Abstract
Phytochelatin (PC) synthesis has been well demonstrated as a major metal tolerance mechanism in Arabidopsis thaliana, whereas its contribution to long-distance element transport especially in monocots remains elusive. Using rice as a cereal model, we examined physiological roles of Oryza sativa phytochelatin synthase 1 (OsPCS1) in the distribution and detoxification of arsenic (As) and cadmium (Cd), two toxic elements associated with major food safety concerns. First, we isolated four different transcript variants of OsPCS1 as well as one from OsPCS2. Quantitative real-time reverse transcription–PCR (RT-PCR) of each OsPCS transcript in rice seedlings suggested that expression of OsPCS1full, the longest OsPCS1 variant, was most abundant, followed by OsPCS2. Heterologous expression of OsPCS variants in PCS-deficient mutants of Schizosaccharomyces pombe and A. thaliana suggested that OsPCS1full possessed PCS activity in response to As(III) and Cd while the activity of other PCS variants was very low. To address physiological functions in toxic element tolerance and accumulation, two independent OsPCS1 mutant rice lines (a T-DNA and a Tos17 insertion line) were identified. The OsPCS1 mutants exhibited increased sensitivity to As(III) and Cd in hydroponic experiments, showing the importance of OsPCS1-dependent PC synthesis for rice As(III) and Cd tolerance. Elemental analyses of rice plants grown in soil with environmentally relevant As and Cd concentrations showed increased As accumulation and decreased Cd accumulation in grains of the T-DNA line. The Tos17 mutant also exhibited the reduced Cd accumulation phenotype. These contrasting effects on As and Cd distribution to grains suggest the existence of at least partially distinct PC-dependent pathways for As and Cd.
Keywords: Arsenic, Cadmium, Food safety, Metal accumulation, Metal tolerance, Rice
Introduction
Plants have transporter proteins for nutrient uptake from soil and distribution to various tissues. Some nutrient transporters also mediate acquisition of non-essential toxic elements such as cadmium (Cd) and arsenic (As) from soil. This is attributable to the chemical similarities between the essential and such non-essential elements and incomplete selectivity of nutrient transporters. As a consequence, plants accumulate Cd and As in their tissues. Phosphate transporter-mediated arsenate [As(V)] transport is a representative example (Shin et al. 2004, Wu et al. 2011, Kamiya et al. 2013). Arsenite [As(III)], another inorganic form of As, is an analog of silicic acid and dominant under the reducing conditions of paddy soil (Zhao et al. 2013). Silicon transporters and channels (Lsi1 and Lsi2) play a major role in As(III) uptake in rice roots (Ma et al. 2008). Plant Cd transport is mainly mediated by a variety of transporters for essential transition metals or cations (Krämer et al. 2007, Uraguchi and Fujiwara 2013). Especially in rice, a series of studies have identified transporters for Cd uptake (OsNramp5), vacuolar sequestration (OsHMA3), xylem loading in roots (OsHMA2) and distribution in shoots (OsLCT1) (Ueno et al. 2010, Miyadate et al. 2011, Uraguchi et al. 2011, Ishikawa et al. 2012, Sasaki et al. 2012, Satoh-Nagasawa et al. 2012). Knock-down or disruption of these transporter genes has achieved a reduction of As or Cd accumulation in rice plants. However, loss of transporter function can adversely affect elemental homeostasis and plant growth in some cases (Mitani-Ueno et al. 2011, Sasaki et al. 2012). Thus, it is important to explore further approaches suitable to ensure human-safe crop production.
Besides transporter proteins, metal ligands are another group of key players in metal transport and homeostasis. In plant cells, two metabolites, mugineic acid (MA) and its precursor nicotianamine (NA), form various metal complexes and facilitate iron (Fe) and zinc (Zn) transport (Haydon and Cobbett 2007, Kobayashi and Nishizawa 2012). Phytochelatins (PCs) represent another major type of metal-chelating ligand in plant cells. In contrast to MA and NA, PCs are predominantly associated with detoxification of non-essential toxic metals and metalloids. PCs are polypeptides with the general structure (γ-Glu-Cys)n-Gly (usually n = 2–7), and are non-ribosomally synthesized from glutathione (GSH) by phytochelatin synthases (PCSs). PCS-dependent PC synthesis in the cytosol is readily triggered by toxic metals and metalloids. In Arabidopsis thaliana, AtPCS1 is crucial for achieving tolerance against the toxic inorganic ions of Cd, As, Hg and Pb (Howden et al. 1995b, Ha et al. 1999, Fischer et al. 2014) as well as against Zn excess (Tennstedt et al. 2009). Another possible function of PCs is a contribution to long-distance element transport in plants. For example, PCs have been detected in the phloem of Brassica napus and implicated in Cd long-distance transport (Mendoza-Cozatl et al. 2008). Analyses of the AtPCS1-deficient A. thaliana mutant cad1-3 and another allele, cad1-6, suggested PC-dependent Zn translocation from roots to shoots under adequate Zn condition (Kühnlenz et al. 2016). cad1-3 also shows reduced root-to-shoot Cd translocation (Chen et al. 2006, Kühnlenz et al. 2016), but exhibits increased As translocation from the root (Liu et al. 2010). These findings in Arabidopsis suggest a potential of PCS to control Cd and/or As accumulation levels in seeds/grains. This question therefore needs to be addressed particularly in cereals.
However, few data derived from the analysis of mutant lines are available on the physiological roles of monocot PCS genes. There are precedent studies on rice PCS genes, but these only examined seed-specific knock-down of rice PCS genes (Li et al. 2007, Das et al. 2017). Only loss-of-function mutant analysis can unravel the contribution of PCSs to the mobility of toxic elements within rice plants. The significance of rice PCS has been strongly suggested by functional analysis of OsABCC1, a rice ortholog of the A. thaliana tonoplast ABC transporters for PC–metal(loid) complexes. OsABCC1 retains a substantial fraction of As in the vacuoles and thereby limits As movement within the plants and especially to the grains (Song et al. 2014).
A major motivation of the present study was to examine physiological roles of PCSs in Cd and As deposition to grains in rice as a cereal model as well as in Cd and As tolerance. We isolated cDNA of OsPCS1 transcript variants and of OsPCS2, and conducted expression and functional analyses. We further identified two independent OsPCS1 mutant rice lines and examined the physiological function of OsPCS1 in Cd and As tolerance. Finally, the element accumulation was compared between the wild-type and OsPCS1 mutant rice grown under hydroponic and soil culture conditions. Importantly, our data show contrasting effects of OsPCS1 on the accumulation of Cd and As in rice grains.
Results
Isolation and quantification of OsPCS transcripts
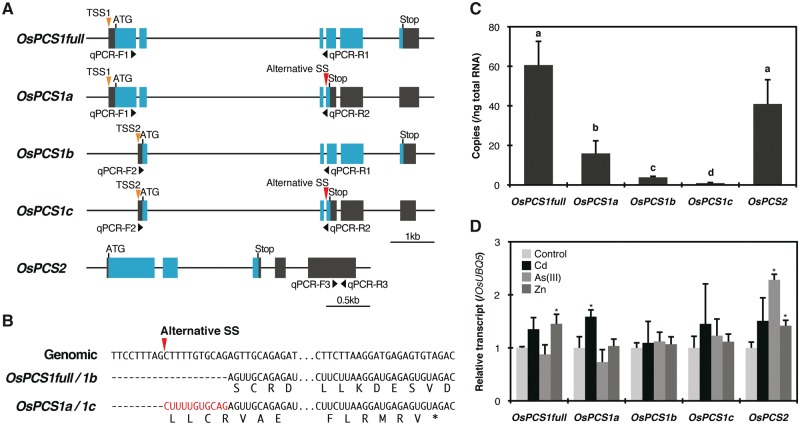
Two PCS genes have been found in the rice genome (Kühnlenz et al. 2014). Os06g0102300 (RAP-DB: http://rapdb.dna.affrc.go.jp/LOC_Os06g01260; MSU Rice Genome Annotation Project: http://rice.plantbiology.msu.edu) was predicted to encode the longest protein similar to AtPCS1. Thus the locus was named OsPCS1, and the second locus Os05g0415200/LOC_Os05g34290, which encodes a shorter protein, was referred to as OsPCS2 (Kühnlenz et al. 2014). Because three different gene models in total were proposed for OsPCS1 by the RAP and MSU databases, we verified the database predictions by isolating cDNA of OsPCS1 transcript variants as well as of OsPCS2 by PCR (Fig. 1A) and by examining their expression using specific primers for each variant (Fig. 1C). Four OsPCS1 transcripts with different coding sequences were detected. The clones corresponding to LOC_Os06g01260.1, Os06t0102300-1/LOC_Os06g01260.2 and Os06t0102300-2 were named hereafter OsPCS1full, OsPCS1a and OsPCS1b, respectively. The OsPCS1 transcript with the shortest coding sequence, not predicted in the databases, was named OsPCS1c. For OsPCS1, two different transcription start sites (TSS1 for OsPCS1full and OsPCS1a and TSS2 for OsPCS1b and OsPCS1c) were apparent as predicted in the databases (Fig. 1A). Furthermore, an alternative splicing site was indicated in exon 3 of OsPCS1a/c, which would add 11 bp to the start of exon 4 of OsPCS1full/b (Fig. 1B). This would result in an early stop codon for OsPCS1a and OsPCS1c. The OsPCS2 sequence was also identical to that of the database. Compared with the length of PCS2 from other higher plants, the coding sequence of OsPCS2 was shorter due to an early stop codon in exon 3 (Fig. 1A).
Fig. 1.
Isolation and characterization of OsPCS genes. (A) Predicted gene structures of OsPCS1 (Os06g0102300/LOC_Os06g01260) and OsPCS2 (Os05g0415200/LOC_Os05g34290). Black and blue boxes represent untranslated regions and coding regions, respectively. Approximate positions of real-time PCR primers, predicted transcription start sites (TSS1 and TSS2) and alternative splicing sites (SS) for OsPCS1 variants are denoted with black, orange and red triangles, respectively. (B) Alternative splicing sites at exon 4/3 of OsPCS1. An additional 11 bp coding sequence for OsPCS1a/1c is indicated in red. (C) Absolute quantification of OsPCS transcripts in rice roots by real-time RT–PCR. Data represent means with the SD of at least three biological replicates. Means sharing the same letter are not significantly different (P < 0.05, Tukey’s HSD). (D) Responses of OsPCS transcripts in rice roots exposed to Cd, As(III) and Zn excess treatment for 3 h. Data represent means with the SD of at least three biological replicates. Asterisks indicate significant differences from control for each variant (*P < 0.05; t-test).
We performed quantitative real-time reverse transcription–PCR (RT–PCR) to verify the abundance of each OsPCS transcript (Fig. 1C). Six specific primers were designed and used in various combinations to distinguish all five OsPCS transcripts (Fig. 1A). Absolute quantification of OsPCS transcripts in rice roots demonstrated that OsPCS1full was the dominant molecule, followed by OsPCS2. The abundance of OsPCS1a was 25% of that of OsPCS1full, while the levels of the other OsPCS1 transcripts were much lower. The public RNA-seq data available in the RAP-DB also indicated that OsPCS1full was the major OsPCS1 variant in various rice tissues (Supplementary Fig. S1). Effects of Cd, As(III) or Zn excess treatment on the transcript abundance were not evident, except that As(III) treatment approximately doubled OsPCS2 expression (Fig. 1D).
Complementation analyses in fission yeast and A. thaliana PCS mutants reveal OsPCS1full as a functional PCS
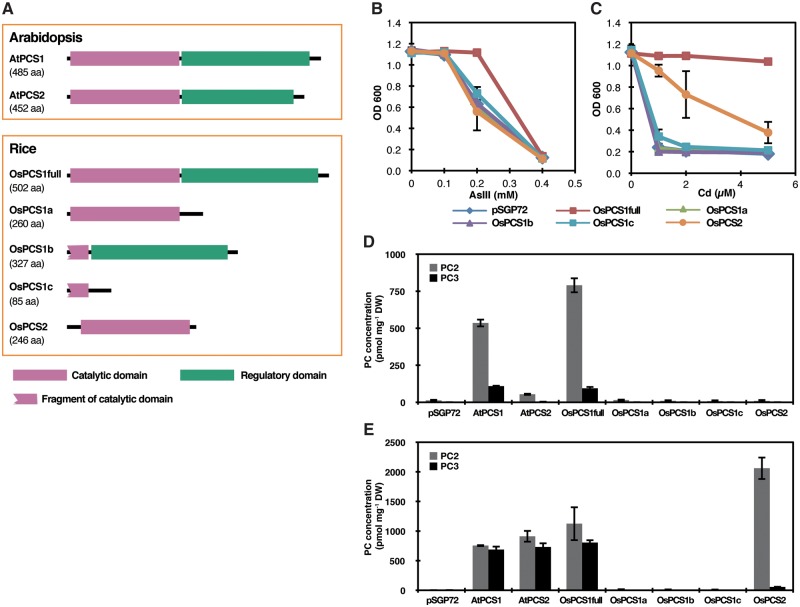
Based on the cDNA sequences, primary structures of the OsPCS proteins were compared with those of AtPCS1 and AtPCS2 (Fig. 2A; Supplementary Fig. S2). Both Arabidopsis PCSs have a structure canonical for higher plant PCS with two major domains (Rea 2012): the N-terminal catalytic domain (pfam05023) and the C-terminal Phytochelatin_C domain of unknown function (pfam09328). Among the predicted rice PCS proteins, OsPCS1full (accession No. LC314599) showed an AtPCS1/2-like structure with the two domains, whereas OsPCS2 (accession No. LC314600) would lack the C-terminal domain. OsPCS1a (accession No. AK071754) and OsPCS1b (accession No. AK071958) would consist of only the N- or C-terminal domain, respectively. OsPCS1c (accession No. LC314598) would contain a very short fragment of the catalytic N-terminal domain.
Fig. 2.
PCS activity of OsPCS variants heterologously expressed in S. pombe Δpcs under As(III) or Cd exposure. (A) Domain structures of AtPCSs and OsPCSs. The N-terminal catalytic domain (pfam05023) and C-terminal Phytochelatin_C domain of unknown function (pfam09328) are indicated. (B and C) Growth of S. pombe Δpcs harboring an empty vector pSGP72 or the cells expressing OsPCS variants when exposed to As(III) (B) or Cd (C). Data represent means with the SD of four independent replicates. (D and E) PC2 and PC3 concentrations in S. pombe Δpcs harboring an empty vector pSGP72 or the cells expressing AtPCS/OsPCS exposed to As(III) (D) or Cd (E). Data represent means with the SD of three independent replicates.
It was shown that C-terminally truncated AtPCS1 maintains substantial Cd-dependent PCS activity, suggesting that the N-terminal domain is sufficient for synthesis of PCs at least in response to Cd (Romanyuk et al. 2006, Kühnlenz et al. 2016). We therefore tested PCS activity of each predicted OsPCS using Schizosaccharomyces pombe as a heterologous expression system. The OsPCS1 variants and OsPCS2 as well as AtPCS1 and AtPCS2 were expressed in the S. pombe PCS knockout strain Δpcs (Clemens et al. 1999). Growth of the cells expressing OsPCSs was first tested under As(III) and Cd exposure. The cells expressing OsPCS1full showed better growth under 0.2 mM As(III) (Fig. 2B) and up to 5 µM Cd (Fig. 2C) than control cells with an empty vector pSGP72. OsPCS2 expression conferred weaker Cd tolerance but did not change As(III) tolerance. Growth of other cell lines was similar to that of the control cells under As(III) and Cd treatments. Subsequently, PC accumulation in the cell lines was quantified. Exposure to As(III) mainly induced drastic PC2 synthesis in the cells expressing OsPCS1full and AtPCS1 (Fig. 2D). In contrast, other OsPCS proteins including OsPCS2 did not show PCS activity. Cd treatment equally induced PC2 and PC3 synthesis mediated by OsPCS1full as well as by the two AtPCS proteins (Fig. 2E). In contrast to As(III), little but significant PC2 synthesis by OsPCS2 was observed in response to Cd exposure. However, far less PC3 was synthesized by OsPCS2.
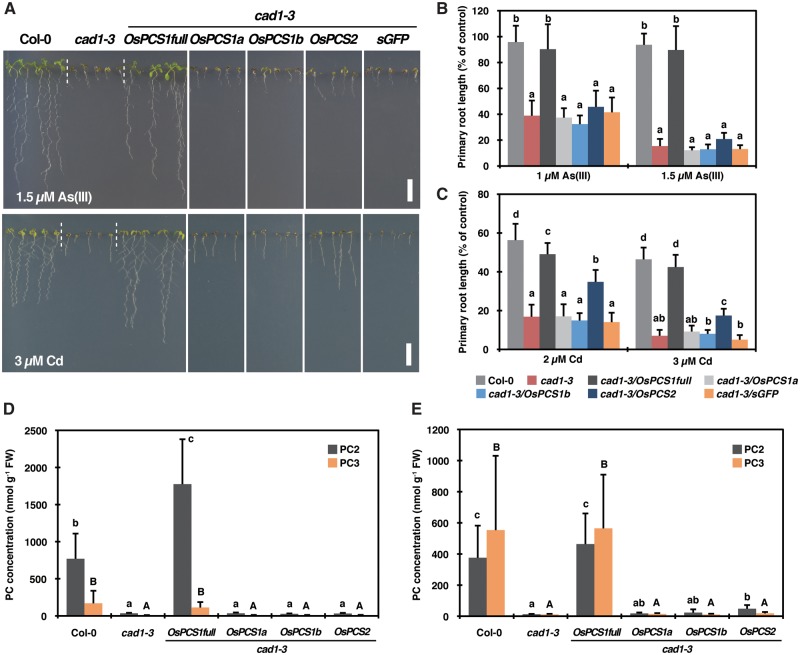
To examine further the PCS activity of predicted OsPCS proteins, in planta complementation was performed using the AtPCS1 null mutant cad1-3 (Howden et al. 1995a). cad1-3 plants were transformed with the vectors carrying the AtPCS1 promoter and each OsPCS coding sequence except for OsPCS1c. The vector carrying sGFP fused with the AtPCS1 promoter served as a control. Three independent T3 homozygous lines were established for each construct, and expression of the OsPCS variants was analyzed by RT–PCR (Supplementary Fig. S3). The transcript of the introduced OsPCS genes was evident in all cad1-3/OsPCS lines except for cad1-3/OsPCS1b line1. Growth of the plants on vertical agar plates was examined under control conditions and in the presence of As(III) or Cd. The concentrations were adjusted so that the stress severely inhibited the growth of cad1-3 but had only a minor effect on Col-0 (Fig. 3A–C). In the absence of metal/metalloid stress, there was no difference in growth between the lines (Supplementary Fig. S4). The As(III)-sensitive phenotype of cad1-3 seedlings was completely rescued by the introduction of OsPCS1full, whereas growth of the plants expressing other OsPCS genes was inhibited by As(III) as strongly as that of cad1-3 (Fig. 3A, B). Similarly, OsPCS1full introduction complemented the Cd-sensitive phenotype of cad1-3, while other OsPCS1 genes did not (Fig. 3A, C). In contrast to As(III), a partial rescue of the cad1-3 phenotype upon Cd exposure was observed in the plants expressing OsPCS2 (Fig. 3A, C). The phenotypes were consistently observed for all independent transgenic lines expressing a particular OsPCS gene (Supplementary Fig. S4). These results suggested OsPCS-mediated PC synthesis in (partially) complemented plants (cad1-3/OsPCS1full and cad1-3/OsPCS2) in response to As(III) or Cd.
Fig. 3.
Complementation assay of OsPCS variants using the AtPCS1 null mutant cad1-3. (A) Phenotypes of Col-0, cad1-3 and cad1-3 transgenic plants expressing OsPCS variants or sGFP under control of the endogenous AtPCS1 promoter grown on medium containing 1.5 µM As(III) or 3 µM Cd for 12 d. Scale bar = 1 cm. (B and C) Relative primary root length of Col-0, cad1-3 and cad1-3 transgenic plants grown on medium containing As(III) (B) or Cd (C) for 12 d. Data represent means with the SD of two independent experiments (n = 13–25). Means sharing the same letter are not significantly different within each treatment (P < 0.05, Tukey’s HSD). (D and E) PC2 and PC3 concentrations in roots of Col-0, cad1-3 and cad1-3 transgenic plants exposed to As(III) (D) or Cd (E). Seven-day-old seedlings were transferred from the control medium to the medium containing 5 µM As(III) or 5 µM Cd and grown for 7 d before harvest. Data represent means with the SD of two independent experiments (n = 3–4). Means sharing the same lower case (PC2) and upper case (PC3) letter are not significantly different (P < 0.05, Tukey’s HSD).
To demonstrate such OsPCS-dependent PC accumulation in planta, PC2 and PC3 concentrations in the transgenic plants exposed to As(III) (Fig. 3D) or Cd (Fig. 3E) were measured. Wild-type-like PC accumulation was observed in plants expressing OsPCS1full under both As(III) and Cd exposure, whereas OsPCS1a and OsPCS1b expression did not enable PC synthesis. A slightly yet significantly elevated PC2 production was observed in Cd-treated cad1-3/OsPCS2 compared with cad1-3 (Fig. 3E). This trend was not observed under As(III) treatment (Fig. 3D). GSH concentrations in the plants were negatively correlated with PC concentrations (Supplementary Fig. S4): Col-0 and cad1-3/OsPCS1full contained less GSH compared with cad1-3 and other transgenic lines under both As(III) (Supplementary Fig. S5A) and Cd exposure (Supplementary Fig. S5B). Taken together with the functional analysis in yeast, in planta complementation experiments showed that OsPCS1full is a fully functional PCS among rice PCSs in response to As(III) and Cd and OsPCS2 possesses a weak Cd-dependent PCS activity.
OsPCS1 mutant plants show increased sensitivity to Cd and As(III) stress
To understand the physiological roles of OsPCS1 in rice, OsPCS1 mutant rice lines were identified (Supplementary Fig. S6A): a T-DNA insertion line (PFG_2D-20992, hereafter ‘T-DNA’ line) and three Tos17 insertion lines (NG5039, NG5045 and NG5071). The Tos17 insertion sites in the three Tos17 insertion lines were confirmed at the same positon of OsPCS1 intron 5 by sequencing. However, these lines appeared to be independent according to their respective secondary Tos17 insertion sites in the genome according to the Tos17 mutant panel database (NG5039, the long arm of Chromosome 7; NG5045, the long arm of Chromosome 6; NG5071, the long arm of Chromosome 12). RT–PCR analysis using primers amplifying a fragment spanning from exon 1 to 6 of OsPCS1full was conducted to estimate the expression level of OsPCS1full in each mutant. The transcript was not detected in the T-DNA line, while relative expression levels among the Tos17 lines varied from 58% to 93% of cv. Nipponbare (hereafter, NB), the wild-type background of the Tos17 lines (Supplementary Fig. S6B). NG5045 was selected for further experiments based on the lowest OsPCS1full expression among the tested Tos17 lines. The second Tos17 insertion of NG5045 is in an intron of a non-protein-coding transcript (Os06g0707800). Quantitative RT–PCR using the primers amplifying an upstream region of the Tos17 or T-DNA insertion further confirmed the reduction of OsPCS1full expression by nearly 50% in NG5045 and down to residual level (<15%) in the T-DNA line, relative to NB and Hwayoung (hereafter HY), the wild type of the T-DNA line, respectively (Supplementary Fig. S6C). Thus, the T-DNA line represents a strong allele of OsPCS1, whereas NG5045 is a weak knock-down line.
The PC concentrations in the mutants exposed to Cd were measured to address whether the mutations affected PCS activity in these plants (Supplementary Fig. S6D). In accordance with the expression levels of OsPCS1 in the mutants, total PC concentration was 60% of that of NB in NG5045 and 30% in the T-DNA line compared with HY. Reduction was more pronounced for PC3 than for PC2.
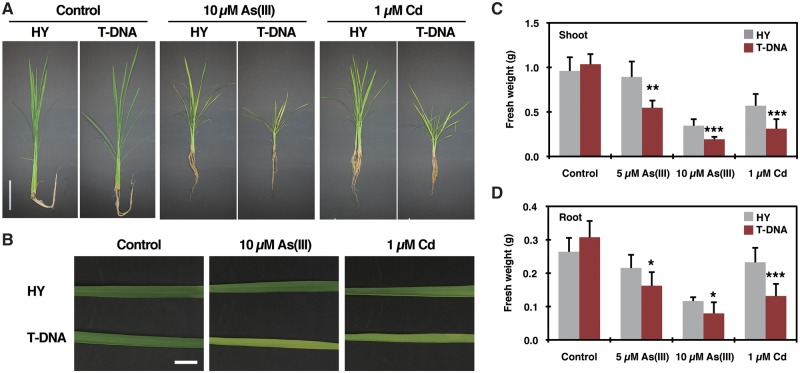
As(III) and Cd tolerance of the mutants as well as the respective wild-type plants were then examined in a hydroponic culture system (Fig. 4; Supplementary Fig. S7). The T-DNA mutant exhibited increased toxicity derived from As(III) or Cd treatment compared with the wild-type HY, while the growth overall under control conditions was comparable between HY and the T-DNA line (Fig. 4A, B). These phenotypes of the T-DNA mutant were further confirmed by measuring biomass. Both shoot (Fig. 4C) and root (Fig. 4D) fresh weight were significantly lower in the T-DNA plants than in HY when exposed to As(III) (5 or 10 µM) or Cd (1 µM). Corresponding yet weaker phenotypes were observed for the second allele NG5045 (Supplementary Fig. S7). The root growth of the Tos17 mutant was significantly more reduced by As(III) and Cd treatments compared with that of NB (Supplementary Fig. S7A, C), while the difference between the mutant and wild-type plants was not significant for the shoot (Supplementary Fig. S7B). Taken together, these results for two independent mutants demonstrate that OsPCS1 plays a major role in achieving tolerance to As(III) and Cd stress in rice plants.
Fig. 4.
Sensitivity assay of the OsPCS1 T-DNA insertion mutant rice under As(III) or Cd stress. (A–C) The wild-type HY and the T-DNA mutant of OsPCS1 were hydroponically grown for 2 weeks without As(III) or Cd addition and then grown further for 3 weeks with or without As(III) or Cd. (A and B) Phenotypes of HY and the T-DNA seedlings (A) and leaf blades (B) after cultivation. Scale bar = 10 cm (A) or 1 cm (B). (C and D) Fresh weight of shoots (C) and roots (D) after cultivation. Data represent means with the SD of two independent experiments (n = 6–10). Asterisks indicate significant differences from the wild-type HY (*P < 0.05, **P < 0.01, ***P < 0.001; t-test).
Distribution of As and Cd is altered in shoot and grains of OsPCS1 mutants
In addition to being essential for Cd and As tolerance, PCS has been implicated in the distribution of Cd, As and Zn in A. thaliana (Chen et al. 2006, Liu et al. 2010, Kühnlenz et al. 2016). We thus examined accumulation patterns of As and Cd in the OsPCS1 mutant plants in comparison with the respective wild-type plants. First, concentrations of As and Cd in shoots and roots obtained from the hydroponic culture were measured (Supplementary Fig. S8). The As concentration in shoots was slightly yet not significantly higher both in NG5045 (P = 0.16, vs. NB) and in T-DNA (P = 0.13, vs. HY) compared with each wild-type line. The As distribution (shoot/root) was significantly higher in the T-DNA line compared with HY. In contrast to As, the Cd concentrations in shoots of the two mutants were significantly lower than in those of the wild-type plants. These results indicated the possibility that disruption of OsPCS1 differentially affects distribution of As and Cd to shoots.
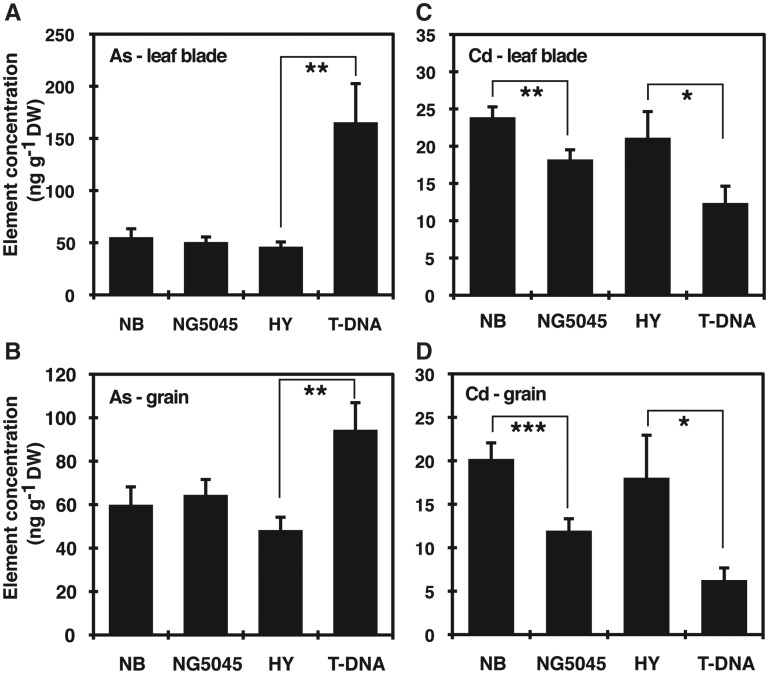
Elemental analyses were further conducted on plant samples obtained from pot experiments. Plants were grown in a commercial nursery soil until grain ripening under flooded conditions for As determination or under intermittent irrigated conditions for Cd determination. The applied concentrations are environmentally relevant (Supplementary Table S2). The flag leaf As concentration was significantly higher in the T-DNA line than the wild-type HY, whereas no difference was found between NG5045 and NB (Fig. 5A). Similar patterns were observed for grain As concentration: As concentration in grains was nearly 2-fold higher in the T-DNA line than in HY, while no significant difference was observed between NG5045 and NB (Fig. 5B). Accumulation patterns of Cd in flag leaves and grains were also well correlated among the tested plants. The Cd concentrations in the leaves of NG5045 and T-DNA were 24% and 41% lower than in the respective wild-type plants (Fig. 5C), and the grains of NG5045 and T-DNA accumulated about 40% and 65% less Cd, respectively, compared with each wild-type line (Fig. 5D). These results suggest that disruption of OsPCS1 function has opposite effects on the accumulation of As and Cd in shoots and grains of rice plants. We further quantified As accumulation in the plants grown under intermittent irrigation conditions (Supplementary Fig. S9). Overall As concentrations in the plants were lower than those in the plants grown under flooded conditions. No significant difference was found in both flag leaf and grain As concentrations between the wild-type plants and mutant plants.
Fig. 5.
As and Cd concentrations in soil-grown OsPCS1 mutant rice and the respective wild-type plants. The wild-type and OsPCS1 mutant rice were grown in pots until grain ripening. After harvest, element concentrations in leaf blade and brown rice were measured by ICP-MS. Plants were grown under flooded or intermittent irrigated conditions for testing As accumulation or Cd accumulation, respectively. (A and B) As concentrations in leaf blade (A) and grain (brown rice) (B). (C and D) Cd concentrations in leaf blade (C) and grain (brown rice) (D). Data represent means with the SD of two independent experiments (n = 8–12). Asterisks indicate significant differences between each wild-type and mutant pair (*P < 0.05, **P < 0.01, ***P < 0.001; t-test).
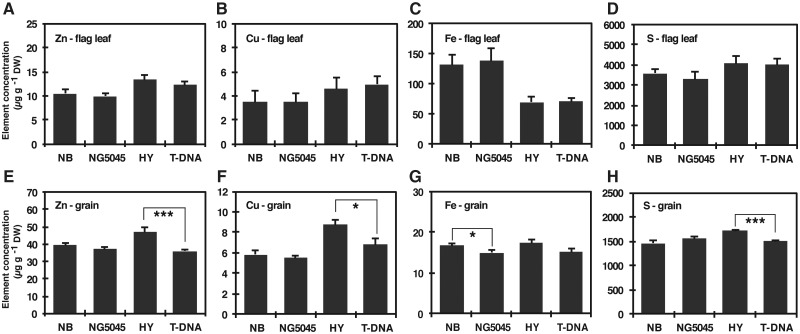
Also, we examined nutrient element concentrations in the same plant samples obtained from the intermittent irrigated cultivation (Fig. 6). In the flag leaves, there was no significant difference for Zn, Cu, Fe and S concentrations between the mutants and respective wild-type plants (Fig. 6A–D). However, the grains of the T-DNA line showed significantly reduced accumulation of Zn, Cu and S compared with HY, whereas such differences were not observed for NG5045 and NB (Fig. 6E, F, H). The grains of NG5045 showed slight but significant reduction of Fe concentration but the T-DNA line did not (Fig. 6G).
Fig. 6.
Nutrient concentrations in the soil-grown OsPCS1 mutant rice and the respective wild-type plants. The wild-type and OsPCS1 mutant rice were grown in pots until grain ripening under intermittent irrigated conditions. After harvest, element concentrations in leaf blade and grain (brown rice) were measured by ICP-MS. (A–D) Zn (A), Cu (B), Fe (C) and S (D) concentrations in flag leaf blade. (E–H) Zn (E), Cu (F), Fe (G) and S (H) concentrations in grain. Data represent means with the SD of two independent experiments (n = 8). Asterisks indicate significant differences between each wild type and mutant pair (*P < 0.05, **P < 0.01; t-test).
Discussion
Structural characteristics and expression of OsPCS gene variants
Like other higher plants, rice has two PCS genes (Kühnlenz et al. 2014). The RAP-DB and MSU database suggested three different transcript variants for OsPCS1 (Os06g0102300 and LOC_Os06g01260 in RAP-DB and MSU, respectively, designated as OsPCS2 in Das et al. 2017 and Hayashi et al. 2017) based on transcript evidence, and a shorter open reading frame (ORF) for OsPCS2 (Os05g0415200/LOC_Os05g34290, designated as OsPCS1 in Das et al. 2017 and Hayashi et al. 2017). Albeit with several differences from these database entries, recent studies isolated OsPCS transcript variants from IR64, an indica cultivar (Das et al. 2017) and the japonica cultivar Koshihikari (Hayashi et al. 2017). However, the genomic situation of OsPCS genes was not fully established. In the present study, we isolated four OsPCS1 variants and the OsPCS2 transcript from the model japonica cultivar NB basically in accordance with the database predictions and established the transcriptional and functional characteristics of the rice PCS genes. Our naming of the OsPCS genes as 1 and 2 was essentially based on the fact that Os06g0102300/LOC_Os06g01260 gives rise to a full-length PCS with higher similarity to the major PCS in A. thaliana, AtPCS1, then to the minor AtPCS2.
First, based on the sequence analyses, we suggest two TSSs and an alternative splicing site at exon 4/3 for OsPCS1, which together result in the generation of four different transcripts (Fig. 1A, B). Three of them (OsPCS1full, 1a and 1b) are identical to the database annotation, while OsPCS1c was not predicted. OsPCS1full was identical to the reported longest clone from the indica cultivar IR64 (accession No. KU670827) (Das et al. 2017). However, our three other OsPCS1 clones are different from the second indica clone (accession No. KU670828) which has a non-spliced intron 1 (Das et al. 2017). Clearly, possible genotypic variation in splicing variants will have to be further examined in various japonica and indica cultivars.
Sequence and functional analyses suggest OsPCS1full as the rice PCS most similar to AtPCS1 with respect to PCS activity: like AtPCS1, OsPCS1full possesses both the N-terminal catalytic and the C-terminal domain implicated in metal activation of the enzyme (Fig. 2A). More importantly, its expression complemented both the S. pombe PCS mutant Δpcs (Fig. 2B–E) and A. thaliana cad1-3 (Fig. 3) by mediating substantial PC synthesis under As(III) and Cd exposure. Other OsPCS1 variants did not show significant PCS activity, probably due to the lack of either the N- or C-terminal domain. We also examined the expression profiles of all OsPCS variants which we isolated (Fig. 1C). Since OsPCS1full showed the highest abundance among OsPCS transcripts in rice plants (Fig. 1C; Supplementary Fig. S1), we suggest that OsPCS1full encoded by Os06g0102300/LOC_Os06g01260 is physiologically the major form of OsPCS1.
A unique feature of OsPCS2, the secondary rice PCS according to our data, is the lack of the C-terminal domain due to an early stop codon in exon 3 of the OsPCS2 gene (Figs. 1A, 2A; Supplementary Fig. S2). The genomic sequence around exon 3 is conserved in the indica cultivar Guangluai4 and the japonica cultivar Koshihikari according to the public short read assembly data (RAP-DB). However, recent studies reported longer OsPCS2 transcripts from cultivars IR64 (Das et al. 2017) and Koshihikari (Hayashi et al. 2017). We therefore examined public RNA-seq data obtained from normally grown NB seedlings (Secco et al. 2013) available in the RAP-DB. We found an indication of an alternative splicing site just before the early stop codon of exon 3 which could result in longer ORFs (Supplementary Fig. S10). Our shorter OsPCS2 variant seems rather abundant in roots, whereas the longer transcript appears dominant in shoots. Another noteworthy feature of OsPCS2 is the expression level close to that of the most abundant OsPCS transcript OsPCS1full (Fig. 1C). In Arabidopsis, AtPCS2 is far less expressed in seedlings compared with AtPCS1 (Cazale and Clemens 2001). With regard to the PCS activity, OsPCS2 is the only clone besides OsPCS1full that showed PCS activity in response to Cd treatment in S. pombe and cad1-3 (Figs. 2E, 3E). No activity of the predicted short form of OsPCS2 was detected in our experiments upon As(III) exposure (Figs. 2D, 3D). In contrast, PCS activity in response to As was reported for the longer OsPCS2 version not covered in our analyses (Hayashi et al. 2017). It has been suggested that different regions of the AtPCS1 C-terminal domain are required for metal-specific activation of PC synthesis (Ruotolo et al. 2004, Kühnlenz et al. 2016). The different responses of the two OsPCS2 variants are another example indicating the importance of the PCS C-terminal domain for determining metal-specific activation.
Roles of OsPCS1 in As(III) and Cd tolerance of rice
PCS-dependent PC synthesis has been well documented as a crucial response of Arabidopsis for detoxifying cytosolic Cd, As, Hg and Pb ions as well as Zn excess (Howden et al. 1995b, Ha et al. 1999, Tennstedt et al. 2009, Fischer et al. 2014). AtPCS1 is the major PCS in Arabidopsis since the AtPCS1 null mutant cad1-3 displays strong hypersensitivity to these elements. While previous reports investigated seed-specific RNA interference (RNAi)-mediated knock-down of OsPCS genes (Das et al. 2017, Li et al. 2007) and another recent study reported a mutant for OsPCS2 (Os05g0415200/LOC_Os05g34290), no physiological data are available for OsPCS1 (Os06g0102300/LOC_Os06g01260), encoding the highly expressed and functional PCS variant.
We identified two independent rice mutants of OsPCS1 (Supplementary Fig. S6) and characterized them to evaluate physiological functions of OsPCS1 with respect to As(III) and Cd tolerance as well as the within-plant mobility of As and Cd. First, the hydroponic experiments demonstrated that OsPCS1 is crucial for As and Cd tolerance of rice seedlings: disruption of OsPCS1 significantly increased sensitivity of rice against As(III) and Cd stresses (Fig. 4; Supplementary Fig. S7). The enhanced sensitivity of the mutants was more evident in the case of As(III) treatment than in the case of Cd, demonstrating the importance of the PC-dependent pathway especially for As(III) detoxification. Our results are in line with the As hypersensitivity phenotype of osabcc1, a rice mutant of the vacuolar ABC transporter OsABCC1 (Song et al. 2014). OsABCC1 is crucial for removing PC–As complexes from the cytosol by vacuolar sequestration, which is the final step of the PC-dependent detoxification process as demonstrated also for Arabidopsis mutants of AtABCC1 and 2 (Song et al. 2010, Song et al. 2014).
The relatively weak phenotype of the OsPCS1 mutants observed under Cd exposure is in contrast to the clear phenotypes under As(III) stress, and again in line with observations on OsABCC1 mutants (Song et al. 2014). Similarly, a recent report on OsPCS2 also indicates its involvement in As tolerance but little contribution to Cd tolerance (Hayashi et al. 2017). As previously discussed (Song et al. 2014), it may be attributable to the contribution of OsHMA3, a vacuolar heavy metal ATPase. Functional OsHMA3 mediates Cd sequestration in the vacuole independent of PCs and thus is an important transporter affecting radial Cd transport in roots and Cd tolerance of rice as well (Ueno et al. 2010, Miyadate et al. 2011, Sasaki et al. 2014). There is no evidence suggesting involvement of OsHMA3 in As transport. Taken together with the OsPCS1 mutant phenotypes reported here, it is suggested that OsPCS1-dependent Cd detoxification and OsHMA3-mediated Cd sequestration independently play an important role in Cd tolerance of rice, while the PC-dependent pathway mediated by OsPCS1 and OsABCC1 rather exclusively accounts for a major fraction of the As detoxification capacity in rice. Moreover, from the report of Hayashi et al. (2017) and our data, it appears that unlike in A. thaliana both PCS genes significantly contribute to As tolerance.
Roles of OsPCS1 in As(III) and Cd distribution within rice plants
In addition to the significant roles of PCS for toxic element detoxification, recent data suggest involvement of PCS in element distribution within plants (Mendoza-CÓzatl et al. 2008, Liu et al. 2010, Kühnlenz et al. 2016). In rice, physiological experiments using different cultivars confirmed PC–As complex formation in the plants grown under non-toxic As exposure conditions (Batista et al. 2014). High-resolution elemental mapping in rice grown with environmentally relevant As concentrations showed co-localization of As and S in vacuoles of nodal phloem parenchyma cells (Moore et al. 2014), suggesting thiol complexation of As in the vacuoles mediated by PCs. Supporting these observations, OsABCC1 mutant analyses suggested OsABCC1-mediated control of long-distance As transport (Song et al. 2014). Co-localization of Cd and S was also reported in rice grown in the presence of environmentally relevant Cd concentrations (Yamaguchi et al. 2012), but functions of PCS related to long-distance Cd transport toward grains had remained elusive.
In this study, using rice mutants of OsPCS1, we examined whether PCS affected the distribution of As and Cd into shoots and grains. The elemental analyses on the hydroponically grown plant samples first indicated contrasting effects of OsPCS1 disruption on As and Cd accumulation in rice shoots: increased As and decreased Cd content of the mutant shoot (Supplementary Fig. S8). This suggested the possibility that OsPCS1-dependent PC synthesis confers As retention in roots but Cd translocation from roots to shoots. The independent soil experiments performed at environmentally relevant concentrations further supported the contrasting effects of OsPCS1 disruption. An increase in As accumulation was evident in grains and leaves of the T-DNA line and a decrease of Cd accumulation in grains and leaves was significant for the two tested mutant lines (Fig. 5). The weak allele NG5045 did not show the As accumulation phenotype, unlike the T-DNA line, but this may be attributable to residual OsPCS1 expression in NG5045 and/or to the secondary OsPCS (Hayashi et al. 2017). The OsPCS1-dependent changes in As and Cd accumulation indicated the physiological significance of OsPCS1 and roles of metal(loid)–PC complexes in the within-plant mobility of these elements, albeit in opposite directions. It should also be noted that the T-DNA line showed reduced accumulation of Zn, Cu and S in the grains (Fig. 6), indicating the possibility that OsPCS1 is also influencing the within-plant mobility of these essential elements.
A recent study on the OsPCS2 mutant showed the increased As accumulation in grains, but no phenotype for Cd was observed (Hayashi et al. 2017). These and our data combined suggest that in fact both OsPCS genes significantly contribute to restricting As mobility within rice plants. This is different from A. thaliana where AtPCS1 is the only PCS isoform controlling metal tolerance and accumulation. A physiological function for AtPCS2 has yet to be found.
In contrast, apparently only OsPCS1, the isoform analyzed in this study, exerts a measurable influence on Cd accumulation. The divergent effects of OsPCS1 disruption on As and Cd accumulation in shoot may be explained again by the relatively large contribution of OsABCC1 to vacuolar transport of As and a comparatively smaller contribution for Cd. The PCS- and OsABCC1-dependent pathway largely contributes to vacuolar sequestration of As in the PC–As complex form in root cells as well as nodal phloem companion cells, resulting in As retention within cells (Song et al. 2014). Lack of OsPCS1 would reduce formation of the PC–As complex, and thereby very probably eliminate the contribution of OsABCC1-mediated vacuolar sequestration of As. This would eventually diminish the As retention capacity of cells and thus enhance As mobility within plants. This hypothesis is basically in line with the one suggested for OsABCC1 mutant phenotypes (Song et al. 2014), and reasonably explains the increased As accumulation in grains of both OsABCC1 and OsPCS1 mutants.
Regarding Cd, OsABCC1 could play a role in the vacuolar sequestration of PC–Cd complexes. However, according to yeast expression experiments, the affinity of OsABCC1 is lower for PC–Cd than for PC–As (Song et al. 2014). Thus unlike the case of As, substantial PC–Cd complex formed in wild-type root cells may not be sequestered in vacuoles, and such escaped Cd would be channeled into long-distance transport pathways. Reduced PCS activity would consequently result in lower mobility of Cd and less accumulation in above-ground tissues, as we observed for leaves and grains. It is also possible that reduced PCS activity in roots diminishes Cd uptake by roots, suggested by reduced Cd concentration in the T-DNA mutant roots (Supplementary Fig. S8D). Formation of PC–Cd complexes in the cytosol may drive Cd uptake into root cells.
An alternative explanation for the contrasting effects on Cd and As accumulation in grains could be that a reduction in As–PC complex concentrations enhances the formation of other, more mobile As complexes, for example with GSH. Such complexes have been detected in plants (Raab et al. 2005). However, we are not aware of data suggesting such higher mobility of As complexes with thiols other than PCs. It will certainly be important to address directly in future studies changes in the speciation of As and Cd caused by a loss of PCS activity.
Potential of PCS for mitigating risks of As and Cd background contamination
Daily consumption of plant-derived foods, especially rice containing trace yet relevant levels of As or Cd, has increasingly been suggested as a potential risk to human health (Tsukahara et al. 2003, Meharg et al. 2009, Gilbert-Diamond et al. 2011, Meharg et al. 2013). For mitigating As and Cd contamination of rice grains, water management control of paddies is a possible approach immediately available in fields. However, there is a clear trade-off relationship between phytoavailability of As and Cd in soil (Arao et al. 2009, Honma et al. 2016): aerobic upland conditions drastically decrease As but increase Cd in soil solution, and, in contrast, flooded reducing conditions have completely opposite effects. Changes in major As and Cd transporters is suggested as another promising approach for reducing toxic element accumulation in grains (Uraguchi and Fujiwara 2013, Clemens and Ma 2016). However, As and Cd do not share the same uptake and distribution pathways (Clemens and Ma 2016) so that it is difficult to establish plants with both low-As and low-Cd phenotypes by manipulating a single transporter gene. Moreover, it is suggested that disruption of such transporter genes harbors a risk of disturbing nutritional homeostasis and impairing plant growth and stress resistance (Ma et al. 2006, Sasaki et al. 2012).
In this study, we establish the foundation for utilization of PCS in controlling both As and Cd partitioning into rice grains. Under environmentally relevant soil conditions, we found significantly decreased Cd accumulation in grains when OsPCS1 is disrupted (Fig. 5). For As, although the OsPCS1 T-DNA insertion line accumulated double the level of As in grains, NG5045, the Tos17 mutant with residual OsPCS1 expression, did not show such increased As accumulation. Taking advantage of the reduced Cd and unaltered As phenotypes of NG5045, it might be possible to achieve mitigation of both Cd- and As-related risks by cultivating NG5045 in fields with rather aerobic conditions which diminish the phytoavailable As fraction in soil solutions. Indeed, our pot experiment with intermittent water management suggests the practical potential of the idea: the Tos17 mutant line showed the reduced grain Cd accumulation attributable to OsPCS1 disruption, and the As levels in NG5045 as well as NB were reduced to <20% of the flooded conditions (Supplementary Fig. S9) due to the relatively aerobic soil condition. It should be noted that NG5045 showed wild-type-like vegetative growth under control conditions (Supplementary Fig. S7) and little alteration of nutrient accumulation (Fig. 6). Performance of the non-transgenic OsPCS1 allele NG5045 especially in regard to As and Cd accumulation as well as yields should be further examined in field conditions. More generally, it appears promising to explore systematically the effects of different PCS alleles on toxic metal accumulation in cereal grains.
Materials and Methods
Plant materials and growth conditions
Rice (Oryza sativa L.) cv. NB was used for the cloning of OsPCS variants and expression analyses. A T-DNA insertion line (PFG_2D-20992) and Tos17 insertion lines (NG5039, NG5045 and NG5071) were obtained from the Rice T-DNA Insertion Sequence Database Center at Pohang University of Science and Technology (POSTECH) and from the Rice Tos17 Insertion Mutant Database at the National Institute of Agrobiological Sciences (NIAS), respectively. Homozygous plants for each line were selected by PCR with specific primers (Supplementary Table S1) and then used for the characterization. NB and HY were used as the wild types of Tos17 and the T-DNA line, respectively.
For hydroponic culture of rice, half-strength Kimura B solution supplemented with 2 mM MES (pH 5.6, KOH) was used (Uraguchi and Fujiwara 2011) and plants were grown under long-day conditions (16 h light/8 h dark, 26°C/23°C). Soil experiments were conducted in a temperature-controlled greenhouse (25–30°C with natural light conditions). A commercial nursery soil (‘Honens nursery soil No. 1’, Honen Agri Co.) containing basal fertilizers was used for the pot experiment.
For Arabidopsis complementation, A. thaliana wild-type Col-0 and the AtPCS1 null mutant cad1-3 were used. Agar plates containing one-tenth modified Hoagland medium (Tennstedt et al. 2009, Fischer et al. 2014) were used for cultivation of Arabidopsis plants [100 µM (NH4)2HPO4, 200 µM MgSO4, 280 µM Ca(NO3)2, 600 µM KNO3, 5 µM Fe-HBED, 1% (w/v) sucrose, 5 mM MES, 1% (w/v) agar, pH 5.7]. Purified agar (Nacalai Tesque) was used for Cd tolerance assay, and Type E agar (Sigma-Aldrich) was used for other experiments. For PC analyses, the following microelements were added to the medium: 4.63 µM H3BO3, 32 nM CuSO4, 915 nM MnCl2, 77 nM ZnSO4 and 11 nM MoO3 (Kühnlenz et al. 2014). Arabidopsis seeds were surface sterilized and sown on agar plates. After 2 d stratification at 4°C, plants were grown vertically in a growth chamber (16 h light/8 h dark, 22°C).
Isolation of OsPCS cDNAs
Total RNA was isolated from NB seedlings using Trizol (Thermo Fisher Scientific). Extracted RNA was reverse-transcribed using a SuperScript First-Strand Synthesis System (Invitrogen) with the oligod(T) primer and random hexamers. The synthesized cDNA was used as a template for isolation of OsPCS cDNAs by PCR using primers designed based on the OsPCS sequences in the databases (Supplementary Table S1). The amplified products were subcloned into the pGEM-T vector (Promega) and sequenced.
Generation of OsPCS-expressing Arabidopsis
A 2,075 bp fragment upstream of the AtPCS1 start codon was amplified from Col-0 genomic DNA with the primers listed in Supplementary Table S1. The obtained fragment spanning the putative AtPCS1 promoter region was ligated into the HindIII and KpnI sites of pTS100 (Uraguchi et al. 2011), which has sGFP within the att cassette of pMDC32 (Curtis and Grossniklaus 2003). The resulting plasmid was named pSUB59. The coding sequences of OsPCS1full, OsPCS1a, OsPCS1b and OsPCS2 were amplified from the pGEM-T vectors harboring each cDNA, and sGFP was amplified from pTS100 using the primers listed in Supplementary Table S1. The respective amplicons were ligated into the KpnI and PacI sites of pSUB59 to obtain pSUB60 (ProAtPCS1-OsPCS1full), pSUB61 (ProAtPCS1-OsPCS1a), pSUB62 (ProAtPCS1-OsPCS1b), pSUB64 (ProAtPCS1-OsPCS2) and pSUB65 (ProAtPCS1-sGFP). The plasmids were introduced into Agrobacterium tumefaciens GV3101::pMP90, which were then used for transformation of cad1-3 plants by the floral dip method (Clough and Bent 1998). Transformants were selected on agar medium containing hygromycin, and T3 homozygous lines were used for the experiments.
Expression analyses
Rice seedlings hydroponically grown for 3 weeks were used for expression analyses of OsPCS genes in the wild-type and mutant plants. To examine responses to metal treatments, 3-week-old NB seedlings were transferred to a hydroponic solution containing 10 µM Cd, As(III) or Zn and treated for 3 h. Total RNA was extracted using Trizol (Thermo Fisher Scientific) from rice seedlings, followed by DNase I treatment (Thermo Fisher Scientific). Obtained RNA was then used for cDNA synthesis by PrimeScript RT Master Mix (TAKARA). Quantitative real-time PCR was performed with iQ SYBR Green Supermix (BioRad). Semi-quantitative RT–PCR was conducted with PrimeSTAR HS DNA Polymerase (TAKARA). OsUBQ5 served as an internal control (Uraguchi et al. 2011). For semi-quantitative RT–PCR, the relative intensity of the obtained bands was quantified by Image J software.
To examine introduced OsPCS expression in the Arabidopsis transformants, Arabidopsis plants grown on Hoagland agar plates for 12 d were subjected to RNA extraction. An RNeasy Plant Mini (QIAGEN) was used for total RNA extraction from the seedlings, and DNase (QIAGEN) treatment was applied during extraction. PrimeScript RT Master Mix (TAKARA) was used for cDNA synthesis, and RT–PCR was performed with GoTaq Green Master Mix (Promega). Elongation factor 1α (At5g60390) served as an internal control. The primer sequences used for expression analyses are listed in Supplementary Table S2.
Heterologous expression of OsPCS variants in Schizosaccharomyces pombe
The S. pombe PCS knockout strain Δpcs (Clemens et al. 1999) heterologously expressing OsPCS variants was used for assaying PCS activity. The OsPCS coding sequences without a stop codon were amplified from the pGEM-T vectors harboring each OsPCS cDNA and respectively ligated into the XhoI and NotI sites of pSGP72, a fission yeast expression vector. The resulting plasmids were used for transformation of Δpcs. Cells carrying the empty vector served as negative control. Cells carrying pSGP72 with AtPCS1 or AtPCS2 (Kühnlenz et al. 2014) were also included in the assay.
Yeast cultivation was carried out as described previously with some modifications (Kühnlenz et al. 2016). Yeast cultivation was carried out at 30°C in Edinburgh minimal medium (EMM). Pre-cultured cells were inoculated to an OD600 = 0.1 in EMM supplemented with 20 mM thiamine and grown overnight. Then cells were washed twice in EMM supplemented with 1 µM thiamine and inoculated at an OD600 = 0.1 in EMM supplemented with 1 µM thiamine in the presence or absence of either Cd or As(III). For growth assay, three different concentrations of As(III) (0.2, 0.4 and 1 mM) and of Cd (1, 2 and 5 µM) were applied and growth of the cells was monitored by measuring OD600 for 24 h. For PC analysis, 10 µM As(III) or 10 µM Cd was applied. After 24 h incubation, cells were harvested, frozen in liquid N2 and lyophilized for PC extraction and analysis.
Phenotyping of the OsPCS-expressing transgenic Arabidopsis lines
For metal tolerance assays, Arabidopsis plants were grown on the agar plates containing Cd (2 or 3 µM CdCl2) or As(III) (1 or 1.5 µM NaAsO2) for 12 d. The plates without supplementation of Cd or As(III) served as controls. Plant growth was assessed by root length measurement at the end of the cultivation.
For PC analyses, Arabidopsis plants were grown on the control plates for 7 d. Uniformly grown seedlings were then transferred to plates containing 5 µM As(III) or Cd and incubated for an additional 7 d. Roots were then separated from shoots and were frozen in liquid nitrogen after fresh weight measurement. Homogenously ground materials were used for further PC analysis as described below.
Phenotyping of OsPCS1 mutant rice
For PC analyses, rice plants were grown hydroponically with half-strength Kimura B medium for 2 weeks. Established seedlings were then transferred to the medium supplemented with 10 µM Cd. Roots were harvested 3 d after transfer and were frozen in liquid nitrogen after fresh weight measurement. Homogenously ground material was used for further PC analysis as described below.
Hydroponic experiments were carried out to examine Cd and As(III) sensitivity and element accumulation of OsPCS1 mutant rice. Two-week-old seedlings grown with half-strength Kimura B medium were exposed to the medium supplemented with 1 µM CdCl2, or 5 or 10 µM NaAsO2 for 3 weeks. The medium without Cd or As(III) addition served as control. The hydroponic solution was renewed every week. Fresh weights of roots and shoots were separately measured at harvest, and roots were subjected to sequential washing procedures: roots were desorbed for 10 min each in ice-cold MilliQ water, 20 mM CaCl2 (twice), 10 mM EDTA (pH 5.7) and MilliQ water. Harvested roots and shoots were dried at 50°C before elemental analysis.
Soil experiments were conducted to investigate elemental profiles of OsPCS1 mutant rice grown in soil containing environmentally relevant levels of Cd and As (Supplementary Table S2). Three-week-old seedlings grown in a nursery box were transferred to 1/5,000 a Wagner pots filled with 2.3 kg of the soil. The mutants were paired with each wild-type plant in a pot and grown until the grain ripening stage. Six pots were set in a plastic container and three or four containers were prepared for each irrigation condition. Plants for determining As accumulation were grown under flooded conditions, whereas intermittent water irrigation was applied for plants subjected to Cd and As determination. Water levels in plastic containers for the flooded condition were maintained to cover the soil surface with 2–3 cm depth. For the intermittent irrigated condition, when the water level was lowered close to the bottom of the plastic containers, plants were watered to the level covering the soil surface (once or twice a week). Flag leaves and panicles were harvested and dried at 50°C before elemental analysis. Dehusked grains (brown rice) were used for acid digestion.
Phytochelatin analysis
All plant material was frozen in liquid nitrogen and ground to a homogenous powder. Schizosaccharomyces pombe samples were frozen in liquid nitrogen and lyophilized. Thiols were extracted and derivatized as described (Kühnlenz et al. 2014). Thiol derivatives were analyzed by HPLC equipped with a fluorescence detector (Kühnlenz et al. 2014, Nishida et al. 2016).
Elemental analysis
Dried plant samples (flag leaves and brown rice) were wet-digested with a mixture of HNO3 and H2O2 as described (Uraguchi et al. 2009, Kühnlenz et al. 2016). Inductively-coupled plasma-optical emission spectrometry (ICP-OES; iCAP 6500, Thermo Fisher Scientific) and inductively-coupled plasma-masss spectrometry (ICP-MS; Agilent 7800, Agilent Technologies) were used for elemental quantification of samples from hydroponic experiments and soil experiments, respectively.
To determine extractable element concentrations in the soil, 1 M HCl extraction was conducted for As determination (Kuramata et al. 2013), and 0.1 M HCl extraction was applied for determination of Cd and other elements (Uraguchi et al. 2009), according to the Agricultural Land-Soil Pollution Prevention Act in Japan. ICP-OES (iCAP7400Duo, Thermo Fisher Scientific) was used for element quantification in soil extract samples.
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the Japan Society for the Promotion of Science [grant Nos. 15H06580 and 16K14873 to S.U.] and the Deutsche Forschungsgemeinschaft [CL 152/7-1 and CL 152/7-2 to S.C.].
Supplementary Material
Acknowledgments
We thank Stephan HÖreth and Natalia Hess for help with ICP and HPLC analyses, respectively, and Silke Matros, Christiane Meinen and Kayoko Aizawa for excellent technical assistance.
Disclosures
The authors have no conflicts of interest to declare.
Glossary
Abbreviations
- EMM
Edinburgh minimal medium
- GSH
glutathione
- HY
Hwayoung
- ICP-MS
inductively-coupled plasma-mass spectrometry
- ICP-OES
inductively-coupled plasma-optical emission spectrometry
- MA
mugineic acid
- NA
nicotianamine
- NB
Nipponbare
- ORF
open reading frame
- PC
phytochelatin
- PCS
phytochelatin synthase
- RT–PCR
real-time reverse transcription–PCR
- TSS
transcription start site
References
- Arao T., Kawasaki A., Baba K., Mori S., Matsumoto S. (2009) Effects of water management on cadmium and arsenic accumulation and dimethylarsinic acid concentrations in Japanese rice. Environ. Sci. Technol. 43: 9361–9367. [DOI] [PubMed] [Google Scholar]
- Batista B.L., Nigar M., Mestrot A., Rocha B.A., Barbosa Junior F., Price A.H.. et al. (2014) Identification and quantification of phytochelatins in roots of rice to long-term exposure: evidence of individual role on arsenic accumulation and translocation. J. Exp. Bot. 65: 1467–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalé A.C., Clemens S. (2001) Arabidopsis thaliana expresses a second functional phytochelatin synthase. FEBS Lett. 507: 215–219. [DOI] [PubMed] [Google Scholar]
- Chen A., Komives E.A., Schroeder J.I. (2006) An improved grafting technique for mature Arabidopsis plants demonstrates long-distance shoot-to-root transport of phytochelatins in Arabidopsis. Plant Physiol. 141: 108–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S., Kim E.J., Neumann D., Schroeder J.I. (1999) Tolerance to toxic metals by a gene family of phytochelatin synthases from plants and yeast. EMBO J. 18: 3325–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S., Ma J.F. (2016) Toxic heavy metal and metalloid accumulation in crop plants and foods. Annu. Rev. Plant Biol. 67: 489–512. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana .Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das N., Bhattacharya S., Bhattacharyya S., Maiti M.K. (2017) Identification of alternatively spliced transcripts of rice phytochelatin synthase 2 gene OsPCS2 involved in mitigation of cadmium and arsenic stresses. Plant Mol. Biol. 94: 167–183. [DOI] [PubMed] [Google Scholar]
- Fischer S., Kühnlenz T., Thieme M., Schmidt H., Clemens S. (2014) Analysis of plant Pb tolerance at realistic submicromolar concentrations demonstrates the role of phytochelatin synthesis for Pb detoxification. Environ. Sci. Technol. 48: 7552–7559. [DOI] [PubMed] [Google Scholar]
- Gilbert-Diamond D., Cottingham K.L., Gruber J.F., Punshon T., Sayarath V., Gandolfi A.J.. et al. (2011) Rice consumption contributes to arsenic exposure in US women. Proc. Natl. Acad. Sci. USA 108: 20656–20660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S.-B., Smith A.P., Howden R., Dietrich W.M., Bugg S., O’Connell M.J.. et al. (1999) Phytochelatin synthase genes from Arabidopsis and the yeast Schizosaccharomyces pombe. Plant Cell 11: 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S., Kuramata M., Abe T., Takagi H., Ozawa K., Ishikawa S. (2017) Phytochelatin synthase OsPCS1 plays a crucial role in reducing arsenic levels in rice grains. Plant J. 91: 840–848. [DOI] [PubMed] [Google Scholar]
- Haydon M.J., Cobbett C.S. (2007) Transporters of ligands for essential metal ions in plants. New Phytol. 174: 499–506. [DOI] [PubMed] [Google Scholar]
- Honma T., Ohba H., Kaneko-Kadokura A., Makino T., Nakamura K., Katou H. (2016) Optimal soil Eh, pH, and water management for simultaneously minimizing arsenic and cadmium concentrations in rice grains. Environ. Sci. Technol. 50: 4178–4185. [DOI] [PubMed] [Google Scholar]
- Howden R., Andersen C.R., Goldsbrough P.B., Cobbett C.S. (1995a) A cadmium-sensitive, glutathione-deficient mutant of Arabidopsis thaliana. Plant Physiol. 107: 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden R., Goldsbrough P.B., Andersen C.R., Cobbett C.S. (1995b) Cadmium-sensitive, cad1 mutants of Arabidopsis thaliana are phytochelatin deficient. Plant Physiol. 107: 1059–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S., Ishimaru Y., Igura M., Kuramata M., Abe T., Senoura T.. et al. (2012) Ion-beam irradiation, gene identification, and marker-assisted breeding in the development of low-cadmium rice. Proc. Natl. Acad. Sci. USA 109: 19166–19171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya T., Islam R., Duan G., Uraguchi S., Fujiwara T. (2013) Phosphate deficiency signaling pathway is a target of arsenate and phosphate transporter OsPT1 is involved in As accumulation in shoots of rice. Soil Sci. Plant Nutr. 59: 580–590. [Google Scholar]
- Kobayashi T., Nishizawa N.K. (2012) Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63: 131–152. [DOI] [PubMed] [Google Scholar]
- Krämer U., Talke I.N., Hanikenne M. (2007) Transition metal transport. FEBS Lett. 581: 2263–2272. [DOI] [PubMed] [Google Scholar]
- Kühnlenz T., Hofmann C., Uraguchi S., Schmidt H., Schempp S., Weber M.. et al. (2016) Phytochelatin synthesis promotes leaf Zn accumulation of Arabidopsis thaliana plants grown in soil with adequate Zn supply and is essential for survival on Zn-contaminated soil. Plant Cell Physiol. 57: 2342–2352. [DOI] [PubMed] [Google Scholar]
- Kühnlenz T., Schmidt H., Uraguchi S., Clemens S. (2014) Arabidopsis thaliana phytochelatin synthase 2 is constitutively active in vivo and can rescue the growth defect of the PCS1-deficient cad1–3 mutant on Cd-contaminated soil. J. Exp. Bot. 65: 4241–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramata M., Abe T., Kawasaki A., Ebana K., Shibaya T., Yano M.. et al. (2013) Genetic diversity of arsenic accumulation in rice and QTL analysis of methylated arsenic in rice grains. Rice 6: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Guo J., Xu W., Ma M. (2007) RNA interference mediated silencing of phytochelatin synthase gene reduce cadmium accumulation in rice seeds. J. Integr. Plant Biol. 49: 1032–1037. [Google Scholar]
- Liu W.-J.J., Wood B.A., Raab A., McGrath S.P., Zhao F.J., Feldmann J. (2010) Complexation of arsenite with phytochelatins reduces arsenite efflux and translocation from roots to shoots in Arabidopsis. Plant Physiol. 152: 2211–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.F., Tamai K., Yamaji N., Mitani N., Konishi S., Katsuhara M.. et al. (2006) A silicon transporter in rice. Nature 440: 688–691. [DOI] [PubMed] [Google Scholar]
- Ma J.F., Yamaji N., Mitani N., Xu X.-Y., Su Y.-H., McGrath S.P.. et al. (2008) Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc. Natl. Acad. Sci. USA 105: 9931–9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meharg A.A., Norton G., Deacon C., Williams P., Adomako E.E., Price A.. et al. (2013) Variation in rice cadmium related to human exposure. Environ. Sci. Technol. 47: 5613–5618. [DOI] [PubMed] [Google Scholar]
- Meharg A.A., Williams P.N., Adomako E., Lawgali Y.Y., Deacon C., Villada A.. et al. (2009) Geographical variation in total and inorganic arsenic content of polished (white) rice. Environ. Sci. Technol. 43: 1612–1617. [DOI] [PubMed] [Google Scholar]
- Mendoza-CÓzatl D.G., Butko E., Springer F., Torpey J.W., Komives E.A., Kehr J.. et al. (2008) Identification of high levels of phytochelatins, glutathione and cadmium in the phloem sap of Brassica napus. A role for thiol-peptides in the long-distance transport of cadmium and the effect of cadmium on iron translocation. Plant J. 54: 249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani-Ueno N., Yamaji N., Zhao F.-J., Ma J.F. (2011) The aromatic/arginine selectivity filter of NIP aquaporins plays a critical role in substrate selectivity for silicon, boron, and arsenic. J. Exp. Bot. 62: 4391–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyadate H., Adachi S., Hiraizumi A., Tezuka K., Nakazawa N., Kawamoto T.. et al. (2011) OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol. 189: 190–199. [DOI] [PubMed] [Google Scholar]
- Moore K.L., Chen Y., van de Meene A.M., Hughes L., Liu W., Geraki T.. et al. (2014) Combined NanoSIMS and synchrotron X-ray fluorescence reveal distinct cellular and subcellular distribution patterns of trace elements in rice tissues. New Phytol. 201: 104–115. [DOI] [PubMed] [Google Scholar]
- Nishida S., Duan G., Ohkama-Ohtsu N., Uraguchi S., Fujiwara T. (2016) Enhanced arsenic sensitivity with excess phytochelatin accumulation in shoots of a SULTR1; 2 knockout mutant of Arabidopsis thaliana (L.) Heynh. Soil Sci. Plant Nutr. 62: 367–372. [Google Scholar]
- Raab A., Schat H., Meharg A.A., Feldmann J. (2005) Uptake, translocation and transformation of arsenate and arsenite in sunflower (Helianthus annuus): formation of arsenic–phytochelatin complexes during exposure to high arsenic concentrations. New Phytol. 168: 551–558. [DOI] [PubMed] [Google Scholar]
- Rea P.A. (2012) Phytochelatin synthase: of a protease a peptide polymerase made. Physiol. Plant. 145: 154–164. [DOI] [PubMed] [Google Scholar]
- Romanyuk N.D., Rigden D.J., Vatamaniuk O.K., Lang A., Cahoon R.E., Jez J.M., Rea P.A. (2006) Mutagenic definition of a papain-like catalytic triad, sufficiency of the N-terminal domain for single-site core catalytic enzyme acylation, and C-terminal domain for augmentative metal activation of a eukaryotic phytochelatin synthase. Plant Physiol. 141: 858–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruotolo R., Peracchi A., Bolchi A., Infusini G., Amoresano A., Ottonello S. (2004) Domain organization of phytochelatin synthase: functional properties of truncated enzyme species identified by limited proteolysis. J. Biol. Chem. 279: 14686–14693. [DOI] [PubMed] [Google Scholar]
- Sasaki A., Yamaji N., Ma J.F. (2014) Overexpression of OsHMA3 enhances Cd tolerance and expression of Zn transporter genes in rice. J. Exp. Bot. 65: 6013–6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A., Yamaji N., Yokosho K., Ma J.F. (2012) Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell 24: 2155–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Nagasawa N., Mori M., Nakazawa N., Kawamoto T., Nagato Y., Sakurai K.. et al. (2012) Mutations in rice (Oryza sativa) heavy metal ATPase 2 (OsHMA2) restrict the translocation of zinc and cadmium. Plant Cell Physiol. 53: 213–224. [DOI] [PubMed] [Google Scholar]
- Secco D., Jabnoune M., Walker H., Shou H., Wu P., Poirier Y.. et al. (2013) Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell 25: 4285–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H., Shin H.-S., Dewbre G.R., Harrison M.J. (2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 39: 629–642. [DOI] [PubMed] [Google Scholar]
- Song W.-Y., Yamaki T., Yamaji N., Ko D., Jung K.-H., Fujii-Kashino M.. et al. (2014) A rice ABC transporter, OsABCC1, reduces arsenic accumulation in the grain. Proc. Natl. Acad. Sci. USA 111: 15699–15704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W.-Y.Y., Park J., Mendoza-CÓzatl D.G., Suter-Grotemeyer M., Shim D., HÖrtensteiner S.. et al. (2010) Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proc. Natl. Acad. Sci. USA 107: 21187–21192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennstedt P., Peisker D., Bottcher C., Trampczynska A., Clemens S. (2009) Phytochelatin synthesis is essential for the detoxification of excess zinc and contributes significantly to the accumulation of zinc. Plant Physiol. 149: 938–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara T., Ezaki T., Moriguchi J., Furuki K., Shimbo S., Matsuda-Inoguchi N.. et al. (2003) Rice as the most influential source of cadmium intake among general Japanese population. Sci. Total Environ. 305: 41–51. [DOI] [PubMed] [Google Scholar]
- Ueno D., Yamaji N., Kono I., Huang C.F., Ando T., Yano M.. et al. (2010) Gene limiting cadmium accumulation in rice. Proc. Natl. Acad. Sci. USA 107: 16500–16505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uraguchi S., Fujiwara T. (2011) Significant contribution of boron stored in seeds to initial growth of rice seedlings. Plant Soil 340: 435–442. [Google Scholar]
- Uraguchi S., Fujiwara T. (2013) Rice breaks ground for cadmium-free cereals. Curr. Opin. Plant Biol. 16: 328–334. [DOI] [PubMed] [Google Scholar]
- Uraguchi S., Kamiya T., Sakamoto T., Kasai K., Sato Y., Nagamura Y. et al. (2011) Low-affinity cation transporter (OsLCT1) regulates cadmium transport into rice grains. Proc. Natl. Acad. Sci. USA 108: 20959–20964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uraguchi S., Mori S., Kuramata M., Kawasaki A., Arao T., Ishikawa S. (2009) Root-to-shoot Cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice. J. Exp. Bot. 60: 2677–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Ren H., McGrath S.P., Wu P., Zhao F.J. (2011) Investigating the contribution of the phosphate transport pathway to arsenic accumulation in rice. Plant Physiol. 157: 498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N., Ishikawa S., Abe T., Baba K., Arao T., Terada Y. (2012) Role of the node in controlling traffic of cadmium, zinc, and manganese in rice. J. Exp. Bot. 63: 2729–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F.J., Harris E., Yan J., Ma J.C., Wu L.Y., Liu W.J.. et al. (2013) Arsenic methylation in soils and its relationship with microbial arsM abundance and diversity, and As speciation in rice. Environ. Sci. Technol. 47: 7147–7154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.