Abstract
Improved prediction of the “most harmful” breast cancers that cause the most substantive morbidity and mortality would enable physicians to target more intense screening and preventive measures at those women who have the highest risk; however, such prediction models for the “most harmful” breast cancers have rarely been developed. Electronic health records (EHRs) represent an underused data source that has great research and clinical potential. Our goal was to quantify the value of EHR variables in the “most harmful” breast cancer risk prediction. We identified 794 subjects who had breast cancer with primary non-benign tumors with their earliest diagnosis on or after 1/1/2004 from an existing personalized medicine data repository, including 395 “most harmful” breast cancer cases and 399 “least harmful” breast cancer cases. For these subjects, we collected EHR data comprised of 6 components: demographics, diagnoses, symptoms, procedures, medications, and laboratory results. We developed two regularized prediction models, Ridge Logistic Regression (Ridge-LR) and Lasso Logistic Regression (Lasso-LR), to predict the “most harmful” breast cancer one year in advance. The area under the ROC curve (AUC) was used to assess model performance. We observed that the AUCs of Ridge-LR and Lasso-LR models were 0.818 and 0.839 respectively. For both the Ridge-LR and Lasso-LR models, the predictive performance of the whole EHR variables was significantly higher than that of each individual component (p<0.001). In conclusion, EHR variables can be used to predict the “most harmful” breast cancer, providing the possibility to personalize care for those women at the highest risk in clinical practice.
Keywords: breast cancer, electronic health records (EHRs), regularized prediction model, least absolute shrinkage and selection operator (Lasso)
1. INTRODUCTION
Breast cancer is still the most common non-skin malignancy affecting women in the United States, disregarding race and ethnicity. To reduce morbidity and mortality from breast cancer, a population based mammography screening protocol is currently recommended with the aim of allowing prevention and early diagnosis. Unfortunately, emerging evidence suggests that mammography for the general population results in over-diagnosis and over-treatment, which may decrease the overall efficacy of breast cancer management on a population level1–3. The need for alternative approaches to improve the overall cost-effectiveness of breast cancer management is duly being acknowledged.
One of the possible alternatives is to predict the “most harmful” breast cancers, which cause the most substantive morbidity and mortality, since it would enable physicians to target intense screening and preventive measures at those women at the highest risk. A series of prediction models have been developed to predict the probability of malignant versus benign tumors, including demographic risk factors4, genetic variants5–7 and imaging features8–11. However, few manuscripts document models to predict the “most harmful” breast cancers. Prior studies have demonstrated that mammography abnormality features could be used to differentiate invasive breast cancer versus ductal carcinoma in situ (DCIS)12–14. DCIS is a non-obligate precursor to subsequent invasive breast cancer, which may remain indolent for many years such that a woman may die of other causes. Invasive breast cancer is more harmful than DCIS. Nevertheless, it is necessary to explore other data sources to predict the “most harmful” breast cancer for the general population, achieving the overall cost-effectiveness of breast cancer management.
Electronic health records (EHRs) are an increasingly common data source for clinical risk prediction, which may yield useful information to improve risk prediction for the “most harmful” breast cancers, thereby improving the overall cost-effectiveness of breast cancer management. EHRs capture and integrate patient data from all aspects of clinical observation, including demographics, history of hospital visits, diagnoses, physiological measurements, and interventions. Although the main purpose of EHRs is to efficiently achieve and manage patient data, secondary use of EHRs is currently being widely explored for various research, among which developing risk prediction models with EHRs has received substantial attention15. Unfortunately, limited research exists on how information from EHRs can successfully be used to improve breast cancer risk prediction15, 16.
This study provides a glimpse into the opportunities of using EHR data to predict the “most harmful” breast cancers. We aim to quantify the value of EHRs in risk prediction for the “most harmful” breast cancers.
2. METHODS
In this study, we investigated the use of various data types in EHRs to predict the “most harmful” breast cancer. We utilized regularization based learning algorithms to build predictive models.
2.1 Subjects
In this study, the subjects were identified from women enrolled in the Marshfield Clinic Health System at Marshfield, WI, who had breast cancer with primary non-benign tumors and their earliest diagnosis on or after 1/1/2004 (total 3,205 women). The primary measure of “harm” in this study was death due to cancer. Since we did not have cause of death available electronically, “cancer death” was defined for analysis as death with: 1) cancer diagnoses within 60 days prior; and/or 2) cancer registry designation of “never disease free”. In the analysis cohort, 245/3,205 (7.6%) showed cancer death with times of cancer death after earliest diagnosis ranging from 0 to 129.5 months (median 31.9 months). Observation times for those not showing cancer death were censored at death (assumed to be from other causes) or date of last contact in the EHR, and these times ranged from 0 to 153.5 months (median 56.1 months).
For this pilot study, we initially collected 400 “most harmful” cases who had the smallest number of months from diagnosis to death (cancer death, death from other causes, or date of last contact). We collected 400 subjects with the “least harmful” breast cancers who had the largest number of months from diagnosis to death. We further found that there were six subjects without an ICD-9 diagnosis code of 174.X (malignant neoplasm of female breast), 233.0 (carcinoma in situ of breast), or C50.X (malignant neoplasm of breast). After these six subjects were removed, we had 395 “most harmful” breast cancer cases and 399 “least harmful” breast cancer cases for analysis.
The Marshfield Clinic Institutional Review Board reviewed and approved this study.
2.2 EHRs and Feature Representation
EHRs were extracted from Marshfield Clinic’s internally developed Research Data Warehouse (RDW). In this study, we extracted the structured EHR components to develop risk prediction models, which includes demographics, diagnoses, medications, laboratory tests and procedures. We also used natural language processing to extract symptoms from Marshfield Clinic’s clinical narratives. The symptoms were mapped to concepts represented as concept unit identifiers (CUIs) and type unit identifiers (TUIs) in the Unified Medical Language System (UMLS) Meta-thesaurus17. The coded CUIs and TUIs were made available to this investigation (Table 1).
Table 1.
EHR data points extracted from the warehouse
| Table name | Data points in each EHR component (partial) |
|---|---|
| demographics | Patient id, date of birth, gender |
| diagnoses | Patient id, age, ICD-9 diagnosis code, diagnosis description |
| medications | Patient id, age, generic code number sequence number, drug name, generic name, dosage, frequency |
| laboratory results | Patient id, age, laboratory id, laboratory description, results, unit id, unit description |
| procedures | Patient id, age, national procedure code, national code type, national code description, national code category |
| symptoms | Patient id, age, concept unique identifier (CUI), type unique identifier (TUI) |
For each subject, we included the age at event for each of the EHR components represented in Table 1. We identified age-at-initial breast cancer diagnosis when an ICD-9 code 174.X, 233.0, or C50.X first appeared in the EHR. We then labeled one year prior to this event as the censor date – the point at which we censored all future data points before we built our feature vector. All EHR data points prior to the censor date was used to predict the “most harmful” breast cancer (Figure 1).
Figure 1.
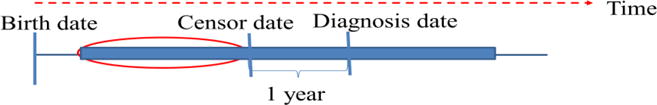
Specification of censor date. EHR data in the red oval were used to predict the “most harmful” breast cancer.
Finally, we mapped important EHR data points prior to the censor date to binary features, which were set to “present” only if the corresponding data points appeared three or more times prior to the censor date to avoid spurious events. Otherwise, the value was set to “not present”. Specifically, for diagnoses, we converted unique ICD-9 diagnosis codes into binary features to predict the “most harmful” breast cancer without consideration of their hierarchical structure (Table 1). For medications, we mapped drug names to binary features. For laboratory results, we generated a set of binary features by combining laboratory test id and lab interpretation of the result (normal, abnormal, high, critical high, low, critical low). For example, the results for a Potassium laboratory test (laboratory test id, 20715) were normal, abnormal, high, critical high, low, or critical low. Six binary features would be created, including 20715normal, 20715abnormal, 20715high, 20715critical_high, 20715low, and 20715critical_low. For procedures, we created binary features using national procedure codes. For symptoms, we created binary features using the concept unique identifiers.
2.3 Statistical Analysis
We first investigated the characteristics of the “most harmful” breast cancers and the “least harmful” breast cancers. Specifically, we examined the distribution difference for breast cancer grade, regional nodes, breast cancer stage, tumor size, estrogen receptors (ER), and progesterone receptors (PR) between two groups. We used the chi-square test to determine whether there was a significant difference.
In this study, we developed two regularized logistic regression models to predict the “most harmful” breast cancer since they are powerful techniques generally used to create parsimonious models to prevent over-fitting in presence of a large number of features. One was the widely used Logistic Regression with ridge regularization (Ridge-LR), which adds a penalty equivalent to the square of the magnitude of coefficients18. The other was Logistic Regression with Lasso (least absolute shrinkage and selection operator) regularization (Lasso-LR), which adds a penalty equivalent to the absolute value of the magnitude of coefficients19–22. Lasso is a regression analysis method that performs variable selection by regularization to enhance the prediction accuracy and interpretability of the prediction model it produces. Regularized logistic regression models provide a natural choice for risk prediction using EHR data since they are high-dimensional and sparse, i.e., patients are described using a large number of features but many have zero values.
For the Ridge-LR model, the model parameters β (regression coefficients) were learned from a training data set by optimizing the following objective function:
where λ was the regularization parameter.
For the Lasso-LR model, the model parameters β were learned by optimizing the following objective function:
where λ was the regularization parameter. Lasso was used to achieve sparsity in the solution such that most coefficients in β were 0.
Prediction models were constructed and evaluated using the Glmnet package23. We selected the tuning parameter λ via 10-fold cross validation method, which divided the data into ten folds of equal size where each fold played the role of testing set whereas the other remaining folds were the training set. For each λ, the predictive performance in terms of the area under the ROC curve (AUC) was calculated for each of the 10 testing sets. After the 10 AUCs were obtained for each λ, we calculated their average. We chose the λ which had the highest average of AUCs. We compared AUCs by using the DeLong method24 implemented in the MATLAB software (MathWorks, Natick, MA). We used a P-value of 0.05 as the threshold for statistical significance testing to assess the difference between the two AUC values.
3. RESULTS
The age range for 395 “most harmful” cases was 29 to 90 years of age (mean = 65.25, standard deviation = 13.87). The age range for 399 “least harmful” cases was 33 to 90 years of age (mean = 64.11, standard deviation = 12.28). We found that the “most harmful” breast cancer cases were likely to have high-grade and advanced-stage tumors. The tumors in the “most harmful” breast cancer cases were likely larger than those in the “least harmful” breast cancers cases. In addition, they would have a higher probability to test negative for ER and PR than those in the “least harmful” breast cancers cases. They also likely tended to test positive for regional nodes. In summary, we found that the distributions of breast cancer grade, regional nodes positive, stage, tumor size, ER, and PR differed significantly between the two groups (p-value < 0.001) (Table 2).
Table 2.
Characteristics of the “most harmful” breast cancers and the “least harmful” breast cancers
| Variables | The “most harmful” cases (N=395) | The “least harmful” cases (N=399) | p-value |
|---|---|---|---|
| Breast cancer grade | p<0.001 | ||
| 1 | 35 (8.86%) | 128 (32.08%) | |
| 2 | 128 (32.41%) | 118 (29.57%) | |
| 3 | 171 (43.29%) | 75 (18.80%) | |
| 4 | 1 (0.25%) | 0 (0%) | |
| unknown | 60 (15.19%) | 78 (19.55%) | |
| Regional nodes | p<0.001 | ||
| negative | 111 (28.10%) | 201 (50.38%) | |
| positive | 156 (39.49%) | 0 (0%) | |
| unknown | 128 (32.41%) | 198 (49.62%) | |
| Breast cancer stage | p<0.001 | ||
| 0 | 8 (2.02%) | 26 (6.51%) | |
| I | 61 (15.44%) | 183 (45.86%) | |
| II | 83 (21.01%) | 0 (0%) | |
| III | 59 (14.94%) | 0 (0%) | |
| IV | 19 (4.81%) | 0 (0%) | |
| unknown | 165 (41.77%) | 190 (47.62%) | |
| Tumor size | p<0.001 | ||
| No mass | 3 (0.76%) | 0 (0%) | |
| Small (<30 mm) | 186 (47.09%) | 292 (73.18%) | |
| Large | 154 (38.99%) | 1 (0.25%) | |
| unknown | 52 (13.16%) | 106 (26.57%) | |
| ER | p<0.001 | ||
| positive | 256 (64.81%) | 285 (71.43%) | |
| negative | 85 (21.52%) | 1 (0.25%) | |
| borderline | 5 (1.27%) | 2 (0.50%) | |
| unknown | 49 (12.41%) | 111 (27.82%) | |
| PR | p<0.001 | ||
| positive | 169 (42.78%) | 250 (62.66%) | |
| negative | 166 (42.03%) | 0 (0%) | |
| borderline | 11 (2.78%) | 35 (8.77%) | |
| unknown | 49 (12.41%) | 114 (28.57%) | |
We observed that there was a total of 28,890 EHR features, including 9,146 diagnosis features, 7,738 symptom features, 5,850 procedure features, 2,838 medication features, and 3,218 laboratory result features (Table 3). For the Ridge-LR model, the AUC was 0.818 using entire EHR features (Figure 2), which was significantly higher than that of each individual EHR component: diagnoses (AUC 0.733, p<0.001), symptoms (AUC 0.779, p<0.001), procedures (AUC 0.763, p<0.001), medications (AUC 0.745, p<0.001), and laboratory results (AUC 0.713, p<0.001). For the Lasso-LR model, the AUC was 0.839 using entire EHR features (Figure 3), which was also significantly higher than that of each individual feature component: diagnoses (AUC 0.761, p<0.001), symptoms (AUC 0.808, p<0.001), procedures (AUC 0.785, p<0.001), medications (AUC 0.744, p<0.001), and laboratory results (AUC 0.754, p<0.001).
Table 3.
Predictive performance of two models and feature selection results
| Feature number | Ridge-LR | Lasso-LR | p (Ridge-LR vs Lasso-LR) | Number of features selected using Lasso-LR | |
|---|---|---|---|---|---|
| diagnoses | 9,146 | 0.733 | 0.761 | 0.0162 | 115 |
| symptoms | 7,738 | 0.779 | 0.808 | 0.169 | 5 |
| procedures | 5,850 | 0.763 | 0.785 | 0.0248 | 36 |
| medications | 2,838 | 0.745 | 0.744 | 0.183 | 18 |
| laboratory results | 3,218 | 0.713 | 0.754 | <0.001 | 63 |
| entire EHR features | 28,890 | 0.818 | 0.839 | 0.203 | 41 |
Figure 2.
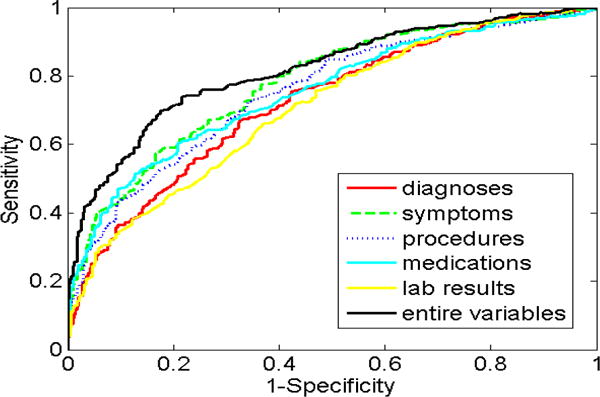
ROC curves for Ridge-LR models.
Figure 3.
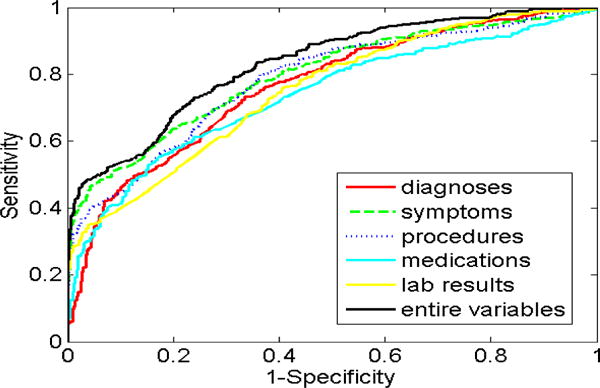
ROC curves for Lasso-LR models.
The difference of predictive performance between the Ridge-LR and the Lasso-LR models was modest using entire EHR features (p=0.203), symptoms (p=0.169), or medications (p=0.183) (Table 3). However, the Lasso-LR model demonstrated significantly higher predictive performance than the Ridge-LR model using diagnoses (p=0.0162), procedures (p=0.0248), or laboratory results (p<0.001).
The Lasso-LR models selected a small set of important features from the original feature sets: 41 vs 28,890 (entire EHR features), 115 vs 9,146 (diagnoses), 5 vs 7,738 (symptoms), 36 vs 5,850 (procedures), 18 vs 2,838 (medications), and 63 vs 3,218 (laboratory results) (Table 3). The ten most important features selected from entire EHR features were described in Table 4. We observed that a history of tobacco use was strongly associated with the “most harmful” breast cancer (47.85%) (p<0.001). These patients who had the “most harmful” breast cancers seldom chose screening mammography (29.37%) but they had a higher probability of recommendation for Computer-Aided Detection (CAD) mammography (18.23%) if they received screening mammography. These patients often had prescriptions generated and transmitted via an e-Prescribing (eRx) system. In contrast, the “least harmful” breast cancer often occurred for woman who chose screening mammography (57.39%), had gynecological examination (43.36%), or exhibited cause of death as an epidemiological consideration such as family cause of death (69.67%) (p<0.001).
Table 4.
The ten most important features selected from entire EHR variables using Lasso-LR
| Ranking | Features | EHR component | Values | the “most harmful” cases (N=395) | the “least harmful” cases (N=399) | p-value |
|---|---|---|---|---|---|---|
| 1 | History of tobacco use | symptoms | present | 189 (47.85%) | 29 (7.27%) | p<0.001 |
| Not present | 206 (52.15%) | 370 (92.73%) | ||||
| 2 | Tobacco use disorder | symptoms | present | 189 (47.85%) | 29 (7.27%) | p<0.001 |
| Not present | 206 (52.15%) | 370 (92.73%) | ||||
| 3 | Mammography, screening, bilateral (two view film study of each breast) | procedures | present | 116 (29.37%) | 229 (57.39%) | p<0.001 |
| Not present | 279 (70.63%) | 170 (42.61%) | ||||
| 4 | Other nonoperative measurements and examinations | diagnoses | present | 191 (48.35%) | 63 (15.79%) | p<0.001 |
| Not present | 204 (51.65%) | 336 (84.21%) | ||||
| 5 | Prescription(s) generated and transmitted via a qualified eRx system | procedure | present | 93 (23.54%) | 0 (0%) | p<0.001 |
| Not present | 302 (76.46%) | 399 (100%) | ||||
| 6 | Normal non-HDL cholesterol | laboratory results | present | 90 (22.78%) | 1 (0.25%) | p<0.001 |
| Not present | 305 (77.22%) | 398 (99.75%) | ||||
| 7 | Hydrocodone acetaminophen | medications | present | 105 (26.58%) | 21 (5.26%) | p<0.001 |
| Not present | 290 (73.42%) | 378 (94.74%) | ||||
| 8 | Cause of death | symptoms | present | 190 (48.10%) | 278 (69.67%) | p<0.001 |
| Not present | 205 (51.90%) | 121 (30.33%) | ||||
| 9 | Computer-Aided Detection (CAD) mammography | procedures | present | 72 (18.23%) | 3 (0.75%) | p<0.001 |
| Not present | 323 (81.77%) | 396 (99.25%) | ||||
| 10 | Gynecological examination | diagnoses | present | 95 (24.05%) | 173 (43.36%) | p<0.001 |
| Not present | 300 (75.95%) | 226 (56.64%) |
4. DISCUSSION
This study provides a glimpse into the opportunities of using EHR feature variables to predict the “most harmful” breast cancer, with the aim of enabling physicians to target intense screening and preventive measures at those women who have the highest risk of breast cancer. Specifically, we developed two regularized prediction models, the Ridge logistic regression and the Lasso logistic regression, to predict the “most harmful” breast cancer one year in advance. The results from both models demonstrated that EHRs could predict the “most harmful” breast cancer, providing the possibility to personalize care for those women at the highest risk in clinical practice.
Lasso based prediction models can select the most important variables from EHRs, offering the potential of collecting novel and important variables from EHRs to improve breast cancer risk prediction. Based on feature selection results, we observed that the patients who were less likely to choose screening mammography would had a higher probability of the “most harmful” breast cancer. The patients who had the “most harmful” breast cancers would have a high tendency to be recommended for CAD mammography if they received screening mammography. These patients often had prescriptions generated and transmitted via an eRx system. They also often used hydrocodone acetaminophen to reduce the pain. Conversely, the patients who had some worrisome epidemiological risk factors, chose regular screening mammography, or had regular gynecological examination would have a high possibility to avoid the “most harmful” breast cancer. These observations align with clinical intuition, which demonstrates that Lasso based prediction model is a powerful tool to identify the important risk factors. In the clinical environment, after breast cancer cases have been identified, these risk factors can be used to categorize the patients who had the highest risk further.
Researchers have strived to explore the value of different sources of patient information to improve breast cancer risk prediction. In this study, we showed that EHRs could be used to predict the “most harmful” breast cancer one year in advance, revealing that EHRs may be another valuable data source to improve breast cancer risk prediction, in addition to demographic risk factors, genetic variants, and breast density. There are a couple of advantages to breast cancer risk prediction using EHRs. EHRs allow researchers to collect data at a fraction of the cost of traditional cohort studies, in which researchers must follow patients for many years and use chart review to obtain predefined data. Additionally, prediction models based on EHRs can be readily implemented in a clinical environment. These advantages would provide unique benefits and opportunities for researchers to accelerate breast cancer risk prediction using EHRs.
The biggest limitation of this study is that we did not reduce the effects of confounding variables when we quantified the association between breast cancer risk and EHR features. For example, history of tobacco use may be one of confounding variables. We found that it was strongly associated with the “most harmful” breast cancer but it may actually only be associated with early death. A future research area would be the use of some techniques to reduce the effects of confounding variables25, 26. In addition, there are several other areas where we could improve this study. We developed risk prediction models based on EHRs collected at a single institution, Marshfield Clinic. External validation using EHRs in other sites or in other EHR systems is required to achieve generalizability. Moreover, we used EHRs to predict the “most harmful” breast cancers. We realize that other data sources could be used to predict breast cancer, including demographic risk factors, genetic variants, and imaging features. It would be interesting to collect these datasets to predict the “most harmful” breast cancer, and compare predictive performance between these datasets and EHRs. Furthermore, the advantage of observing changes of patient records with time is a strength of EHRs. We largely disregarded these temporal trends by pulling all events together prior to the censor date. In the future, we will explore methods to add the characteristics of these temporal trends into statistical models27.
5. CONCLUSIONS
Electronic health records can be used to predict the “most harmful” breast cancer, providing the possibility to personalize care for those women at the highest risk in clinical practice. In addition, Lasso based prediction models have the potential to lead to substantial improvements due to their capability of selecting important prediction variables when a large amount of electronic health record derived variables are used to predict the “most harmful” breast cancer.
Acknowledgments
The authors acknowledge the support of NIH grants U54AI117924, K24CA194251 and the NIH NCATS grant (UL1TR000427). We also acknowledge support from the University of Wisconsin Madison Office of the Vice Chancellor for Research and Graduate Education with funding from the Wisconsin Alumni Research Foundation and the University of Wisconsin Carbone Comprehensive Cancer Center (P30CA014520).
References
- 1.Feig S. Overdiagnosis of breast cancer at screening is clinically insignificant. Acad Radiol. 2015;22:961–966. doi: 10.1016/j.acra.2015.01.020. [DOI] [PubMed] [Google Scholar]
- 2.Helvie M, Chang J, Hendrick R, et al. Reduction in late-stage breast cancer incidence in the mammography era: Implications for overdiagnosis of invasive cancer. Cancer. 2014;120(17):2649–56. doi: 10.1002/cncr.28784. [DOI] [PubMed] [Google Scholar]
- 3.Ong M, Mandl K. National expenditure for false-positive mammograms and breast cancer overdiagnoses estimated at $4 billion a year. Health Aff. 2015;34(4):576–83. doi: 10.1377/hlthaff.2014.1087. [DOI] [PubMed] [Google Scholar]
- 4.Gail M, Brinton L, Byar D, et al. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J Natl Cancer Inst. 1989;81(24):1879–86. doi: 10.1093/jnci/81.24.1879. [DOI] [PubMed] [Google Scholar]
- 5.Gail MH. Discriminatory accuracy from single-nucleotide polymorphisms in models to predict breast cancer risk. J Natl Cancer Inst. 2008;100(14):1037–41. doi: 10.1093/jnci/djn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gail MH. Value of adding single-nucleotide polymorphism genotypes to a breast cancer risk model. J Natl Cancer Inst. 2009;101(13):959–63. doi: 10.1093/jnci/djp130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wacholder S, Hartge P, Prentice R, et al. Performance of common genetic variants in breast-cancer risk models. N Engl J Med. 2010;362(11):986–93. doi: 10.1056/NEJMoa0907727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burnside ES, Davis J, Chhatwal J, et al. Probabilistic computer model developed from clinical data in national mammography database format to classify mammographic findings. Radiology. 2009;251(3):663–72. doi: 10.1148/radiol.2513081346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burnside ES, Liu J, Wu Y, et al. Comparing mammography abnormality features to genetic variants in the prediction of breast cancer in women recommended for breast biopsy. Acad Radiol. 2016;23:62. doi: 10.1016/j.acra.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chhatwal J, Alagoz O, Lindstrom MJ, et al. A logistic regression model based on the national mammography database format to aid breast cancer diagnosis. AJR Am J Roentgenol. 2009;192(4):1117–27. doi: 10.2214/AJR.07.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Y, Alagoz O, Ayvaci MU, et al. A comprehensive methodology for determining the most informative mammographic features. J Digital Imaging. 2013;26(5):941–7. doi: 10.1007/s10278-013-9588-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ayvaci MU, Alagoz O, Chhatwal J, et al. Predicting invasive breast cancer versus DCIS in different age groups. BMC Cancer. 2014;14:584. doi: 10.1186/1471-2407-14-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo JY, Baker JA, Kornguth PJ, et al. Predicting breast cancer invasion with artificial neural networks on the basis of mammographic features. Radiology. 1997;203(1):159–63. doi: 10.1148/radiology.203.1.9122385. [DOI] [PubMed] [Google Scholar]
- 14.Weaver DL, Vacek PM, Skelly JM, et al. Predicting biopsy outcome after mammography: what is the likelihood the patient has invasive or in situ breast cancer? Ann Surg Oncol. 2005;12(8):660–73. doi: 10.1245/ASO.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Goldstein B, Naver A, Pencina M, et al. Opportunities and challenges in developing risk prediction models with electronic health records data: a systematic review. J Am Med Inform Assoc. 2017;24(1):198–208. doi: 10.1093/jamia/ocw042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y, Burnside ES, Cox J, et al. Breast cancer risk prediction using electronic health records. IEEE International Conference on Healthcare Informatics (ICHI) 2017:224–228. [Google Scholar]
- 17.Bodenreider O. The Unified Medical Language System(UMLS): integrating biomedical terminology. Nucleic Acids Research. 2004;32:267–270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Cessie S, Van Houwelingen J. Ridge estimators in logistic regression. Appl Statist. 1992;41(1):191–201. [Google Scholar]
- 19.Fan J, Wu Y, Yuan M, et al. Structure-leveraged methods in breast cancer risk prediction. J Machine Learning Research. 2016;17:1–15. [PMC free article] [PubMed] [Google Scholar]
- 20.Tibshirani R. Regression shrinkage and selection via the lasso. J R Statist Soc B. 1996;58:267–288. [Google Scholar]
- 21.Tibshirani R, Saunders M, Rosset S, et al. Sparsity and smoothness via the fused lasso. J R Statist Soc B. 2005;67:91–108. [Google Scholar]
- 22.Yuan M, Lin Y. Model selection and estimation in regression with grouped variables. J R Statist Soc B. 2006;68:49–67. [Google Scholar]
- 23.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Statistical Software. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 24.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–45. [PubMed] [Google Scholar]
- 25.Pourhoseingholi M, Baghestani A, Vahedi M. How to control confounding effects by statistical analysis. Gastroenteral Hepatol Bed Bench. 2012;5(2):79–83. [PMC free article] [PubMed] [Google Scholar]
- 26.Jager K, Zoccali C, MacLeod A, et al. Confounding: what it is and how to deal with it. Kidney International. 2008;73:256–260. doi: 10.1038/sj.ki.5002650. [DOI] [PubMed] [Google Scholar]
- 27.Henderson R, Diggle P, Dobson A. Joint modeling of longitudinal measurements and event time data. Biostat Oxf Engl. 2000;1(4):465–480. doi: 10.1093/biostatistics/1.4.465. [DOI] [PubMed] [Google Scholar]


