Abstract
As a stress-inducible natural killer (NK) cell ligand, B7H6 plays a role in innate tumor immunosurveillance and is a fairly tumor selective marker expressed on a variety of solid and hematologic cancer cells. Here, we describe the isolation and characterization of a new family of single chain fragment variable (scFv) molecules targeting the human B7H6 ligand. Through directed evolution of a yeast surface displayed non-immune human-derived scFv library, eight candidates comprising a single family of clones differing by up to four amino acid mutations and exhibiting nM avidities for soluble B7H6-Ig were isolated. A representative clone re-formatted as an scFv-CH1-Fc molecule demonstrated specific binding to both B7H6-Ig and native membrane-bound B7H6 on tumor cell lines with a binding avidity comparable to the previously characterized B7H6-targeting antibody, TZ47. Furthermore, these clones recognized an epitope distinct from that of TZ47 and the natural NK cell ligand NKp30, and demonstrated specific activity against B7H6-expressing tumor cells when expressed as a chimeric antigen receptor (CAR) in T cells.
Keywords: Antibody, B7H6, cancer, chimeric antigen receptor, immunotherapy, single chain fragment variable
Introduction
The immune system relies upon an intricate network of intracellular communication via the specific expression and activation of numerous cell markers to defend the host against perceived threats. One such marker, B7H6, is a stress-inducible immunostimulatory ligand for the activating natural killer (NK) cell surface receptor, NKp30 (Brandt et al., 2009), and serves to initiate NK cell-mediated cytokine release and cytotoxicity. Cells from various tissue types have been found to express this surface antigen (Ag) in the settings of cancer (Brandt et al., 2009; Fiegler et al., 2013), infection (Matta et al., 2013; Zou et al., 2015; Koumbi et al., 2016) and autoimmune diseases (Salimi et al., 2016), suggesting a native role of B7H6 expression in NK cell immunosurveillance of various pathological states.
Two key features render B7H6 an especially attractive target for tumor immunotherapy: its tumor specificity and broad expression across multiple tumor tissue types. Normal tissue cells lacked B7H6 expression as measured by qRT-PCR analysis of B7H6 mRNA on a normal tissue array (Brandt et al., 2009) and by microarray analysis of gene expression (Shyamsundar et al., 2005; Brandt et al., 2009). In contrast, varying levels of B7H6 expression were found in many cancer cell lines (Brandt et al., 2009; Fiegler et al., 2013; Cao et al., 2015) and primary tumor samples from patients with both solid and hematologic malignancies, including a range of lymphomas, leukemias, breast cancers, ovarian cancers, sarcomas and brain tumors (Brandt et al., 2009; Fiegler et al., 2013; Wu et al., 2015a, b).
Given this tumor specificity and broad potential applicability, there has been considerable interest in the significance and utility of B7H6-targeting molecules in cancer and inflammatory disease immunotherapy and diagnosis (Brandt et al., 2010; Cerwenka and Moldenhauer, 2014; Vivier and Matta, 2014; Choi et al., 2015; Pierres et al., 2015). Furthermore, a recent study suggests that current tumor therapeutics may already indirectly leverage the B7H6-NKp30 axis to promote anti-tumor immunity; treatment with chemotherapy, radiation therapy, hyperthermia and immunotherapy, were all demonstrated to upregulate B7H6 expression on tumor cells, subsequently enhancing tumor sensitivity to NKp30-mediated NK cell cytolysis (Cao et al., 2015). Thus, even if a subject's primary tumor does not natively express B7H6, combination therapy with conventional anti-tumor therapeutics to induce B7H6 expression may render tumor cells susceptible to B7H6-targeted therapies, increasing the applicability of anti-tumor strategies targeting B7H6. In addition, researchers have proposed monitoring soluble B7H6 levels in sepsis patients as a prognostic marker (Matta et al., 2013), suggesting utility for B7H6-binding reagents beyond malignancy applications.
To demonstrate the anti-tumor potential of targeting B7H6, we have shown the successful application of B7H6-targeting molecules in bispecific T cell engaging molecules (BiTEs™) and chimeric antigen receptor (CAR) T cells in both in vitro and in vivo tumor models (Zhang et al., 2012; Wu et al., 2015a, b). Beyond specific targeting and efficacy against B7H6-expressing tumors, B7H6-specific CAR T cells generated a robust immune response against additional tumor cell markers that facilitated tumor eradication after rechallenge with an otherwise identical, but non-B7H6-expressing tumor cell line in in vivo mouse models (Wu et al., 2015a, b). Early evaluations of B7H6-targeting CAR T cells used NKp30 extracellular domains to target B7H6-expressing tumor cells, but suffered from self-reactivity against peripheral blood mononuclear cells (PBMCs) and immature dendritic cells (iDCs) expressing other NKp30 ligands (Zhang et al., 2012). A subsequent approach employing TZ47, an antibody generated by mouse immunization, increased B7H6 specificity and demonstrated little to no self-reactivity (Wu et al., 2015a, b).
Given these promising pre-clinical data, we sought to expand the repertoire of B7H6-targeting domains. Humanized versions of TZ47 (Choi et al., 2015) have previously been described resulting from an effort to decrease the risk of immunogenicity associated with the administration of murine antibodies (Presta, 2008). Here, we report generation of a new clonal family of fully human B7H6-targeting scFvs, determine their avidity and specificity for both soluble and membrane-expressed antigen, localize their binding to a novel B7H6 epitope, and demonstrate their cytotoxicity in the form of CAR T cells.
Materials and Methods
Production of soluble proteins (B7H6-Ig, NKp30-Ig, TZ47 and PB11 scFv-CH1-Fc)
Plasmids for Human Embryonic Kidney (HEK) cell expression of the two soluble B7H6-Ig and NKp30-Ig fusion proteins were generated by fusing the extracellular domain of B7H6 or NKp30 to a mouse IgG2a Fc domain in pCMV expression vectors. For TZ47, a pCMV expression vector encoding the heavy chain (HC) sequence followed by a T2A ‘self-cleaving peptide’ sequence (Carey et al., 2009) and the light chain (LC) sequence was generated for simultaneous expression of both HC and LC. To create an expression plasmid for PB11 scFv-CH1-Fc, the VH region of a human VRC01 Ab HC expression vector (pCMVR-VRC8552) was replaced with the PB11 scFv sequence. The PB11 scFv-CH1-Fc thus contains all CH1-CH3 domains, differing from traditional scFv-Fcs which lack the CH1 domain.
HEK cells were transfected with expression plasmids using polyethyleneimine and recombinant proteins were purified from cell supernatants as previously described (Choi et al., 2015). Briefly, HEK-293F cells at a density of 106 cells/mL were transfected with expression plasmids at a concentration of 1.0 mg/L each and cultured for 5–7 days. Cell cultures were spun down at 9000 g for 15 min and supernatants were purified via Protein A resin-based affinity chromatography: five column volumes (CV) of PBS were applied to equilibrate a pre-packed Protein A column, followed by cell supernatant to bind Fc-containing proteins to the column, another five CVs of PBS to wash excess unbound protein from the column, and finally five CVs of 100 mM glycine, pH 2.7 to elute proteins from the column. Eluates were then buffer-exchanged into PBS using Amicon-10 K (NKp30-Ig) or −30 K (B7H6-Ig, TZ47, PB11 scFV-CH1-Fc) ultracentrifugation tubes and purified by size exclusion chromatography to separate aggregates from functional protein.
Directed evolution of PB scFvs by yeast surface display
Yeast library preparation and sequencing
A previously described non-immune human scFv library (Feldhaus et al., 2003) served as the initial g1.0 population. To monitor diversity, plasmids were extracted from the library using the ZymoPrep Yeast Plasmid Miniprep II Kit (Zymo Research) according to the manufacturer's protocol. Recovered plasmids were then transformed into highly comptent ElectroSHOX E. coli and 8–12 bacterial clones were sent for bacterial colony sequencing.
Library selection
Directed evolution of the yeast-displayed scFv library was carried out according to a previously published protocol (Chao et al., 2006). To induce yeast libraries to express scFvs, pre-induction cultures were started at OD of 0.2 in growth media (30°C, shaking at 250 rpm) and induced at mid-log phase (OD of 1.0) by transferring cells to induction media for 16–48 h at 30°C with shaking. For each round of selection, a minimum number of cells equal to 10 times the theoretical diversity were used for both pre-induction cultures and sort samples to increase the likelihood of sampling the complete diversity available.
Magnetic-activated cell sorting of yeast libraries
Magnetic bead-based sorting of yeast libraries was conducted as described in a previously published protocol (Ackerman et al., 2009). Briefly, Streptavidin-coated magnetic beads (DynaBeads M-270) were washed 5x with PBSF (PBS + 0.1% BSA) prior to an overnight incubation with saturating amounts of biotinylated B7H6-Ig (50 μL of 100 nM biotinylated B7H6-Ig per μL of beads) or negative control normal mouse IgG (MsIgG, Jackson Laboratories) as determined through bead titration experiments. Both B7H6-Ig and normal Mouse IgG were chemically biotinylated on primary amines for capture on Streptavidin beads. After conjugation with biotinylated proteins, beads were washed 5× with PBSF to remove any excess soluble protein prior to co-incubation with yeast libraries for 1.5 h at 4°C. In general, each MACS round included two negative selections using bare SA beads and biotinylated MsIgG-coated beads, prior to one positive selection using biotinylated B7H6-Ig-coated beads. After each round of selection, beads were washed 1x with 1 mL PBSF for 30 min at 4°C to separate truly bead-bound yeast from those that were simply trapped between beads (wash fraction). Bead-bound and washed yeast were counted separately but recombined in subsequent rounds of selection. After each round of selection for B7H6-binders, the B7H6-Ig-bead binding population was resuspended in 1 mL of growth media and various dilutions were plated to obtain diversity estimates. After plating, the 1 mL of resuspended B7H6-Ig-bead binding yeast libraries were grown out in 5 mL of growth media overnight.
Fluorescence -activated cell sorting of yeast libraries
For each round of FACS, at least 10 times the estimated population diversity, or a minimum of 10 million, induced yeast expressing scFvs on their cell surface were stained. First, cells were spun down at 3000 g for 5 min and washed three times with PBSF to remove cellular debris. For some selections, a blocking step was employed in which cells were incubated with 500 nM normal mouse IgG for 15 min to block mouse Fc binders prior to addition of B7H6-Ig. Primary stains comprised an incubation with Chicken anti-cMyc tag Ab (Gallus) to monitor full-length expression and either B7H6-Ig or biotinylated B7H6-Ig antigen at concentrations ranging from 2 nm to 2.9 μM depending upon the stringency and stage of selection. After primary incubation, cells were washed 3× with PBSF. Fluorescent staining with secondary reagents included a 30–60 min incubation with goat anti-chicken Abs, goat anti-mouse Abs, or streptavidin secondary staining reagents conjugated to various combinations of AlexaFluor(AF)-488, AF-647, FITC or APC (all from Life Technologies). For most FACS rounds, a gate was drawn for the top 1% of cells with enhanced Ag Binding:Expression signal and sorted on a Sony iCyt Sorter. For selection of g2.5, a low stringency gate was applied to separate Ag-binders from non-binders in g2.4. for collection and clonal selection.
Library diversification
Plasmids recovered from the g1.7 population using the ZymoPrep Yeast Plasmid Miniprep II Kit (Zymo Research) served as the template for an error-prone PCR based on a salt-based method (Fromant et al., 1995) to introduce two to four random mutations per scFv for new diversity in generation 2.0, as previously described (Grimm et al., 2015). Libraries were created by the electroporation of scFv PCR products and digested pCT- vector sharing homology at 5′ and 3′ ends into EBY100 yeast as previously described (Chao et al., 2006). Transformed cells were grown in 500 mL growth media at 30°C with shaking at 250 rpm, and were passaged at least once prior to induction. Library diversity was estimated by counting colonies from plated dilutions of the transformed yeast mixture on selective media.
Avidity assays
Fluorescent staining of B7H6-expressing cells for avidity KD measurements
Fluorescent staining of cells was performed as described previously (Choi et al., 2015). Briefly, 96-well plates containing 2.5 × 105 cells/well of RMA and RMA-B7H6 cells were washed 3× with PBSF before a primary incubation for 1 h with varying concentrations of PB11 scFv-CH1-Fc ranging from 0 to 1000 nM. Cells were then washed 3× with PBSF before incubation with Anti-Human-AF647 secondary Ab and Calcein Violet to stain for live cells for 20 min. Live cell gates were drawn based on FSC vs. SSC profiles and positive Calcein Violet staining, and only this population was used for determination of mean fluorescence intensity (MFI) values. MFI values are geometric means of fluorescence intensities measured for each well using a Miltenyi Biotech MACSQuant Flow Cytometer.
BioLayer interferometry for avidity KD measurements
BioLayer Interferometry measurements of avidity KD were obtained using the ForteBio Octet RED96 system as previously described (Choi et al., 2015). Briefly, anti-human CH1-coated biosensor tips were ‘activated’ by incubation in 83 nM PB11 scFv-CH1-Fc for 10 min to load tips. Binding to soluble B7H6-Ig and negative control NKp30-Ig were assessed by allowing PB11-loaded tips to equilibrate in PBS + 0.1% Tween-20 (PBST) for 5 min, followed by association steps in 200 μL of each antigen at concentrations ranging from 0 to 500 nM for 10 min, and dissociation steps in 200 μL of PBST buffer for 5 min.
Competitive binding assays
Competitive binding assays were carried out essentially identically to the fluorescent staining of B7H6-expressing cells described above, with the modification of co-primary incubations with combinations of PB11 and either TZ47 or NKp30-Ig at 500 nM concentrations each.
Tumor cell lines and cell culture
All murine RMA cell line variants were cultured in Complete RPMI media, which was made by supplementing RPMI 1640 with 10% heat-inactive fetal bovine serum, 10 mM HEPES, 0.1 mM non-essential amino acids, 1 mM sodium pyruvate, 100 U/mL penicillin, 100 μg/mL streptomycin and 50 μM of 2-mercaptoethanol. Cell lines were incubated under 5% CO2 at 37°C. The murine cell line RMA and a variant RMA-B7H6 engineered to express human B7H6 were previously generated (Wu et al., 2015a, b). RMA cell lines expressing Chimera 1 and 2 were generated by transducing RMA cells with dualtropic virus produced by viral packaging PT67 cells containing the chimeric B7H6. The cell lines underwent G418 selection at concentration of 1 mg/mL. To monitor cell viability in cytotoxicity assays, RMA and RMA-B7H6 cell lines were transduced with a dualtropic virus containing a PPyre9-GFP fusion gene followed by Puromycin selection at concentration of 2 μg/mL.
Construction and transduction of CAR T cells
Construction of CARs
The TZ47-CAR was constructed previously (Wu et al., 2015a, b). Furthermore, the TZ47-CAR was fused to a Furin cleavage site-SGSG-T2A sequence followed by a gene encoding a truncated mouse CD19 and cloned into the pFB-neo retroviral vector. The PB6-CAR was generated by using the pFB-neo-TZ47-CAR-MsCD19 as a template. DNA sequence of the TZ47 was excised and the PB6 scFv was subcloned into this vector. All PCR reactions were carried out using high-fidelity DNA polymerase Phusion (New England BioLabs, Ipswich, MA USA) and DNA primers purchased from IDT DNA (Coralville, Iowa).
Retroviral production
Before transfection, 2.5 × 106 293T cells were plated on a 10 cm plate in 10 mL of Complete Dulbecco's modified Eagle's (DMEM) media to produce ecotropic virus. Complete DMEM media is made with DMEM with a high glucose concentration (4.5 g/L) supplemented with the same supplements as the Complete RPMI 1640 media. Cells were then transfected with 20 μg of CAR plasmid and 10 μg of Ψeco packaging plasmid using X-fect (Clontech) according to the manufacturer's protocol. At 48 h post transfection, media was harvested and filtered through a 0.45 μm filter before storage at −80°C. To generate stable virus packaging cell lines, ecotropic virus made from 293T cells was used to infect PT67 cells followed by G418 selection (1 mg/mL) for 7 days. To make stable cell lines which produce ecotropic virus, dualtropic virus generated from PT67 cells was used to infect E86 cells followed by G418 selection (1 mg/mL). All cell lines were cultured under 5% CO2 at 37°C.
Retroviral transduction
Spleens were extracted from C57BL/6 mice purchased from either the National Cancer Institute or Jackson Laboratories. Mice were 7–12 weeks old at the start of experiments. All animal experiments were conducted under the approval of Dartmouth College's Institution Animal Care and Use Committee. Spleens were processed and activated with Concavalin A, as previously described (Wu et al., 2015a, b). After activation, T cells were transduced with retrovirus containing either the PB6-CAR, TZ47-CAR, or vector only (Mock) based on a spin transduction protocol previously described (Wu et al., 2015a, b). CAR T cells underwent G418 selection for 3 days at 0.5 mg/mL then were histopaque enriched to isolate live cells. Mouse T cells were cultured in Complete RPMI 1640 media with 25 U/mL of recombinant IL-2 under 5% CO2 atmosphere at 37°C.
Flow cytometry analysis of CAR T cells
Mouse CAR T cells were counted, washed, centrifuged, and resuspended in PBS containing 2% w/v of FBS. 105 CAR T cells were incubated with antibodies for mouse CD3ɛ (145-2c11 Biolegend) at 2 μg/mL, mouse CD4 (Clone GK1.5 eBioscience) at 5 μg/mL, and mouse CD19-phycoerythrin (PE) 2 μg/mL (Biolegend) in a total volume of 100 μL of PBS containing 2% at 4°C for 25 min. After incubation, cells were washed and resuspended in 200 μL of PBS containing 2% w/v FBS. Samples were analyzed on an Accuri C6 Cytometer (BD biosciences, San Jose, CA, USA).
CAR T cell activity assays
Cytotoxicity assay
Tumor cell lines of RMA and RMA-B7H6 expressing the PPyre9-GFP gene were plated at 5 × 104 cells per well in a 96-well plate. RMA and RMA-B7H6 cell lines were plated in round bottom or flat bottom plates, respectively. CAR T cells were added at various T cell effector to target ratios of 5:1, 1:1, 0.2:1. Cells were co-cultured at 37°C for 24 h followed by addition of 50 μL of luciferin (200 μg/mL) (Promega) and incubated at 37°C for 30 min before analyzing luminescence.
Cytokine production by T cells
About 1 × 105 CAR T cells and target RMA, RMA-B7H6, RMA Chimera 1 or RMA Chimera 2 tumor cells were co-incubated at an effector to target cell ratio of 1:1 in a 96-well plate for 24 h at 37°C. Cell-free medium was collected and analyzed for mouse IFN-γ production by ELISA (Biolegend).
Results
Isolation and characterization of yeast-displayed PB scFvs
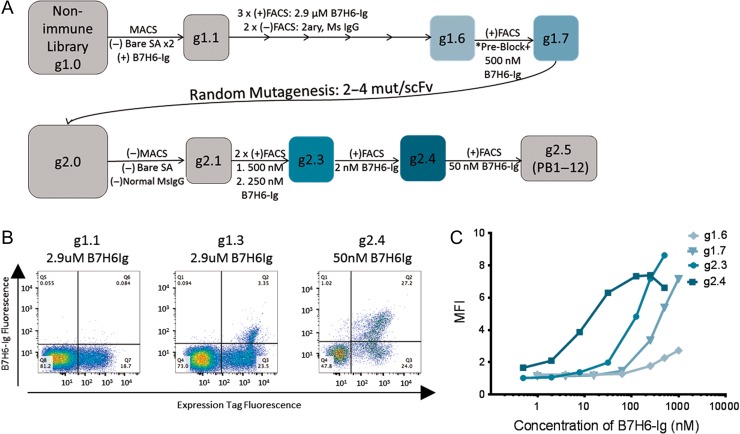
B7H6-targeting scFvs were evolved from a previously described yeast surface display non-immune human antibody fragment library (Feldhaus et al., 2003) using a soluble fusion protein consisting of the B7H6 extracellular domain and a mouse IgG2a Fc domain (B7H6-Ig) as bait. A summary of the evolutionary strategy is shown in Fig. 1. First, a magnetic bead-based approach was used (Ackerman et al., 2009), where each round included two sequential negative selections against binders to ‘bare’ streptavidin beads and normal Mouse IgG-coated beads followed by one positive selection for binders to B7H6-Ig-coated beads. After magnetic selection to narrow down the diversity/search space from ~10^9 to ~10^7, libraries were primarily evolved using fluorescence-activated cell sorting (FACS) with gradually decreasing concentrations of bivalent B7H6-Ig (Fig. 1B). Additionally, in some rounds, negative selections were conducted to eliminate secondary reagent binders and blocking steps with normal Mouse IgG were incorporated to reduce the likelihood of enriching Ig Fc binders. When library diversity was estimated to be on the order of a dozen variants and titration staining suggested that clones had not yet reached nanomolar avidity (Fig. 1C), the population was diversified by error-prone PCR. The resulting population (g2.0) was further selected using 1 round of MACS and 4 rounds of FACS resulting in g2.4. A final non-stringent, binary sort to separate binders from non-binders yielded g2.5, from which PB clones were selected.
Fig. 1.
Directed Evolution of PB clones from a non-immune human scFv library. (A) A series of magnetic-activated cell sorting (MACS) and fluorescence-activated cell sorting (FACS) selections were used, both including both positive (+) selections of Ag-binders and negative (−) selections against reagent binders such as secondary (2ary) Ab, streptavidin bead (Bare SA) and Mouse IgG Fc binders. Generation names x.y represent the number of x) diversification steps and y) selections after diversification. Pre-Block indicates a blocking step against the mouse IgG Fc tag using 500 nM normal msIgG prior to staining with B7H6-Ig. Concentrations of B7H6-Ig used for selections are indicated under selection arrows. Boxes indicate generations for which titration curves of antigen were tested and shown in (C). PB clones were isolated from g2.5. (B) Representative FACS plots for a subset of improving generations are shown. Binding to B7H6-Ig (y-axis) and expression tag antibodies (x-axis) are shown. (C) Titration curves depict the MFI of library generations stained with B7H6-Ig antigen at a range of concentrations to compare binding avidities.
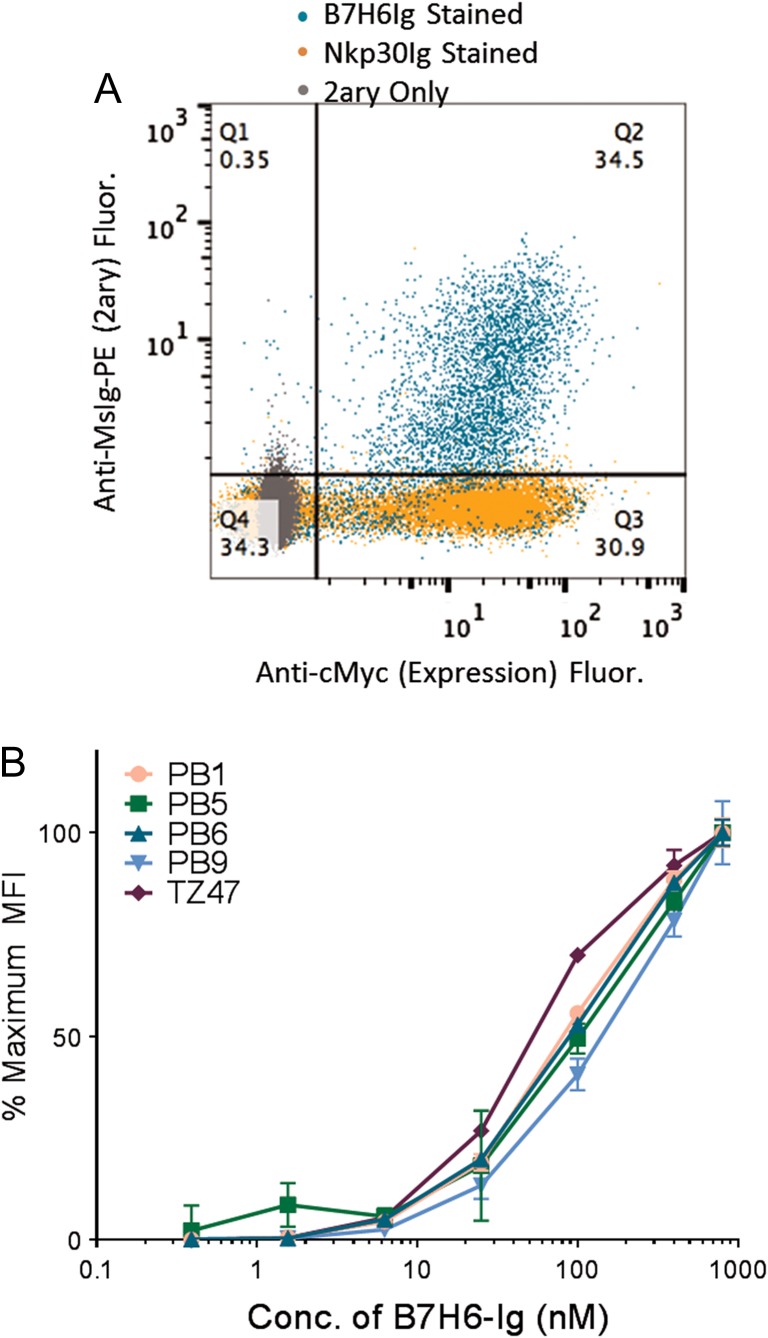
To monitor diversity, clones from libraries were sequenced at g1.7 prior to the diversification step, and after the final selection of g2.5 containing nanomolar avidity binders. Whereas sequence diversity existed at g1.7, all eight full-length scFv clones sequenced from g2.5 were determined to be related based on high sequence similarity. The PB clonal family was thus defined as scFvs sharing IgH-V1 and -J3/4 and IgL V2 and -J7/3 gene usage with identical CDR3 lengths, as predicted by VBASE2 (Retter et al., 2005). To ensure that B7H6-Ig-binders bound to the extracellular B7H6 domain of the fusion protein bait, libraries were tested for binding to secondary reagents and an NKp30-Ig fusion protein containing the same Ig Fc (Fig. 2A). Clones representing four distinct protein sequences (PB1, 5, 6 and 9) were tested for binding to soluble B7H6-Ig, revealing binding avidities comparable to the previously isolated murine anti-B7H6 Ab TZ47 (Fig. 2B). Quantitative estimates of binding avidity KD values for individual clones ranged from 10 nM to >100 nM, depending upon the batch of B7H6-Ig antigen used for each run, potentially due to some instability of the antigen at storage conditions (4°C or freeze-thaw) and/or glycosylation differences as B7H6 contains six potential N-linked glycosylation sites (Li et al., 2011). However, within each individual run using a single batch of B7H6-Ig, PB scFvs bound with similar avidities to B7H6 differing from each other by at most three-fold.
Fig. 2.
Characterization of Yeast-Displayed PB Clones. (A) Population g2.4 was stained with B7H6-Ig, Nkp30-Ig and secondary only using an anti-MsIg-PE secondary for fluorescence read-out (y-axis). Yeast were also stained for the cMyc expression tag (x-axis). (B) A range of B7H6-Ig antigen concentrations were used to stain yeast-displayed PB clones and TZ47. MFI values were normalized to represent the % of the maximum signal observed at the highest concentration tested. Error bars indicate the standard deviation of three technical replicates and relative trends are representative of three experimental replicates.
Specificity and avidity of a soluble PB scFv-CH1-Fc
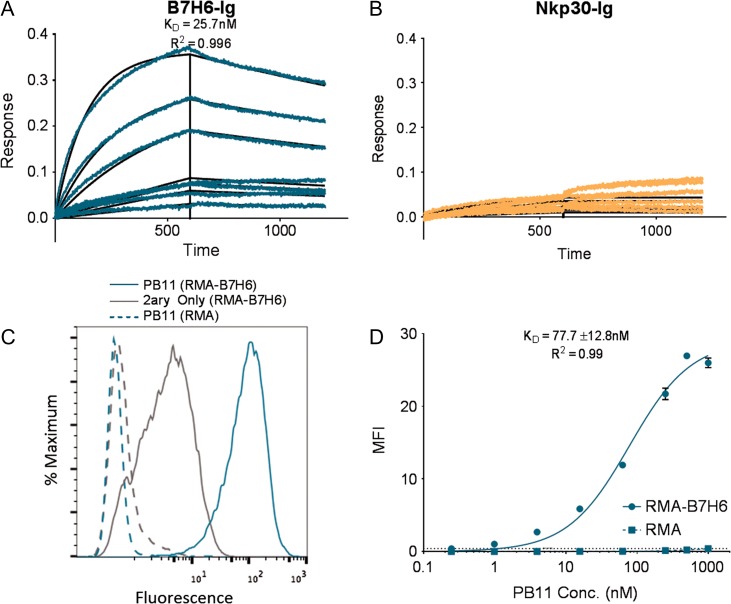
To further confirm that yeast-displayed clones specifically recognized tumor antigen B7H6, the PB11 scFv was cloned into a pCMV-based antibody HC expression vector, replacing the variable heavy domain with the scFv to generate PB11 scFv-CH1-Fc constructs for expression in HEK cells. By BioLayer Interferometry, PB11 scFv-CH1-Fc bound soluble B7H6-Ig with avidity KD of 25.7 nM (Fig. 3A), which was comparable to the 290 nM KD reported for the previously isolated mouse antibody TZ47 (Choi et al., 2015). In contrast, PB11 did not bind to the negative control NKp30-Ig molecule bearing the same mouse Ig portion (Fig. 3B), demonstrating specific binding to the B7H6 extracellular domain of soluble B7H6-Ig antigen. To determine whether the PB family of clones were able to recognize native membrane-bound B7H6 antigen, the soluble PB11 scFv-CH1-Fc was tested for binding to the mouse lymphoma RMA cell line and RMA-B7H6 cells transduced to express human B7H6 on the cell surface. PB11 specifically bound B7H6-expressing cells with a similar avidity (KD ~ 78 nM) as recombinant antigen, and demonstrated undetectable binding to the background cell line lacking B7H6 expression (Fig. 3C and D). Although some background fluorescence was observed for secondary reagent binding to RMA-B7H6 cells which has previously been observed (Choi et al., 2015), a marked shift in fluorescence signal was observed for PB11 scFv-CH1-Fc staining of RMA-B7H6 cells that was not observed in RMA cells, confirming B7H6-specific binding, and demonstrating recognition of this Ag in a cellular context.
Fig. 3.
Binding characterization of soluble PB11 scFv-CH1-Fc. (A and B) Kinetic binding curves for the binding of soluble PB11 scFv-CH1-Fc to (A) B7H6-Ig and (B) Nkp30-Ig (negative control) were obtained by BioLayer Interferometry tested at a range of antigen concentrations from 0 to 500 nM. (C) Histograms depicting the staining of B7H6-expressing cells (RMA-B7H6, solid lines) and B7H6-null cells (RMA, dashed lines) with 500 nM PB11 scFv-CH1-Fc or secondary only. (D) Titration curves generated by staining B7H6-expressing cells (RMA-B7H6, solid) or B7H6-null cells (RMA, dashed) with varying concentrations of PB11 scFv-CH1-Fc ranging from 0 to 1000 nM. Error bars depict standard deviations of the MFI values from triplicate stains.
Activation and cytotoxicity of PB-based CAR T cells
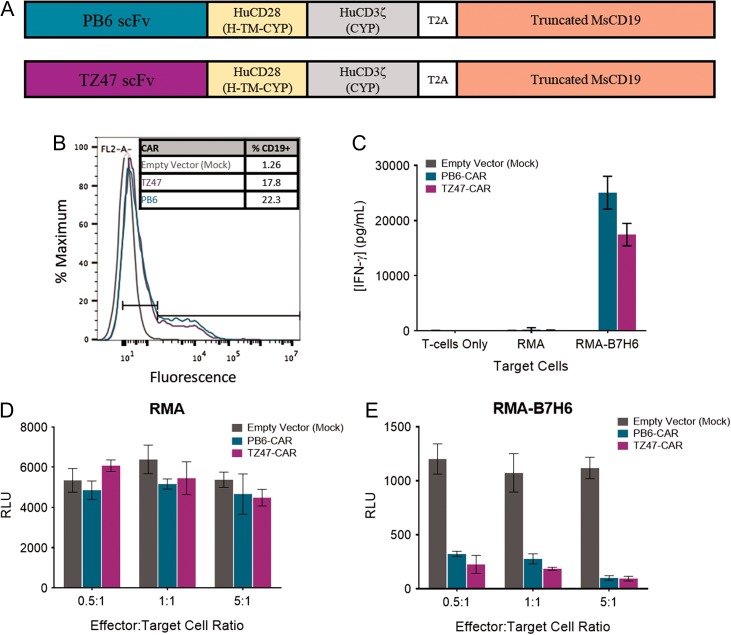
Given this promising characterization of the PB scFv family, PB6, the clone with the highest relative avidity (Supplementary Fig. S1 and Supplementary Table S1), was incorporated into a CAR construct (Fig. 4A). This CAR contained the PB6 scFv fused to human CD28 hinge, transmembrane, and cytoplasmic domains, followed by the human CD3ζ signaling domain. After the CAR sequence, a T2A ‘self-cleaving peptide’ sequence (Carey et al., 2009) was added followed by a sequence encoding a truncated mouse CD19 to allow co-expression of the CAR and the truncated mouse CD19, which serves as a surrogate marker for transduction efficiency and CAR expression as demonstrated previously in T cells transduced with similar CAR constructs (Wu et al., 2015a, b). To test functional activity of the PB6-CAR, mouse T cells were transduced with viruses containing this PB6-CAR construct. As controls, T cells were also transduced with the murine mAb-based CAR (TZ47-CAR) or a vector only control (Mock). As demonstrated by similar CD19 expression levels after transduction, T cells appeared to express the PB6-CAR at a level similar to that observed for the TZ47-CAR (Fig. 4B).
Fig. 4.
PB6-CAR T cell Activity. (A) Schematic diagrams of PB6- and TZ47-CAR constructs. H = Hinge, TM = Transmembrane domain, CYP = cytoplasmic domain. (B) Histograms of fluorescence signal after staining with anti-CD19-PE detection reagents for CD19 expression. (C) In vitro activation of CAR T cells measured by IFN-γ release in response to RMA-B7H6 target cells, negative control RMA cells, and no target cells for T cells transduced with empty vector, PB6 and TZ47 CARs. (D and E) Tumor cell cytotoxicity of CAR T cells measured by luciferase reporter assays in which target RMA (D) or RMA-B7H6 (E) cells express luciferase as a marker of viability. Empty vector, PB6 and TZ47- CAR T cells were incubated with target cells at effector:target cell ratios of 0.5, 1 and 5:1. In all graphs, error bars indicate standard deviations.
To determine whether PB6-CAR T cells were active against B7H6-expressing cells, PB6-CAR T cells were co-cultured with target RMA and RMA-B7H6 cells and tested for T cell activation and cytotoxicity. Demonstrating specific activation, PB6-CAR T cells were observed to secrete IFN-γ only when co-cultured with RMA-B7H6 cells (Fig. 4C). Demonstrating specific cytotoxicity, PB6-CAR T cells induced tumor cell toxicity of RMA-B7H6 cells and not RMA cells over a range of effector to target cell ratios (Fig. 4D and E). Together, these data demonstrate that the PB6-CAR can be expressed on T cells to induce a potent anti-tumor response in vitro in the presence of B7H6-positive target cells.
Epitope characterization of the PB scFv family
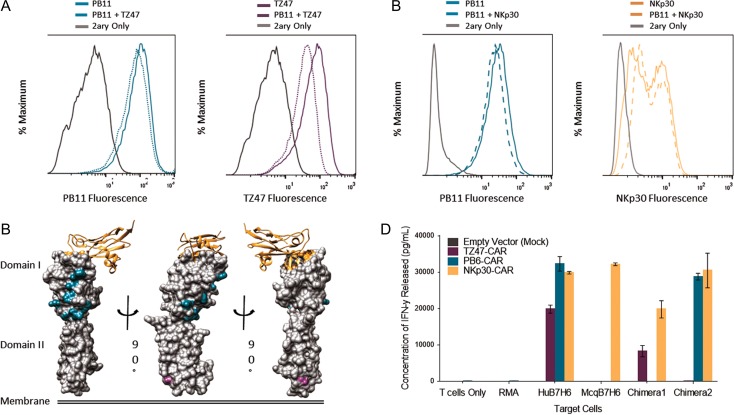
After confirming that PB scFvs successfully target B7H6-expressing tumor cells, studies to further characterize the binding mode by which PB scFvs recognize the B7H6 antigen were pursued via competitive binding assays and chimeric antigen design. First, competitive binding experiments demonstrated that PB11 could bind B7H6-expressing cells simultaneously with TZ47 and NKp30 (Fig. 5A and B). Additionally, TZ47 and NKp30 did not compete with each other for binding to B7H6 (data not shown). The lack of competition among these three proteins suggests that PB11 recognizes an epitope distinct from those of TZ47 and NKp30.
Fig. 5.
Epitope Characterization of TZ47 and PB scFvs. (A and B) Histograms depicting results of competitive staining of PB11 scFv-CH1-Fc with TZ47 (A) and NKp30-Ig (B) on RMA-B7H6 cells. Solid colored lines indicate staining with 500 nM of a single reagent, dotted colored lines with an equimolar mixture of reagents, and gray lines with secondary reagents (2ary) only. Results are presented for PB11, TZ47, and NKp30-Ig. (C) Structural representations of B7H6 are shaded by positions at which Human B7H6 residues differed and were replaced by Macaque B7H6 residues in Chimera 1 (Domain I) and Chimera 2 (Domain II). NKp30 is also shown as a ribbon diagram, with B7H6 residues <5 Å away highlighted in orange. (D) CAR T cell activation in response to Human, Macaque, and Chimeric Hu-Mcq B7H6 expressing RMA cells is shown, in addition to controls for non-transduced RMA cells and no target cells. Colors indicate T cells that were transduced with empty vector, TZ47-CAR, PB6-CAR and NKp30-CAR. Error bars depict the standard deviation of three technical replicates and are representative of two biological replicates.
To localize the PB epitope, an ortholog-based chimeragenesis approach was used based on the finding that PB6-CAR T cells were not activated against cells expressing the Macaque B7H6-ortholog, despite nearly 80% sequence identity to the extracellular domain of Human B7H6. Two chimeric Macaque-Human B7H6 variants were made by swapping out portions of the human B7H6 sequence for that of the macaque ortholog (Fig. 5C). Epitopes were localized via CAR T cell activation assays against chimera-expressing target cells. Activation of NKp30-CAR T cells by both chimeras suggested successful expression and folding of chimeric antigens on the surface of target cells. In contrast, PB6-CAR and TZ47-CAR T cells lost activation upon co-incubation with Chimera 1 and Chimera 2 with replacements in Domain I and Domain II, respectively, but maintained activity against the alternate chimera (Fig. 5D), supporting the specific contribution of distinct chimeric residues within each variant to PB6 and TZ47 binding. Thus, the PB scFv family targets an epitope localized in Domain I which is distinct from both the TZ47 epitope found within Domain II and the NKp30 binding site in Domain I.
Discussion
The PB family of scFvs provides a novel means through which to target B7H6-expressing tumor cells, enabling opportunities to focus and direct various types of immunotherapies in addition to the CAR T cell application described here. Since these scFvs were evolved from a human non-immune library and included only one round of mutagenesis targeting two to four mutations per scFv, they are presumed to pose a decreased risk of immunogenicity than murine antibodies such as TZ47. As with all antibodies, however, it cannot be ruled out that immunogenic epitopes may have been introduced during affinity maturation to either relatively quiescent human framework regions or within the complementarity determining regions (Harding et al., 2010). This possibility could be explored through experimental immunogenicity studies or computational epitope predictors (reviewed in Jawa et al., (2013)), and various deimmunization (Jones et al., 2009; Parker et al., 2010) or tolerization (De Groot et al., 2013) strategies may be employed to mitigate risk, if necessary.
Although established protocols have been written on isolating antigen binders from human scFv libraries (Chao et al., 2006), the specific decisions involved for the selection strategy—e.g. the type of selection, the amount of antigen, the gating strategy, the incorporation of blocking agents, the timing of diversification steps—remain somewhat subjective. Many modifications to the selection strategy described here could have potentially increased the efficiency with which binders were isolated, including earlier incorporation of blocking steps or alternating the use of diverse secondary reagents to prevent the enrichment of clones specific for secondary reagents and the eventual need for negative selections. Additional modifications could have changed the diversity of resulting clones after selection: for example, a less dramatic decrease in antigen concentration used in g2 selecting the g2.3 population may have resulted in a greater diversity of final clones isolated from the library. Given the extent of diversity observed at g1.7, the restriction of sequence diversity likely happened at this stage. Thus, additional B7H6-binding clonal families likely could have been evolved from the starting library employing even slightly modified selection strategies, potentially resulting in isolation of clones with additional epitope specificities.
Avidity considerations
All PB clones isolated from generation 2.5 exhibited similar binding avidities to bivalent B7H6-Ig, with binding avidity KD values of approximately 10–150 nM across yeast-displayed scFv titration stains and soluble scFv-CH1-Fc binding assays, including measurements by BLI and titration staining of antigen-expressing tumor cells. Besides incorporation into Ab-based therapies such as CAR T cells, whether PB clones may offer clinical benefit as monoclonal Ab therapy remains an open question. While this avidity KD range indicates weaker binding than the low nM-pM affinity typical of many engineered monoclonal Ab therapeutics, it suffices for CAR T cell activation in vitro where both CARs and B7H6 are expressed multivalently on the surface of T cells and tumor cells, respectively. In fact, typical immune effector cell surface receptors have KD values for their ligands that can span the range from 100 nM to 1 μM.
Among mAb therapies, the optimal affinity and avidity range to balance tumor penetration (Thurber et al., 2008), cell internalization (Ackerman et al., 2008) and tumor specificity (reviewed in Rudnick and Adams (2009)) remains to be determined. Similarly, how the affinity of extracellular targeting domains impact CAR therapy is unclear. Previous work has attempted to elucidate how the affinity of an scFv affects CAR T cell efficacy with respect to cytotoxicity and cytokine production (Guest et al., 2005; Chmielewski et al., 2011; Haso et al., 2013; Hudecek et al., 2013; Liu et al., 2015; Lynn et al., 2016). However, many of these studies utilized separately generated scFvs with different affinities targeting the same antigen, which may not recognize the same epitope (Guest et al., 2005; Hudecek et al., 2013; Lynn et al., 2016). Epitope differences among scFvs studied confound determination of the specific role of the affinity of an scFv on CAR efficacy in these studies. One major advantage of affinity maturation through yeast display is the ability to isolate scFv families such as the PB family described here that target the same epitope with a range of affinities. Isolation of such scFv families will enable better controlled experiments to assess the role of affinity of scFvs used in CAR constructs while eliminating extraneous factors such as binding modes. Therefore, use of the PB family to study CAR affinities can further expand on earlier investigations of affinity-activity relationships and may enable a more fine-tuned approach to the optimal design of CAR constructs.
Epitope considerations
We demonstrated specific activation of PB-based CAR T cells against B7H6-expressing tumor cells and recognition of an epitope distinct from those of the previously isolated TZ47 Ab and the native receptor NKp30. Furthermore, these two scFvs target epitopes localized on two different domains of B7H6. Targeting of these different regions may have implications in potency of CAR T cells.
Studies have sought to optimize CAR designs to investigate how CAR construction affects T cell activity. The length of the extracellular domain of a CAR has been shown to influence CAR T cell functional output and in vivo therapeutic efficacy (Guest et al., 2005; Haso et al., 2013; Hudecek et al., 2013, 2015), which may be due to target epitope accessibility or the orientation of effector and target cell membranes. For example, TZ47's membrane proximal epitope in Domain I may be more challenging to access on the surface of target cell membranes than the PB family's membrane distal epitope. Alternatively, the greater inter-membrane distance expected between PB-based CAR T cell and target tumor cell membranes may be less favorable resulting in less efficient activation (James et al., 2008). These dynamics may play a more critical role in CAR T cell efficacy when targeting tumors expressing low amounts of antigen, as CAR T cells require a minimum amount of receptor-ligand interactions to induce activity (Watanabe et al., 2015). Future studies comparing equivalent avidity PB-, TZ47- and NKp30-based CAR T cells targeting distinct epitopes may enable systematic studies into such spatial contributions to CAR T cell potency.
Conclusions
These studies have demonstrated that PB-based CARs in mouse T cells are specifically activated and induce cytotoxicity against mouse tumor cell lines expressing human B7H6 in vitro. Though beyond the scope of this study, we are pursuing studies to evaluate the efficacy of PB-based CARs in human T cells and in vivo using mouse models. Future studies comparing the efficacy of TZ47 and PB family B7H6-specific scFvs will expand the knowledge of how to tune CARs to interact with their binding partner to induce specific functional T cell responses. Ideally, given the success of TZ47-based CARs in in vivo models (Wu et al., 2015a, b) and the comparable binding and activation of TZ47- and PB-based CARs, we anticipate that PB scFvs may offer clinical utility as CAR T cell therapy in the future.
Supplementary Material
Supplementary data
Supplementary data are available at Protein Engineering, Design and Selection online.
Conflict of interest
C.K.H., M.E.A. and C.L.S. are co-inventors on a patent application on the PB family of scFvs. C.L.S. is also an inventor on patent applications related to targeting B7H6. This work is managed in compliance with the policies of Dartmouth College.
Funding
This work was supported in part by the National Institutes of Health [R01 CA164178 to C.L.S. and F30 AI122970 to C.K.H.]; the National Institutes of Health Center of Biomedical Research Excellence [8P30GM103415 to M.E.A. and C.L.S.]; and The Elmer R. Pfefferkorn and Allan U. Munck Education and Research Fund at Dartmouth [to M.E.A. and C.L.S].
References
- Ackerman M., Levary D., Tobon G., Hackel B., Orcutt K.D. and Wittrup K.D. (2009) Biotechnol. Prog., 25, 774–783. doi:10.1002/btpr.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman M.E., Pawlowski D. and Wittrup K.D. (2008) Mol. Cancer Ther., 7, 2233–2240. doi:10.1158/1535-7163.MCT-08-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt C.S., Baratin M., Yi E.C. et al. (2009) J. Exp. Med., 206, 1495–1503. doi:10.1084/jem.20090681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt C.S., Kennedy J.J., Xu W., Yi E.C., Fox B.A. and Gao Z. (2010). Anti-zB7H6 antibody-drug conjugates US 7858759 B2. https://www.google.com/patents/US7858759.
- Cao G., Wang J., Zheng X., Wei H., Tian Z. and Sun R. (2015) J. Biol. Chem., 290, 29964–29973. doi:10.1074/jbc.M115.674010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B.W., Markoulaki S., Hanna J., Saha K., Gao Q., Mitalipova M. and Jaenisch R. (2009) Proc. Natl. Acad. Sci. U. S. A., 106, 157–162. doi:10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerwenka A. and Moldenhauer G. (2014). B7-h6 therapeutically active monoclonal antibody against b7-h6 polypeptide PCT/EP2012/067637. https://www.google.com/patents/US20140341915.
- Chao G., Lau W.L., Hackel B.J., Sazinsky S.L., Lippow S.M. and Wittrup K.D. (2006) Nat. Protoc., 1, 755–768. doi:10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- Chmielewski M., Hombach A.A. and Abken H. (2011) Gene. Ther., 18, 62–72. doi:10.1038/gt.2010.127. [DOI] [PubMed] [Google Scholar]
- Choi Y., Hua C., Sentman C.L., Ackerman M.E. and Bailey-Kellogg C. (2015) mAbs, 7, 1045–1057. doi:10.1080/19420862.2015.1076600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groot A.S., Terry F., Cousens L. and Martin W. (2013) Expert Rev. Clin. Pharmacol, 6, 651–662. doi:10.1586/17512433.2013.835698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldhaus M.J., Siegel R.W., Opresko L.K. et al. (2003) Nat. Biotechnol., 21, 163–170. doi:10.1038/nbt785. [DOI] [PubMed] [Google Scholar]
- Fiegler N., Textor S., Arnold A., Rolle A., Oehme I., Breuhahn K., Moldenhauer G., Witzens-Harig M. and Cerwenka A. (2013) Blood, 122, 684–693. doi:10.1182/blood-2013-02-482513. [DOI] [PubMed] [Google Scholar]
- Fromant M., Blanquet S. and Plateau P. (1995) Anal. Biochem., 224, 347–353. doi:10.1006/abio.1995.1050. [DOI] [PubMed] [Google Scholar]
- Grimm S.K., Battles M.B. and Ackerman M.E. (2015) PLoS ONE, 10, e0117227 doi:10.1371/journal.pone.0117227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest R.D., Hawkins R.E., Kirillova N. et al. (2005) J. Immunother., 28, 203–211. [DOI] [PubMed] [Google Scholar]
- Harding F.A., Stickler M.M., Razo J. and DuBridge R.B. (2010) mAbs, 2, 256–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haso W., Lee D.W., Shah N.N. et al. (2013) Blood, 121, 1165–1174. doi:10.1182/blood-2012-06-438002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudecek M., Lupo-Stanghellini M.T., Kosasih P.L., Sommermeyer D., Jensen M.C., Rader C. and Riddell S.R. (2013) Clin. Cancer Res., 19, 3153–3164. doi:10.1158/1078-0432.CCR-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudecek M., Sommermeyer D., Kosasih P.L., Silva-Benedict A., Liu L., Rader C., Jensen M.C. and Riddell S.R. (2015) Cancer Immunol. Res., 3, 125–135. doi:10.1158/2326-6066.CIR-14-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James S.E., Greenberg P.D., Jensen M.C., Lin Y., Wang J., Till B.G., Raubitschek A.A., Forman S.J. and Press O.W. (2008) J. Immunol., 180, 7028–7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawa V., Cousens L.P., Awwad M., Wakshull E., Kropshofer H. and De Groot A.S. (2013) Clin. Immunol., 149, 534–555. doi:10.1016/j.clim.2013.09.006. [DOI] [PubMed] [Google Scholar]
- Jones T.D., Crompton L.J., Carr F.J. and Baker M.P. (2009) Methods Mol. Biol., 525, 405–423. doi:10.1007/978-1-59745-554-1_21xiv. [DOI] [PubMed] [Google Scholar]
- Koumbi L., Pollicino T., Raimondo G., Kumar N., Karayiannis P. and Khakoo S.I. (2016) Ann. Gastroenterol., 29, 348–357. doi:10.20524/aog.2016.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang Q. and Mariuzza R.A. (2011) J. Exp. Med., 208, 703–714. doi:10.1084/jem.20102548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Jiang S., Fang C. et al. (2015) Cancer Res., 75, 3596–3607. doi:10.1158/0008-5472.CAN-15-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn R.C., Feng Y., Schutsky K., Poussin M., Kalota A., Dimitrov D.S. and Powell D.J. Jr (2016) Leukemia, 30, 1355–1364. doi:10.1038/leu.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matta J., Baratin M., Chiche L. et al. (2013) Blood, 122, 394–404. doi:10.1182/blood-2013-01-481705. [DOI] [PubMed] [Google Scholar]
- Parker A.S., Zheng W., Griswold K.E. and Bailey-Kellogg C. (2010) BMC. Bioinformatics, 11, 180 doi:10.1186/1471-2105-11-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierres M., Vivier E. and Baratin M. (2015). Monoclonal antibodies that bind b7h6 and uses thereof PCT/IB2010/003411. https://www.google.com/patents/EP2510011B1.
- Presta L.G. (2008) Curr. Opin. Immunol., 20, 460–470. doi:10.1016/j.coi.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Retter I., Althaus H.H., Munch R. and Muller W. (2005) Nucleic Acids Res., 33, D671–D674. doi:10.1093/nar/gki088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnick S.I. and Adams G.P. (2009) Cancer Biother. Radiopharm., 24, 155–161. doi:10.1089/cbr.2009.0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimi M., Xue L., Jolin H., Hardman C., Cousins D.J., McKenzie A.N. and Ogg G.S. (2016) J. Immunol., 196, 45–54. doi:10.4049/jimmunol.1501102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyamsundar R., Kim Y.H., Higgins J.P. et al. (2005) Genome. Biol., 6, R22 doi:10.1186/gb-2005-6-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber G.M., Schmidt M.M. and Wittrup K.D. (2008) Adv. Drug Deliv. Rev., 60, 1421–1434. doi:10.1016/j.addr.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier E. and Matta J. (2014). Google Patents.
- Watanabe K., Terakura S., Martens A.C. et al. (2015) J. Immunol., 194, 911–920. doi:10.4049/jimmunol.1402346. [DOI] [PubMed] [Google Scholar]
- Wu M.R., Zhang T., DeMars L.R. and Sentman C.L. (2015. a) Gene. Ther., 22, 675–684. doi:10.1038/gt.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.R., Zhang T., Gacerez A.T., Coupet T.A., DeMars L.R. and Sentman C.L. (2015. b) J. Immunol., 194, 5305–5311. doi:10.4049/jimmunol.1402517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Wu M.R. and Sentman C.L. (2012) J. Immunol., 189, 2290–2299. doi:10.4049/jimmunol.1103495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y., Bao J., Pan X., Lu Y., Liao S., Wang X., Wang G. and Lin D. (2015) PLoS ONE, 10, e0134568 doi:10.1371/journal.pone.0134568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.