Abstract
Study Objectives
The current definition of sleep arousals neglects to address the diversity of arousals and their systemic cohesion. Autonomic arousals (AA) are autonomic activations often associated with cortical arousals (CA), but they may also occur in relation to a respiratory event, a leg movement event or spontaneously, without any other physiological associations. AA should be acknowledged as essential events to understand and explore the systemic implications of arousals.
Methods
We developed an automatic AA detection algorithm based on intelligent feature selection and advanced machine learning using the electrocardiogram. The model was trained and tested with respect to CA systematically scored in 258 (181 training size/77 test size) polysomnographic recordings from the Wisconsin Sleep Cohort.
Results
A precision value of 0.72 and a sensitivity of 0.63 were achieved when evaluated with respect to CA. Further analysis indicated that 81% of the non-CA-associated AAs were associated with leg movement (38%) or respiratory (43%) events.
Conclusions
The presented algorithm shows good performance when considering that more than 80% of the false positives (FP) found by the detection algorithm appeared in relation to either leg movement or respiratory events. This indicates that most FP constitute autonomic activations that are indistinguishable from those with cortical cohesion. The proposed algorithm provides an automatic system trained in a clinical environment, which can be utilized to analyze the systemic and clinical impacts of arousals.
Keywords: autonomic arousals, heart rate variability, electrocardiography, neural networks
Statement of Significance
Autonomic arousals have been postulated to be related to the cardiovascular and neurocognitive dysfunction associated with sleep-disordered breathing, independent of their relation to cortical arousals. However, most studies to date have explored experimentally induced autonomic arousals only in healthy/control populations. We developed an arousal detector that can learn the complex decision-making patterns of human scorers through application of state-of-the-art machine learning to the polysomnogram’s RR tachogram in a population-based sample, with a variety of sleep disorders. The finding that many autonomic arousals were associated with leg movement or breathing events, despite a lack of an electroencephalographic correlates, suggests that future explorations of the pathophysiologic consequences of autonomic arousals may help us to improve management of patients with sleep-disordered breathing.
Introduction
Arousals in sleep are naturally occurring microevents, which reflect the reversibility of sleep. Despite their vital function, arousals have been found to be associated with the pathophysiology of several sleep disorders [1]. The American Academy of Sleep Medicine (AASM) states that scoring of arousals must be attained through electroencephalographic (EEG) analysis and cannot be based on alternative biosignals alone [2]. However, this definition neglects to address the diversity of arousals and their systemic cohesion. In fact, experimentally induced sleep fragmentation has shown evidence of autonomic arousals (AA) without EEG correlates [3, 4]. Moreover, in studies intentionally limiting the number of cortical arousals (CA), AA were sufficient to diminish the restorative value of sleep in healthy individuals [5]. Moreover, in clinical populations, obstructive breathing events and snoring have been associated with AA, even in the absence of CA [6, 7]. To date, a complete analysis of the physiological impacts of naturally occurring AA in a clinical setting is still missing. Before this can be achieved, however, there is an unmet need for agreeing on the manifestations of AA and a clear definition of them.
Analysis of heart rate variability (HRV) has been used for decades to assess changes in the autonomic nervous system (ANS), which maps the balance between the parasympathetic nervous system (PNS) and sympathetic nervous system (SNS). HRV has been used as the primary marker of the systemic manifestation of CA [1, 4, 7–11] and has the potential to shed new light on the systemic impacts of arousals and sleep fragmentation, through more meaningful associations with clinical outcomes. Furthermore, it is widely known that manual scoring of arousals has low interscorer agreement [11], and it is time-consuming and expensive. There is an unmet need for the development of automatic systems to substitute or assist human scorers in the scoring process such that a consistent, objective, and fast analysis can be achieved.
Very few attempts at automatic detection of HRV-based arousals have been made. Basner et al. addressed this problem by presenting a semiautomatic electrocardiographic (ECG)–based arousal detection algorithm [10]. The algorithm could detect 68% of CA; however, as mentioned above, the criteria for CA may be insufficient for capturing all the electrophysiological changes that define an arousal. Furthermore, the algorithm was trained and tested in a setup where external stimuli were introduced to provoke arousal; therefore, it is uncertain how well it would behave in a clinical environment. Finally, the RR tachogram required editing by visual inspection; thus, the algorithm was only semiautomatic.
The study of Pillar et al. and Pillar et al. of Refs. 12 and 13 introduced and elaborated, respectively, on the use of a rule-based method to detect AA using peripheral arterial tonometry (PAT) recordings from participants suffering from obstructive sleep apnea [12, 13]. A correlation of 0.82 and 0.87, respectively, was achieved between such events and CA. Although the method is demonstrated in a clinical environment, only people with obstructive sleep apnea were considered, thereby limiting its application in broader populations, e.g. people with leg movements. Furthermore, the method relies on PAT recordings, which are not routinely performed, thereby limiting its applications.
The purpose of this study is to use the existing gold-standard diagnostic method—the polysomnogram (PSG) and manually or automatically scored sleep stage data—to develop an automatic detection algorithm for the detection of AA in a clinical setting, i.e. from participants suffering from a variety of sleep disorders and heart diseases. We adapted the approach of modeling and detecting autonomic behavior during CA [10, 12], since CA have shown significant correlation with autonomic activations and their cohesion occurs with consistent onset [4, 9, 10]. It is important to note that even without an EEG change that is sufficient for scoring an arousal by the AASM criteria [2], EEG spectral power density has been noted to change in association with the physiologically important sympathetic surges that cause AA [5, 7]. The detection algorithm performance will be measured against the current gold standard of EEG-based arousals (according to the AASM criteria), but will also be compared to other physiologically relevant sleep-disorder phenomena, to determine whether ANS analysis may provide an alternative, complementary metric of sleep health.
Methods
Datasets
As none of the databases available had all the required annotations to allow the development of both an automatic ectopic beat and arousal detection algorithms, two distinct databases were used. Sample size and demographics information for both databases are presented in Table 1.
Table 1.
Demographics of participants in databases
| Database | N (male/female) | Age: mean (SD) | BMI: mean (SD) |
|---|---|---|---|
| MITDB | 20 (9/11) | 61 (17.5) | — |
| WSC | 258 (138/120) | 65 (7) | 31.7 (7) |
The MIT BIH arrhythmia database (MITDB) was chosen to develop a functional ectopic beat detection algorithm. The MITDB includes a subset of forty-six 30 min recordings from over 4000 long-term Holter recordings that were collected between 1975 and 1979 by the Beth Israel Hospital Arrhythmia Laboratory [14]. The 23 first recordings, i.e. 100–124, are considered to represent usual variations in heart rhythm encountered at a routine arrhythmia clinic. Twenty of these 23 recordings were used for the development, training, and testing of an ectopic beat detection algorithm, and three were excluded due to the presence of paced beats. Inclusion criteria were limited to focus on the most common types of ectopic beats, i.e. atrial premature beats (APB) and ventricular premature beats (VPB).
The Wisconsin Sleep Cohort (WSC) [15] was used to develop, train, and test our new arousal detection algorithm. The WSC is a longitudinal study of population-based sample of randomly selected Wisconsin state employees, a subset of who are suffering from a variety of sleep pathologies ranging from normal to severe cases. These recordings have all been annotated by either of two specialized medical personnel for sleep stages, respiratory events, leg movement events, and arousals according to the AASM criteria [2]. Furthermore, a subgroup of 306 randomly selected recordings from the WSC were annotated with arousal subgroup to indicate if they appeared spontaneously or in response to a respiratory or a leg movement event. Forty-eight recordings were excluded from the study if more than 50% of the ECG-channel had signal loss. The remaining 258 recordings were included for AA algorithm training and testing. Therefore, the participants included in this study were randomly selected and were not excluded based on any medication or medical comorbidity. The University of Wisconsin–Madison Health Sciences and Stanford University Institutional Review Boards approved the study (Stanford #19207).
Signal acquisition
The MITDB provides ECG data in lead 2 configuration along with another lead of varying types. Clearly, more ECG leads improve classification; however, the WSC recordings follow standard PSG requirements provided by the AASM [2]; thus, only lead 2 configuration was used from both databases.
The ECG signal in the WSC was digitized with a sampling frequency, fs, of 200 Hz, whereas in the MITDB, fs = 360 Hz. In the WSC, Grass Comet montages were used with a pre-bandpass filter of 0.3–35 Hz. In the MITDB, a pre-filter of 0.1–100 Hz was used.
Automatic RR Tachogram Extraction Algorithm
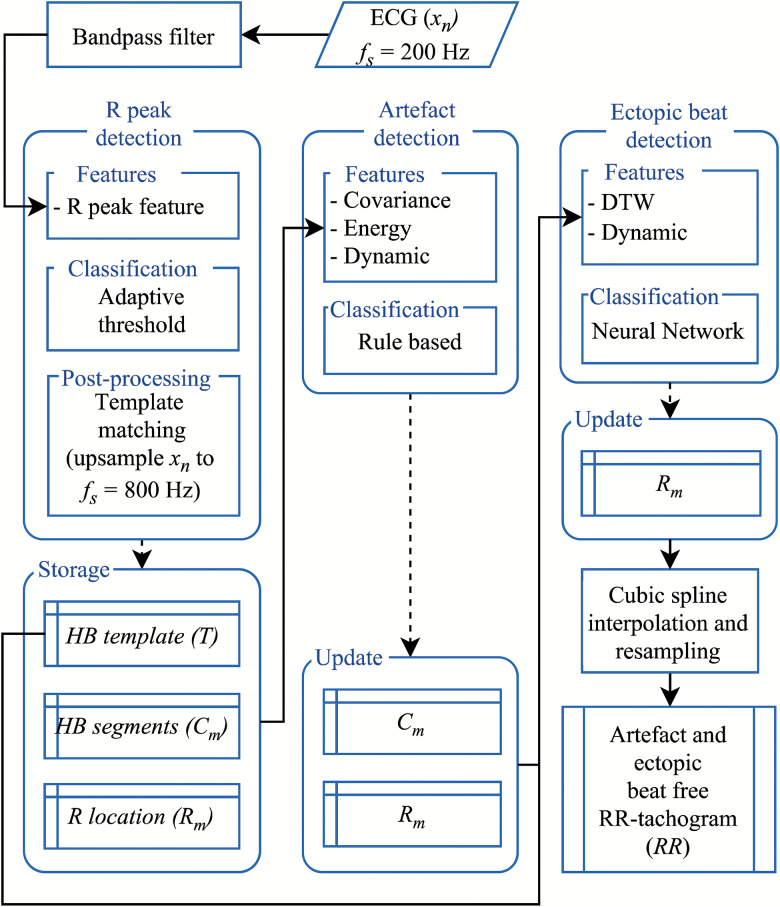
The RR tachogram directly presents information on duration and variation between heart beats, i.e. the HRV, and contains the necessary information to identify AA. Our automatic RR tachogram extraction algorithm includes an initial bandpass filtering of the ECG signal followed by three processing submodules: an R peak detection, an artifact detection, and an ectopic beat detection module. An artifact-free and ectopic beat–free RR tachogram was then extracted by cubic spline interpolation and resampling. The first block receives the bandpass filtered ECG signal, xn, and the output of the algorithm module was the RR tachogram (Figure 1). Specifics of the automatic RR tachogram extraction algorithm can be found in Supplementary Material.
Figure 1.
RR tachogram extraction algorithm overview. Input, xn, is the ECG signal, which is initially bandpass filtered and then processed in the three blocks: R peak detection, artifact detection, and ectopic beat detection. Output from the R peak detection block is used to store variables that contain dynamic information, Rm, and morphological information for each heartbeat, HB segments, Cm, which is used to design a HB template, T. Based on this information, each Cm is firstly evaluated in the artifact detection block, updated, and then evaluated in the ectopic beat block. The final output is the RR tachogram extracted from the updated Rm, fs: samling frequency.
Automatic Arousal Detection System
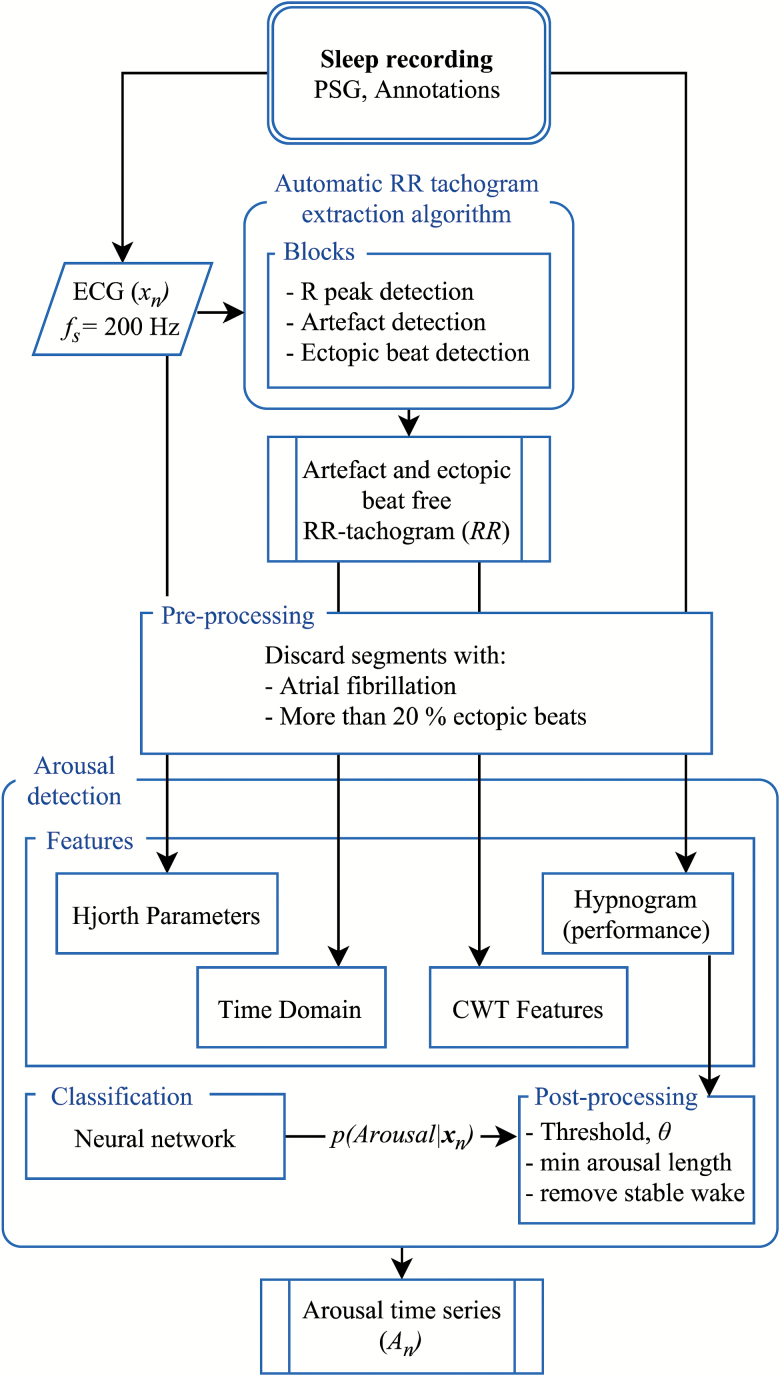
Our automatic arousal detection system includes preprocessing, feature extraction, classification, and postprocessing of features from three modalities: the ECG, the RR tachogram, and hypnogram. An overview of the automatic arousal detection algorithm is presented in Figure 2.
Figure 2.
Arousal detection algorithm overview. From the sleep recording, the PSG, the following is inputted for preprocessing and then feature extraction in the arousal detection block: the ECG, xn, the RR tachogram, RR (Figure 1), and hypnogram. A neural network is then trained and tested on recordings from the WSC. The output is the posterior probability of an arousal, p(Arousal|xn). In the postprocessing step, a threshold is used to categorize the posterior probability into binary categories: arousal or no arousal. Finally, wake stages are removed. fs: samling frequency.
Preprocessing
All modalities considered for feature extraction are first processed using a preprocessing module, where segments are evaluated for removal if the heart rhythm is too unstable. Criteria for an unstable heart rhythm were inspired by Kleiger et al. [16] and any 30 s segment contained more than 20% ectopic beats (found as described in the previous section) and/or atrial fibrillation (AF) were discarded and were not evaluated for feature extraction. Detection of AF was carried out using the algorithm of Petrenas et al. [17]. All other segments were included for feature extraction.
Feature extraction
In all prior work, HRV features have been developed for screening purposes. Consequently, they were extracted from long time segments, often 5 min or longer, and compared between different groups of participants [18]. CA introduce activations of the autonomic system that occur spontaneously. The HRV features we seek must therefore be adapted to have a good time resolution so that we could capture abrupt shifts in autonomic balance. For this reason, we selected a sliding window with overlap, such that a time resolution of 1 s was achieved. Specifically, the SDNN feature (Table 2) was extracted from 0 to 30 s and assigned to time bin 15, and from 1 to 31 s and assigned to time bin 16, etc. Table 2 shows features extracted from the ECG, RR tachogram, and hypnogram, along with window lengths that have been used to compute them. References to original work(s) where they were used for sleep-related detection tasks are provided. All features were interpolated to match the 1 s time bins used for classification.
Table 2.
Arousal detection features
| Type | Feature | Number |
|---|---|---|
| Time domain | , 18 SDNN, 18 SDSD, 18 RMSSD 18 | 1–4 |
| Range(RR), 18,19 MAD(RR) 18,19 | 5–6 | |
| 18,19 | 7–11 | |
| MSLDshort(RR), MSLDlong(RR), LR, 10 LRback, 10 | 12–15 | |
| Frequency domain | HF, 18 LF, 18 VLF, 18 TP, 18 all from CTW | 16–19 |
| Hjorth parameters | Activity, 20 mobility, 20 complexity20 | 20–22 |
| Sleep stage | p(wake), p(NREM), p(REM) | 23–25 |
Features used for arousal detection. Features 1–11 were computed for 30 s windows. Features 12–13 were computed for local windows of length 5 s and 15 s and global windows of length 30 s and 180 s, respectively. Features 14–15 were computed on a beat-to-beat basis. Features 16–25 were calculated in 1 s bins.
RR = RR interval; = mean; SDNN = standard deviation of RR; SDSD = standard deviation of RR differences; RMSSD = root mean square of successive RR differences; MAD = mean absolute difference; MSLD = median signed local difference; LR = likelihood ratios; HF = high frequency; LF = low frequency; VLF = very low frequency; TP = total power; CTW = continuous wavelet transformation; NREM = non-rapid eye movement; REM = rapid eye movement.
CWT features
Spectral features of the RR tachogram approximate the balance of ANS [18]. Inclusion of these features was important, since arousals are associated with an activation of the SNS. Spectral features were extracted with the continuous wavelet transformation (CWT), using the morlet wavelet, since it has been shown to map changes in ANS well [19]. Ten levels of decomposition using two number of voices per octave created a satisfactory frequency resolution to fit usual frequency bands used for HRV analysis, which is very low frequency (VLF): 0.0033–0.04 Hz, low frequency (LF): 0.04–0.15 Hz, high frequency: 0.15–0.4 Hz, and total power (TP): 0.0033–0.4 Hz [18].
Hjorth parameters
The definition of arousals clearly states that during rapid eye movement (REM) sleep, arousal scoring must be accompanied by muscle activation [2]. Muscle activations may introduce movement artifacts, which inflict and corrupt recorded signals during PSG. This apparent movement artifact can, however, also be utilized as a descriptive feature. In this study, Hjorth parameters were introduced to extract artifactual movements from the ECG signal. Hjorth parameters are a set of nonlinear parameters that describe a different degree of signal complexity: activity is the variance of the signal; mobility can be interpreted as the mean frequency; and complexity is the change of mean frequency [20].
Sleep stages
Sleep stages were annotated in 30 s epochs and were translated into features by the one-hot representation. This representation has been widely used in machine learning and works by translating a categorical class into a set of numerical parameters by assigning 1 for the present category and 0 for all other categories [21]. The AASM definition clearly distinguishes arousals by sleep stages, which makes sleep stages a natural choice to include as a feature [2]. In recent years, HRV features have shown to be useful for the classification of sleep stages [22, 23]. We elected to collect all non-rapid eye movement (NREM) stages in one feature, firstly because most HRV feature-based sleep stage classification algorithms are not sufficiently performing to distinguish the different NREM sleep stages, and secondly because the arousal definition provided by the AASM does not distinguish arousals emerging from different NREM sleep stages.
Time domain
Time domain features serve to give contextual information about variability and dynamics of the heart rate [18]. Apart from traditional time domain features (Number 1–11, Table 2), four additional features were extracted (Number 12–15, Table 2), the mean average deviation (MAD), and a novel feature developed and assigned with the name median signed local deviation (MSLD), characterized by
where is the median operator, index descriptions of RRsmall and RRlarge indicate window size used to extract RR intervals, and RR intervals from a smaller window are compared with a larger window (window sizes are presented in Table 2). These features give a signed estimation of local deviations from the median, such as the tachycardia-bradycardia seen during AA, while keeping the sign to allow heart rate increases and decreases to be distinguished.
Likelihood ratios (LR) were developed to detect tachycardia associated with AA. Two versions of the LR feature were extracted, one as calculated in [10], and the other implemented to work in the opposite direction of the RR time series. The latter allowed for a detection of bradycardia often following the tachycardia associated with an arousal.
Transformation and Normalization
Relative changes rather than absolute changes must be calculated within participants, as one participant might have a naturally higher baseline heart rate than another. The logarithmic operator can be used to transform the features from absolute to relative values,
Normalization of features is important to develop generic detection algorithms that can work on big data set with participants having different physiological states and various medical conditions. A min–max normalization was deemed an inappropriate choice, since it is very sensitive to noise. Thus, soft normalization was performed for each participant, using 0.1 and 0.9 quantiles,
All features but sleep stage features were log transformed and normalized.
Classification
Neural networks are powerful machine learning tools that can learn high dimensional patterns using nonlinear transformation. A feed forward neural network (FFNN) was considered for the classification task of detecting AA. A nonrecurrent neural network was considered sufficient, since temporal information was already incorporated in the features.
Architecture
For each time bin, the input was a vector of 25 features. This input was fed to a single hidden layer with bias and tanh activation function, which served as the active part for the nonlinear transformation. An output layer served to assign a posterior probability to each of two classes, given by the softmax activation function. The hidden layer contains a number of hidden units (HU) that each allows for a nonlinear transformation of the data. Naturally, more HU will lead to a more flexible model. Three different models were trained and evaluated with N = [50, 200, 500] HU. Cross entropy is useful for classification problems and was used as loss function, since it gives penalty that increases exponentially the further away the output probability is from the target class. Weights were optimized using the scaled conjugated gradient, which is a fast, automatic back-propagation solution that avoids user-dependent settings [24].
Participants and regularization
The 258 participants from the WSC (Table 1) were randomly divided into a training set (181 participants, 70%) and test set (77 participants, 30%). Nonoverlapping segments of 10 min of nonwake periods were extracted from each participant. These were shuffled between participants and collected into minibatches of size 10 in both the training and test set. To ensure model generalization and to avoid over-fitting to the training set, data regularization was introduced by batch normalization and by using an early stopping criterion, where the training was stopped if the test error did not improve through 20 iterations. Furthermore, generalization was strengthened by the large amount of data in this study.
Targets
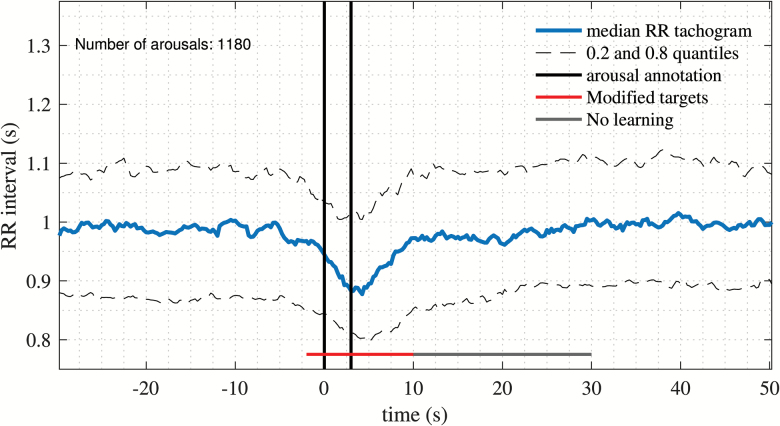
Targets must include part of the signal of interest to detect. Autonomic activations have been found to begin prior to and to last longer than, associated CA, with duration of autonomic activation varying with CA duration [9]. All annotations provided by specialized medical personal were of the arbitrary minimum 3 s duration in concordance with the AASM [2] and were localized at the beginning of a CA. To capture autonomic activations associated with CA, arousal annotations from 12 randomly selected participants were extracted, comprising 1180 arousals. The median of all events along with the 0.2 and 0.8 quantiles was calculated and is presented in Figure 3. From this figure, it is possible to extract information on median heart rate responses during CA. Clearly, this response lasts longer in the RR tachogram than what the annotation captures. By visual inspection and assuming that the annotations are localized at the beginning of an arousal, targets were designed to begin 2 s prior to the beginning of the annotation and to end 10 s after the annotation stopped (Figure 3, red horizontal line). Furthermore, 20 s following an arousal were not considered in the loss function (Figure 3, grey horizontal line). This can be justified, since some AA last longer than this fixed target size. There is no benefit from penalizing the model from such events, if they are in fact genuine autonomic activations.
Figure 3.
RR tachogram at time-locked arousals. Median, 0.2 and 0.8 quantiles of RR tachogram time-locked to annotated arousals from 12 randomly selected participants, comprising 1180 arousal events. The targets used for the loss function are indicated with a red line and has a 15 s duration, followed by a 20 s window not included to update the loss function in order to prevent overpenalization of the model.
Postprocessing
The output from the neural network is the probability of an arousal occurring at each time bin, given by
Arousals occur in events of various lengths. A threshold was fixed to segment the output into a binary vector of events,
The threshold will be determined based on the performance metrics discussed in a later section.
Arousal length
Different minimum arousal lengths are considered to control the minimum arousal intensity required to be considered an AA.
Removal of stable wake periods
Arousal events are transitions from sleep towards wakefulness; thus, periods of stable wake are not of interest. Wake periods lasting more than 1 min were therefore partly removed by retaining the first 30 s and removing the rest. This was done to enable the capturing of arousals in the transition towards wake. In this study, wake periods were removed by using hypnogram; a step that could easily be replaced later by an automatic sleep stage classifier.
Validation
To evaluate the performance of a time series event detection algorithm, it only makes sense to focus on events that have been annotated or detected by the model, and not areas where no such events are present. To validate the performance, precision (P+) and sensitivity (Se) were considered. Se describes the fraction of annotated arousals that have been detected, whereas P+ describes the fraction of detected arousals which are indeed annotated.
Both Se and P+ are important performance metrics. The F-score combines these and is given by
where β can be chosen to put more emphasis on either Se or P+. There exists no formula for the optimal choice of β; it should be chosen based on the application of the model. Some false positives (FP) are expected, since prior knowledge indicates that AA may occur without concomitant cortical activation; thus, β was chosen to emphasize Se, thereby reducing influence of FP. A value of was chosen to evaluate the performance of the model. All manually scored arousals are arbitrarily annotated to last the minimum 3 s, required in the AASM scoring guidelines [2]. The autonomic response to CA might not be at the exact same location as the annotation. A window starting 2 s before the beginning of an annotation and 10 s after the end was considered in the evaluation process to search for an autonomic response corresponding to the modified target presented in Figure 3.
Results
Network
The log-likelihood of three different FFNN models with 50, 200, and 500 HU performed with test error 0.0871, 0.0866, and 0.0867, respectively. It seemed appropriate to select the model containing 200 HU, since a less complex model (50 HU) scored a higher test error, indicating that the model was not flexible enough, whereas the more complex model (500 HU) scored a similar test error, suggesting no further improvement in performance.
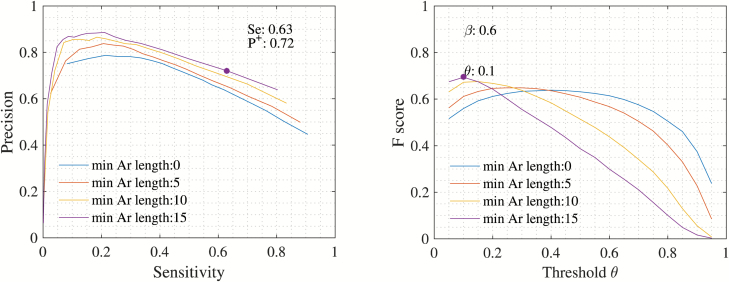
Optimal postprocessing parameters
The F-score was used to determine an optimal threshold and an optimal arousal duration. Figure 4 (right) shows the F-score for different threshold and minimum arousal duration. The highest F-score of 0.70 was achieved with a threshold of and minimum arousal length of 15 s.
Figure 4.
Performance metrics. Performance metrics displayed for different minimum arousal lengths (in seconds) indicated by the different colors. Left: Precision, P+, vs. Sensitivity, Se. Right: F-score and threshold, θ. The threshold for the final model is chosen from these performance metrics and is indicated with a dot. As shown, a threshold of θ = 0.1 and minimum arousal length of 15 s is chosen.
Performance
Using optimal postprocessing parameters, performance metrics resulted in P+ = 0.72 and Se = 0.63 (Figure 4, left). From Figure 4, it is clear that P+ improved significantly when removing short duration arousals, indicating that increasing minimum arousal length removes more FP than true positives (TP).
Figure 5 shows a scatterplot of performance metrics for every participant used in the test set. It is noted that most points are gathered in the upper right quadrant. An observation is that no participants have 100% Se or P+. Assuming a perfect model where all AA are identified, the former case (Se) suggests that no participant has a perfect correlation between CA and AA, whereas the latter case (P+) indicates that some AA occur without cortical activations.
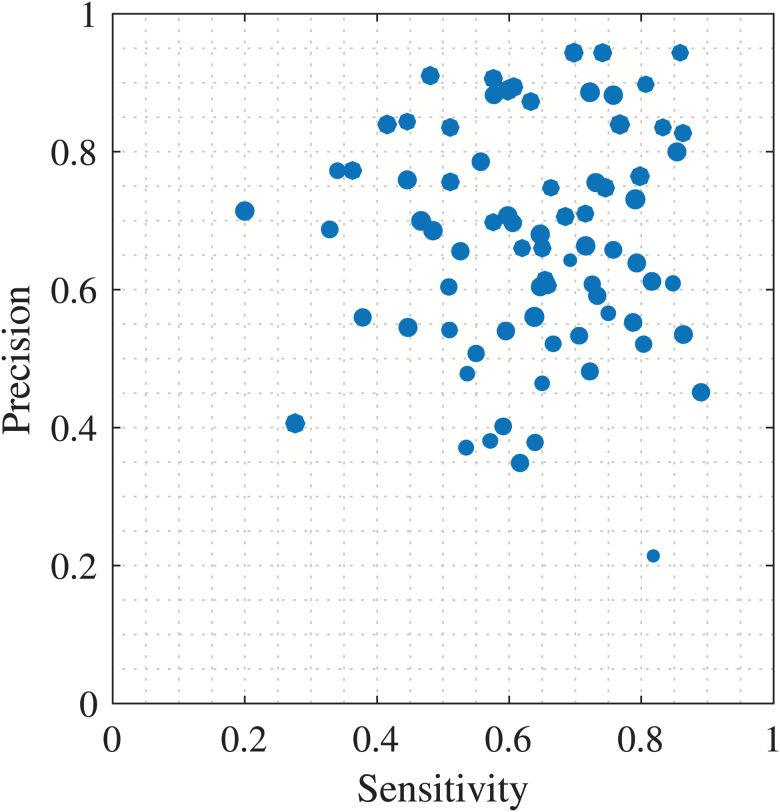
Figure 5.
Performance by participant. Scatterplot of performance metrics shown for every participant used in the test set (77 participants). Sizes of scatter-dots are relative to the number of arousals present per participant.
Sleep stages
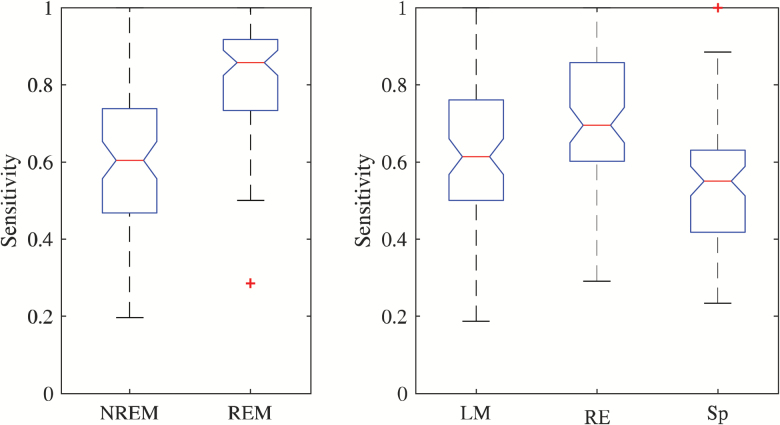
Boxplots of Se with respect to arousal sleep stage are shown in Figure 6 (left). The model performs very well in REM sleep compared with NREM. This suggests that autonomic activations in REM sleep introduce larger heart rate changes than in other stages.
Figure 6.
Arousal subtype. Left: Sensitivity of model with respect to sleep stage. Right: Sensitivity with respect to arousal subtype, i.e. leg movement (LM), respiratory event (RE), and spontaneous (Sp). The red line is the median and the edge of the boxes represents the 25 and 75 percentiles. Whiskers indicate nonoutlier extremes. Red crosses are outliers.
Arousal type
Boxplots of Se with respect to arousal subtype are shown in Figure 6 (right). Clearly, arousals in response to a leg movement or respiratory event are better detected in comparison to spontaneous events, a result that might be explained by the fact that respiratory events and leg movement events are already associated with tachycardia independent of CA [25, 26]. In this case, these events might lead to larger impacts on the heart rate compared with spontaneous arousals.
False positive
It was of interest to explore the 28% FP that were present with the chosen post-processing parameters. Of these, 38% fell into a leg movement event, and 43% fell into a respiratory event. This confirms that autonomic activations occur without concomitant cortical stimulation.
Discussion
This study presents an algorithm that, when provided with raw PSG data and manually or automatically scored sleep stage data, allows for automatic detection of AA by adapting the approach of modeling and detecting autonomic behavior during CA, and choosing the post-processing parameters such that the influence of FP events is reduced, thereby providing for a multivariate definition of AA. The algorithm was trained and tested on recordings from the WSC and included participants suffering from a variety of sleep and cardiac disorders. The detection algorithm includes a module for automatic extraction of an artefact- and ectopic-beat-free RR tachogram. Ectopic beats were detected by development of a model trained and tested on ECG recordings from the MITDB. Using CA as gold-standard, arousals were detected with P+ = 0.72 and Se = 0.63. The postprocessing parameters were chosen to put more emphasis on Se, thereby reducing influence of FP.
P + = 0.72 shows that 28% of AA occur without sufficient cohesive CA. It is well-known that both leg movement and respiratory events introduce tachycardia that appears with and without concomitant cortical responses [25, 26]. In this study, we found that 38% of FP appeared in relation to leg movement events, and 43% appeared in relation to respiratory events. This indicates that most (81%) FP are likely genuine AA triggered without associated CA, although it may also reflect the fact that the current definition of arousals does not capture the full spectrum of EEG disturbances that may occur (e.g. duration and change of frequency). Alternatively, it may be that the reticular activation systems present dynamic monitoring-activations throughout the night in response to physiologic disruptions to sleep continuity, which do not necessarily involve a full-blown CA. This is fundamentally important to the assessment of sleep disorders by PSG, suggesting that manually scored, EEG-limited arousal scoring may not fully capture the adverse impact of sleep disorders.
Se = 0.63 shows that 37% of CA do not have sufficient cohesive AA. This is clearly a consequence of the chosen postprocessing parameters, as a Se = 0.9 can be achieved, just by changing the minimum duration limit to 0 s and (Figure 4). This also confirms the correlation between AA and CA intensity, i.e. duration [9]. However, choosing these parameters will consequently increase the amount of FP, achieving P+ = 0.45. Conveniently, as explained above, most FP appear in cohesion to a respiratory or leg movement event; hence, they are likely genuine autonomic activations. The high proportion of CA that did not have an associated AA points to a potentially distinct physiologic phenomenonology, though, as mentioned, a more liberal parameter threshold results in much higher correlation, at the risk of increased FP. Toward this end, an exploration of clinically validating the overlapping CA and AA, as well as those that did not have cohesion, will further our understanding of the interaction between these phenomena. Furthermore, this may highlight that the arbitrarily chosen CA threshold of 3 s of EEG frequency changes, which was meant to ensure sufficient inter-rater reliability in scoring, may not prove to be optimal for defining a CA.
From a clinical standpoint, these findings are highly relevant. The finding that arousals of various types correlate significantly with clinical outcomes, most notably daytime sleepiness [11], suggests a need for better methods of arousal detection. Furthermore, the differential impact evidenced by objective worsening of daytime sleepiness induced by isolated AA points to a potentially hidden pathophysiology of sleep disorders [5]. Additionally, the pathophysiologic consequences of the SNS surges associated with sleep disruptions bear a clear connection to the cardiovascular morbidity and mortality of common sleep disorders, such as obstructive sleep apnea [11] and periodic limb movements of sleep [11]. In fact, recent evidence from studies targeting subphenotypes of obstructive sleep apnea has indicated that comorbid medical issues (including hypertension, diabetes, and cardiovascular disease) are more probable in otherwise asymptomatic patients [27], highlighting a role for more robust PSG analyses in the identification of clinically relevant sleep perturbations such as AA.
Basner et al. proposed a similar system, reporting a Se = 0.68, P+ = 0.64, and specificity, Sp = 0.95 [10]. Reporting Sp in arousal detection results in model interpretation difficulties, due to a large class imbalance between event (Arousal) and no event (No arousal). Basner et al. solved the imbalance problem by randomly selecting control arousals such that they had no overlap with arousal scorings, wake epochs, and signal loss (including a safety margin of 60 s), and so the ratio between actual arousals and control arousals was 0.5. However, this design choice biases the model towards areas with stable sleep conditions and neglects to address areas with natural variability. In general, the design choices are very different for the two systems, which makes it difficult to compare. Basner et al. tested their algorithm using only 56 participants and used external stimuli to provoke arousals, which makes it uncertain how well it models naturally occurring arousals. They tested their model using healthy participants, not suffering from heart diseases or sleep disorders. On the other hand, they reported performance by not excluding wake epochs. This contrasts with the proposed algorithm that was tested using the WSC, a population-based study known to include participants having various sleep disorders and arrhythmias [15]. Furthermore, it was tested in a clinical environment on naturally occurring arousals, but FP during wake epochs were removed. Overall, Basner et al. present a simple system, which is easy to reproduce, and only needs 2 min of manual editing for removing signal loss. The presented algorithm is automatic, but more complex, as model flexibility was prioritized to capture the diversity of AA in a clinical study. One could argue that this work should at least be as good as the system presented by Basner et al., since their algorithm was included as input feature. Ultimately, better performance is reported by tuning the postprocessing parameters presented in Figure 4, e.g. a Se = 0.8, P+ = 0.64.
The study of Pillar et al. and Pillar et al. of Refs. 12 and 13 introduced a rule-based method to detect AA using PAT recordings [12, 13]. A correlation of 0.82 and 0.87, respectively, was achieved between such events and CA. No information about P+ was provided, which makes it difficult to compare with our own performance metrics. It is noted that their method relies on PAT recordings, which are not routinely performed, thereby limiting its applications.
All previous detection algorithms rely on rule-based classification systems [10, 12], which are limited by their static design. On the other hand, supervised machine learning models, e.g. neural networks, have the capabilities to learn the complex patterns of human scorers in a clinical setting. Recent years have shown that machine learning approaches used in sleep medicine can provide reliable classification in other areas such as sleep stage classification [22, 23]. In this study, we used a FFNN that was trained to detect AA. A FFNN treats every input as new and has no memory of the context in the time series. To compensate for this, temporal information was incorporated into the features. Alternatively, a recurrent neural network could have been implemented, providing the temporal context through the network itself.
Limitations
Feature selection in this model was unlikely affected by individual-specific or infrequent autonomic phenomena (e.g. Traub–Hering–Mayer waves) or low-intensity, naturally occurring autonomic fluctuations (e.g. respiratory sinus arrhythmia), due to incorporation of both time- and frequency-domain features of HRV into our model parameters. If any of these phenomena significantly contributed to autonomic arousals across individuals and events, they could have been selected as influential by the algorithm.
The performance of the algorithm is calculated using the hypnogram as input features so that periods with stable wake could be removed. In recent years, HRV features have proven useful for sleep stage classification [22, 23], and it would be of interest to include such a classifier as opposed to manual annotations in later implementations, potentially allowing for a more physiologic staging system than the classically constrained 30 s epoch. Although the algorithm is automatic and work without manual removal of artefacts and common ectopic beats and arrhythmias in the ECG signal, it cannot be used if only the ECG signal is available, since the outcome of the algorithm still depends on visual scoring of the EEG. However, with the abundance of automated, sleep-staging algorithms that are being developed (including one from our lab), this process is likely to be part of an automated analysis pipeline.
Analysis of HRV is affected by the presence of ectopic beats and arrhythmias. A study on the clinical utility of HRV stresses that analysis of HRV requires normal sinus rhythm and reasonable signal quality, and that AF and ectopic complexes preclude its use [16]. In this study, AF and ectopic beats were accounted for by using models trained on recordings from participants with different physiological conditions than participants from the WSC. There is a need for scoring the heart beats and rhythms in the recordings from the WSC to optimally identify sinus rhythm.
As with other physiologic consequences of sleep disturbances (e.g. CA and desaturations), the AA themselves were not able to differentiate between distinct types of sleep disturbances (e.g. respiratory events, leg movements, CA, and spontaneous). Further work would be needed to analyze if AA occurring spontaneously, or in association with CA, leg movements, or respiratory events have differential implications for sleep recuperation and long-term health.
The proposed detection algorithm was evaluated in relation to CA annotations as there is no gold standard for AA [2]. Figure 3 shows that most CA lead to autonomic activations, justifying the use of CA as a gold standard. Still, the few CA that did not have cohesive AA, as well as the AA that occur without cohesive CA will cause incorrect penalization of the model. Moreover, the postprocessing parameters, i.e. threshold of and minimum arousal length of 15 s, were chosen based on optimizing the F-score, consequently biasing the model towards longer CA and longer AA without cohesive CA. Ultimately, tuning these model parameters should not be based on presence of CA but rather on an outcome measure such as sleepiness, daytime function, increased blood pressure, and cardiovascular morbidity and mortality. Ultimately, the manifestation, i.e. the duration and intensity, of AA should be based on their physiological impacts for them to be considered as isolated events of importance. Furthermore, to uncover the complete autonomic response during AA different biosignals that have shown to capture autonomic activity should be considered in the analysis, such as, but not limited to PAT [12, 13], pulse plethysmography, pulse transit time, and electrodermal activity [1].
In conclusion, the presented algorithm shows good performance when considering that more than 80% of the FP found by the detection algorithm appeared in relation to either leg movement events or respiratory events, indicating that most FP constitute autonomic activations that are indistinguishable from those with cortical cohesion. The proposed algorithm provides an automatic system trained in a clinically relevant environment, which can be utilized to analyze the systemic and clinical impacts of arousals.
Supplementary Material
Supplementary material is available at SLEEP online.
Funding
This work was supported by gifts to the Stanford Sleep Center, grants from the Lundbeck Foundation, Technical University of Denmark, and Danish Center for Sleep Medicine. L.S. and J.C. were supported by NIH T32 HL110952. P.E.P. was supported by R01 HL062252 and UL1 RR025011. Instrumentation provided by grants S10 RR021086 and S10 OD010569. The funding institutes played no role in the design and conduct of the study; no role in the collection, management, analysis, or interpretation of data; and no role in the preparation, review, or approval of the manuscript.
Conflict of interest statement. M.O. has nothing to disclose. L.D.S. reports grants from National Institutes of Health (NIH), during the conduct of the study. J.C. has nothing to disclose. P.E.P. reports grants from NIH, during the conduct of the study; personal fees from ResMed, outside the submitted work. P.J.J. has nothing to disclose. E.M. reports grants from NIH, during the conduct of the study; grants and personal fees from Jazz Pharmaceutical, personal fees from Actelion, grants from The Lundbeck Foundation, other from ALPCO, other from Federal Trade Commission, grants from GSK, other from Novo Nordisk, grants from Sunovion, grants from Merck, other from Resmed, other from INC, other from Google/Verily, other from Balance, other from Airweave, other from Flamel, outside the submitted work. H.B.D.S. has nothing to disclose.
Work Performed: Stanford Center for Sleep Sciences and Medicine, Stanford University, Palo Alto, CA and Department of Electrical Engineering, Technical University of Denmark, Kongens Lyngby, Denmark
References
- 1. Halász P, et al. The nature of arousal in sleep. J Sleep Res. 2004; 13(1): 1–23. [DOI] [PubMed] [Google Scholar]
- 2. Berry RB, et al. The American Academy of Sleep Medicine. The AASM Manual for the Scoring of Sleep and Associated Events: Rules, terminology and Technical Specifications, Version 2.3. wwww.aasmnet.org. Darien, Illinois: American Academy of Sleep Medicine; 2016. [Google Scholar]
- 3. Pitson D, et al. Changes in pulse transit time and pulse rate as markers of arousal from sleep in normal subjects. Clin Sci (Lond). 1994; 87(2): 269–273. [DOI] [PubMed] [Google Scholar]
- 4. Catcheside PG, et al. Noninvasive cardiovascular markers of acoustically induced arousal from non-rapid-eye-movement sleep. Sleep. 2002; 25(7): 797–804. [DOI] [PubMed] [Google Scholar]
- 5. Martin SE, et al. The effect of nonvisible sleep fragmentation on daytime function. Am J Respir Crit Care Med. 1997; 155(5): 1596–1601. [DOI] [PubMed] [Google Scholar]
- 6. Lofaso F, et al. Arterial blood pressure response to transient arousals from NREM sleep in nonapneic snorers with sleep fragmentation. Chest. 1998; 113(4): 985–991. [DOI] [PubMed] [Google Scholar]
- 7. Rees K, et al. Arousal responses from apneic events during non-rapid-eye-movement sleep. Am J Respir Crit Care Med. 1995; 152(3): 1016–1021. [DOI] [PubMed] [Google Scholar]
- 8. Lombardi C, et al. Autonomic arousals in sleep related breathing disorders: a link between daytime somnolence and hypertension?Sleep. 2009; 32(7): 843–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Azarbarzin A, et al. Relationship between arousal intensity and heart rate response to arousal. Sleep. 2014; 37(4): 645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Basner M, et al. An ECG-based algorithm for the automatic identification of autonomic activations associated with cortical arousal. Sleep. 2007; 30(10): 1349–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bonnet MH, et al. The scoring of arousal in sleep: reliability, validity, and alternatives. J Clin Sleep Med. 2007; 3(2): 133–145. [PubMed] [Google Scholar]
- 12. Pillar G, et al. Autonomic arousal index: an automated detection based on peripheral arterial tonometry. Sleep. 2002; 25(5): 543–549. [PubMed] [Google Scholar]
- 13. Pillar G, et al. An automatic ambulatory device for detection of AASM defined arousals from sleep: the WP100. Sleep Med. 2003; 4(3): 207–212. [DOI] [PubMed] [Google Scholar]
- 14. Goldberger AL, et al. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation. 2000; 101(23): E215–E220. [DOI] [PubMed] [Google Scholar]
- 15. The Wisconsin Sleep Cohort [Online]. Available from: //pophealth.wisc.edu/Research/WSC. Accessed January, 22 2018.
- 16. Kleiger RE, et al. Heart rate variability: measurement and clinical utility. Ann Noninvasive Electrocardiol. 2005; 10(1): 88–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Petrėnas A, et al. Low-complexity detection of atrial fibrillation in continuous long-term monitoring. Comput Biol Med. 2015; 65: 184–191. [DOI] [PubMed] [Google Scholar]
- 18. Task Force of The European Society of Cardiology and The North American Society of Pacing and Electrophysiology. Guidelines Heart rate variability, Standards of measurement, physiological interpretation, and clinical use. Eur Heart J. 1996; 17: 354–381. [PubMed] [Google Scholar]
- 19. Neto OP, et al. Morlet wavelet transforms of heart rate variability for autonomic nervous system activity. Appl Comput Harmon Anal. 2016; 40(1): 200–206. [Google Scholar]
- 20. Hjort B. EEG analysis based on time domain properties. Electroenceph Clin Neurophysiol. 1970; 29: 306–310. [DOI] [PubMed] [Google Scholar]
- 21. Harris D, et al. Digital Design and Computer Architecture. Waltham, MA: Elsevier; 2012. [Google Scholar]
- 22. Yilmaz B, et al. Sleep stage and obstructive apneaic epoch classification using single-lead ECG. Biomed Eng Online. 2010; 9: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mendez MO, et al. Sleep staging from heart rate variability: time-varying spectral features and hidden Markov models. Int J Biomed Eng Technol. 2010; 3(3): 246–263. [Google Scholar]
- 24. Møller MF. A scaled conjugate gradient algorithm for fast supervised learning. Neural networks 1993; 6: 525–533. [Google Scholar]
- 25. Yang CK, et al. Heart rate response to respiratory events with or without leg movements. Sleep. 2006; 29(4): 553–556. [DOI] [PubMed] [Google Scholar]
- 26. Winkelman JW. The evoked heart rate response to periodic leg movements of sleep. Sleep. 1999; 22(5): 575–580. [DOI] [PubMed] [Google Scholar]
- 27. Ye L, et al. The different clinical faces of obstructive sleep apnoea: a cluster analysis. Eur Respir J. 2014; 44(6): 1600–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.