Abstract
Detection of Salmonella is very important to minimize the food safety risk. In this study, the recombinant PagC protein and PagC antibody were prepared and coupled with immunomagnetic beads (IMBs) to capture Salmonella cells from pork and milk samples. And then the SYBR Green qualitative PCR was developed to detect the pathogenic Salmonella. The results showed that the PagC polyclonal antiserum is of good specificity and the capture rate of 0.1 mg IMBs for Salmonella tended to be stable at the range of 70–74% corresponding to the concentrations between 101 and 104 CFU/mL. The method developed demonstrated high specificity for the positive Salmonella samples when compared to non-specific DNA samples, such as Escherichia coli, Staphylococcus aureus, Yersinia enterocolitica, and Yersinia pseudotuberculosis. The limit of detection of this assay was 18 CFU/mL. Detection and quantitative enumeration of Salmonella in samples of pork or milk shows good recoveries of 54.34% and 52.07%. In conclusion, the polyclonal antibody of recombinant PagC protein is effective to capture Salmonella from detected samples. The developed pagC antibody IMBs-qPCR method showed efficiency, sensitivity and specificity for 30 Salmonella detection, enabling detection within 10 h, which is a promising rapid method to detect Salmonella in emergency.
Keywords: Salmonella, IMBs-qPCR, pagC gene, Detection
Introduction
Salmonella species have been found to be responsible for food-borne illnesses such as typhoid and paratyphoid fevers, gastroenteritis, pneumonia and bacteraemia. The major sources of salmonellosis outbreaks include meat, poultry, fish/seafood, eggs and processed food.1, 2 Food-borne salmonellosis is a huge public health burden not only in the developing countries but also in industrialized countries, leading to high rates of enteric diseases, hospitalizations and even deaths worldwide each year.3, 4, 5 As one of the most important food borne pathogens, Salmonella infects over 160,000 individuals in the EU annually, with a morbidity rate of 35 cases per 100,000.6 Salmonella was the second laboratory-confirmed etiologic agent account for 229 (30%) reported food poisoning outbreaks in the United States7 and the annual economic cost of Salmonella infections is at $2.4 billion.8 In Hong Kong, Salmonella spp. account for 22% of foodborne disease outbreaks during 2000–2004.9
Detection of pathogenic Salmonella in various food matrixes is very important to minimize the food safety risk. Conventional culture detection methods are time-consuming and labour-intensive, as it takes 3–7 days including selective enrichment steps for positive results, which is inconvenient for rapid detection in food industry. Immunology-based methods involving antigen–antibody bindings have been widely used for the detection of food-borne pathogens. Immunomagnetic bead separation (IMS) techniques use antibodies against the outer membrane protein (OMP) of Salmonella attached to a magnetic microbeads. These immunomagnetic beads can combine and concentrate Salmonella bacteria in detection samples and then be used in subsequent detection assay, such as the technique of real-time quantitative PCR (qPCR), ELISA, fluorescence immunoassay and flow cytometry (FCM),10, 11, 12, 13, 14 which may improve the efficiency and sensitivity, and decrease the non-specificity.15, 16, 17, 18
PagC protein expressed in most pathogenic Salmonella is a 188-aminoacid outer membrane protein coded in chromosome of Salmonella which is known to be with strong immunogenicities and immunoprotection.19, 20 PagC is a protein regulated by phoP-phoQ involved in bacterial virulence and macrophage survival of Salmonella typhimurium,21 and played an essential role in the serum resistance of S. enterica serovar choleraesuis.22, 23 Furthermore, the prevalence of pagC was significantly higher (p < 0.01) among the isolates from the diseased pigs than in isolates from the healthy pigs.22 The expression of PagC is activated by conditions that mimic acidified macrophage phagosomes.24 The recombinant PagC outer membrane protein is a promising tool for the serum resistance upon Salmonella.20
It has been shown that real-time PCR methods are very effective to detect Salmonella organisms after pre-enrichment of foods.25 To avoid the false-positive results from the detection of dead cells, a TaqMan quantitative real-time RT-PCR (qRT-PCR) assay to assay the invA mRNA level of Salmonella was developed.26 However, the invA mRNA levels vary with temperature, manure or culture conditions.27 Different from the monoclonal antibody against Salmonella somatic antigen, the polyclonal antibodies of recombinant OMP antigen keep the strong bacterial recognition ability, good antigen-antibody reactivity and multi-pathogenic isolate compatibility, but a lower cross-reactivity as polyclonal antibody against whole bacterial antigen do. In our study, an alternative rapid method combining SYBR Green qPCR and immunomagnetic beads coated with specific PagC antibody was developed to quantify Salmonella cells in complex food matrix contaminated.
Methods
Bacterial strains and culture conditions
The bacterial strains and plasmids used in this study are described in Table 1. All Yersinia strains were cultured in Luria–Bertani (LB) broth (DingGuo, Beijing, China) at 28 °C for 14 h, and other strains were incubated at 37 °C in LB with gentle shaking. All of the bacterial strains were supplemented with glycerol (final concentration of 25%) and stored in a refrigerator at -80 °C until use.
Table 1.
Key bacterial strains used in this study.
| Bacterial strains | Source (origin) | Detection of pagC genomic DNA |
|---|---|---|
| Salmonella abortusequi | CVCC 514 | + |
| Sahnonella anatum | CMCC 50135 | + |
| Salmonella blegdam | CMCC 50764 | + |
| Sahnonella bonariensis | CMCC 50149 | + |
| Salmonella chittagong | CMCC 50844 | + |
| Salmonella choleraesuis | CMCC 50019 | + |
| Salmonella choleraesuis | CMCC 50020 | + |
| Salmonella choleraesuis | CMCC 50306 | + |
| Salmonella choleraesuis | CVCC 2146 | + |
| Salmonella deversoir | CMCC 50883 | + |
| Salmonella djakarta | CMCC 50884 | + |
| Salmonella eallinarum | CMCC 50770 | + |
| Salmonella grumpensis | CMCC 50887 | + |
| Salmonella jodhpur | CMCC 50902 | + |
| Salmonella milwaukee | CMCC 50861 | + |
| Salmonella paratyphi A | CMCC 50503 | + |
| Salmonella paratyphi B | CMCC 50605 | + |
| Salmonella paratyphi B | CMCC 50681 | + |
| Salmonella poona | CMCC 50136 | + |
| Salmonella salinatis | CMCC 50830 | + |
| Salmnonella telaviv | CMCC 50062 | + |
| Salmonella typhi | CMCC 50407 | + |
| Salmonella typhi | CMCC 50420 | + |
| Salmonella typhi | CMCC 50521 | + |
| Salmonella typhi | CMCC 50522 | + |
| Salmonella typhi | CMCC 50532 | + |
| Salmonella typhi | CMCC 50533 | + |
| Salmonella typhi | CMCC 50534 | + |
| Salmonella typhi | CMCC 50607 | + |
| Salmonella typhi | CMCC 50618 | + |
| Salmonella typhi | CMCC 50619 | + |
| Salmonella typhi | CMCC 50621 | + |
| Salmonella typhi | CMCC 50663 | + |
| Salmonella typhi | CMCC 50639 | + |
| E. coli top10 | TansGen, Corp, Beijing, China | − |
| Staphylococcus aureus | PNY1, human isolate | − |
| Yersinia enterocolitica | CMCC 55075 | − |
| Yersinia pseudotuberculosis | CMCC 53520 | − |
Expression and purification of recombinant PagC protein
The gene pagC was PCR-amplified from the DNA of Salmonella using primers harbouring restriction sites (underlined): pagC-F (5′-CGGGATCCAGCGTTT TGGTTGTAAATG-3′) and pagC-R (5′-CCGCTCGAGTCAGA AACGGTATCCAAYT-3′). The PCR conditions initial denaturation at 94 °C for 6 min followed by the 5 cycles of denaturation at 94 °C for 30 s, annealing at 54 °C for 30 s and extension at 72 °C for 35 s and then 25 cycles of denaturation at 94 °C for 30 s, annealing at 52 °C for 30 s and extension at 72 °C for 35 s; finally, extension at 72 °C for 6 min. The 1% agarose gel with ethidium bromide (0.5 μg/mL) was used to analyze the amplicons. The PCR product was cloned into the TA cloning vector pUC-T Simple (CWBIO Corp, Beijing, China) to generate pUC-pagC. The BamHI–XhoI fragments of pUC-pagC were ligated with the same sites of pET28a to produce pET28a-pagC. Plasmid pET28a-pagC in Escherichia coli Top10 (TransGen Biotech, Beijing, China) producing recombinant PagC was sequenced (Genewiz Corp., Beijing, China) and no frame shift or other mutations in the coding sequence of PagC were detected. Expression and purification of recombinant PagC were performed by using procedures described as our previously report.28 Equal amounts of protein were subjected to SDS-PAGE in 12% polyacrylamide gel with Mini-Protean (BioRad) apparatus as previously described.29 Protein concentrations were determined by using Bicinchoninic acid (BCA) protein assay, with BSA (Solarbio Corp, Beijing, China) as a standard.
Generation of polyclonal antisera of recombinant PagC protein
Approximately 1 mg of highly purified PagC obtained from E. coli BL21(DE3) (TransGen Biotech, Beijing, China) was used for production of polyclonal antisera. The SPF Rabbit purchased from Tianjin Laboratory Animal Center were maintained in SPF (Specific pathogen-free) facilities and was immunized subcutaneously following standard immunization protocols with 1 mg of recombinant protein emulsified with equal volume of mineral oil adjuvant. Animal experiments were performed on 6-month-old rabbit in compliance with regulations by Tianjin University Institutional Animal Care and Use Committee. The immunization protocol consists of three subcutaneous injections at 2-week intervals. The rabbit was taken blood from vein 4 days after the third immunization and PagC polyclonal antisera were purified by ammonium sulfate precipitation.30 For immunostaining of PagC protein, proteins were transferred to a PVDF membrane and labelled with rabbit anti-PagC antiserum diluted at 1:1000 and HRP goat anti-rabbit IgG antibody (CWBIO Corp, Beijing, China) diluted at 1:5000, as described previously.31 The concentration of purified anti-PagC polyclonal antibodies was 5.35 mg/mL, determined by BCA protein assay. Western blotting was performed and HRP goat anti-rabbit IgG antibody (CWBIO Corp, Beijing, China) was used as previously described.29
Preparation of anti-Salmonella immunomagnetic beads
Affimag PSC magnetic beads (mean diameter of 500 nm, 10 mg/mL) were purchased from BaseLine Technology Co. Ltd. (Tianjin, China). Uncoated magnetic beads were washed three times with 0.01 M phosphate buffered saline (PBS, pH 6.0) and then resuspended in 500 μL of freshly made 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide (5 mg/mL EDC) and Nhydroxysuccinimide (5 mg/mL NHSS) solution in 0.01 M PBS (pH 6.0). After vortex mixing, activation was carried out at 37 °C for 30 min in orbital shaker. After incubation, the beads were washed three times with 0.01 M PBS containing Tween 20 (PBST, pH 7.4) and resuspended in PBST solution. Purified PagC antibodies were added gradually with gentle mixing and placed on the shaker at 37 °C for 3 h. The antibody-conjugated beads were washed three times with PBST. The coated IMBs were then blocked by incubation with 1 mL 1% BSA in 0.01 M PBS at 37 °C for 30 min and were washed three times with PBST. Finally, the IMBs were resuspended in 10 mg/mL in 0.01 M PBS (pH 7.4) with 0.1% BSA and stored at 4 °C until use.
Optimization of immunomagnetic separation
The capture capacity of the IMBs against Salmonella was studied for four coupling rates between anti-PagC antibodies and magnetic beads (10, 50, 100, and 200 μg/mg), six volumes of IMBs (0.005, 0.01, 0.02, 0.05, 0.1and 0.2 mg), and a series of concentrations (100–106 CFU/mL).
Protocol of immunomagnetic separation and specificity test
Salmonella typhi 50618 cultures were grown to a concentration of 109 CFU/mL and serially diluted to 103–104 CFU/mL in sterile PBS buffer (0.65 M NaCl, 0.01 M phosphate buffer, pH 7.2). The immunomagnetic beads were mixed and captured for 30 min at room temperature, and then separated by magnet device (BaseLine Technology Co. Ltd., Tianjin, China) according to the manufacture's instruction. The supernatant was collected and spread on Bismuth Sulfite (BS; Beijing Land Bridge Technology Co. Ltd., Beijing, China) Agar for bacterial enumeration after appropriate dilution. The IMBs-bacteria conjunctures were used for DNA extraction (DNALyse Amplification Kit; CWBIO Corp, Beijing, China). All the BS agar plates were incubated at 37 °C for 24–48 h for observation. Triplets of each sample were plated. Further, Salmonella choleraesuis CMCC 50306, E. coli top10, Staphylococcus aureus PNY1-3 were cultured in LB at 37 °C, 150 rpm for 12 h. Yersinia enterocolitica CMCC 55075 and Yersinia pseudotuberculosis CMCC 53520 were cultured in LB at 28 °C for 14 h and diluted to 103–104 CFU/mL in PBS. IMBs capture and plating were done as described above to test the specificity of the anti-PagC IMBs.
Preparation of samples for scanning with electron microscope
S. typhi CMCC 50618 cultures were grown to a concentration of 109 CFU/mL and serially diluted to 103–104 CFU/mL in PBS. IMBs capture was done as described above, and the IMBs-Salmonella conjunctures were diluted 30 times and fixed on glass slide by air-drying. All the samples were sputter-coated with platinum using E1045 Pt-coater (Hitachi High-technologies CO., Japan), following by imaging under an S-4800 field emission scanning electron microscope (SEM, Hitachi High-technologies CO., Japan) at an acceleration voltage of 3 keV. Bare IMBs and bare S. typhi CMCC 50618 were used as controls.
Quantification of Salmonella by qPCR
A pair of primers (F: 5′-AGCGTTTTGGTTGTA AATG-3′, R: 5′-CCTGGCTGTCTCCATATAAG-3′, amplified length:180 bp) was designed in highly conserved region of the PagC gene (Oligo 6.0) for specific detection of Salmonella. Salmonella strains were grown at 37 °C for 12 h in LB and subjected to serial 10-fold dilutions of different concentrations. Genomic DNA (20 μL) was extracted from 1 mL of each certain culture as templates that would be used to generate the standard curve of qPCR. The IMBs-combined Salmonella was used for DNA extraction as described above.
The qPCR assay was performed using 7500 Real-time PCR system (Applied Biosystems, Foster City, CA, USA) and TransStart Top Green qPCR Super Mix (TransGen Biotech, Beijing, China). The reaction mixture contained 2 × TransStart Top Green qPCR Super Mix (12.5 μL), 0.4 μL of 10 μM forward and reverse primers, 0.4 μL of passive reference dye (TransGen Biotech, Beijing, China), 2 μL of DNA template and sterile double distilled water in a final volume of 20 μL. The qPCR thermal profile for pagC gene was as follows: initial denaturation at 95 °C for 4 min, followed by 40 cycles at 95 °C for 20 s, 53 °C for 30 s and 70 °C for 30 s.
Further, E. coli top10, S. aureus PNY1-3 were cultured in LB at 37 °C for 12 h. Then Y. enterocolitica CMCC 55075 and Y. pseudotuberculosis CMCC 53520 were cultured in LB at 28 °C for 14 h and all cultures were diluted to 105 CFU/mL in PBS, with DNA of each bacterial strain as template to test the specificity of the qPCR method. Besides, a mixture of all qPCR reagents containing 2 μL of sterile distilled water instead of DNA template served as no template control. The results of the qPCR method were considered negative if the fluorescent signal did not increase within 40 cycles.
To evaluate the specificity of the qPCR method targeting pagC gene in presence of background DNA of nonspecific bacteria, 107 CFU/mL of E. coli top10, S. aureus PNY1-3, Y. enterocolitica CMCC 55075 were spiked in triplicate with reference strain (S. typhi CMCC 50618) to generate serially 10 folds diluted cultures from 102 to 107 CFU/mL DNA template of genomic DNA (200 μL) was extracted from 1 mL of each spiked samples as described above. Detection and quantification of S. typhi CMCC 50618 in the presence of non-specific DNA was performed by qPCR described previously.
Immunomagnetic separation (IMS) of Salmonella in food samples
Fresh pork and whole milk was purchased from a local grocery store and used for rapid detection of Salmonella. Salmonella cultures were decimally diluted in PBS for the further addition of milk pork and beef samples. The mixture of 25 g of fresh pork or 25 mL whole milk and 225 mL of BPW was homogenized in stomacher for 2 min respectively. Serial dilutions of Salmonella were added to final concentrations of 100–101 CFU/mL, followed by enrichment culture for 4 h to increase the concentration of the target bacteria of at 37 °C, 150 rpm. After enrichment, 1 mL of each sample was collected, mixed with 0.1 mg of IMBs and transferred to the magnet device (BaseLine Technology Co. Ltd., Tianjin, China) according to the manufacture's instruction. The IMB bacteria conjugates from the capture phase were washed 2 times with PBS and suspended with 1.5 mL of PBS. A 200 μL aliquot of each suspension was used for enumeration, and the rest was used for DNA extraction.
Bacterial recovery was evaluated by plating appropriately diluted supernatant onto BS agar (BS; Beijing Land Bridge Technology Co. Ltd., Beijing, China) as above. Three replications of the study were performed in triplicate.
Statistics
The statistical significance of the results was determined with help of GraphPad Prism (version 5.0a; San Diego, CA), and results are expressed as the mean ± standard error of the mean. Statistical significance was determined by a Student's t-test, two-way ANOVA for multiple comparisons. Probability values less than 0.05 (p < 0.05) were considered statistically significant.
Results
The gene pagC is widely expressed among various Salmonella strains and the polyclonal antiserum produced by recombinant PagC is of good specificity
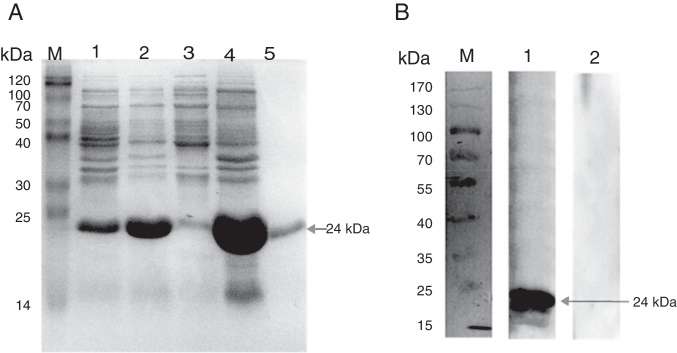
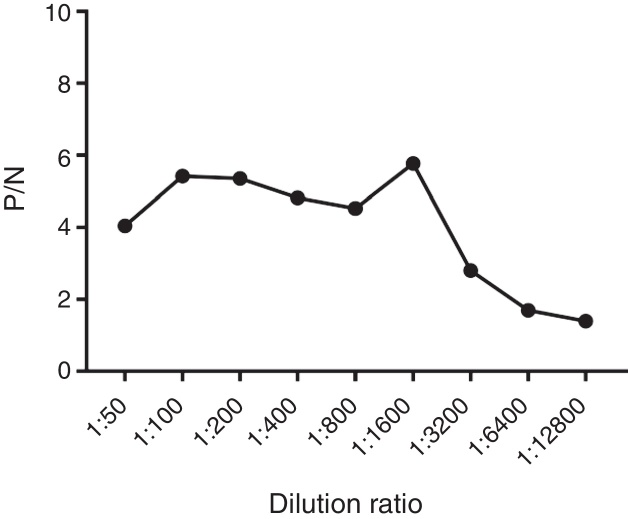
To determine the prevalence of pagC, 34 Salmonella strains was evaluated by PCR analysis using pagC-specific primers, which demonstrated that all of the Salmonella strains tested contained the pagC gene (Table 1). To determine the antigenicity of pagC, full-length histidine-tagged pagC recombinant protein was produced in E. coli and purified for preparation of PagC-specific antiserum. Recombinant PagC had a molecular mass of approximately 24 kDa as determined by SDS-PAGE (Fig. 1A), consistent with the calculated molecular mass based on the deduced amino acid sequence of PagC. The good specificity of the polyclonal antiserum with the recombinant PagC protein as shown in Fig. 1B. The ELISA antibody titre of PagC-specific polyclonal antiserum was determined as 1:3200 (Fig. 2).
Fig. 1.
Expression of the recombinant protein PagC and Western blot analysis. (A) Proteins were separated on 12% acrylamide gel and stained with Coomassie blue. Lane M, molecular mass markers (Blue Plus II Protein Marker, TransGen Biotech, Beijing, China); lane 1, cells carrying the vector with the insert before IPTG induction; lane 2, total cellular protein after IPTG induction; lane 3, supernatant of cells ultrasound pyrolysis products after IPTG induction; lane 4, sediment of cells ultrasound pyrolysis products after IPTG induction; lane 5, the purified protein. (B) Western blot analysis using Goat anti Rabbit HRP-IgG monoclonal antibody. Lane M, molecular mass markers (PageRuler Prest Protein Ladder, Fermentas, Shanghai, China); lane 1, western blot analysis using antiserum to recombinant protein; lane 2, western blot using pre-immune serum.
Fig. 2.
The ELISA titre detection of PagC-specific polyclonal antibodies.
Capture capacity and specificity of IMBs
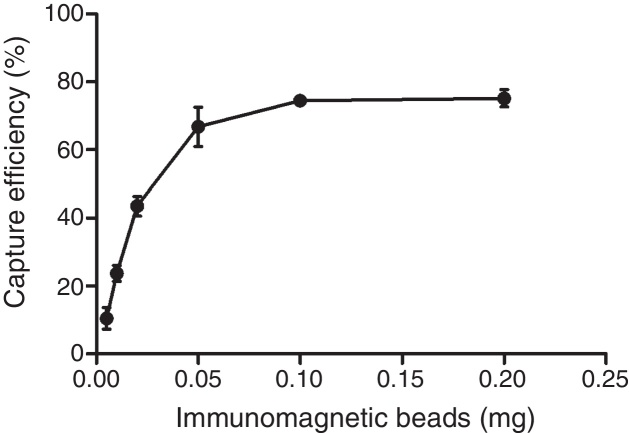
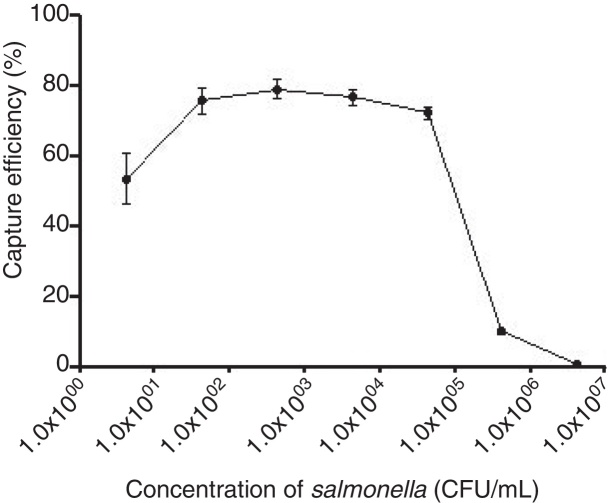
The optimum ratio between anti-PagC polyclonal antibody and magnetic beads was determined in preliminary experiment by preparing PAb in different concentrations from 10 to 200 μg per 1 mg of magnetic beads (data not shown). The IMBs coated by 100 μg of PAb per 1 mg of magnetic beads were optimum. The capture efficiency (CE) of IMBs was defined as the percentage of the total bacteria combined with IMBs, which was based on counting the unbound bacteria left in the supernatant on Agar plating. It showed that when bacteria of approximately 104 CFU/mL were captured, the capture efficiency was 10.48 ± 3.16%, 23.71 ± 2.34%, 43.52 ± 2.88%, 66.76 ± 5.77%, 74.48 ± 1.67% and 75.14 ± 2.54% respectively for 0.005 mg, 0.01 mg, 0.02 mg, 0.05 mg, 0.1 mg and 0.2 mg. The CE of 0.1 mg of IMBs came to the highest 74.48% and higher IMBs volumes did not change the CE of IMBs (Fig. 3). The CE of 0.1 mg of IMBs against 1 mL 4.2 × 100 –4.2 × 106 CFU/mL of S. typhi CMCC 50618 for 30 min of immunoreaction time was 53.46 ± 7.19%, 75.74 ± 3.74%, 79.05 ± 2.61%, 76.62 ± 2.25%, 72.13 ± 1.65%, 9.52 ± 0.97% and 0.87 ± 0.61% respectively. It tended to be stable at the range of 70–74% corresponding to the concentrations between 101 and 104 CFU/mL (Fig. 4). The image of IMBs-Salmonella conjunctures under electron microscope is shown in Fig. 5. IMBs appear to form much larger aggregation of dozens of beads due to the binding of many Salmonella bacteria.
Fig. 3.
The changes of capture efficiency to S. typhi in different immunomagnetic beads content conditions.
Fig. 4.
The capture efficiency of the 0.1 mg IMBs to S. typhi cultures in different concentrations (CFU/mL).
Fig. 5.
The combination of IMBs and S. typhi CMCC 50618 in SEM. (A) Bare IMBs. IMBs appear to be small aggregation of a couple of beads; (B) S. typhi CMCC 50618 only. (C) S. typhi CMCC 50618 was captured by IMBs. IMBs appear to be large aggregation of dozens of beads, which helps the binding of bacteria.
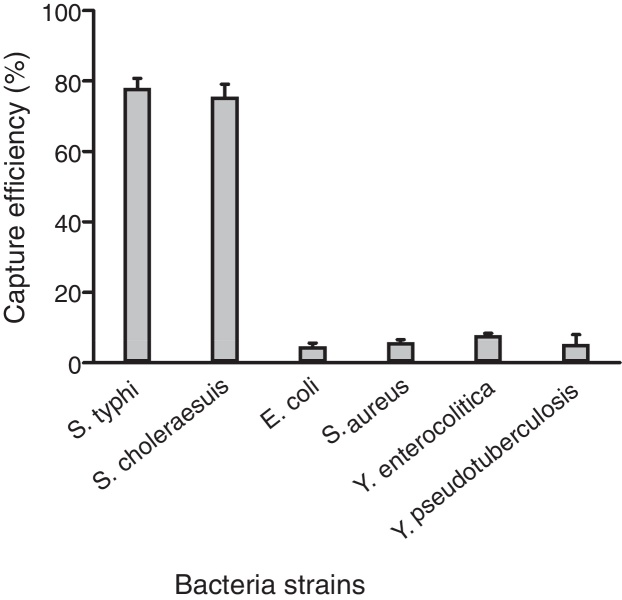
However, the capture efficiency for S. choleraesuis CMCC 50306, E. coli top10, S. aureus PNY1-3, Y. enterocolitica CMCC 55075, and Y. pseudotuberculosis CMCC 53520 strains was determined to be 75.65%, 6.46%, 7.90%, 6.69%, and 4.74% respectively for concentrations of 3 × 104 CFU/mL (Fig. 6). The comparison of CEs of Salmonella strains with that of non-target bacteria shows good specificity of the polyclonal antibody coated magnetic beads.
Fig. 6.
The specificity of IMBs with selected bacterial strains. The capture efficiency of different bacteria with 0.1 mg of IMBs.
Estimation of Lower of Dosage (LOD), selectivity and specificity of the IMBs-qPCR assay
A standard curve was established by using ten serial dilutions of S. typhi CMCC 50618 ranging from 1.8 × 102 CFU to 1.8 × 108 CFU per-mililiter. The assay could detect lowest 18 CFU/mL linearly (Amplification efficiency: 101.526 percent, y = -0.3061x + 12.818, R2 = 0.995).
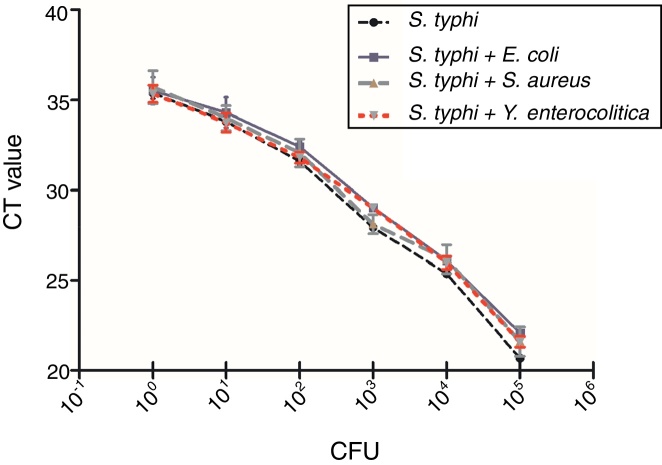
A total number of 39 samples including 34 Salmonella strains, 4 non-targeted bacterial strains (E. coli top10, S. aureus PNY1-3, Y. enterocolitica CMCC 55075, Y. pseudotuberculosis CMCC 53520) and 1 blank has been tested for the selectivity and specificity of the IMS-qPCR method. It has been observed that all of the 34 Salmonella strains were positive for the pagC gene while no amplicon band was detected in non-targeted bacterial strains. Further, presence of background DNA from different non-specific bacteria including E. coli top10, S. aureus PNY1-3, and Y. enterocolitica CMCC 55075 has no impact on the specificity of the Salmonella specific qPCR method (Fig. 7). The variation in Ct values of Salmonella vs that in existence of non-specific DNA from different bacteria was not significant by analysis of Student's t-test (p>0.05). Which shows that qPCR method developed in this study is of good specificity and screening for the detection of Salmonella strains.
Fig. 7.
Influence of background flora on detection and quantitative enumeration of S. typhi though qPCR. Mean CT values (n = 3) ± SD. No amplification was observed in no template control (2 μL sterile water instead of DNA in qPCR reaction). Abbreviation: CFU, colony forming units.
Detection and quantitative enumeration of Salmonella in milk and pork samples
The fresh pork and whole milk samples were artificially contaminated with 100–101 CFU/mL S. typhi and then analyzed with the IMBs-qPCR method after incubation in enrichment broth. Salmonella reached 104 CFU/mL after enrichment in BPW for 5 h. The recovery of IMBs for Salmonella in fresh pork and milk samples for the IMBs-qPCR method was 54.34% and 52.07%, respectively (Table 2).
Table 2.
Capture efficiency analysis of IMBs in food samples.
Abbreviation: CFU, colony forming units; BPW buffered peptone water; CE, capture efficiency.
a: total numbers of Salmonella in 1 mL food samples after 5 h enrichment in BPW. b: total numbers of Salmonella in 1 mL culture medium after 30 min capture at 37 °C.
CE (%) = (a − b)/a × 100%, capture efficiency of IMBs.
Discussion
It shows that real-time PCR methods are very effective to detect Salmonella organisms. The real-time PCR coupled with either immunomagnetic separation (PCR-IMS) was proved to be the effective and less costly method for the detection of both healthy and sanitizer-injured Salmonella on mung bean sprouts after pre-enrichment of foods and immunomagnetic separation steps.32, 33 The PagC is a 188-aminoacid outer membrane protein expressed in Salmonella chromosome,34, 35, 36 and has an extensive distribution among Salmonella enterica including S. typhi, Salmonella paratyphi,37, 20 S. typhimurium38, 22 and S. choleraesuis.23 Studies have found that PagC is a promising role in the development of new vaccines with strong immunogenicities and immunoprotection.23, 19, 20
The use of immunomagnetic separation (IMS) for efficient capture of bacteria from food has received wide attention.39, 40, 41 In our study, a method for detection of Salmonella in food samples that uses PagC antibody coated with magnetic beads for immunomagnetic separation (IMS) associated with qPCR assay targeting the gene pagC was developed. All the 34 pathogenic Salmonella strains examined in this study have been detected with pagC gene by PCR and the recombinant PagC protein had a molecular mass of approximately 24 kDa. ELISA and immunoblotting assays revealed that the PagC protein polyclonal antibody prepared by recombinant PagC protein was of high titre and specificity.
Commercially available immunomagnetic separation (IMS) techniques for rapid Salmonella detection including Dynal BeadRetriever, Pathatrix Auto, KingFisher Flex and BioVeris M1 M Platform showed that the IMS methods were generally effective in improving the recovery from sample matrices, and are more rapid and specific than cultural methods.42, 43 Our result showed that the PagC immunomagnetic separation (IMS) were specific and effective in collecting Salmonella spp. cells from pre-enrichment medium, and the sensitivity of Salmonella detection could be enhanced by IMS, particularly in primary animal production with much higher amounts of substances.25 The nano- (300 nm) and micro- (2.8 μm) sized magnetic particles modified with anti-Salmonella antibody are effective to pre-concentrate the bacteria from the samples throughout an immunological reaction.44 The diameter of the Affimag PSC magnetic beads used in this study was 500 nm, which had high ratio of surface area to volume and showed good stability in solution. Coated with specific immune polyclonal antibodies, the anti-PagC IMBs scan specifically attach to a target strain and exhibited good capture efficiency for Salmonella at 74.48% with a detection limit of about 18 CFU/mL. Non-specific capture rates were lower than 10%.
The final concentration of the Salmonella in food samples reached 104 CFU/mL after 4 h of enrichment in BPW before capture. With IMBs-based qPCR assay we are able to rapidly detect Salmonella from fresh pork and whole milk with a recovery of 54.34% and 52.07% lower than the capture of the same concentration of Salmonella pure bacteria (74%). The low recovery is most probably due to the complex tissues and matrices in the food samples that disturbed the magnetic separation and lead to loss of capture magnetic beads.42 Thus eliminating the complex tissues and matrices before immunomagnetic separation may help improve the capture efficiency. Furthermore, specificity of the antibody conjugated on the magnetic beads is a significant factor to the capture rates.45 Thus increasing the IMBs-based in the treated food sample and screening the highly specific monoclonal antibody of PagC should be benefit to improve the capture efficiency.
The traditional agar plate cultivation method to detect Salmonella in food sample is a time consuming and laborious process that takes several days, and has limitations in accuracy, specificity and sensitivity.46 So, there is an urgent need for development of rapid and cost-effective alternative methods for monitoring of Salmonella. Some nano- and micro-scaled and aptamers based biosensors for detection of pathogen or disease have been developed,47, 48, 49 the method of IMBs-qPCR based on PagC protein is of good efficiency and specificity, allowing “real-time” detection of Salmonella. The whole process of detection Salmonella from food samples needs no more than 10 h, which significantly reduces the time of detection for Salmonella in laboratories compared with traditional cultured method, which is a promising and rapid tool to detect Salmonella in case of emergency.
Author contributions
Conceived and designed the experiments: JH H. Performed the experiments: Y L, H D, JS W, J C, ZX L, DP H. Analyzed the data: J C, JH H. Contributed reagents/materials/analysis tools: JH H. Wrote the paper: J C, JH H.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 31272540) and the underprop project of Tianjin Science and Technology committee (No. 16YFZCNC00640).
Associate Editor: Luis Augusto Nero
References
- 1.Mcguinness S., Mccabe E., O’Regan E. Development and validation of a rapid real-time PCR based method for the specific detection of Salmonella on fresh meat. Meat Sci. 2009;83(3):555–562. doi: 10.1016/j.meatsci.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Feasey N.A., Dougan G., Kingsley R.A., Heyderman R.S., Gordon M.A. Invasive non-typhoidal Salmonella disease: an emerging and neglected tropical disease in Africa. Lancet. 2012;379(9835):2489–2499. doi: 10.1016/S0140-6736(11)61752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunkley K.D., Callaway T.R., Chalova V.I. Foodborne Salmonella ecology in the avian gastrointestinal tract. Anaerobe. 2008;15(1–2):26–35. doi: 10.1016/j.anaerobe.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Hur J., Jawale C., Lee J.H. Antimicrobial resistance of Salmonella isolated from food animals: a review. Food Res Int. 2012;45(2):819–830. [Google Scholar]
- 5.Callaway T., Edrington T., Anderson R., Byrd J., Nisbet D. Gastrointestinal microbial ecology and the safety of our food supply as related to Salmonella. J Anim Sci. 2008;86(14 suppl):E163–E172. doi: 10.2527/jas.2007-0457. [DOI] [PubMed] [Google Scholar]
- 6.Helwigh B., Korsgaard H. European Food Safety Authority; 2007. The Community Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents, Antimicrobial Resistance and Foodborne Outbreaks in the European Union in 2006. [Google Scholar]
- 7.Woh P.Y., Thong K.L., Behnke J.M., Lewis J.W., Zain S.N.M. Characterization of nontyphoidal Salmonella isolates from asymptomatic migrant food handlers in Peninsular Malaysia. J Food Prot. 2017:1378–1383. doi: 10.4315/0362-028X.JFP-16-342. [DOI] [PubMed] [Google Scholar]
- 8.Golan E. 2014. USDA Economic Research Service-Calculating the Cost of Foodborne Illness. [Google Scholar]
- 9.Cheung P.Y., Kai M.K. Salmonella in food surveillance: PCR, immunoassays, and other rapid detection and quantification methods. Food Res Int. 2012;45(2):802–808. [Google Scholar]
- 10.Hibi K., Abe A., Ohashi E. Combination of immunomagnetic separation with flow cytometry for detection of Listeria monocytogenes. Anal Chim Acta. 2006;573–574(1):158–163. doi: 10.1016/j.aca.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 11.Power M., Shanker S., Sangster N., Veal D. Evaluation of a combined immunomagnetic separation/flow cytometry technique for epidemiological investigations of Cryptosporidium in domestic and Australian native animals. Vet Parasitol. 2003;112(1):21–31. doi: 10.1016/s0304-4017(02)00414-4. [DOI] [PubMed] [Google Scholar]
- 12.Hibi K., Mitsubayashi K., Fukuda H. Rapid direct determination using combined separation by prepared immunomagnetic and flow cytometry of Flavobacterium psychrophilum. Biosens Bioelectron. 2007;22(9):1916–1919. doi: 10.1016/j.bios.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 13.Yazdankhah S.P., Sølverød L., Simonsen S., Olsen E. Development and evaluation of an immunomagnetic separation-ELISA for the detection of Staphylococcus aureus thermostable nuclease in composite milk. Vet Microbiol. 1999;67(2):113–125. doi: 10.1016/s0378-1135(99)00035-8. [DOI] [PubMed] [Google Scholar]
- 14.Zhu P., Shelton D.R., Li S. Detection of E. coli O157: H7 by immunomagnetic separation coupled with fluorescence immunoassay. Biosens Bioelectron. 2011;30(1):337–341. doi: 10.1016/j.bios.2011.09.029. [DOI] [PubMed] [Google Scholar]
- 15.Taskila S., Tuomola M., Ojamo H. Enrichment cultivation in detection of food-borne Salmonella. Food Control. 2012;26(2):369–377. [Google Scholar]
- 16.Cudjoe K.S., Hagtvedt T., Dainty R. Immunomagnetic separation of Salmonella from foods and their detection using immunomagnetic particle (IMP)-ELISA. Int J Food Microbiol. 1995;27(1):11–25. doi: 10.1016/0168-1605(94)00134-r. [DOI] [PubMed] [Google Scholar]
- 17.Hagren V., von Lode P., Syrjälä A. An 8-hour system for Salmonella detection with immunomagnetic separation and homogeneous time-resolved fluorescence PCR. Int J Food Microbiol. 2008;125(2):158–161. doi: 10.1016/j.ijfoodmicro.2008.03.037. [DOI] [PubMed] [Google Scholar]
- 18.Moreira Â.N., Conceição F.R., Conceição Rd.C.S. Detection of Salmonella Typhimurium in raw meats using in-house prepared monoclonal antibody coated magnetic beads and PCR assay of the fimA gene. J Immunoass Immunochem. 2007;29(1):58–69. doi: 10.1080/15321810701735096. [DOI] [PubMed] [Google Scholar]
- 19.Yang T.-C., Ma X.-C., Liu F. Screening of the Salmonella paratyphi A CMCC 50973 strain outer membrane proteins for the identification of potential vaccine targets. Mol Med Rep. 2012;5(1):78–83. doi: 10.3892/mmr.2011.587. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J., Fan X., Ge Y., Yan J., Sun A. Distribution of Salmonella paratyphi A pagC gene and immunoprotective effect of its recombinant expressed products. Zhejiang da xue xue bao Yi xue ban. 2013;42(2):171–176. doi: 10.3785/j.issn.1008-9292.2013.02.007. 231. [DOI] [PubMed] [Google Scholar]
- 21.Miller S.I., Kukral A.M., Mekalanos J.J. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci U S A. 1989;86(13):5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamang M.D., Gurung M., Nam H.M. Antimicrobial susceptibility and virulence characteristics of Salmonella enterica Typhimurium isolates from healthy and diseased pigs in Korea. J Food Prot. 2014;77(9):1481–1486. doi: 10.4315/0362-028X.JFP-14-084. [DOI] [PubMed] [Google Scholar]
- 23.Nishio M., Okada N., Miki T., Haneda T., Danbara H. Identification of the outer-membrane protein PagC required for the serum resistance phenotype in Salmonella enterica serovar Choleraesuis. Microbiology. 2005;151(3):863–873. doi: 10.1099/mic.0.27654-0. [DOI] [PubMed] [Google Scholar]
- 24.Kitagawa R., Takaya A., Ohya M. Biogenesis of Salmonella enterica serovar typhimurium membrane vesicles provoked by induction of PagC. J Bacteriol. 2010;192(21):5645–5656. doi: 10.1128/JB.00590-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braun S.D., Methner U. Comparison of DNA isolation methods and detection of Salmonella spp. from animal faeces and dust using invA real-time PCR. Berl Munch Tierarztl Wochenschr. 2011;124(5–6):177–185. [PubMed] [Google Scholar]
- 26.Gonzalez-Escalona N., Hammack T.S., Russell M. Detection of live Salmonella sp. cells in produce by a TaqMan-based quantitative reverse transcriptase real-time PCR targeting invA mRNA. Appl Environ Microbiol. 2009;75(11):3714–3720. doi: 10.1128/AEM.02686-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia R., Baelum J., Fredslund L., Santorum P., Jacobsen C.S. Influence of temperature and predation on survival of Salmonella enterica serovar Typhimurium and expression of invA in soil and manure-amended soil. Appl Environ Microbiol. 2010;76(15):5025–5031. doi: 10.1128/AEM.00628-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang H.Z., Deng H., Shen M.L. Superantigenicity analysis of staphylococcal enterotoxins SElK and SElQ in a mouse model. RSC Adv. 2015;5(38):29684–29692. [Google Scholar]
- 29.Kothari H., Kumar P., Singh N. Prokaryotic expression, purification, and polyclonal antibody production against a novel drug resistance gene of Leishmania donovani clinical isolate. Protein Expr Purif. 2006;45(1):15–21. doi: 10.1016/j.pep.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Emmrich F. Handbook of experimental immunology: edited by D.M. Weir (co-editors L.A. Herzenberg, C. Blackwell and Leonore A. Herzenberg), Blackwell Scientific Publications, 1986. £180.00 (for all four volumes) ISBN 0 632 01499 7 Vol. 1 Immunochemistry £75.00 (XXV) Immunol Today. 1986;7(12):384–385. [Google Scholar]
- 31.Miki T., Okada N., Danbara H. Two periplasmic disulfide oxidoreductases, DsbA and SrgA, target outer membrane protein SpiA, a component of the Salmonella pathogenicity island 2 type III secretion system. J Biol Chem. 2004;279(33):34631–34642. doi: 10.1074/jbc.M402760200. [DOI] [PubMed] [Google Scholar]
- 32.Zheng Q., Miks-Krajnik M., Yang Y., Lee S.M., Lee S.C., Yuk H.G. Evaluation of real-time PCR coupled with immunomagnetic separation or centrifugation for the detection of healthy and sanitizer-injured Salmonella spp. on mung bean sprouts. Int J Food Microbiol. 2016;222:48–55. doi: 10.1016/j.ijfoodmicro.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 33.Notzon A., Helmuth R., Bauer J. Evaluation of an immunomagnetic separation-real-time PCR assay for the rapid detection of Salmonella in meat. J Food Prot. 2006;69(12):2896–2901. doi: 10.4315/0362-028x-69.12.2896. [DOI] [PubMed] [Google Scholar]
- 34.Gunn J.S., Alpuche-Aranda C.M., Loomis W.P., Belden W.J., Miller S.I. Characterization of the Salmonella typhimurium pagC/pagD chromosomal region. J Bacteriol. 1995;177(17):5040–5047. doi: 10.1128/jb.177.17.5040-5047.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pulkkinen W., Miller S. A Salmonella typhimurium virulence protein is similar to a Yersinia enterocolitica invasion protein and a bacteriophage lambda outer membrane protein. J Bacteriol. 1991;173(1):86–93. doi: 10.1128/jb.173.1.86-93.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J., Fan X., Ge Y., Yan J., Sun A. Distribution of Salmonella paratyphi A pagC gene and immunoprotective effect of its recombinant expressed products. J Zhejiang Univ (Med Sci) 2013;42(2):171–176. doi: 10.3785/j.issn.1008-9292.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 37.Parkhill J., Dougan G., James K. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413(6858):848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 38.Mohamed T., Zhao S., White D.G., Parveen S. Molecular characterization of antibiotic resistant Salmonella Typhimurium and Salmonella Kentucky isolated from pre- and post-chill whole broilers carcasses. Food Microbiol. 2014;38:6–15. doi: 10.1016/j.fm.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 39.Amoako K.K., Thomas M.C., Kong F. Rapid detection and antimicrobial resistance gene profiling of Yersinia pestis using pyrosequencing technology. J Microbiol Methods. 2012;90(3):228–234. doi: 10.1016/j.mimet.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 40.Tatavarthy A., Peak K., Veguilla W. An accelerated method for Isolation of Salmonella enterica Serotype Typhimurium from artificially contaminated foods, using a short preenrichment, immunomagnetic separation, and xylose-lysine-desoxycholate agar (61X method) J Food Prot. 2009;72(3):583–590. doi: 10.4315/0362-028x-72.3.583. [DOI] [PubMed] [Google Scholar]
- 41.Kanki M., Seto K., Kumeda Y. Simultaneous immunomagnetic separation method for the detection of Escherichia coli O26, O111, and O157 from food samples. J Food Prot. 2014;77(1):15–22. doi: 10.4315/0362-028X.JFP-13-225. [DOI] [PubMed] [Google Scholar]
- 42.Lee K.M., Runyon M., Herrman T.J., Phillips R., Hsieh J. Review of Salmonella detection and identification methods: aspects of rapid emergency response and food safety. Food Control. 2015;47:264–276. [Google Scholar]
- 43.Bushon R.N., Likirdopulos C.A., Brady A.M. Comparison of immunomagnetic separation/adenosine triphosphate rapid method to traditional culture-based method for E. coli and enterococci enumeration in wastewater. Water Res. 2009;43(19):4940–4946. doi: 10.1016/j.watres.2009.06.047. [DOI] [PubMed] [Google Scholar]
- 44.Brandao D., Liebana S., Campoy S., Alegret S., Isabel Pividori M. Immunomagnetic separation of Salmonella with tailored magnetic micro and nanocarriers. A comparative study. Talanta. 2015;143:198–204. doi: 10.1016/j.talanta.2015.05.035. [DOI] [PubMed] [Google Scholar]
- 45.Xiong Q., Cui X., Saini J.K. Development of an immunomagnetic separation method for efficient enrichment of Escherichia coli O157:H7. Food Control. 2014;37(1):41–45. [Google Scholar]
- 46.Melo A.M., Alexandre D.L., Furtado R.F. Electrochemical immunosensors for Salmonella detection in food. Appl Microbiol Biotechnol. 2016;100(12):5301–5312. doi: 10.1007/s00253-016-7548-y. [DOI] [PubMed] [Google Scholar]
- 47.Pashazadeh P., Mokhtarzadeh A., Hasanzadeh M., Hejazi M., Hashemi M., Guardia M.D.L. Nano-materials for use in sensing of Salmonella infections: recent advances. Biosens Bioelectron. 2016;87:1050. doi: 10.1016/j.bios.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 48.Zhu X., Li J., He H., Huang M., Zhang X., Wang S. Application of nanomaterials in the bioanalytical detection of disease-related genes. Biosens Bioelectron. 2015;74:113–133. doi: 10.1016/j.bios.2015.04.069. [DOI] [PubMed] [Google Scholar]
- 49.Mouammine A., Lanois A., Pagès S. Ail and PagC-related proteins in the entomopathogenic bacteria of Photorhabdus genus. PLOS ONE. 2014;9(10) doi: 10.1371/journal.pone.0110060. e110060. [DOI] [PMC free article] [PubMed] [Google Scholar]