mean values ± standard deviation (S.D). Statistical significance was determined by one way ANOVA with Bonferroni correction. Significance was set at the level of P<0.05. P values are expressed as * P<0.05, ** P<0.01, and ***, ### or ˆˆˆ P<0.001.
Supplementary Material
Acknowledgments
The work was supported by grants from the NIH (DC008603, DC008603-S1 and DC014748 to Y.W.L.).
Reference List
- Andrade LR, Lins U, Farina M, Kachar B, Thalmann R. Immunogold TEM of otoconin 90 and otolin - relevance to mineralization of otoconia, and pathogenesis of benign positional vertigo. Hear Res. 2012;292:14–25. doi: 10.1016/j.heares.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade LR. Evidence for changes in beta- and gamma-actin proportions during inner ear hair cell life. Cytoskeleton (Hoboken) 2015;72:282–291. doi: 10.1002/cm.21227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anniko M. Development of otoconia. Am J Otolaryngol. 1980;1:400–410. doi: 10.1016/s0196-0709(80)80021-4. [DOI] [PubMed] [Google Scholar]
- Anniko M, Wenngren BI, Wroblewski R. Aberrant elemental composition of otoconia in the dancer mouse mutant with a semidominant gene causing a morphogenetic type of inner ear defect. Acta Otolaryngol. 1988;106:208–212. doi: 10.3109/00016488809106427. [DOI] [PubMed] [Google Scholar]
- Anniko M, Ylikoski J, Wroblewski R. Microprobe analysis of human otoconia. Acta Otolaryngol. 1984;97:283–289. doi: 10.3109/00016488409130990. [DOI] [PubMed] [Google Scholar]
- Baloh RW, Honrubia V, Jacobson K. Benign positional vertigo: clinical and oculographic features in 240 cases. Neurology. 1987;37:371–378. doi: 10.1212/wnl.37.3.371. [DOI] [PubMed] [Google Scholar]
- Banfi B, Malgrange B, Knisz J, Steger K, Dubois-Dauphin M, Krause KH. NOX3, a superoxide-generating NADPH oxidase of the inner ear. J Biol Chem. 2004;279:46065–46072. doi: 10.1074/jbc.M403046200. [DOI] [PubMed] [Google Scholar]
- Calzada Audrey P, Lopez Ivan A, Ishiyama Gail, Ishiyama Akira. Otolithic Membrane Damage in Patients with Endolymphatic Hydrops and Drop Attacks. Otology & Neurotology. 2012 doi: 10.1097/MAO.0b013e318271c48b. Ref Type: In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans MR, Peterson JM, Wong GW. Mammalian Otolin: a multimeric glycoprotein specific to the inner ear that interacts with otoconial matrix protein Otoconin-90 and Cerebellin-1. PLoS ONE. 2010;5:e12765. doi: 10.1371/journal.pone.0012765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror AA, Politi Y, Shahin H, Lenz DR, Dossena S, Nofziger C, Fuchs H, Hrabe de AM, Paulmichl M, Weiner S, Avraham KB. Calcium oxalate stone formation in the inner ear as a result of an Slc26a4 mutation. J Biol Chem. 2010;285:21724–21735. doi: 10.1074/jbc.M110.120188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Amraoui A, Cohen-Salmon M, Petit C, Simmler MC. Spatiotemporal expression of otogelin in the developing and adult mouse inner ear. Hear Res. 2001;158:151–159. doi: 10.1016/s0378-5955(01)00312-4. [DOI] [PubMed] [Google Scholar]
- Everett LA, Belyantseva IA, Noben-Trauth K, Cantos R, Chen A, Thakkar SI, Hoogstraten-Miller SL, Kachar B, Wu DK, Green ED. Targeted disruption of mouse Pds provides insight about the inner-ear defects encountered in Pendred syndrome. Hum Mol Genet. 2001;10:153–161. doi: 10.1093/hmg/10.2.153. [DOI] [PubMed] [Google Scholar]
- Ferrary E, Tran Ba HP, Roinel N, Bernard C, Amiel C. Calcium and the inner ear fluids. Acta Otolaryngol Suppl. 1988;460:13–17. doi: 10.3109/00016488809125130. [DOI] [PubMed] [Google Scholar]
- Gross EM, Ress BD, Viirre ES, Nelson JR, Harris JP. Intractable benign paroxysmal positional vertigo in patients with Meniere's disease. Laryngoscope. 2000;110:655–659. doi: 10.1097/00005537-200004000-00022. [DOI] [PubMed] [Google Scholar]
- Huang L, Gitschier J. A novel gene involved in zinc transport is deficient in the lethal milk mouse. Nat Genet. 1997;17:292–297. doi: 10.1038/ng1197-292. [DOI] [PubMed] [Google Scholar]
- Hughes CA, Proctor L. Benign paroxysmal positional vertigo. Laryngoscope. 1997;107:607–613. doi: 10.1097/00005537-199705000-00010. [DOI] [PubMed] [Google Scholar]
- Hughes I, Saito M, Schlesinger PH, Ornitz DM. Otopetrin 1 activation by purinergic nucleotides regulates intracellular calcium. Proc Natl Acad Sci U S A. 2007;104:12023–12028. doi: 10.1073/pnas.0705182104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurle B, Ignatova E, Massironi SM, Mashimo T, Rios X, Thalmann I, Thalmann R, Ornitz DM. Non-syndromic vestibular disorder with otoconial agenesis in tilted/mergulhador mice caused by mutations in otopetrin 1. Hum Mol Genet. 2003;12:777–789. doi: 10.1093/hmg/ddg087. [DOI] [PubMed] [Google Scholar]
- Igarashi M, Saito R, Mizukoshi K, Alford BR. Otoconia in young and elderly persons: a temporal bone study. Acta Otolaryngol Suppl. 1993;504:26–29. doi: 10.3109/00016489309128117. [DOI] [PubMed] [Google Scholar]
- Ignatova EG, Thalmann I, Xu B, Ornitz DM, Thalmann R. Molecular mechanisms underlying ectopic otoconia-like particles in the endolymphatic sac of embryonic mice. Hear Res. 2004;194:65–72. doi: 10.1016/j.heares.2004.03.019. [DOI] [PubMed] [Google Scholar]
- Jang YS, Hwang CH, Shin JY, Bae WY, Kim LS. Age-related changes on the morphology of the otoconia. Laryngoscope. 2006;116:996–1001. doi: 10.1097/01.mlg.0000217238.84401.03. [DOI] [PubMed] [Google Scholar]
- Jones SM, Erway LC, Bergstrom RA, Schimenti JC, Jones TA. Vestibular responses to linear acceleration are absent in otoconia-deficient C57BL/6JEi-het mice. Hear Res. 1999;135:56–60. doi: 10.1016/s0378-5955(99)00090-8. [DOI] [PubMed] [Google Scholar]
- Jones SM, Erway LC, Johnson KR, Yu H, Jones TA. Gravity receptor function in mice with graded otoconial deficiencies. Hear Res. 2004;191:34–40. doi: 10.1016/j.heares.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Karlberg M, Hall K, Quickert N, Hinson J, Halmagyi GM. What inner ear diseases cause benign paroxysmal positional vertigo? Acta Otolaryngol. 2000;120:380–385. doi: 10.1080/000164800750000603. [DOI] [PubMed] [Google Scholar]
- Kim E, Hyrc KL, Speck J, Lundberg YW, Salles FT, Kachar B, Goldberg MP, Warchol ME, Ornitz DM. Regulation of cellular calcium in vestibular supporting cells by Otopetrin 1. J Neurophysiol. 2010 doi: 10.1152/jn.00525.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss PJ, Knisz J, Zhang Y, Baltrusaitis J, Sigmund CD, Thalmann R, Smith RJ, Verpy E, Banfi B. Inactivation of NADPH oxidase organizer 1 Results in Severe Imbalance. Curr Biol. 2006;16:208–213. doi: 10.1016/j.cub.2005.12.025. [DOI] [PubMed] [Google Scholar]
- Kozel PJ, Friedman RA, Erway LC, Yamoah EN, Liu LH, Riddle T, Duffy JJ, Doetschman T, Miller ML, Cardell EL, Shull GE. Balance and hearing deficits in mice with a null mutation in the gene encoding plasma membrane Ca2+-ATPase isoform 2. J Biol Chem. 1998;273:18693–18696. doi: 10.1074/jbc.273.30.18693. [DOI] [PubMed] [Google Scholar]
- Legan PK, Lukashkina VA, Goodyear RJ, Kossi M, Russell IJ, Richardson GP. A targeted deletion in alpha-tectorin reveals that the tectorial membrane is required for the gain and timing of cochlear feedback. Neuron. 2000;28:273–285. doi: 10.1016/s0896-6273(00)00102-1. [DOI] [PubMed] [Google Scholar]
- Legan PK, Richardson GP. Extracellular matrix and cell adhesion molecules in the developing inner ear. Semin Cell Dev Biol. 1997;8:217–224. doi: 10.1006/scdb.1997.0145. [DOI] [PubMed] [Google Scholar]
- Lim DJ. Otoconia in health and disease. A review. Ann Otol Rhinol Laryngol Suppl. 1984;112:17–24. doi: 10.1177/00034894840930s404. [DOI] [PubMed] [Google Scholar]
- Lundberg YW, Xu Y, Thiessen KD, Kramer KL. Mechanisms of otoconia and otolith development. Dev Dyn. 2015;244:239–253. doi: 10.1002/dvdy.24195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg YW, Zhao X, Yamoah EN. Assembly of the otoconia complex to the macular sensory epithelium of the vestibule. Brain Res. 2006;1091:47–57. doi: 10.1016/j.brainres.2006.02.083. [DOI] [PubMed] [Google Scholar]
- Marcus DC, Wangemann P. Cochlear and Vestibular Function and Dysfunction. In: Alvarez-Leefmans F Javier, D E., editors. Physiology and Pathology of Chloride Transporters and Channels in the Nervous System-From molecules to diseases. San Diego: Academic Press; 2009. pp. 421–433. [Google Scholar]
- Marino G, Fernandez AF, Cabrera S, Lundberg YW, Cabanillas R, Rodriguez F, Salvador-Montoliu N, Vega JA, Germana A, Fueyo A, Freije JM, Lopez-Otin C. Autophagy is essential for mouse sense of balance. J Clin Invest. 2010;120:2331–2344. doi: 10.1172/JCI42601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama E, Herbomel P, Kawakami A, Takeda H, Nagasawa H. Otolith matrix proteins OMP-1 and Otolin-1 are necessary for normal otolith growth and their correct anchoring onto the sensory maculae. Mech Dev. 2005;122:791–803. doi: 10.1016/j.mod.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Nakano Y, Longo-Guess CM, Bergstrom DE, Nauseef WM, Jones SM, Banfi B. Mutation of the Cyba gene encoding p22phox causes vestibular and immune defects in mice. J Clin Invest. 2008;118:1176–1185. doi: 10.1172/JCI33835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogun OA, Buki B, Cohn ES, Janky KL, Lundberg YW. Menopause and benign paroxysmal positional vertigo. Menopause. 2014;21:886–889. doi: 10.1097/GME.0000000000000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paffenholz R, Bergstrom RA, Pasutto F, Wabnitz P, Munroe RJ, Jagla W, Heinzmann U, Marquardt A, Bareiss A, Laufs J, Russ A, Stumm G, Schimenti JC, Bergstrom DE. Vestibular defects in head-tilt mice result from mutations in Nox3, encoding an NADPH oxidase. Genes Dev. 2004;18:486–491. doi: 10.1101/gad.1172504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau A, Legan PK, Richardson GP. Tectorin mRNA expression is spatially and temporally restricted during mouse inner ear development. J Comp Neurol. 1999;405:271–280. [PubMed] [Google Scholar]
- Ross MD, Peacor D, Johnsson LG, Allard LF. Observations on normal and degenerating human otoconia. Ann Otol Rhinol Laryngol. 1976;85:310–326. doi: 10.1177/000348947608500302. [DOI] [PubMed] [Google Scholar]
- Salt AN, Inamura N, Thalmann R, Vora A. Calcium gradients in inner ear endolymph. Am J Otolaryngol. 1989;10:371–375. doi: 10.1016/0196-0709(89)90030-6. [DOI] [PubMed] [Google Scholar]
- Salvinelli F, Firrisi L, Casale M, Trivelli M, D'Ascanio L, Lamanna F, Greco F, Costantino S. Benign paroxysmal positional vertigo: diagnosis and treatment. Clin Ter. 2004;155:395–400. [PubMed] [Google Scholar]
- Schuknecht HF. Positional vertigo: clinical and experimental observations. Trans Am Acad Ophthalmol Otolaryngol. 1962;66:319–332. [PubMed] [Google Scholar]
- Schuknecht HF. Cupulolithiasis. Arch Otolaryngol. 1969;90:765–778. doi: 10.1001/archotol.1969.00770030767020. [DOI] [PubMed] [Google Scholar]
- Simmler MC, Cohen-Salmon M, El-Amraoui A, Guillaud L, Benichou JC, Petit C, Panthier JJ. Targeted disruption of otog results in deafness and severe imbalance. Nat Genet. 2000;24:139–143. doi: 10.1038/72793. [DOI] [PubMed] [Google Scholar]
- Sollner C, Schwarz H, Geisler R, Nicolson T. Mutated otopetrin 1 affects the genesis of otoliths and the localization of Starmaker in zebrafish. Dev Genes Evol. 2004;214:582–590. doi: 10.1007/s00427-004-0440-2. [DOI] [PubMed] [Google Scholar]
- Takemura T, Sakagami M, Nakase T, Kubo T, Kitamura Y, Nomura S. Localization of osteopontin in the otoconial organs of adult rats. Hear Res. 1994;79:99–104. doi: 10.1016/0378-5955(94)90131-7. [DOI] [PubMed] [Google Scholar]
- Takumida M, Harada Y. Development of the utricular macula in the mouse. Arch Otorhinolaryngol. 1984;241:9–15. doi: 10.1007/BF00457911. [DOI] [PubMed] [Google Scholar]
- Takumida M, Zhang DM. Electron probe X-ray microanalysis of otoconia in the guinea pig inner ear. ORL J Otorhinolaryngol Relat Spec. 1997;59:187–192. doi: 10.1159/000276938. [DOI] [PubMed] [Google Scholar]
- Thalmann I, Hughes I, Tong BD, Ornitz DM, Thalmann R. Microscale analysis of proteins in inner ear tissues and fluids with emphasis on endolymphatic sac, otoconia, and organ of Corti. Electrophoresis. 2006;27:1598–1608. doi: 10.1002/elps.200500768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohse H, Takagi Y, Nagasawa H. Identification of a novel matrix protein contained in a protein aggregate associated with collagen in fish otoliths. FEBS J. 2008;275:2512–2523. doi: 10.1111/j.1742-4658.2008.06400.x. [DOI] [PubMed] [Google Scholar]
- Verpy E, Leibovici M, Petit C. Characterization of otoconin-95, the major protein of murine otoconia, provides insights into the formation of these inner ear biominerals. Proc Natl Acad Sci U S A. 1999;96:529–534. doi: 10.1073/pnas.96.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kowalski PE, Thalmann I, Ornitz DM, Mager DL, Thalmann R. Otoconin-90, the mammalian otoconial matrix protein, contains two domains of homology to secretory phospholipase A2. Proc Natl Acad Sci U S A. 1998;95:15345–15350. doi: 10.1073/pnas.95.26.15345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZM, Zhang SZ, Liu XJ, Chen X, Ji F, Chen AT, Yang WY, Han DY. Benign paroxysmal positioning vertigo related to inner ear disorders. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2007;42:821–825. [PubMed] [Google Scholar]
- Xu Y, Zhang H, Yang H, Zhao X, Lovas S, Lundberg YW. Expression, functional and structural analysis of proteins critical for otoconia development. Dev Dyn. 2010;239:2659–2673. doi: 10.1002/dvdy.22405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Zhao X, Xu Y, Wang L, He Q, Lundberg YW. Matrix recruitment and calcium sequestration for spatial specific otoconia development. PLoS ONE. 2011;6:e20498. doi: 10.1371/journal.pone.0020498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Jones SM, Yamoah EN, Lundberg YW. Otoconin-90 deletion leads to imbalance but normal hearing: a comparison with other otoconia mutants. Neuroscience. 2008;153:289–299. doi: 10.1016/j.neuroscience.2008.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yang H, Yamoah EN, Lundberg YW. Gene targeting reveals the role of Oc90 as the essential organizer of the otoconial organic matrix. Dev Biol. 2007;304:508–524. doi: 10.1016/j.ydbio.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaenepoel I, Mustapha M, Leibovici M, Verpy E, Goodyear R, Liu XZ, Nouaille S, Nance WE, Kanaan M, Avraham KB, Tekaia F, Loiselet J, Lathrop M, Richardson G, Petit C. Otoancorin, an inner ear protein restricted to the interface between the apical surface of sensory epithelia and their overlying acellular gels, is defective in autosomal recessive deafness DFNB22. Proc Natl Acad Sci U S A. 2002;99:6240–6245. doi: 10.1073/pnas.082515999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.