Abstract
BACKGROUND
The use of clinical samples and circulating cell free DNA (cfDNA) collected from liquid biopsies for diagnostic and prognostic applications in cancer is burgeoning, and improved methods that reduce the influence of excess wild-type (WT) portion of the sample are desirable. Here we present enrichment of mutation-containing sequences using enzymatic degradation of wild-type DNA (WT-DNA). Mutation enrichment is combined with high-resolution-melting (HRM) performed in multiplexed closed-tube reactions, as a rapid, cost-effective screening tool prior to targeted re-sequencing.
METHODS
We developed a homogeneous, closed-tube approach to utilize a double-strand DNA-specific nuclease (DSN) for degradation of WT-DNA at multiple targets simultaneously. The No-Denaturation Nuclease-assisted Minor Allele Enrichment with Probe-Overlap (ND-NaME-PrO) employs WT-oligonucleotides overlapping both strands on putative DNA targets. Under conditions of partial denaturation (DNA-breathing), the oligonucleotide-probes enhance double-stranded-DNA-specific nuclease digestion at the selected targets, with high preference towards WT over mutant DNA. To validate ND-NaME-PrO we employed multiplexed-HRM, digital PCR and Miseq targeted re-sequencing of mutated genomic DNA and cfDNA.
RESULTS
Serial dilution of KRAS mutation-containing DNA demonstrates mutation enrichment by 10–120-fold and detection of allelic-fractions down to 0.01%. Multiplexed ND-NaME-PrO combined with multiplexed PCR-HRM demonstrated mutation scanning of 10–20 DNA amplicons simultaneously. ND-NaME-PrO applied on cfDNA from clinical samples enables mutation enrichment and HRM-scanning over 10 DNA targets. cfDNA mutations were enriched up to ~100-fold, average ~25-fold, and identified via targeted re-sequencing.
CONCLUSIONS
Closed-tube homogeneous ND-NaME-PrO combined with multiplexed-HRM is a convenient approach to efficiently enrich for mutations on multiple DNA-targets and enabling pre-screening prior to targeted re-sequencing.
Keywords: Mutation detection, mutation enrichment, high resolution melting, targeted re-sequencing, nuclease-assisted minor-allele enrichment
Introduction
There is mounting evidence that tumor mutations identified in cfDNA can potentially act as a powerful liquid biopsy based diagnostic tool (1–5). Clinical studies indicate the use of cfDNA to complement (6) or replace (2) tissue biopsies. Rare mutations identified in cfDNA via digital droplet PCR (ddCR) or massively parallel sequencing (MPS) can lead to changes in clinical practice (7). Despite its promise, technical hurdles persist. The limited amount of cfDNA obtained from a standard blood draw and the excess amount of circulating wild-type (WT) DNA are persistent issues that often compromise the diagnostic results. A number of approaches have been described to reduce excess WT DNA (8), thereby facilitating detection of mutated DNA, via the use of polymerase chain reaction (PCR) (9–19). Use of PCR based mutation enrichment on multiple DNA targets simultaneously has also been reported (20, 21). However, the approach is technically demanding and requires extensive optimization which becomes more difficult as the number of multiplexed targets increases (22). Enzymatic approaches using endonucleases to degrade WT DNA, on the other hand, can be highly parallel but these are restricted to sequences recognized by the enzyme used (23–27). Accordingly, mutation enrichment prior to highly parallel processes like targeted re-sequencing remain difficult to implement.
To circumvent these hurdles we recently developed Nuclease-Assisted Minor-Allele enrichment using Overlapping Probes (NaME-PrO) (28), an enzymatic approach to remove WT-DNA from multiple DNA targets selected at-will, prior to DNA-amplification, following which current genomic analysis processes remain substantially unchanged. After DNA denaturation, the temperature is reduced to allow addition of a thermostable double-strand-DNA-specific nuclease (DSN) and mutation-overlapping oligonucleotide-probes that guide nuclease digestion to the selected WT-DNA sequences (28). NaME-PrO can be applied to numerous DNA targets in parallel and provides mutation enrichment up to several hundred-fold depending on conditions applied, resulting in mutation detection of allelic frequencies of 0.01% or less. Despite these advantages, the requirement of adding DSN enzyme after an initial denaturation step prevents application in a homogeneous format that can be performed on multiple clinical samples in parallel using commonly available laboratory equipment, eg a 96-sample PCR machine. Indeed, manual tube opening and addition of reagents in the pre-PCR setting introduces a major risk for cross-contamination, a critical concern for the reliable detection of rare mutations.
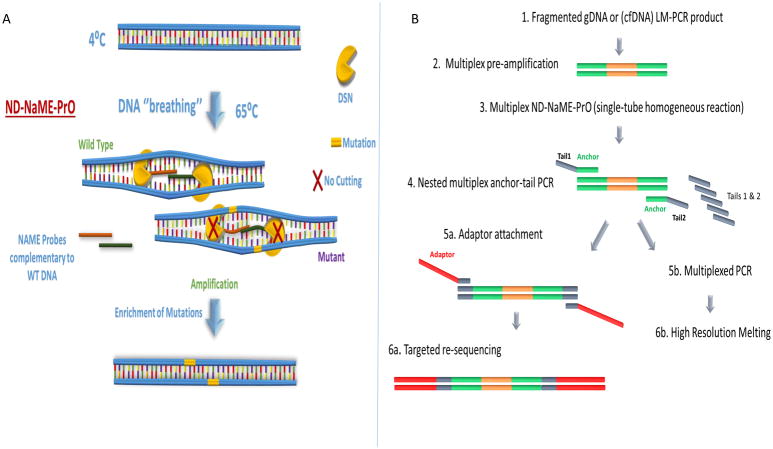
Here we introduce a modified approach via which NaME-PrO is applied in a homogeneous closed-tube format using partial denaturation at 65°C and without a denaturation step at 98°C (No-Denaturation NaME-PrO or ND-NaME-PrO). The modified technique (Fig. 1A) utilizes the strong preference of a thermostable duplex-specific nuclease (DSN) towards digestion of dsDNA over single stranded DNA (29). Local DNA mismatches reduce greatly DSN digestion (30), thereby enabling oligonucleotides matching WT DNA on top and bottom strands to direct DSN digestion and discriminate between mutation and WT sequences (28). ND-NaME-PrO utilizes a natural property of the dsDNA to spontaneously locally and partially denature, below DNA melting temperatures Tm (DNA ‘breathing’) (31). Our data indicate that, under appropriate conditions, DNA breathing allows the overlapping oligonucleotide probes to transiently invade the double-stranded structure and bind the partially denatured DNA strands (Fig. 1A), thereby guiding and enhancing DSN digestion of WT sequences despite absence of complete denaturation of the parent strands. ND-NaME-PrO provides a key practical advantage over the originally described NaME-PrO as it can be performed using a common 96-well thermocycler at high throughput fashion and with reduced risk for contamination.
Figure 1.
Concept and workflow for No-Denaturation Nuclease-assisted Minor-allele Enrichment using PRobe-Overlap, ND-NaME-PrO. A. ND-NaME-PrO the spontaneous partial denaturation of dsDNA (DNA ‘breathing’) at elevated temperatures remaining below DNA melting temperatures, allowing overlapping probes to bind to complementary target DNA strands. Double strand specific nuclease DSN is then digesting fully matched templates while mismatched sequences remain substantially undigested, thereby resulting to mutation enrichment upon subsequent amplification. It should be noted that dsDNA not targeted by probes also becomes digested, but at a much lower rate than targeted DNA (vide infra). B. Sample preparation workflow for multiplexed ND-NaME-PrO followed by targeted re-sequencing. The workflow includes an initial multiplex amplification from genomic DNA or cfDNA followed by nested multiplex anchor-tail PCR for a target panel. The anchor-tail-PCR product is then enriched for mutations at multiple positions via multiplexed ND-NaME-PrO, followed by multiplexed HRM-scanning and MiSeq sequencing of HRM-positive samples.
We further demonstrate the use of single-tube, multiplexed nested PCR followed by high-resolution melting, HRM scanning (32, 33), of 10–20 DNA targets combined with mutation enrichment. This process can be adopted as a rapid pre-screening tool to identify presence or absence of mutations on multiple targets simultaneously, prior to conducting demanding targeted re-sequencing. By combining ND-NaME-PrO with multiplexed PCR-HRM we amplify mutation-enriched targets and interrogate them for mutations prior to library formation for targeted re-sequencing. This novel process is applied in mutation containing genomic DNA and circulating DNA from clinical cancer samples.
Materials and Methods
Cell Lines and clinical samples
Human genomic DNA was extracted from SW480 (ATCC CCL-228™, KRAS mutation p.G12V, c.35G>T) commercial cell line. DNA extraction was performed with the DNeasy™ Blood and Tissue kit (Qiagen) following the manufacturer’s protocol. A standard reference DNA (Horizon Discovery HD728) was used to provide genomic DNA containing multiple mutations. Human genomic DNA (Promega) was used as a wild type control DNA. Serial dilution DNA mixtures of wild type and mutant DNA were prepared to obtain 5%, 1%, 0.3%, 0.1%, 0.03% and 0.01% mutation abundance. Cell-free circulating DNA (cfDNA) samples obtained under Institutional IRB approval from breast cancer patients were provided by the Broad Institute. Tumor biopsies and matched blood samples were collected from patients with metastatic breast cancer consented to Dana-Farber Cancer Institute IRB protocol #05-246.
Shearing of Genomic DNA
Genomic DNA shearing was performed with the use of dsDNA Shearase Plus (Zymo Research) in a total 10μl reaction (1x dsDNA Shearase Plus Reaction Buffer, 100ng of genomic DNA and 1 unit of dsDNA Shearase Plus enzyme). Genomic DNA was quantified with Qubit 3.0 fluorometer (Life Technologies).
Pre-amplification PCR
PCR reactions targeting KRAS exon 2 were prepared in a final volume of 25 μl (10ng of genomic DNA, 1× GoTaq buffer (Promega), 400nM of each primer, 200μM of each of the 4 dNTPs (BioLine), 1.25U of GoTaq Polymerase (Promega) and 10× LC Green). The primer sequences (KRAS-2-F1 and KRAS-2-R1) are depicted in Supplementary Table 1. The reaction was performed on a SmartCycler real-time PCR system (Cepheid). PCR protocol included an initial denaturation step at 98°C for 2 minutes followed by 40 cycles of denaturation at 98°C for 10 seconds, annealing at 58°C for 20 seconds and elongation at 72°C for 10 seconds.
ND-NAME-PrO
The single-plex ND-NaME-PrO was performed on the pre-amplified PCR products for KRAS exon 2. The 10-μl reaction contained 1μl of 1000-fold diluted KRAS PCR products, 0.75 X DSN buffer, 0.375 X GoTaq buffer, 200 nm of each overlapping probes (KRAS-sense-1 and KRAS-antisense-2 in Supplementary Table 2). The reaction setup was prepared on ice and mixed well. Then 1 μl of 1 unit/μl of DSN enzyme (Evrogen) was added in each reaction tube followed with brief vortex and centrifugation. Also, a No-DSN control sample (No DSN enzyme) was included and assessed in parallel with the rest of the samples. Then the PCR tubes were put on to a pre-heated thermocycler at 65°C. The incubation program was 65°C for 20 minutes, followed by an enzyme deactivation step at 95°C for 2 minutes. ND-NAME-PrO was followed by a nested PCR using KRAS-2-F2 and KRAS-2-R2 primers (sequences in Supplementary Table 1), following the same condition as the pre-amplification PCR described above (1μl of ND-NaME-PrO or No-DSN control product was used as DNA input for this reaction). In selected experiments, the activity of DSN enzyme on double stranded DNA was quantified by performing real time PCR before and after digestion, using nested KRAS primers (34).
Droplet digital PCR for mutation abundance validation
Droplet digital PCR (ddPCR) reaction was prepared as previously described (28) in order to quantify the mutation abundance of DNA samples before and after ND-NaME-PrO mutation enrichment. Primer and probe sequences are depicted in Supplementary Table 1. PCR was performed using Eppendorf Mastercycler EP gradient thermocycler (Eppendorf) and the reaction plate was transferred to a QX100 droplet reader (Bio-Rad) for endpoint reading. Quantification analysis and calculation of the ratios of positive events for a given channel (FAM or HEX) was performed with Quanta soft software (Bio-Rad) to calculate the mutation abundance.
Targeted re-sequencing (MiSeq) sample preparation using nested multiplexed anchor-tail PCR reactions
Thirty nanograms fragmented genomic DNA samples with serially diluted mutated DNA or cell free circulating DNA (cfDNA) were initially amplified for 10 cycles via ligation mediated PCR (LM-PCR) using common linkers (adaptor and primer sequences shown in Supplementary Table 3) as per instructions of the NEBNext® Ultra™ II DNA Library Prep kit (New England Biolabs). Then 30ng of LM-PCR product were transferred into a multiplex pre-amplification reaction using Ion AmpliSeq™ Cancer Hotspot Panel v2 (Thermo Fisher Scientific) or customized targets panel (primer sequences in Supplementary Table 3). Multiplex pre-amplification products were purified with Agencourt® AMPure XP (Beckman Coulter) and 1.5ng of product input was included in 10μl ND-NaME-PrO reactions with 200nM for each probe in the multiplex probe pool and 1 unit of DSN. The incubation was at 65°C for 20min followed by 95°C for 2 min. A No-treatment control sample with the same DNA input but without probes or DSN was run in parallel. Then 5μL of the ND-NaME-Pro or No-treatment product was added into 25μl multiplex ‘anchor-tail’ PCR reactions to further amplify the intended target sequences (primer sequences and PCR protocols are in Supplementary Table 4 and 5). The anchor-tail PCR reactions comprise a low concentration (1–10nM) of primers that are nested to the first set of primers and contain a gene-specific portion and common oligonucleotide ‘tails’. In addition, a high concentration (200nM) of the forward and reverse oligonucleotide tails is added to the same reaction. The anchor-tail PCR product was purified with Agencourt® AMPure XP and 5μl of product were added into a final adaptor-PCR for library preparation using NEBNext® Ultra™ II Q5® Master Mix and NEBNext® Multiplex Oligos for Illumina (New England Biolabs). The PCR protocol followed is depicted in Supplementary Table 5. The libraries were then purified with Agencourt® AMPure XP and delivered to the Center for Cancer Computational Biology at the Dana-Farber Cancer Institute to perform Illumina MiSeq sequencing. Libraries with ligated Illumina adapters were assessed for DNA quality and quantity on Agilent bio-analyzer, and then pooled together into a single tube prior to MiSeq sequencing. Data analysis was conducted using a MiSeq Reporter software and the alignment sequencing data were loaded into Integrative Genome Viewer 2.3 (IGV, Broad Institute) using human genome hg19 as reference.
Whole-exome sequencing and analysis
Whole-exome sequencing of tumor, germline, and cell-free DNA samples was performed using the Nextera Rapid Capture Exome Kit, with the exception that cell-free DNA libraries were first constructed using the Kapa Hyper Prep Kit. MuTect (35) was used to identify the somatic mutations in the whole-exome sequencing data.
Multiplex High resolution melting (HRM) analysis
The multiplex anchor-tail PCR products were diluted 100-fold into H2O and 1μl was transferred into 25μl-multiplex-anchor-tail-PCR reactions using the targets selected for each sample. Each sample was run in duplicates and wild-type samples were run in quadruplicates. The PCR protocol is depicted in Supplementary Table 5. 10μl of the 25μl-multiplex-anchor-tail-PCR products were then transferred to a 96-well plate and 20μl of mineral oil was added to each well. HRM was performed on a 96-well LightScanner®system (Idaho Technology). All experiments were independently replicated at least three times for assessing the reproducibility of results.
Results
Single DNA target ND-NaME-PrO
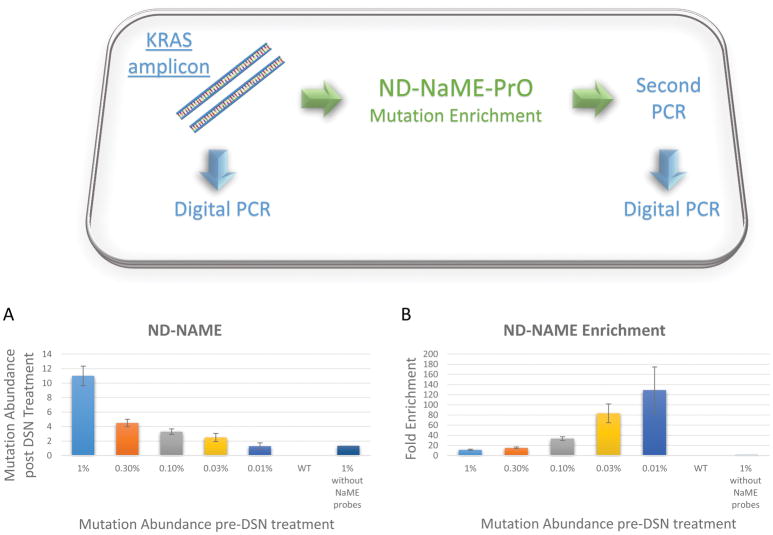
To examine ND-NaME-PrO efficiency in removing WT DNA and enriching mutations, a serial dilution experiment with decreasing KRAS exon 2 mutation abundance into WT DNA were assessed (1%, 0.3%, 0.1%, 0.03% and 0.01%), Figure 2A. Pre-amplification of KRAS was first conducted on ~100 ng fragmented genomic DNA containing various KRAS mutation abundances, then ND-NaME-PrO was conducted to diluted PCR products. The mutation abundance, before and after ND-NaME-PrO, was derived via droplet digital PCR. Mutation enrichment ranging from 10 to over 120-fold enrichment was obtained. Although it is understdood that, at mutation abundances as low as 0.01%, Poisson statistics can lead to significant variability in the number of mutant copies, the data indicate a trend for more pronounced mutation enrichment for lower initial mutation abundance, Figure 2B. The dependence of mutation enrichment on probe concentration is depicted in Supplementary Figure 1. Mutation enrichment increases with probe concentration with a maximum reached at about 200nM. In the absence of probes, double stranded DNA digestion proceeds but there is no mutation enrichment, Supplementary Figure 1. Mutation enrichment was then examined, at a single mutation abundance of 0.3%, for different DSN enzyme incubation temperatures during ND-NaME-PrO, Supplementary Figures 2A and 2B. The mutation enrichment is pronounced for temperatures ranging from 60°C-70°C, while there is little enrichment outside this range of temperatures. As demonstrated in Supplementary Figure 2C using real time PCR to examine threshold differences after DSN digestion, the lack of enrichment outside the 60°-70°C temperature range is not due to DSN inactivity. Also, the DSN digestion efficiency in the presence or absence of NaMe probes are demonstrated in Supplementary Figure 2D and 2E. The data is consistent with the scheme proposed in Figure 1, where transient DNA denaturation (‘breathing’) allows probe binding to fully-matched sequence positions and site-specific DNA digestion by DSN. As temperature decreases DNA breathing is also expected to decrease along with site-specific probe binding, while at temperatures higher than 70°C probe Tm is exceeded and DNA binding is reduced. It should be appreciated that since DNA remains double-stranded during ND-NaME-PrO, all DNA molecules are digested to some extent. However, the data indicate that at temperatures where DNA is expected to undergo ‘breathing’, WT-DNA targets addressed by NaME-PrO probes become digested faster than mutated DNA or DNA not targeted by probes. Supplementary Figure 2D indicates that, when probes are present, WT KRAS is almost completely eliminated, as assessed by the threshold of a real time PCR reaction applied to the ND-NaME-PrO product (the threshold is equal to that of a No-Template-Control reaction run in parallel). In contrast, when probes are omitted, DNA undergoes only modest DSN digestion as assessed by the PCR threshold compared to no-DSN control. Further, in the presence of mutated KRAS plus probes, the digestion is also modest and the PCR reaches threshold earlier since there are mismatches at the probe-binding positions. Accordingly, the data are consistent with ND-NaME-PrO causing modest general degradation of the DNA sample, with significantly increased degradation at wild-type DNA sites addressed by probes (Supplementary Figure 2E).
Figure 2.
Single-plex ND-NaME-PrO from PCR products. A. Mutation abundance obtained after single-plex ND-NaME-PrO for KRAS (c.35G>T, p.G12V) on serially decreasing mutated KRAS-containing DNA diluted in WT KRAS-containing DNA. Mutation abundance was assessed via droplet digital PCR following ND-NaME-PrO. B. Mutation enrichment obtained for KRAS mutations after ND-NaME-PrO ranged from 10-fold to 120-fold for 1% and 0.01% respectively.
Finally, ND-NaME-PrO was also performed directly from sheared genomic DNA without prior amplification. Mutation enrichment was observed, Supplementary Figure 3A, B, however the enrichment was lower than that obtained from a PCR product. Also, the lowest mutation abundance detectable was about 0.3%. 100ng of genomic DNA was used as starting material in this experiment, corresponding to 10,000–30,000 amplifiable genomic DNA copies. Since during ND-NaME-PrO a proportion of mutated DNA molecules are also digested one potential explanation for the difference between the results obtained when starting from genomic DNA versus a PCR product is the limited number of mutated molecules in the former case. Accordingly, in all subsequent experiments ND-NaME-PrO was performed following an initial PCR amplification step.
Multiplexed ND-NaME-PrO followed by multiplex-HRM
To perform multiplex ND-NaME-PrO followed by single-tube multiplexed HRM and Miseq sequencing we followed the sample preparation workflow depicted in Figure 1b. Fragmented genomic DNA samples with serially decreasing mutation abundances (5%, 2.5%, 1%, 0.3% and 0.1%) were first amplified via LM-PCR, targeted multiplexed PCR and multiplexed anchor-tail-PCR to obtain common tails at each amplicon matching the Illumina adaptors used for targeted re-sequencing (MiSeq). The anchor-tail-PCR products were then diluted and processed via ND-NaME-PrO performed in a multiplexed single tube reaction using probes targeting multiple DNA targets. The ND-NaME-PrO products were then added to adaptor-PCR reactions including Illumina adaptors and sample indexes, to yield sequencing libraries. No-treatment control samples were also run in parallel throughout the process, by omitting ND-NaME-PrO. Before conducting MiSeq targeted re-sequencing, multiplexed single-tube HRM was performed for rapid pre-screening prior to time and expense-demanding sequencing.
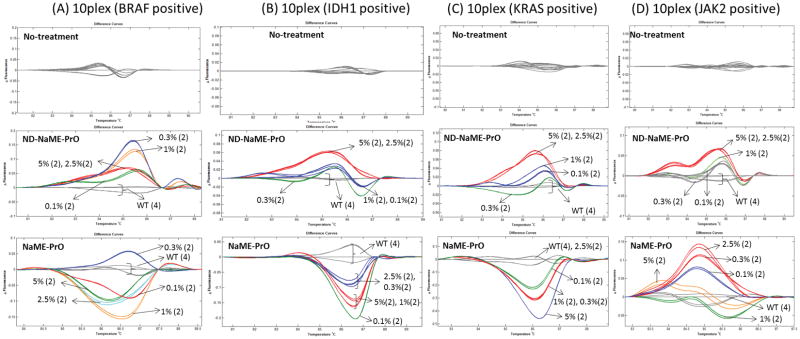
To validate the use of multiplexed HRM for mutation scanning prior to conducting targeted re-sequencing, 10-plex PCR reactions where just one of 10 DNA targets was mutated were first tested via HRM, using serially decreasing mutation dilutions. Four different genes were tested in this approach, in separate experiments, BRAF, IDH1, KRAS, and JAK2, Figure 3A–D. While it was not possible to discriminate the mutations from WT samples in the no-treatment samples, in the presence of ND-NaME-PrO the presence of a mutated target was clearly evident, and the detection limit was 0.1%–1%, depending on the target gene. When ND-NaME-PrO was replaced with NaME-PrO which includes a denaturation step prior to initiating DSN digestion (28), similar results were observed indicating the equivalence of the two approaches. In another test, a 20plex-HRM was conducted, where 8 targets contained serially decreasing mutation abundances (5%, 2.5%, 1%, 0.3% and 0.1%). HRM was not able to distinguish mutant DNA from wild-type DNA in the no-treatment samples, Supplementary Figure 4. When ND-NaME-PrO was conducted, the mutant DNA melting profiles were clearly distinguished from WT DNA, presumably as a result of mutation enrichment. Mutation abundance down to 0.1% was detectable via multiplexed-HRM mutation scanning in this case. Taken together, these data demonstrate the potential of using multiplexed HRM in combination with mutation enrichment on 10–20 targets, where mutations in any one target can be identified prior to sequencing.
Figure 3.
Multiplexed ND-NaME-PrO followed by 10-plex-PCR and multiplexed HRM scanning of amplicons containing a single mutated target at decreasing dilutions. A. BRAF gene, B. IDH1 gene, C. KRAS gene and D. JAK2 gene. Top four panels, samples run in parallel while omitting DSN treatment. Middle four panels, samples treated via ND-NaME-PrO (homogeneous, closed tube process). Bottom four panels, samples treated via NaME-PrO (non-homogenous process). All Mutation samples were assessed in duplicate (2) and wild type controls in quadruplicate (4).
Multiplexed ND-NaME-PrO-HRM and Miseq sequencing on circulating DNA from cancer patients
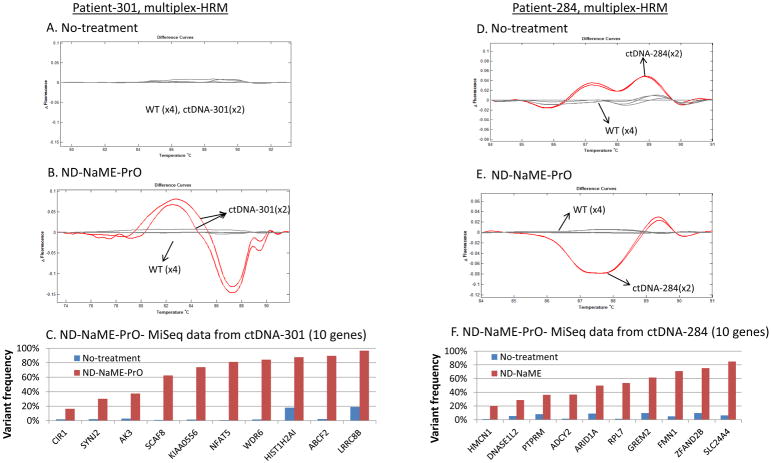
To perform ND-NaME-PrO-HRM-Miseq on circulating DNA from plasma obtained from two metastatic breast cancer patients (#301 and #284), we selected two samples where exome sequencing had been performed on matched tumor biopsies (36, 37). We selected ten mutations that were confidently detected by WES of the tumor biopsies. These were detected at random, without consideration on whether they were potential driver mutations or passenger mutations, to assess our ability to enrich any tumor mutation in a customizable manner. Primers and probes corresponding to these ten mutated DNA target regions were designed for each sample (Supplementary Tables 1 and 4). The workflow depicted on Figure 1b was then applied. In samples that contained multiple mutations, multiplexed HRM in the absence of ND-NaME-PrO could differentiate melting profiles from WT samples run in parallel, in just one of two samples (Figure 4a, 4d). The differentiation of melting profiles in HRM is important as it can distinguish WT from clinical samples. When mutation enrichment via ND-NaME-PrO was applied, melting profile differentiation from WT was evident in both samples (Figure 4b, 4e). Upon MiSeq sequencing the presence of mutations was demonstrated in all 10 targets for both samples. While in the no-treatment samples the mutations were close to the noise limit (1–2%), in the ND-NaME-PrO treated samples mutations were enriched in all ten targets and were clearly evident (Figure 4c, 4f). The mutation enrichment generated via ND-NaME-PrO is variable, Figure 4. Mutations having lower original mutation abundance are enriched the most (Supplementary Figure 5; also Figure 2) providing for very sensitive detection of low-level mutations. Overall, following multiplex ND-NaME-PrO, cfDNA mutations from metastatic breast cancer samples were enriched by up to 98-fold with an average of 24-fold.
Figure 4.
ND-NaME-PrO-HRM-MiSeq performed in circulating tumor DNA (ctDNA) containing mutations in 10 different targets obtained from metastatic breast cancer patients. (A, D) No-treatment samples without DSN were processed in parallel comparison. (B, E) Multiplexed mutation scanning pre-screening performed via multiplexed High Resolution Melting (HRM). (C, F). Variant frequency at the 10 mutated targets on cfDNA as derived via targeted re-sequencing (Miseq) for no-treatment samples (blue bars) and ND-NaME-treated samples (red bars).
DISCUSSION
We previously, reported on a homogeneous, closed tube approach to reduce or eliminate WT DNA from multiple targets of interest based on the preferential digestion of double-stranded, mismatch-free DNA by a double strand specific nuclease, DSN. The originally described approach (28) involved DNA denaturation followed by cooling, open-tube addition of DSN enzyme and contamination-prone sample manipulations. Additionally, we have shown that if the DNA is single stranded, such as in bisulfite treated DNA, DNA denaturation is not required (38). The present work reveals that this process also works with double stranded DNA in a closed-tube format that can be performed on a standard PCR machine. We observed that, under certain temperature, probe and enzyme concentration conditions, partly denatured DNA enables WT-DNA-matching oligonucleotide probes to transiently bind their target and enable local DSN digestion. Consistent with previous reports (28, 29), probes fully matching their DNA targets are digested by DSN at a substantially higher rate than mismatch-containing targets, thereby leading to preferential WT elimination. The thermal stability properties of the crab hepato-pancreas derived nuclease DSN (29) enables enzymatic activity at 65°C which matches the Tm of the oligonucleotide probes used in this work. Additional double strand DNA specific nucleases have been reported (39), and these can also be potentially adapted to produce mutation enrichment in a similar manner by matching oligonucleotide probe Tm, denaturation conditions (buffer) and enzyme incubation temperature.
The ability to enrich multiple targets simultaneously for mutations lead to a novel adaptation of HRM for multiplexed mutation scanning. While HRM-genotyping has been used for multiplexing up to 3–4 targets (40), to our knowledge multiplexing of multiple amplicons for HRM-scanning has not been reported. Mutation scanning via HRM provides a straightforward, low-cost method for identifying DNA variations on single PCR amplicons (32, 33) and it may be performed prior to Sanger sequencing to avoid screening of non-informative wild type samples. Application of HRM scanning with next generation sequencing platforms required two improvements; adaptation of HRM-scanning to a multiplexed format following nested multiplexed PCR (Figure 1B), and boosting HRM sensitivity via mutation enrichment, to ensure low level mutations present in clinical samples like cfDNA are not missed. Indeed, since for single PCR amplicons the HRM scanning sensitivity is of the order of ~5% (33, 41), multiplexed HRM with 10 amplicons might not be sensitive enough to detect a mutation in just one of the amplicons, unless the mutation is present at high level. Here, combining mutation enrichment via ND-NaME-PrO with nested multiplexed PCR and HRM resulted in sensitive HRM-scanning for single mutations over 10 targets simultaneously (Figure 3). This development opens up the possibility of filtering out uninformative samples and concentrating targeted re-sequencing effort and resources to just the mutation-containing samples.
Supplementary Material
Supplementary Table 1. Primers and fluoresence probes for PCR and droplet digital PCR.
Supplementary Table 2. Overlapping probes for ND-NaME-PrO assay
Supplementary Table 3. Adaptor and primers for LM-PCR, and Multiplex PCR primers for cfDNA
Supplementary Table 4. Multiplex anchor-PCR primers
Supplementary Table 5. PCR protocols
Acknowledgments
The present work was partially supported by a Bridge Award from Dana Farber Harvard Cancer Center and the Kock Institute for Integrative Cancer Research at MIT; also by SPORE Grant # P50CA168504; and by an ACT-NOW Fund at Dana-Farber Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health/NCI.
References
- 1.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, Kinzler KW, Vogelstein B, Diaz LA., Jr Circulating mutant DNA to assess tumor dynamics. Nature medicine. 2008;14:985–90. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thierry AR, Mouliere F, El Messaoudi S, Mollevi C, Lopez-Crapez E, Rolet F, Gillet B, Gongora C, Dechelotte P, Robert B, Del Rio M, Lamy PJ, Bibeau F, Nouaille M, et al. Clinical validation of the detection of kras and braf mutations from circulating tumor DNA. Nature medicine. 2014;20:430–5. doi: 10.1038/nm.3511. [DOI] [PubMed] [Google Scholar]
- 3.Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE, Shrager JB, Loo BW, Jr, Alizadeh AA, Diehn M. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nature medicine. 2014;20:548–54. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, Antonarakis ES, Azad NS, Bardelli A, Brem H, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, Diaz LA, Jr, Goodman SN, David KA, Juhl H, Kinzler KW, Vogelstein B. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A. 2005;102:16368–73. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwaederle M, Husain H, Fanta PT, Piccioni DE, Kesari S, Schwab RB, Patel SP, Harismendy O, Ikeda M, Parker BA, Kurzrock R. Use of liquid biopsies in clinical oncology: Pilot experience in 168 patients. Clinical cancer research : an official journal of the American Association for Cancer Research. 2016;22:5497–505. doi: 10.1158/1078-0432.CCR-16-0318. [DOI] [PubMed] [Google Scholar]
- 7.Roschewski M, Dunleavy K, Pittaluga S, Moorhead M, Pepin F, Kong K, Shovlin M, Jaffe ES, Staudt LM, Lai C, Steinberg SM, Chen CC, Zheng J, Willis TD, et al. Circulating tumour DNA and ct monitoring in patients with untreated diffuse large b-cell lymphoma: A correlative biomarker study. The Lancet Oncology. 2015;16:541–9. doi: 10.1016/S1470-2045(15)70106-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milbury CA, Li J, Makrigiorgos GM. Pcr-based methods for the enrichment of minority alleles and mutations. Clin Chem. 2009;55:632–40. doi: 10.1373/clinchem.2008.113035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, Smith JC, Markham AF. Analysis of any point mutation in DNA. The amplification refractory mutation system (arms) Nucleic acids research. 1989;17:2503–16. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun X, Hung K, Wu L, Sidransky D, Guo B. Detection of tumor mutations in the presence of excess amounts of normal DNA. Nat Biotechnol. 2002;20:186–9. doi: 10.1038/nbt0202-186. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Wang L, Mamon H, Kulke MH, Berbeco R, Makrigiorgos GM. Replacing pcr with cold-pcr enriches variant DNA sequences and redefines the sensitivity of genetic testing. Nature medicine. 2008;14:579–84. doi: 10.1038/nm1708. [DOI] [PubMed] [Google Scholar]
- 12.Wu LR, Wang JS, Fang JZ, Evans ER, Pinto A, Pekker I, Boykin R, Ngouenet C, Webster PJ, Beechem J, Zhang DY. Continuously tunable nucleic acid hybridization probes. Nat Methods. 2015;12:1191–6. doi: 10.1038/nmeth.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guha M, Castellanos-Rizaldos E, Liu P, Mamon H, Makrigiorgos GM. Differential strand separation at critical temperature: A minimally disruptive enrichment method for low-abundance unknown DNA mutations. Nucleic acids research. 2013;41:e50. doi: 10.1093/nar/gks1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milbury CA, Li J, Makrigiorgos GM. Ice-cold-pcr enables rapid amplification and robust enrichment for low-abundance unknown DNA mutations. Nucleic acids research. 2011;39:e2. doi: 10.1093/nar/gkq899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.How Kit A, Mazaleyrat N, Daunay A, Nielsen HM, Terris B, Tost J. Sensitive detection of kras mutations using enhanced-ice-cold-pcr mutation enrichment and direct sequence identification. Human mutation. 2013;34:1568–80. doi: 10.1002/humu.22427. [DOI] [PubMed] [Google Scholar]
- 16.Galbiati S, Brisci A, Lalatta F, Seia M, Makrigiorgos GM, Ferrari M, Cremonesi L. Full cold-pcr protocol for noninvasive prenatal diagnosis of genetic diseases. Clin Chem. 2011;57:136–8. doi: 10.1373/clinchem.2010.155671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Milbury CA, Li C, Makrigiorgos GM. Two-round coamplification at lower denaturation temperature-pcr (cold-pcr)-based sanger sequencing identifies a novel spectrum of low-level mutations in lung adenocarcinoma. Human mutation. 2009;30:1583–90. doi: 10.1002/humu.21112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Berbeco R, Distel RJ, Janne PA, Wang L, Makrigiorgos GM. S-rt-melt for rapid mutation scanning using enzymatic selection and real time DNA-melting: New potential for multiplex genetic analysis. Nucleic acids research. 2007;35:e84. doi: 10.1093/nar/gkm403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy DM, Bejar R, Stevenson K, Neuberg D, Shi Y, Cubrich C, Richardson K, Eastlake P, Garcia-Manero G, Kantarjian H, Ebert BL, Mike Makrigiorgos G. Nras mutations with low allele burden have independent prognostic significance for patients with lower risk myelodysplastic syndromes. Leukemia. 2013;27:2077–81. doi: 10.1038/leu.2013.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castellanos-Rizaldos E, Richardson K, Lin R, Wu G, Makrigiorgos MG. Single-tube, highly parallel mutation enrichment in cancer gene panels by use of temperature-tolerant cold-pcr. Clin Chem. 2015;61:267–77. doi: 10.1373/clinchem.2014.228361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mauger F, Daunay A, Deleuze JF, Tost J, How-Kit A. Multiplexing of e-ice-cold-pcr assays for mutation detection and identification. Clin Chem. 2016;62:1155–8. doi: 10.1373/clinchem.2016.258830. [DOI] [PubMed] [Google Scholar]
- 22.Milbury CA, Correll M, Quackenbush J, Rubio R, Makrigiorgos GM. Cold-pcr enrichment of rare cancer mutations prior to targeted amplicon resequencing. Clin Chem. 2012;58:580–9. doi: 10.1373/clinchem.2011.176198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parry JM, Shamsher M, Skibinski DO. Restriction site mutation analysis, a proposed methodology for the detection and study of DNA base changes following mutagen exposure. Mutagenesis. 1990;5:209–12. doi: 10.1093/mutage/5.3.209. [DOI] [PubMed] [Google Scholar]
- 24.Haliassos A, Chomel JC, Grandjouan S, Kruh J, Kaplan JC, Kitzis A. Detection of minority point mutations by modified pcr technique: A new approach for a sensitive diagnosis of tumor-progression markers. Nucleic acids research. 1989;17:8093–9. doi: 10.1093/nar/17.20.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward R, Hawkins N, O’Grady R, Sheehan C, O’Connor T, Impey H, Roberts N, Fuery C, Todd A. Restriction endonuclease-mediated selective polymerase chain reaction: A novel assay for the detection of k-ras mutations in clinical samples. Am J Pathol. 1998;153:373–9. doi: 10.1016/S0002-9440(10)65581-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bielas JH, Loeb LA. Quantification of random genomic mutations. Nat Methods. 2005;2:285–90. doi: 10.1038/nmeth751. [DOI] [PubMed] [Google Scholar]
- 27.Gu W, Crawford ED, O’Donovan BD, Wilson MR, Chow ED, Retallack H, DeRisi JL. Depletion of abundant sequences by hybridization (dash): Using cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol. 2016;17:41. doi: 10.1186/s13059-016-0904-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song C, Liu Y, Fontana R, Makrigiorgos A, Mamon H, Kulke MH, Makrigiorgos GM. Elimination of unaltered DNA in mixed clinical samples via nuclease-assisted minor-allele enrichment. Nucleic acids research. 2016;44:e146. doi: 10.1093/nar/gkw650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shagin DA, Rebrikov DV, Kozhemyako VB, Altshuler IM, Shcheglov AS, Zhulidov PA, Bogdanova EA, Staroverov DB, Rasskazov VA, Lukyanov S. A novel method for snp detection using a new duplex-specific nuclease from crab hepatopancreas. Genome research. 2002;12:1935–42. doi: 10.1101/gr.547002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shao ZM, Wu J, Shen ZZ, Nguyen M. P53 mutation in plasma DNA and its prognostic value in breast cancer patients. Clin Cancer Res. 2001;7:2222–7. [PubMed] [Google Scholar]
- 31.Fei J, Ha T. Watching DNA breath one molecule at a time. Proceedings of the National Academy of Sciences. 2013;110:17173–4. doi: 10.1073/pnas.1316493110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liew M, Pryor R, Palais R, Meadows C, Erali M, Lyon E, Wittwer C. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin Chem. 2004;50:1156–64. doi: 10.1373/clinchem.2004.032136. [DOI] [PubMed] [Google Scholar]
- 33.Reed GH, Wittwer CT. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem. 2004;50:1748–54. doi: 10.1373/clinchem.2003.029751. [DOI] [PubMed] [Google Scholar]
- 34.Amicarelli G, Shehi E, Makrigiorgos GM, Adlerstein D. Flag assay as a novel method for real-time signal generation during pcr: Application to detection and genotyping of kras codon 12 mutations. Nucleic acids research. 2007;35:e131. doi: 10.1093/nar/gkm809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lennon NJ, Adalsteinsson VA, Gabriel SB. Technological considerations for genome-guided diagnosis and management of cancer. Genome Med. 2016;8:112. doi: 10.1186/s13073-016-0370-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adalsteinsson VA, Love JC. Towards engineered processes for sequencing-based analysis of single circulating tumor cells. Curr Opin Chem Eng. 2014;4:97–104. doi: 10.1016/j.coche.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, Song C, Ladas I, Fitarelli-Kiehl M, Makrigiorgos GM. Methylation-sensitive enrichment of minor DNA alleles using a double-strand DNA-specific nuclease. Nucleic acids research. 2016 doi: 10.1093/nar/gkw1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nilsen IW, Overbo K, Jensen Havdalen L, Elde M, Gjellesvik DR, Lanes O. The enzyme and the cdna sequence of a thermolabile and double-strand specific dnase from northern shrimps (pandalus borealis) PloS one. 2010;5:e10295. doi: 10.1371/journal.pone.0010295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seipp MT, Pattison D, Durtschi JD, Jama M, Voelkerding KV, Wittwer CT. Quadruplex genotyping of f5, f2, and mthfr variants in a single closed tube by high-resolution amplicon melting. Clin Chem. 2008;54:108–15. doi: 10.1373/clinchem.2007.097121. [DOI] [PubMed] [Google Scholar]
- 41.Milbury CA, Li J, Makrigiorgos GM. Cold-pcr-enhanced high-resolution melting enables rapid and selective identification of low-level unknown mutations. Clin Chem. 2009;55:2130–43. doi: 10.1373/clinchem.2009.131029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Primers and fluoresence probes for PCR and droplet digital PCR.
Supplementary Table 2. Overlapping probes for ND-NaME-PrO assay
Supplementary Table 3. Adaptor and primers for LM-PCR, and Multiplex PCR primers for cfDNA
Supplementary Table 4. Multiplex anchor-PCR primers
Supplementary Table 5. PCR protocols