Abstract
The combination of yeast surface display and flow cytometric analyses and selections is being used with increasing frequency to alter specificity of macromolecular recognition, including both protein–protein and protein–nucleic acid interactions. Here we describe the use of yeast surface display and cleavage-dependent flow cytometric assays to increase the specificity of an engineered meganuclease. The re-engineered meganuclease displays a significantly tightened specificity profile, while binding its cognate target site with a slightly lower, but still sub-nanomolar affinity. When incorporated into otherwise identical megaTAL protein scaffolds, these two nucleases display significantly different activity and toxicity profiles in cellulo. The structural basis for reprogrammed DNA cleavage specificity was further examined via high-resolution X-ray crystal structures of both enzymes. This analysis illustrated the altered protein–DNA contacts produced by mutagenesis and selection, that resulted both in altered readout of those based and a necessary reduction in DNA binding affinity that were necessary to improve specificity across the target site. The results of this study provide an illustrative example of the potential (and the challenges) associated with the use of surface display and flow cytometry for the retargeting and optimization of enzymes that act on nucleic acid substrates in a sequence-specific manner.
Keywords: crystal structure, genome engineering, meganuclease, protein DNA recognition, protein engineering
Introduction
Engineering and redesign of the recognition specificity of a DNA-binding protein is a very challenging area of research and development (Romano Ibarra et al., 2016). Whereas considerable progress has been made in engineering protein–protein recognition (Procko et al., 2014; Hsia et al., 2016), engineering of protein–nucleic acid recognition remains difficult (Thyme et al., 2016). This disparity is partially attributable to the differing composition of these two types of molecular interfaces, with the latter involving a large number of buried hydrogen bonds, solvent molecules and counterions, and highly distortable nucleotide base pairs that are challenging to model and compute.
MegaTALs are single-chain (i.e. monomeric) DNA-binding proteins that are used (in a manner similar to TAL effector nucleases, zinc-finger nucleases and CRISPR/Cas9 nucleases) for targeted genome modification. MegaTALs are comprised of the fusion of an N-terminal TAL DNA-binding domain and a C-terminal meganuclease (Boissel et al., 2013; Boissel and Scharenberg, 2015). Both components contribute to specificity and activity: the TAL effector region via sequence-specific binding at the 5′ end of the DNA target site, and the meganuclease via additional sequence-specific DNA binding and cleavage across 22 additional base pairs at the 3′ end of the target site. MegaTALs often display elevated gene modification activity relative to the isolated meganuclease (Takeuchi et al., 2011; Boissel et al., 2013; Wang et al., 2014; Romano Ibarra et al., 2016). Optimal specificity is dependent on an appropriate balance of relative DNA binding affinities by the two components of the protein, such that cleavage requires the simultaneous engagement of the TAL effector and the meganucleases on the two ends of the DNA target (Boissel et al., 2013). Properly engineered and optimized megaTALs can be extremely efficient gene targeting systems in primary human T-cells (Boissel et al., 2013; Sather et al., 2015; Romano Ibarra et al., 2016)
The creation of a gene-targeting megaTAL requires reprogramming of the DNA binding and cleavage specificity of its corresponding meganuclease. This process is challenging to accomplish due to the extensive, non-modular nature of the protein–DNA interface (Stoddard, 2014). The engineering of a meganuclease to create an alternative DNA cleavage specificity profile is accompanied by multiple technical requirements: (i) generation and efficient screening of large combinatorial protein libraries harboring randomized amino acid substitutions at many simultaneous positions, (ii) sampling as large of a fraction of these protein constructs as possible, (iii) filtering out destabilized and non-functional constructs at each stage of the engineering and selection process and (iv) carrying out redesign in a sequential, iterative manner across the molecular interface, resulting in the assembly of active redesigned protein constructs.
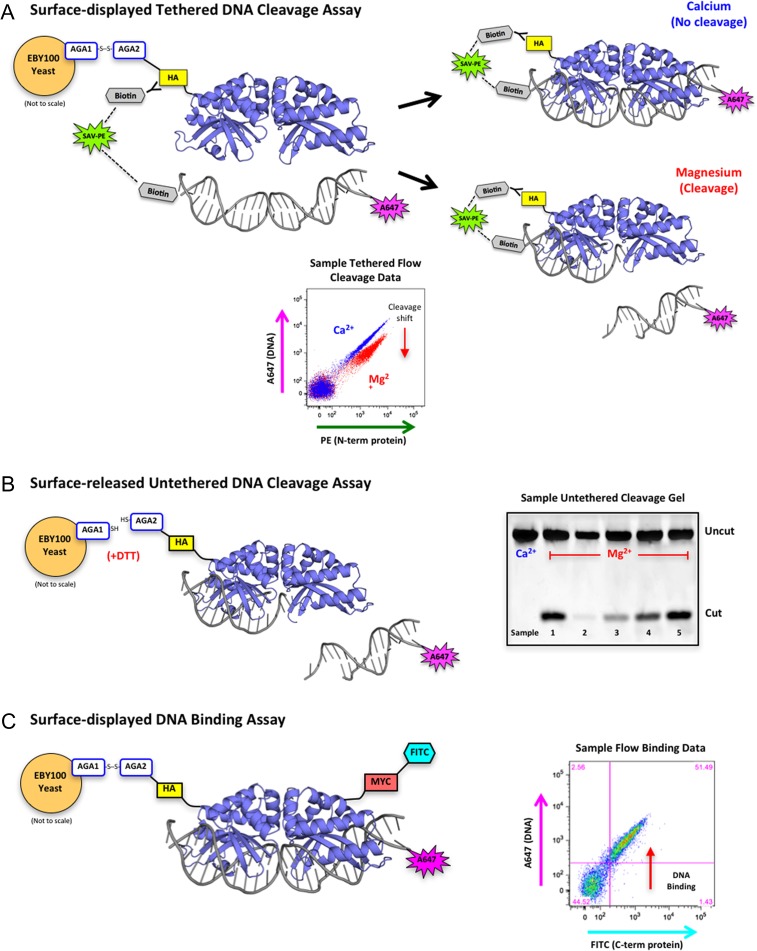
Several methods have been described for redesign of DNA recognition by meganucleases, including in cellulo activity screens allowing visible readout of nuclease-induced cleavage or recombination activity (Arnould et al., 2006; Doyon et al., 2006; Chen et al., 2009), purely in vitro translation and activity screens where formation of cleaved DNA products is directly coupled to the nuclease reading frame (Takeuchi et al., 2014), and flow-cytometric readout of DNA target cleavage by enzymes displayed on yeast surface (Baxter et al., 2013). Each method has its own particular advantages; none are absolutely ideal. The use of yeast surface display combined with flow-cytometric sorting for selection of DNA cleavage activity has several useful features: it combines ease of library construction and cloning, moderately high throughput screening, and reliable filtering and removal of significantly destabilized constructs at each stage of selection (Fig. 1).
Fig. 1.
Overview of yeast surface display and flow cytometric assays. (A) Schematic of the tethered flow cytometric cleavage assay. A meganuclease is expressed on the surface of yeast with an N-terminal HA epitope tag. The HA tag is stained with a biotinylated anti-HA antibody and used to create a physical tether between the protein and a fluorescently labeled DNA target substrate via a fluorescent streptavidin-phycoerythrin (SAV-PE) bridge. In the presence of calcium, the DNA is able to bind but not cleave (blue diagonal population on the sample flow data plot). In the presence of magnesium, the DNA may be cleaved (red population in sample flow plot), which releases the fluorescent tag and produces a drop in A647 signal (cleavage shift designated by the red arrow). The ratio of A647 signal in the calcium vs. magnesium samples is used to quantitate cleavage activity against a given DNA target substrate. (B) Schematic of the surface-released untethered cleavage assay. The meganuclease is released from the surface of the yeast with dithiothreitol (DTT) and the A647-labeled DNA substrate is free-floating in solution with no tethering. This assay requires the enzyme to both bind and cleave the DNA substrate before cleavage can occur. The cleaved products are separated on an acrylamide gel and visualized by the fluorescence of the A647 tag on the DNA substrate. The sample untethered cleavage gel displays cleaved products from five unique samples with varying levels of cleavage. (C) Schematic of the flow cytometric DNA binding assay. The meganuclease is expressed on the surface of yeast, stained with an anti-Myc-FITC antibody to detect full-length protein expression (the Myc epitope tag is located at the C-terminal end of the protein), and incubated with varying concentrations of untethered DNA target substrate. Cells with both FITC and A647 signal (see sample flow binding plot) are expressed on the yeast surface and have bound the DNA substrate
This article describes the use of surface display and flow cytometry to improve the DNA cleavage specificity of an engineered meganuclease and to create a megaTAL protein scaffold that targets a conserved sequence in the HIV pol gene (coding for the viral integrase enzyme). The ultimate goal of this work was to (i) test the ability of our engineering strategy to improve the specificity profile of an initial engineered meganuclease construct; (ii) explore the balance of DNA affinity and base-pair discrimination that results in improved specificity; and (iii) examine the structural basis for improved specificity during iterative rounds of protein engineering.
Materials and Methods
Nomenclature
Per published standards established for restriction and homing endonucleases (Roberts et al., 2003), the engineered meganucleases described in this article are officially named as ‘I-OnuI-e-vHIVInt_vX’. ‘I-OnuI’ refers to the parental wild-type meganuclease and ‘e-vHIVInt’ indicates that the enzyme is an engineered variant created to cleave a viral sequence in the HIV integrase reading frame. ‘vX’ refers to the version of the enzyme being described, ‘v1’ indicates the original engineered ‘version 1’ of the enzyme used in initial experiments, while ‘v2’ indicates the subsequent version of the same enzyme produced by additional rounds of selection and engineering. Those full names are provided in depositions of sequences, specificities and structures into both the REBASE nuclease database (Roberts et al., 2015) and into the RCSB structural database (Rose et al., 2017). However, in the text of this article, a shorter naming convention is used for convenience, corresponding to ‘eOnu_HIVInt_vX’. For megaTAL constructs harboring either of the engineered nuclease scaffolds, a suffix is added that indicates the number of repeats in the TAL effector DNA binding region (i.e. ‘eOnu_HIVInt_v1_7.5mT’ indicates a megaTAL constructed from a TALE harboring 7.5 DNA-matched TAL repeats tethered to ‘version 1’ of the engineered meganuclease.)
Overview
As illustrated in Fig. 1, the basis for reprogramming the DNA cleavage specificity of a meganuclease, described in this work, is the use of yeast surface display coupled with flow cytometry. This approach allows us to screen protein libraries for desired DNA cleavage activities and specificities, as well as to assay the activity of individual protein constructs at the end of each round of selections. High-throughput screening of enzyme libraries for cleavage activity (and subsequent analysis of the activity of individual enzymes) uses a tethered system (Fig. 1A) in which each individual enzyme construct is physically linked to its DNA target substrate (to ensure that the capture of yeast cells expressing active constructs is not confounded by ‘cross-talk’ between neighboring cells). Subsequent to the isolation and validation of individual active enzyme constructs, the system can then be used to assay DNA cleavage in trans (by liberating the enzyme from the yeast surface and conducting traditional untethered DNA cleavage assays; Fig. 1B) or to assay DNA binding affinity, by titrating and staining yeast cells harboring displayed enzymes constructs with increasing concentrations of untethered fluorescently labeled DNA targets; Fig. 1C.
Plasmids
Plasmids eOnu_HIVInt_v1 and eOnu_HIVInt_v1_7.5mT were obtained from Dr Sandrine Boissel and Dr Andrew Scharenberg (Seattle Children's Research Institute), and Dr Jordan Jarjour (Pregenen Inc.) and have been previously described in detail (Sedlak et al., 2016). Each plasmid, in the backbone of a lentiviral vector, expresses the BFP fluorescent reporter protein and an I-OnuI derived meganuclease re-engineered to recognize a 22-nucleotide sequence in the HIV pol gene. Plasmid eOnu_HIVInt_v1_7.5mT encodes a fusion of a TAL effector domain and a meganuclease, termed a megaTAL, as previously described (Boissel et al., 2013; Boissel and Scharenberg, 2015). This megaTAL contains 7.5 DNA-binding repeat domains fused to the meganuclease.
For yeast-based assays of endonuclease cleavage activity, the eOnu_HIVInt_v1 protein coding sequence was cloned into the pETCON yeast surface expression vector (Addgene #41 522). The pETCON vector incorporates an N-terminal hemagglutinin (HA) epitope tag and a C-terminal Myc tag to allow for fluorescent antibody staining in flow cytometric assays and cell sorting. A modified version of this vector, containing the I-OnuI protein scaffold, was also used in all of the yeast libraries.
The second re-engineered version of the eOnu_HIVInt_v1 meganuclease, termed eOnu_HIVInt_v2, was used as a stand-alone meganuclease in the pETCON vector in all of the yeast-based assays and then cloned into the same megaTAL scaffold vector as eOnu_HIVInt_v1, containing an identical 7.5 central repeat domains and termed eOnu_HIVInt_v2_7.5mT.
HIV plasmids pDHIV3 and pDHIV3-GFP were provided by Dr Vicente Planelles and have been previously described (Andersen et al., 2006).
Yeast transformation and protein surface display for DNA-binding and cleavage selections and assays
Constructs in the pETCON yeast surface display vector were transformed into frozen competent EBY100 Saccharomyces cerevisiae (Invitrogen) using the lithium acetate method (Gietz and Schiestl, 2007). Single yeast colonies were grown in selective culture media (SC) + 2% w/v glucose at 30°C with shaking overnight. For the induction of surface display, cells from the SC + glucose culture were transferred to SC + 2% raffinose + 0.1% glucose media at an initial density of 24 million cells/μL and grown at 30°C with shaking until reaching a density of 80–100 million/mL. Cells were then washed with water and transferred to SC + 2% galactose media at a density of 20 million/mL for overnight induction on the benchtop (room temperature with no shaking).
Labeled DNA target substrates for yeast binding and cleavage assays
Double-stranded DNA oligonucleotide target substrates for yeast flow cytometry and in vitro cleavage assays were prepared as previously described (Baxter et al., 2013, 2014). The 54-base pair double-stranded oligonucleotide substrates were generated by PCR using Platinum Taq High Fidelity DNA polymerase (Invitrogen) with biotin- and AlexaFluor647-labeled primers (5′-/5Biosg/TCAGCACAGCACTACG-3′ and 5′-/5Alex647N/TGGACACGACTTGAGC-3′, IDT) and the following single-stranded template: 5′-TGGACACGACTTGAGCAATGGCAGTATTCATCCACAATCGTAGTGCTGTGCTGA-3′ (IDT). Leftover contaminating single-stranded template and/or primers were removed with a 6-h Exonuclease I (NEB) digest at 37°C followed by purification on G-100 Sephadex (GE Healthcare) filter plate columns. The final labeled substrates were analyzed for purity on a 15% acrylamide gel.
Flow cytometric cleavage assays using surface-displayed endonucleases
Two million induced yeast cells per sample were washed with a ‘yeast staining buffer’ (YSB): 180 mM KCl, 10 mM NaCl, 0.2% bovine serum albumin (BSA), 0.1% galactose and 10 mM HEPES, pH 7.5. The cells were then stained in a 50 μL volume with 1:250 biotinylated anti-HA antibody (BioLegend, BIOT-101L) and a 1:100 anti-Myc-FITC antibody (ICL Labs, CMYC-45F) for 2 h at 4°C. Staining of the C-terminal Myc epitope tag confirms successful full-length surface expression of the desired protein. The fluorescently labeled DNA target substrates were pre-conjugated at 40 nM concentration with 5 nM streptavidin-phycoerythrin (SAV-PE) (BD-Biosciences, 554 061) in 50 μL volume with a higher-salt YSB buffer (additional 400 mM KCl). The DNA mixes were then incubated with the stained yeast cells to form the physical biotin-streptavidin tether between the DNA substrates and the N-termini of the surface-expressed proteins. High-salt YSB buffer served to discourage undesired binding of tethered DNA to enzymes expressed on neighboring cells. The cells with tethered DNA substrates were washed with an ‘oligo cleavage buffer’ (OCB): 250 mM KCl, 10 mM NaCl, 5 mM K-glutamate, 0.05% BSA and 10 mM HEPES, pH 7.4. Next, the cells were divided equally between two wells and incubated with 50 mM OCB + 5 mM CaCl2 (supporting DNA binding with no cleavage) or MgCl2 (supporting cleavage activity) at 37°C for 20 min. After transfer of the samples back to YSB, the cells were run on a BD LSRII cytometer (BD Biosciences), collecting signal from the FITC, PE and APC (for the A647 fluorophore) channels, and the data was analyzed with FloJo software (Tree Star, Inc.). See Supplementary Fig. S1a for a description of our analysis and quantification of the flow cleavage data and the Supplementary Data for samples of the raw flow cytometric cleavage plots. We performed at least two biological replicates of each flow cleavage experiment, using separately induced yeast cultures for each replicate. The replicates with the best surface expression and corresponding highest signal-to-noise were presented in the main figures.
‘One-off’ cleavage specificity profile determinations using surface-displayed endonucleases
A647-labeled DNA target substrates were generated with each position of the 22 bp HIVInt target sequence systematically substituted with each of the three alternate bases. This created a set of 66 unique ‘one-off’ target substrates. The tethered flow cytometric cleavage assay (described above) was performed in 96-well plate format with surface-expressed meganuclease for each of the 66 one-off DNA target substrates. The resulting quantified cleavage shift against each one-off target was arranged in bar-graph format relative to cleavage of the wildtype base at each position. See the Supplementary Data for samples of the raw flow cytometric plots used to generate the one-off profiles.
Flow cytometric DNA binding assays using surface-displayed endonucleases
The flow cytometric binding assay uses the same A647-labeled DNA substrates as the tethered flow cleavage assay, but the DNA remains free in solution with no tethering. The 50 000 yeast cells with surface-expressed HIV-Intv1 or HIV-Intv2 meganuclease were used per condition in a 96-well format. The cells were washed with OCB buffer supplemented with 5 mM CaCl2 to allow for binding of the DNA without cleavage. The cells were then stained for 2 h in 50 μL OCB + 5 mM CaCl2 with anti-Myc-FITC antibody (stains the C-term of the expressed protein) and DNA substrate ranging from 0 to 50 nM concentration (0, 1, 2.5, 5.0 , 7.5, 10, 25, 50, 75, 100, 250, 500 and 750 pM, 1, 1.25, 2.5, 5, 7.5, 10, 25 and 50 nM). Cells were then washed twice with OCB + CaCl2 and run on a BD Fortessa X50 cytometer (BD Biosciences), collecting signal from the FITC and APC (for the A647 fluorophore) channels. The resulting data were analyzed with FloJo software (Tree Star, Inc.). See Supplementary Fig. S1b for a description of our analysis and quantification of the flow binding data and the Supplementary Data for the raw flow cytometric binding plots.
Partially randomized meganuclease library design, generation and selection
A homology model of the eOnu_HIVInt_v1 meganuclease was generated using the I-OnuI crystal structure as template (PDB ID 3QQY). Inspection of this model allowed us to identify eight amino acid positions located directly over base pairs −8, −7 and −6 of the DNA target sequence for engineering experiments intended to improve DNA cleavage specificity at those base pairs (Fig. S2). Protein residue positions 26, 28, 30, 42, 44, 70, 80 and 82 (numbering based on the structure of wild-type I-OnuI) were considered for randomization. The sidechains at these positions are pointing downward from the central portion of the beta sheet forming the primary DNA-contacting surface, an area of the protein which is completely conserved across all I-OnuI family meganuclease structures (Lambert et al., 2016). Each position was designated for either full variation (NNS codon), limited variation (to a subset of appropriate DNA-contacting amino acids), or in the case of Arg42, constrained to a single residue. The total number of theoretically possible amino acid sequences in this library was calculated to be 12.4 million (NNS codon = all 20 possible amino acids plus a STOP codon). Using previously described methods (Baxter et al., 2014), the insert DNA encoding the meganuclease ORF was created through assembly PCR using Accuprime Pfx polymerase (Thermo Fisher Scientific) with variation introduced by ultramers (IDT DNA) containing degenerate codons at each of the altered positions. The library was cloned into the pETCON vector through homologous recombination inside EBY100 yeast after transformation with amplified insert DNA and open I-OnuI scaffold pETCON vector. The transformed library culture was diluted and plated, with each surviving colony representing a successful transformant. Under the assumption that each colony contained a unique sequence, we obtained sufficient numbers of post-transformation colonies to represent at least 3-fold coverage of the theoretical library variation.
Transformed yeast were cultured and induced for surface expression of the meganuclease library. A scaled-up version of the tethered flow cytometric cleavage assay (described above) was performed with 50 million induced yeast in 400 μL. Two million stained and DNA substrate-tethered cells were incubated with calcium for use as a no-cleavage control (for the purpose of drawing sort gates), while the remaining cells were incubated in buffer containing magnesium to allow for cleavage to occur. The yeast cells were run over a BD FACS Aria II instrument, with sort gates chosen to collect only those cells demonstrating a shift in A647 fluorescence (indicating cleavage of the tethered DNA substrate). Two subsequent rounds of sorting were performed, with a 45-min digest (37°C) for the first sort and a 30-min digest (37°C) for the second sort. An additional 100 mM KCl was added to the cleavage buffer for the second sort to help discourage the activity and selection of cells expressing variants with compromised DNA binding affinity. Only those cells with the largest A647 shifts were collected in the second sort (see Supplementary Fig. S3 for screenshots and additional explanation of the actual gates used for sorting).
Human cell culture
HEK 293 T cells (ATCC# CRL-3216) were cultured in DMEM supplemented with 10% FBS. SupT1 CD4+ T cells (ATCC# CRL-1942) were grown in RPMI supplemented with 10% FBS.
Gene disruption analysis in HEK 293 T cells
HEK 293 T cells were seeded in a 12-well plate at 2.5 × 105 cells/well. Each well was co-transfected with 0.5 μg of an env-defective replication deficient HIV target plasmid that expresses GFP (pDHIV3-GFP) and 1ug of the eOnu_HIVInt_v1_7.5mT or eOnu_HIVInt_v2_7.5mT engineered megaTAL plasmid, both with and without Trex2. Trex2 is a DNA end-processing exonuclease whose activity increases the chance of repair by mutagenic NHEJ (Certo et al., 2012). Transfection was performed using polyethylenimine (PEI) as previously described (Sedlak et al., 2016). Media was changed 18 h after transfection, and at three days post transfection, the cells were harvested and gDNA was extracted using the Qiagen DNeasy Blood and Tissue kit per manufacturer's instructions. The genomic DNA was used for determination of mutation rates with both a T7 mismatch cleavage assay (see below) and sequencing analysis.
T7 endonuclease enzymatic assay of genomic target disruption
Genomic DNA was extracted from cells using the Qiagen DNeasy Blood and Tissue kit (see above). We analyzed on-target cleavage activity by amplifying the region containing the meganuclease target site (AATGGCAGTATTCATCCACAAT) from the genomic DNA using the primers Int_Forward: TAGCAGGAAGATGGCCAGTA and Int_Reverse: TCCTGTATGCAGACCCCAAT (Supplementary Table S1). The T7 mismatch cleavage assay (Vouillot et al., 2015) was performed using T7 endonuclease I (New England Biolabs), following the manufacturer's instructions. The resulting cleavage bands were visualized on a 3% agarose gel and quantified using ImageJ software (Guschin et al., 2010; Schneider et al., 2012). Off-target cleavage analysis was performed in the same way with alternate regions of DNA amplified using the primers listed in Supplementary Table S2.
Production of HIV
DHIV3 is a replication-deficient env-deficient viral clone derived from NL4-3. Virus was made by transfecting HEK 293 T cells with pDHIV3 (Bosque and Planelles, 2011) and pLET-LAI (a CXCR4 tropic envelope expressing plasmid), using a previously described protocol (De Silva Feelixge et al., 2016). Viral supernatant was harvested 72 h post transfection, filtered using a 0.45 μm filter, and titered using the TZM-BL assay as previously described (Montefiori, 2009).
Lentiviral vector production
To make lentiviral vectors, the transfer plasmids eOnu_HIVInt_v1, eOnu_HIVInt_v1_7.5mT or eOnu_HIVInt_v2_7.5mT were transfected into HEK 293 T cells. The cells were co-transfected with the packaging plasmid psPAX2 and the VSV-G envelope pMDG2. Cell growth media was changed 18 h post transfection. The viral supernatant was collected 72 h post transfection and filtered using a 0.45 μm filter. The lentiviral vectors were concentrated by centrifugation for 8 h at 8000 g. The resulting pellet was re-suspended in media concentrating the virus ×100. The lentiviruses containing the enzymes were titered by infecting SupT1 CD4+ T cells and performing flow cytometry 3 days post infection to assess transduction levels using BFP as a proxy for enzyme transduction.
Gene disruption analysis in SupT1 CD4+ T cells
SupT1 CD4+ T cells were seeded into six well plates (200 000 cells/well) and infected with the CXCR4 tropic DHIV3 virus at a multiplicity of infection (MOI) of 2 infectious units/cell. At 24 h post infection with DHIV3, cells were infected with 20 μL of the HIV-specific engineered meganuclease or megaTAL in a VSV-G pseudotyped lentiviral vector concentrated to ×100. The efficiency of transduction was measured at 72 h post infection by flow cytometry using BFP expression as a surrogate for meganuclease or megaTAL expression. We used the p24 assay to measure the efficiency of DHIV transduction or GFP for DHIV3-GFP (De Silva Feelixge et al., 2016). At 72 h post-lentivirus transduction, genomic DNA was extracted from the SupT1 CD4+ T cells and used for quantification of mutation rates using sequence analysis.
Prediction and analysis of off-target sites in the human genome
We used the web-based off-target site prediction program PROGNOS (Fine et al., 2014) to identify the most closely matched sites in the human genome to the 22 base pair target sequence of our eOnu_HIVInt meganuclease. We performed the T7 mismatch cleavage assay (Vouillot et al., 2015) for the 10 most closely matched sites and a yeast-based flow cytometric cleavage assay for the top 30 sites. We also amplified a subset of these genomic loci and performed Illumina sequencing, as previously described (De Silva Feelixge et al., 2016), as an alternate method for detecting off-target cleavage activity by the meganuclease.
In vitro cell toxicity analysis
Toxicity in SupT1 cells was assessed after transduction with a stand-alone HIV-specific meganuclease or megaTAL (eOnu_HIVInt_v1_7.5mT or eOnu_HIVInt_v2_7.5mT). At 72 h post infection, we stained the cells with Annexin V and Propidium Iodide (PI) using an apoptosis detection kit (BD Biosciences) and performed flow cytometric analysis for cell viability. First, we normalized for transduction efficiency by gating on BFP expressing cells as a proxy for transduction by the HIV-specific engineered meganuclease or megaTALs. We then assessed the percentage of BFP-expressing cells that were stained for Annexin V and PI or either of these cell viability markers alone. Cells in the early apoptotic stage are Annexin V positive while cells in the late apoptotic phase should be positive for both PI and Annexin V. The number of stained cells was used to estimate the percentage of cell viability in each experimental condition. Our measure of cell viability was calculated as the percentage of cells that were negative for both Annexin V and PI stains. Untreated SupT1 CD4+ T cells were used as an control for background cell death. Flow-cytometric analysis was performed using a LSRII flow cytometer (BD Biosciences) and FloJo software version 10.0.8 (TreeStar).
Next generation sequencing and sequence analysis
Purified PCR products described above were diluted to 0.5ng/μL and next-generation sequencing library preparation was performed using quarter-volume reactions of Nextera XT (Illumina) and 14 cycles of PCR amplification with dual-indexed adapters following the manufacturer's protocol (De Silva Feelixge et al., 2016; Sedlak et al., 2016). Reactions were cleaned using 1.0× Ampure XP beads (Beckman Coulter) and quantitated on a Qubit 3.0 fluorometer (Life Technologies) and a Bioanalyzer 2100 (Agilent Technologies). Samples were pooled and sequenced with an Illumina MiSeq system.
Raw reads were pre-processed using tools from the Galaxy suite (Goecks et al., 2010) and trimmed using Trimmomatic (Bolger et al., 2014) and Cutadapt (Martin, 2011) to remove adapter contaminants and low-quality regions (Q < 28) at the 3′ and 5′ ends. Any remaining reads shorter than 35 nucleotides were discarded. Trimmed reads were mapped to the reference sequence using Bowtie2 (Langmead and Salzberg, 2012) and exported for further analysis. Variant analysis was performed using a custom script that incorporated functions from the Rsamtools, ShortRead and Biostrings packages in R/Bioconductor (Gentleman et al., 2004; Morgan et al., 2009; R Core Team, 2013). Aligned reads that completely overlapped the meganuclease HIV target site or a predicted off-target site were scanned for insertions and deletions and the length and position of each indel was recorded. The percentage of reads containing each mutation was tabulated after discarding singletons (mutations that were only detected in a single read) and the overall mutation rate computed for each sample. Finally, we used a Chi-square test of proportions to determine whether the mutation rate for each sample was significantly higher than the corresponding untreated controls.
Statistical analysis
For cell toxicity analysis in SupT1 CD4+ T cells, we performed a one-way analysis of variance (ANOVA) to detect differences between the treatment conditions. We also performed a two-sided t-test analysis to determine P values for pairwise comparison between treatment groups using Stata 13 (Stata corp).
Recombinant protein expression and purification
Both eOnu_HIVInt_v1 and eOnu_HIVInt_v2 meganuclease open reading frames were cloned into the pET21d tagless bacterial expression vector and transformed into BL21(DE3) RIL cells (Agilent Technologies). Protein expression was induced with 0.2 mM IPTG and the bacteria were incubated overnight at 16°C with shaking. The recombinant protein was purified over a heparin column followed by size exclusion chromatography. The heparin column buffer contained 25 mM Tris pH 7.5 with a sodium chloride (NaCl) gradient of 200–1000 mM. The proteins eluted from the column at ~600 mM NaCl. Size exclusion chromatography was performed in a buffer containing 25 mM Tris pH 7.5, 200 mM NaCl, and 5% glycerol.
Crystallization, structure determination and refinement
Crystals of the two meganucleases in this study were grown in the presence of the same double-stranded DNA oligo substrate (IDT DNA):
Top strand 5′-GGGAATGGCAGTATTCATCCACAATG-3′
Bottom strand 5′-CCATTGTGGATGAATACTGCCATTCC-3′
eOnu_HIVInt_v1 crystals grew with 50 μM protein and 80 μM DNA substrate in 19% w/v PEG 2000, 6% w/v PEG-MME 8000, 100 mM BIS-TRIS pH 8.0 and 200 mM NaCl with 6 mM CaCl2 present in the protein solution. The crystals were cryoprotected with 20% ethylene glycol.
eOnu_HIVInt_v2 crystals grew with 65 μM protein and 100 μM DNA substrate in 0.2 M ammonium sulfate, 0.1 M HEPES pH 7.0 and 22.5% w/v polyethylene glycol 3350 with 6 mM CaCl2 present in the protein solution. The crystals were cryoprotected with 20% sucrose, through a step-wise soaking procedure.
Data was collected at the Advanced Light Source (Lawrence Berkeley National Laboratory, Berkeley, CA) on Beamline 5.0.2. with a Pilatus detector. Datasets were processed with HKL2000 (Otwinowski et al., 1997) and the structures were solved by molecular replacement with the wildtype I-OnuI structure (PDB ID 3QQY) as search model. Phasing and refinement were performed using the PHENIX software suite (Adams et al., 2010). The eOnu_HIVInt_v1 structure was deposited into the RCSB protein database with the PDB ID 5V0Q and the eOnu_HIVInt_v2 structure with PDB ID 5T8D. The structure of 5T8D was recently described for the purpose of comparing the mechanism of shifting specificity between multiple redesigned variants of the same meganuclease scaffold (Werther et al., 2017). All of the engineering and activity studies and data in this article, including the structure of 5V0Q, are otherwise novel.
Results
Please refer to the first paragraph of ‘Materials and Methods’ for a description of the endonuclease name and nomenclature conventions and for a brief overview of the engineering approach referenced below.
Initial versions of an engineered HIV-specific megaTAL cleave the intended target sequence
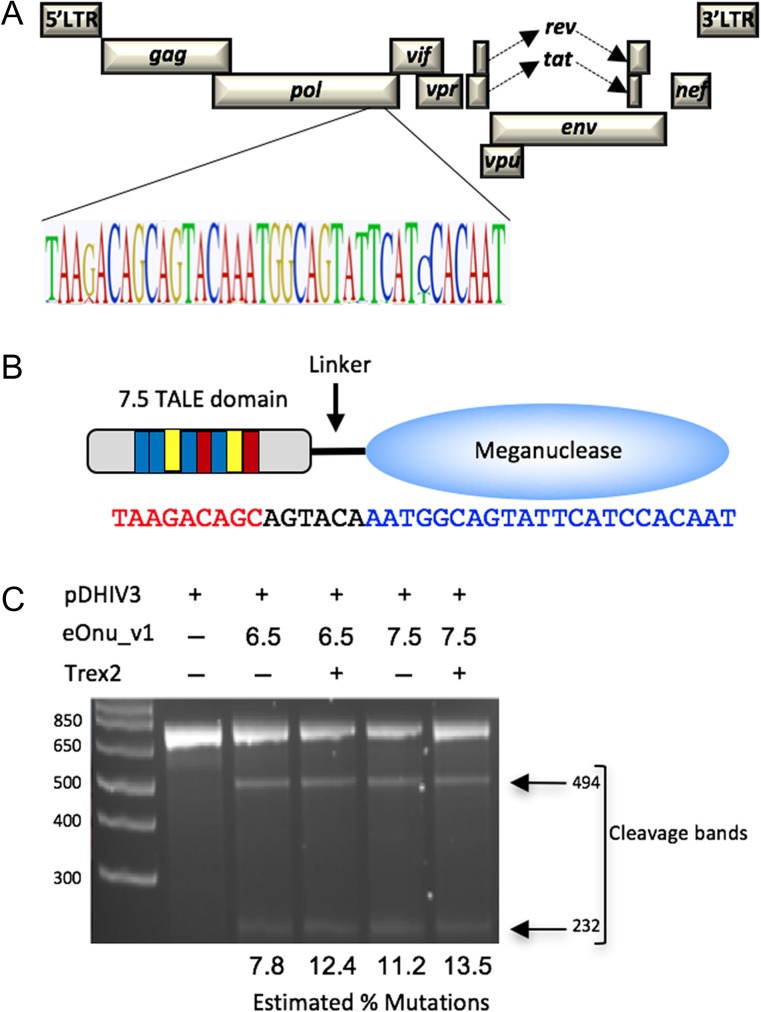
A previously engineered version of an HIV-specific meganuclease, ‘eOnu_HIVInt_v1’ (for ‘engineered I-OnuI targeting a site in HIV Integrase, version 1’) recognizes a 22 base pair target sequence in the HIV pol gene (Sedlak et al., 2016). This sequence is located in the viral genomic region encoding the catalytic domain of HIV-1 integrase and is highly conserved across multiple HIV-1 variants (Fig. 2A). Mutations in this catalytic core domain of HIV-1 integrase have been shown to lead to the production of non-viable viral progeny (Cannon et al., 1994). To improve the DNA binding affinity and specificity of this engineered enzyme, the meganuclease was fused to a TAL effector domain using previously described methods (Boissel and Scharenberg, 2015) to generate two megaTALs: eOnu_HIVInt_v1_6.5mT and eOnu_HIVInt_v1_7.5mT. These megaTALs contain 6.5 and 7.5 DNA-contacting TAL repeats, respectively, which recognize an additional 7 or 8 base pairs (including a ‘T’ preceding the TAL recognition site), increasing the overall length of the megaTAL recognition sequence to 29 or 30 base pairs (Fig. 2B).
Fig. 2.
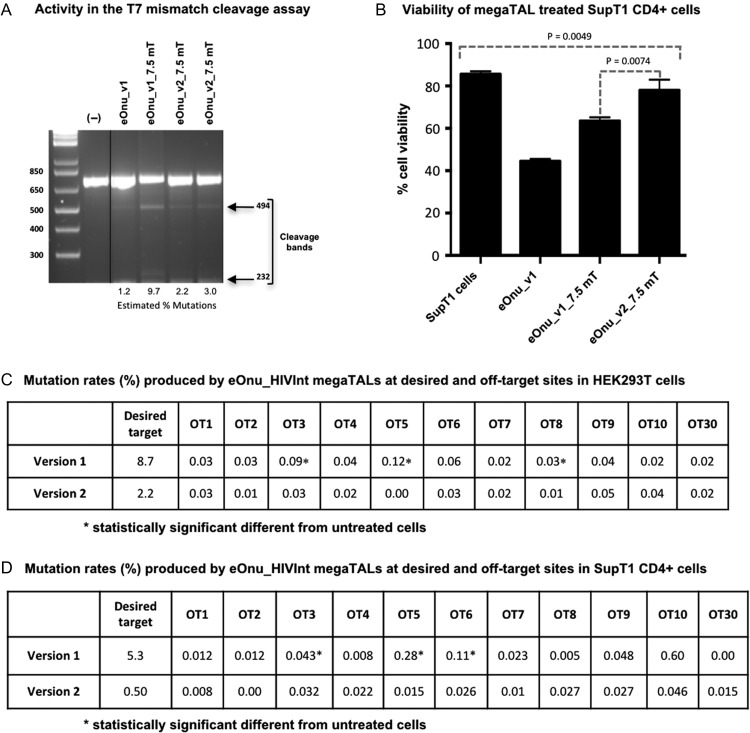
Target sequence and initial characterization of an HIV-specific engineered meganuclease. (A) The engineered I-OnuI meganuclease (eOnu_HIVInt) targets a highly conserved site in the HIV provirus, as illustrated by a Logo plot of the target region using 2635 sequences from the Los Alamos National Laboratory HIV database. (B) Schematic of the eOnu_HIVInt megaTAL enzyme and its bipartite DNA target site, with the TAL effector portion binding the sequence shown in red text and the meganuclease binding the sequence in blue text. (C) Gel from a T7 mismatch cleavage assay demonstrating activity at the desired HIVInt target sequence by eOnu_HIVInt_v1 megaTALs (with either 6.5 or 7.5 RVDs) in the presence or absence of Trex2. The estimated percentage of mutated targets is designated below each lane
Cleavage of the desired target sequence by these megaTALs is expected to introduce insertions and/or deletions (indels) at the target site due to the mutagenic nature of the non-homologous end-joining (NHEJ) DNA repair pathway (Lieber, 2010). Therefore, we could detect cleavage activity in transfected cells using a T7 endonuclease mismatch cleavage assay (Vouillot et al., 2015). We observed cleavage bands after digestion with T7 endonuclease (Fig. 2C), suggesting megaTAL activity at the desired target sequence. We quantified these bands using ImageJ software (Schneider et al., 2012) and obtained mutation rates for eOnu_HIVInt_v1_6.5mT of 7.8 and 12.14% in the absence or presence of Trex2, respectively. For eOnu_HIVInt_v1_7.5mT, we obtained mutation rates of 11.2 (−Trex2) or 13.5% (+Trex2). Based on its higher relative cleavage activity, and from previous work in our lab (Sedlak et al., 2016), we chose to move forward with eOnu_HIVInt_v1_7.5mT for subsequent experiments.
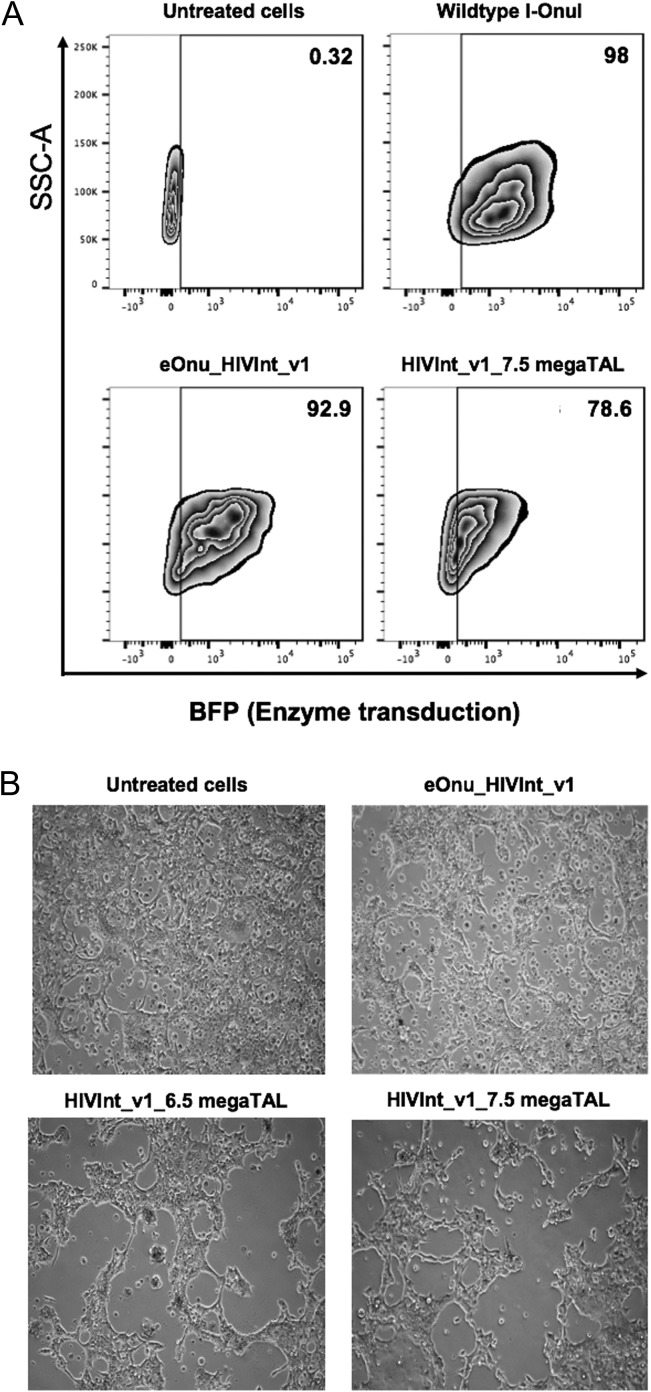
Next, we wanted to demonstrate activity of the eOnu_HIVInt_v1 megaTAL on integrated HIV proviral sequences in SupT1 CD4+ T cells. We had previously established that 20 μL of a ×100 concentration of VSV-G pseudotyped lentiviral vector would give us nearly 100% transduction of the SupT1 CD4+ T cells (Fig. 3A). At 72 h post lentivirus transduction, we extracted genomic DNA from the SupT1 CD4+ T cells and performed Illumina sequencing of PCR amplicons containing the HIVInt target sequence. From this sequence analysis, the mutation frequency at the target site in megaTAL-treated SupT1 CD4+ cells was calculated to be 5.3%.
Fig. 3.
Toxicity of megaTALs in cell culture. (A) Transduction of SupT1 CD4+ T cells with megaTALs in a VSV-G pseudotyped lentiviral vector. For each condition, 200 000 SupT1 CD4+ T cells were infected with 20uL of ×100-concentrated VSV-G pseudotyped lentiviral vectors in the presence of 4 mg/mL polybrene and spinoculated at 1200 g for 60 min. Media was changed 18 h post-infection and flow cytometry was performed 72 h post-infection to determine the transduction efficiency using BFP expression as a proxy for enzyme transduction. The transduction efficiency decreases with the addition of the TAL effector domain; 98% transduction for wildtype I-OnuI meganuclease, 92.9% for eOnu_HIVInt_v1 meganuclease, and 78.6% with HIVInt_v1_7.5 megaTAL. (B) Toxicity associated with treatment of 293 T cells with the eOnu_HIVIntv1 meganuclease or megaTALs. Images taken 72 h post-transfection show widespread cell death in the cells transfected with megaTAL enzyme compared to untreated cells
Initial versions of engineered HIV-specific megaTALs are associated with elevated cell toxicity
In the transfection experiments in HEK293T cells and infection experiments in SupT1 CD4+ cells, we observed significant cell toxicity in all assayed enzyme conditions, as evidenced by cell retraction from culture plates and cell loss (Fig. 3B). This suggested a possible lack of specificity by the endonuclease for the desired viral target and the presence of off-target cleavage activity. Although there was observable toxicity in all the experimental conditions, there was much less toxicity with the meganuclease alone and more toxicity seen in the cells transfected with the megaTALs (with either 6.5 RVDs or 7.5 RVDs). We hypothesized that the increased toxicity with the megaTALs is likely due to increased overall cleavage activity of the enzyme in the megaTAL format.
The initial version of the engineered meganuclease domain exhibits poor base pair discrimination across the 5′ half of its DNA target site
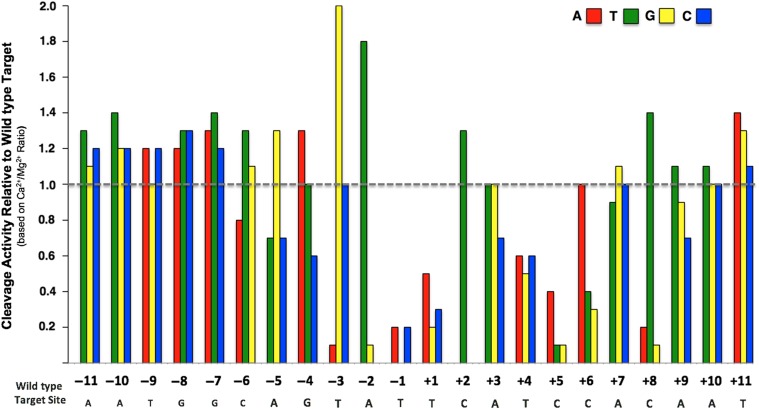
Based on these results, we decided to examine the off-target cleavage activity of the eOnu_HIVInt_v1 enzyme and its corresponding megaTAL. First, we determined the ‘one-off’ cleavage specificity profile for the stand-alone meganuclease using the same yeast surface display and flow cytometry assay as described in Fig. 1A. In this assay, each individual position of the 22 bp DNA target sequence is substituted with each of the three alternative base pairs, creating a set of 66 unique DNA target substrates. The resulting specificity profile quantitates the ability of the meganuclease to cleave a series of double-stranded DNA target substrates, each of which contains a single base pair change from the desired wild type target.
The one-off specificity data illustrates the positions across the meganuclease target sequence where single base pair changes are tolerated by the enzyme (Fig. 4). As expected from previous studies of wild-type meganucleases (Lambert et al., 2016), the eOnu_HIVInt_v1 enzyme is most specific across the ‘central four’ base pair positions of its target sequence (corresponding to positions −2 to +2, when the positions in the 22 base pair target are numbered sequentially from −11 to +11). Also expected was reduced specificity at the outer-most positions of the target sequence, where the wild type I-OnuI enzyme makes very few base-specific contacts to the bound DNA (Takeuchi et al., 2011). However, we also observed a significant lack of specificity across several additional positions within the 5′ (i.e. the ‘left’ or ‘minus’) half-site of the target sequence, where the enzyme appears to tolerate (or in some cases, actually prefer) any of the three alternate bases relative to the original base pair in the desired HIV DNA target site.
Fig. 4.
One-off specificity profile for the eOnu_HIVInt_v1 meganuclease using the tethered flow cytometric cleavage assay. Each bar represents the ability of the eOnu_HIVInt_v1 meganuclease to cleave a ‘one-off’ target substrate, relative to its activity against the wild type target sequence (designated by the gray dotted line). Cleavage activity is quantified as the ratio of A647 signal in calcium vs. magnesium conditions. Activity against each target substrate was measured by the tethered flow cytometric cleavage assay with a 20 min digest at 37°C. The wild type target sequence is shown in gray text below the graph. Each alternative nucleotide base is represented by color: adenine (red), thymine (green), guanine (yellow) and cytosine (blue). We performed at least two biological replicates of this specificity profile, using separately induced yeast cultures for each experiment. For the sake of clarity, we have not included errors bars. Variation in surface expression between replicates leads to differences in signal-to-noise and large errors bars, even when representing relative activity values. We present here the replicate with the highest surface expression and corresponding cleanest signal. See Supplementary Fig. S4a for an additional replicate of this experiment
Low specificity at individual base pair positions correlates with cleavage activity against off-target sites in the human genome
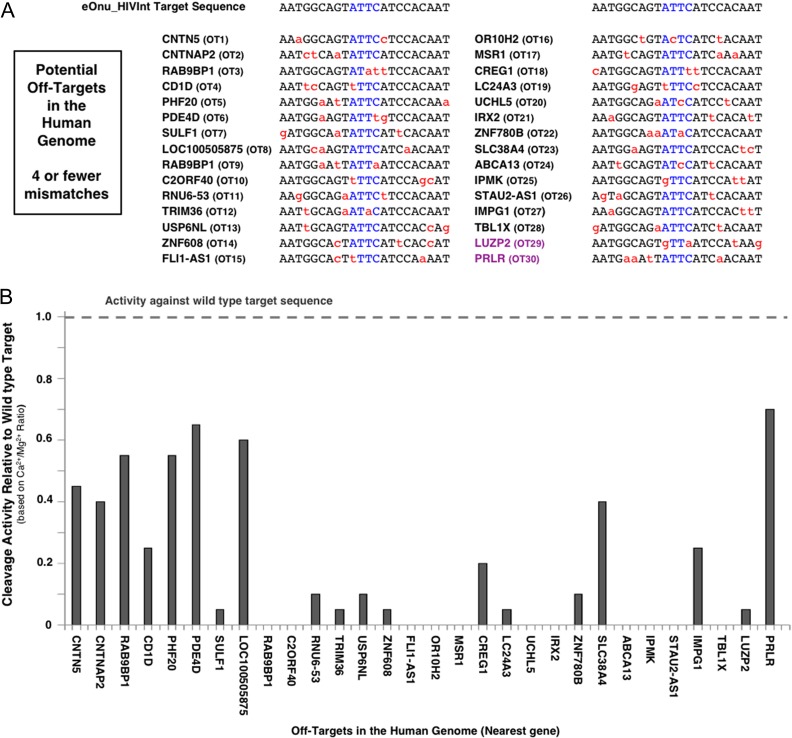
We therefore hypothesized that the relatively low specificity of the eOnu_HIVInt_v1 meganuclease within the 5′ half of its target site might be related to the cell death observed in our initial cellular assays, due to off-target cleavage activity. To examine this hypothesis, we looked for evidence of cleavage activity at predicted off-target sequences in the human genome, using the specificity profile described above as a guide. We used the PROGNOS online off-target prediction tool (Fine et al., 2014) to identify the most closely matched off-target sites in the human genome. From the output of that analysis, we selected 30 potential off-target sites containing two to four base pair mismatches relative to the desired eOnu_HIVInt meganuclease target sequence (Fig. 5A). We generated A647-labeled DNA target substrates for each of these 30 off-target sequences and performed the tethered flow-cytometric cleavage assay using surface-expressed eOnu_HIVInt_v1 meganuclease enzyme. The activity of this enzyme against each human off-target sequence was quantified and plotted relative to cleavage of the wild type target (Fig. 5B).
Fig. 5.
Potential off-target sites in the human genome. (A) Predicted off-target sequences in the human genome (PROGNOS) for the eOnu_HIVInt_v1 meganuclease. We examined activity at the 30 closest genomic off-target sequences (denoted as ‘OT##’), defined as having four or fewer nucleotide mismatches (shown in red) from the desired HIVInt target site. The central four nucleotides of the meganuclease target sequence are shown in blue. The majority of the off-target sites are located in intergenic or intronic regions; the identity of the nearest human coding sequence to each site is noted with the corresponding human gene name. Two target sites are resident within a gene's coding sequence and are colored purple. (B) Double-stranded DNA substrates were generated for each of the 30 predicted off-target sequences and tested with the tethered flow-cytometric cleavage assay. Activity is presented as cleavage of each off-target sequence relative to cleavage of the desired HIVInt target sequence
Cleavage activity was evident at 19 of the 30 off-target sites, with five sites (RAB9BP1, PHF20, PDE4D, LOC100505875 and PRLR) cleaved at levels >50% that of the intended HIVInt target. A closer analysis of the 30 predicted off-targets, compared to the ‘one-off’ specificity profile described above, revealed that the most highly cleaved off-target sites contain mismatches that are fully tolerated by the eOnu_HIVInt_v1 enzyme (Supplementary Fig. S5). Therefore, we further hypothesized that undesired cleavage activity at these off-target sites might be reduced by increasing the specificity of the meganuclease across the 5′ half of the target site. This half of the DNA target is contacted and recognized by the enzyme's N-terminal domain, implying that additional rounds of partially randomizing mutagenesis and selections intended to increase specificity in the N-terminal region of the enzyme would be required.
Engineering of the eOnu_HIVInt_v1 meganuclease domain for improved cleavage specificity
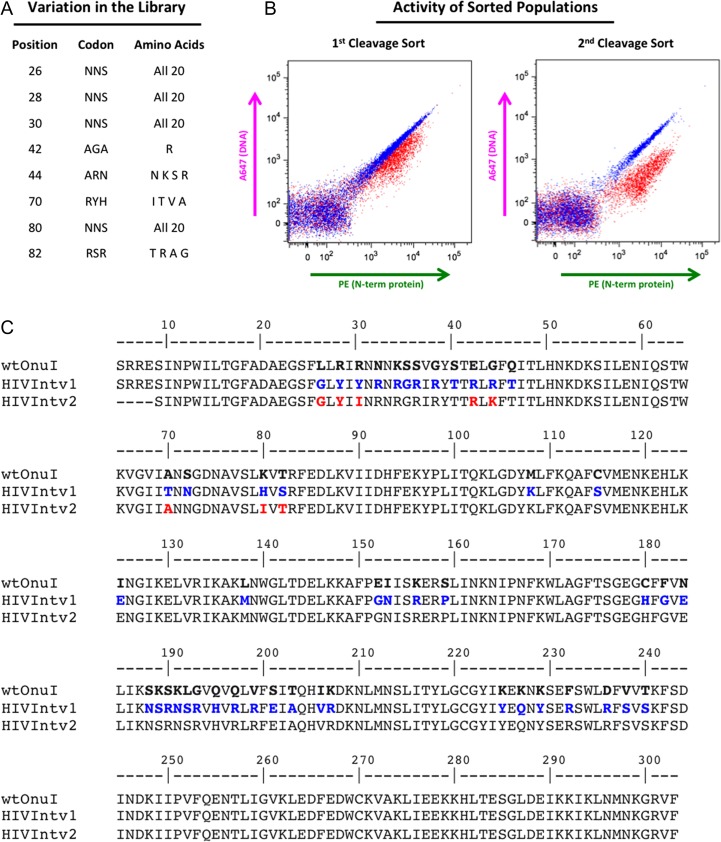
Examination of our set of predicted human off-target sites indicated that specificity at the largest number of highly tolerated mismatches might be improved through tightening of specificity at positions −8, −7 and −6 of the meganuclease target sequence. To accomplish this, we designed a library of meganuclease variants for expression on the surface of yeast and selection using fluorescence-activated cell sorting (FACS) in combination with our tethered flow-cytometric cleavage assay. We generated a homology model of the eOnu_HIVInt_v1 meganuclease based on the I-OnuI structure (PDB ID 3QQY) with the SWISS-MODEL homology modeling server (Arnold et al., 2006). From the homology model, we identified the amino acid residues making contacts to base pair positions −8, −7 and −6 of the target sequence. These sidechains are pointing downward from the central portion of the beta sheet forming the primary DNA-contacting surface (Fig. S2), an area of the protein which is completely conserved across all I-OnuI family meganuclease structures (Lambert et al., 2016). We selected the following residues for variation in our library: complete variation (all 20 possible amino acids) at positions 26, 28, 30 and 80, and limited variation at position 44 (N, K, S, R), position 70 (I, T, V, A) and position 82 (T, R, A, G). The sidechains included at positions of limited variation were chosen from successful substitutions in previous engineering libraries (data not shown) and limited in order to keep our theoretical library size manageable (12.4 million possible amino acid sequences). We chose to keep position 42 constrained to arginine due to its contact to the guanine base at −8 (Fig. 6A).
Fig. 6.
eOnu_HIVInt meganuclease engineering. (A) Eight amino acid positions were considered for variation in our eOnu_HIVInt engineering library. The codon used and variation introduced at each position is listed. (B) Cleavage of the desired HIVInt target sequence by sorted yeast populations is maintained and improved from the first to the second library sorting step, as illustrated by the magnitude of A647 shifts in the tethered flow cytometric cleavage assay. (C) Amino acid sequences are shown for wild type I-OnuI (the parent engineering scaffold), the original eOnu_HIVInt_v1 meganuclease (HIVIntv1), and our further engineered eOnu_HIVInt_v2 meganuclease (HIVIntv2). Numbering of residues matches that of the original I-OnuI crystal structure (PDB ID 3QQY). Blue text indicates altered residues between wild type I-OnuI and eOnu_HIVInt_v1 and red text highlights the eight amino acid positions considered for variation (contacting base pairs −8, −7 and −6 of the target sequence) in the engineering of eOnu_HIVInt_v2
We generated our library insert using assembly PCR and transformed the insert and open pETCON vector into yeast for homologous recombination. The transformed yeast were induced for surface expression of the variant meganucleases, and cells expressing active enzyme were identified using the tethered flow cytometric cleavage assay. Cells with FITC signal (indicating successful expression of the full-length protein) and demonstrating a drop in A647 signal (indicating cleavage of the tethered DNA substrate) were selected in two sequential rounds of sorting (see Supplementary Fig. S3 for the actual gates used during sorting).
The sorted populations were cultured, re-induced for expression, and re-tested for activity using the same flow cytometric cleavage assay. The resulting cleavage shifts demonstrated active enzyme and a clear improvement in cleavage activity from the first to the second sorted population (Fig. 6B). Plasmid DNA was extracted from the second sort population, and individual clones were isolated and sequenced. Of the 24 clones sequenced, 7 were unique. These seven candidate clones were tested individually in the flow cytometric tethered cleavage assay against the desired HIVInt target sequence and each of the top four most tolerated off-target sequences (PHF20, PDE4D, LOC100505875 and PRLR) (Supplementary S6a and b). Off-target RAB9BP1 was excluded from this analysis because its mismatches are located outside of the −8−7−6 base pair window expected to be affected by our engineering strategy (Supplementary Fig. S5). Candidates were considered promising if they showed activity against the desired target sequence and reduced activity against the off-target sequences (by visual inspection of cleavage shifts from the tethered flow cleavage assay).
In a direct comparison to the original eOnu_HIVInt_v1 enzyme, all of the redesigned clones appeared to be less active against the desired target (smaller magnitude drop in A647 signal), but many also showed a distinct reduction in activity against the off-target sequences. The three best clones from the tethered flow cytometric assay were also tested in a complementary non-tethered in vitro cleavage assay (as illustrated in Fig. 1B), which requires both binding and cleavage of a labeled DNA substrate in order to see a shifted band in an acrylamide gel. In the non-tethered assay, we observed shifted bands for the original eOnu_HIVInt_v1 enzyme (with both desired and off-target substrates), but only one of the redesigned variants bound and cleaved a single target substrate (one of the undesired off-targets). No cleavage shifts were observed for the desired wildtype target sequence (Supplementary Fig. S7). This result suggests that all of the redesigned variants demonstrated a decrease in overall DNA binding affinity, as evidenced by their inability to bind a non-tethered substrate. In the end, a single redesigned clone (#202) was chosen based on good activity against the desired target in the tethered flow cytometric cleavage assay and reduced cleavage of off-target sequences in both the flow cytometric and non-tethered in vitro cleavage assays. We named this re-engineered variant eOnu_HIVInt_v2 for ‘engineered I-OnuI targeting a site in HIV Integrase, version 2.’ Of the eight positions included in our −8, −7 and −6 redesign library, five amino acid changes were incorporated in the new enzyme (Fig. 6C).
The redesigned meganuclease displays an unexpected increase in cleavage specificity across the entire target site and decreased cleavage of most human off-target sites
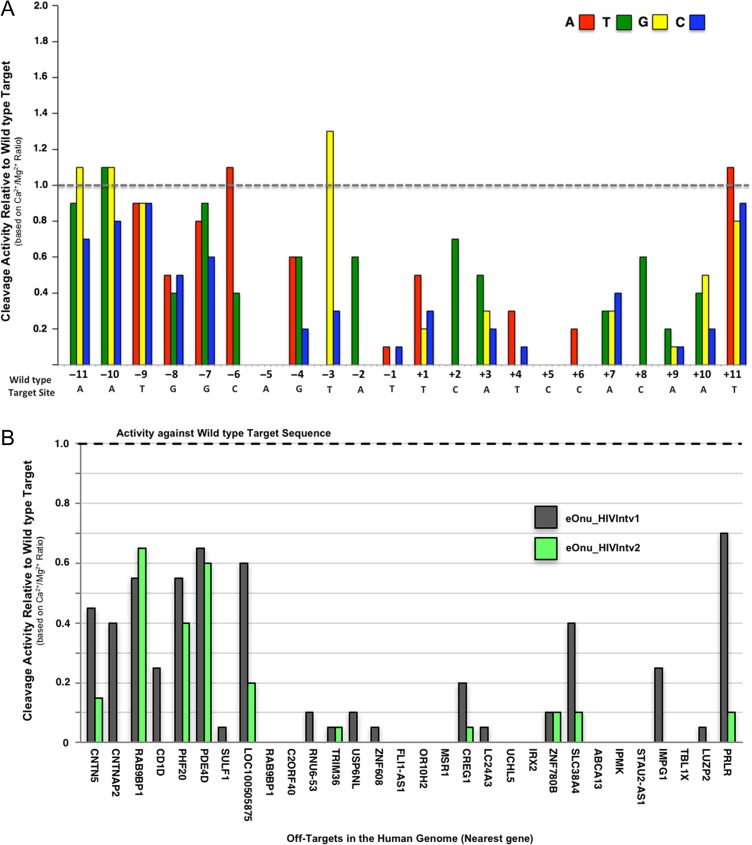
Next, we generated another ‘one-off’ cleavage specificity profile for the re-engineered eOnu_HIVInt_v2 meganuclease (Fig. 7A) using the same set of 66 target substrates assayed for the original enzyme. The specificity profile for the eOnu_HIVInt_v2 meganuclease shows significant improvement over the original eOnu_HIVInt_v1 enzyme. Within the re-engineered −8, −7 and −6 base pair window, only one alternative base is tolerated as well as the wild type base (adenine at position −6), as opposed to 8 of 9 in the eOnu_HIVInt_v1 enzyme (Fig. 4). Strikingly, the specificity at adjacent position −5 has also increased, no longer tolerating any alternate base at this position, and the overall specificity profile, relative to eOnu_HIVInt_v1, is considerably improved.
Fig. 7.
Increased specificity of the re-engineered eOnu_HIVInt_v2 meganuclease. (A) A one-off specificity profile for the re-engineered eOnu_HIVInt_v2 meganuclease was generated with the tethered flow cytometric cleavage assay. Each bar represents the ability of surface-expressed eOnu_HIVInt_v2 meganuclease to cleave a ‘one-off’ target substrate, relative to its activity against the wild type target sequence (designated by the gray dotted line). Activity is quantified as the ratio of A647 signal in calcium vs. magnesium conditions. The wild type target sequence is shown in gray text below the graph. Each alternative nucleotide base is represented by color: adenine (red), thymine (green), guanine (yellow) and cytosine (blue). See Supplementary Fig. S4b for a second replicate of this experiment. (B) The improved specificity re-engineered eOnu_HIVInt_v2 meganuclease was assayed with the tethered flow cytometric cleavage assay against DNA substrates containing each of the 30 predicted human off-target sequences. Activity is presented as cleavage of each off-target sequence relative to cleavage of the desired HIVInt target sequence
The improvement in cleavage specificity of the eOnu_HIVInt_v2 meganuclease was further demonstrated by testing the re-engineered enzyme against each of the 30 predicted human off-target sites in the tethered flow cytometric cleavage assay. In our previous test of the original eOnu_HIVInt_v1 enzyme, we observed various levels of activity against 19 of the 30 predicted off-target sequences. Using the improved specificity eOnu_HIVInt_v2 enzyme in an identical assay, all measurable activity has been eliminated for 9 of these 19 previously tolerated targets (Fig. 7B). Activity against the top four most tolerated off-targets is noticeably reduced for targets LOC100505875 and PRLR, with a slight reduction in cleavage of off-target PHF20. Not surprisingly, activity against off-target PDE4D remains unchanged, as the adenine base at position −6 of this target is still fully tolerated by the eOnu_HIVInt_v2 enzyme (Fig. 7A).
The overall DNA binding affinity of the redesigned meganuclease is decreased about 3× (but is still sub-nanomolar); its DNA binding specificity is unchanged
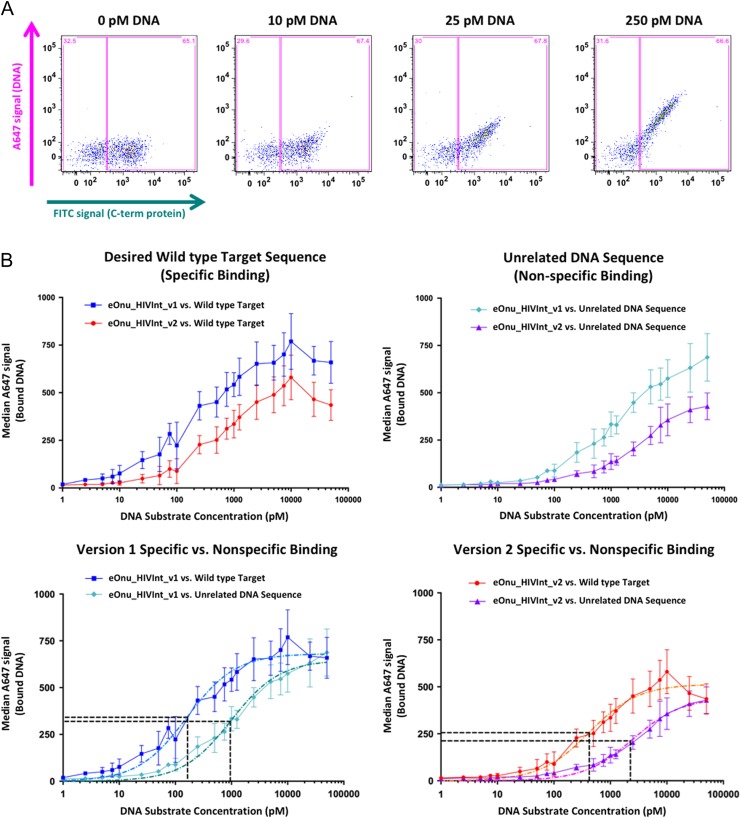
We decided to further investigate the observations described above by directly comparing the DNA binding affinity of the original eOnu_HIVInt_v1 and improved-specificity eOnu_HIVInt_v2 meganucleases in a flow cytometric DNA binding assay (as illustrated in Fig. 1C). The two enzymes were again expressed on the surface of yeast and incubated with a range of A647-labeled DNA substrate concentrations. A plot of FITC signal (C-term of protein) vs. A647 signal (DNA) illustrates the amount of DNA substrate bound by the enzyme with increasing substrate concentration (Fig. 8A). The assay was performed with DNA substrates containing either the desired HIVInt target sequence (5′-AATGGCAGTATTCATCCACAAT-3′) or an unrelated target sequence corresponding to the target site recognized by an entirely different wild-type meganuclease, I-SmaMI (5′- TATCCTCCATTATCAGGTGTAC-3′) (Fig. 8B).
Fig. 8.
Specific vs. non-specific binding using a flow cytometric DNA binding assay. (A) Sample raw data from the flow cytometric binding assay. Cells are incubated with anti-Myc-FITC antibody (to stain for full-length protein expressed on the surface of yeast) and increasing concentrations of A647-labeled DNA target substrate. Gates (pink boxes) are drawn to separate the expressing and non-expressing cell populations, and DNA binding is quantified by measuring the median A647 signal from the expressing gate (DNA substrate bound by the enzyme after washing the cells). (B) Specific vs. non-specific DNA binding by eOnu_HIVInt version 1 and version 2 meganucleases. Median A647 signal (bound DNA) is plotted vs. increasing DNA substrate concentration for version 1 and version 2 meganucleases against the desired wild type DNA target sequence (specific binding, upper left graph) and an unrelated DNA target sequence (non-specific binding, upper right graph). Next, the same data is represented with specific vs. non-specific binding compared for each version of the enzyme separately (lower left and lower right graphs). Colors and shapes for the various datasets are indicated as follows: Binding of the original eOnu_HIVInt_v1 meganuclease to its wild type target (blue squares), binding of the redesigned eOnu_HIVInt_v2 meganuclease to its wild type target (red circles), binding of the version 1 enzyme to an unrelated DNA sequence (cyan diamonds), and binding of the version 2 enzyme to an unrelated DNA sequence (purple triangles). Dashed lines in the lower graphs represent curve fitting of the data (light blue, dark teal, orange, and magenta lines) and horizontal black lines indicate the Y and X values used for estimation of the specificity index for each version of the eOnu_HIVInt meganuclease (concentration of substrate at ‘half-max’). The numerical values determined by the curve fitting are as follows: HIVInt_v1 specific binding (light blue dashed curve) Ymax = 680, Y1/2_max = 340, X1/2_max = 163, HIVInt_v1 non-specific binding (dark teal dashed curve) Ymax = 638, Y1/2_max = 319, X1/2_max = 954. HIVInt_v2 specific binding (orange dashed curve) Ymax = 511, Y1/2_max = 256, X1/2_max = 421, HIVInt_v2 non-specific binding Ymax = 422, Y1/2_max = 211, X1/2_max = 2258. Error bars are shown for the standard deviation of three replicates, performed on independently induced yeast cultures on separate days. The A647 signal is strongly influenced by enzyme expression levels on the surface of yeast for each induced culture (varying for each replicate, as illustrated by the errors bars).
In this assay, we observed a slightly decreased binding affinity towards the desired ‘on-target’ DNA sequence by the redesigned eOnu_HIVInt_v2 enzyme (the KD towards that target being increased by ~3-fold, from 160 to 420 nM, ±~20 nM). Both versions of the meganuclease display similar abilities to discriminate between the desired target sequence vs. the unrelated DNA sequence—corresponding to an ‘specificity index’ of binding (calculated as the ratio of KD-unrelated/KD-OnTarget) of ~5 for both enzymes (Fig. 8B, lower panels).
A new megaTAL containing the redesigned meganuclease domain retains cleavage activity at desired HIVInt target sequences
The redesigned meganuclease, eOnu_HIVInt_v2, was fused to the same TAL effector domain used previously (containing 7.5 TAL repeats) to create a new version of the HIV-specific megaTAL, eOnu_HIVInt_v2_7.5mT. The new megaTAL was tested side-by-side with the original eOnu_HIVInt_v1_7.5mT enzyme in our cellular cleavage assays, starting with HEK293T cells. From the T7 mismatch cleavage assay, we obtained cleavage bands in all the enzyme conditions, suggesting cleavage of the desired HIV DNA sequences by all enzymes tested (Fig. 9A). From a sequence analysis of the same extracted gDNA, we calculated mutation rates of 8.7% with the original megaTAL eOnu_HIVInt_v1_7.5mT, and 2.2% with the improved-specificity megaTAL eOnu_HIVInt_v2_7.5mT.
Fig. 9.
Cleavage activity of meganucleases in HEK293T cells and SupT1 CD4+ cells. (A) T7 mismatch cleavage assay for meganuclease and megaTAL activity in HEK293T cells. Cleavage bands are visible in all treated lanes, suggesting active enzymes are creating mutations at the desired target site. Data for the eOnu_HIVInt_v2_7.5mT enzyme was performed in duplicate. The negative control lane contains DNA from untreated HEK293T cells. A duplicate negative control lane was removed from this image for simplicity (indicated by vertical black line). (B) Cell viability in endonuclease-treated SupT1 CD4+ cells. Cells were transduced with 20ul of 100x concentrated VSV-G pseudotyped lentiviral vectors containing either eOnu_HIVInt_v1_7.5mT, eOnu_HIVInt_v2_7.5mT or eOnu_HIVInt_v1. Three days post transduction, the cells were stained with PI and Annexin V and cell viability was assessed using flow cytometry. Cells in the early apoptotic stage are Annexin V positive while cells in the late apoptotic phase are positive for both PI and Annexin V. Cells that are considered viable are negative for both PI and Annexin V stains. (C) Table of percent cleavage activity of megaTALs eOnu_HIVInt_v1_7.5mT and eOnu_HIVInt_v2_7.5mT in HEK293T cells. HEK293T cells were co-transfected with plasmids containing the megaTALs and DHIV3 plasmids. Three days post transfection, DNA was extracted and Illumina sequencing was performed to quantify mutation rates at both the desired HIVInt target site and predicted human genomic off-target sites. Statistically significant percentages are designated with asterisks. (D) Table of percent cleavage activity of megaTALs eOnu_HIVInt_v1_7.5mT and eOnu_HIVInt_v2_7.5mT in SupT1 CD4+ T cells. SupT1 CD4+ T cells were first infected with DHIV3 and then 24 h later infected with the megaTALs in a VSV-G lentiviral vector concentrated to ×100. Three days post infection, DNA was extracted and Illumina sequencing was performed to quantify mutation rates at both desired and off-target sites. Statistically significant percentages are designated with asterisks
We also used sequence analysis to determine the mutation rates at the target HIV sequence in SupT1 CD4+ T cells treated with either eOnu_HIVInt_v1_7.5mT or the redesigned megaTAL eOnu_HIVInt_v2_7.5mT. In SupT1 CD4+ T cells treated with eOnu_HIVInt_v2_7.5mT, we calculated mutation rates of 0.5% compared to mutation rates of 5.3% with eOnu_HIVInt_v1_7.5mT.
Cellular toxicity and off-target mutation rates are reduced when employing the new megaTAL eOnu-HIVInt_v2_7.5mT
We next examined the in vitro toxicity of the HIVInt-specific engineered meganucleases and megaTALs in SupT1 CD4+ T cells. We infected the cells with the original stand-alone meganuclease eOnu_HIVInt_v1, the original megaTAL eOnu_HIVInt_v1_7.5mT, or the improved-specificity megaTAL eOnu_HIVInt_v2_7.5mT. Viability in cells treated with the original stand-alone meganuclease eOnu_HIVInt_v1 was 42%, vs. 64.8% viability in cells treated with eOnu_HIVInt_v1_7.5mT and 78.2% viability in cells treated with the improved-specificity megaTAL eOnu_HIVInt_v2_7.5mT. The % cell viability in our untreated control was 85.4% (Fig. 9B). The overall difference in cell viability after treatment with the various endonucleases is statistically significant using a two-tailed t-test (P = 0.0049). The difference in cell viability between eOnu_HIVInt_v1_7.5mT (64.8%) and eOnu_HIVInt_v2_7.5mT (78.2%) is also statistically significant (P = 0.0074).
Next, we compared off-target cleavage activity of the megaTALs at predicted off-target sites in both HEK293T cells and SupT1 CD4+ T cells using Illumina sequencing of PCR amplicons. For this comparison, we assayed 11 of the 30 predicted off-target site regions (the first ten as well as the final off-target site, which encompasses all of the predicted off-target sequences cleaved at ≥0.5 relative activity). In HEK293T cells, the calculated mutation rates in cells treated with eOnu_HIVInt_v1_7.5mT at off-target sites RAB9BP1 (0.09%, P = 0.0146), PHF20 (0.12%, P = 0.0096), and LOC100505875 (0.03%, P = 0.0130) were statistically significantly increased compared to untreated cells (Fig. 9C). In contrast, the cells treated with eOnu_HIVInt_v2_7.5mT showed mutation rates ranging from 0.00 to 0.05%, with no statistically significant differences from the untreated cells.
In SupT1 CD4+ T cells treated with eOnu_HIVInt_v1_7.5mT, the calculated mutation rates at predicted off-target sites RAB9BP1 (0.043% P = 0.023), PHF20 (0.28% P = 1.4 × 10−9) and PDE4D (0.11% P = 0.00048) are statistically significantly increased compared to untreated cells. With the improved specificity megaTAL, eOnu_HIVInt_v2_7.5mT, mutation rates at the predicted off-target sites ranged from 0 to 0.046%, and there were again no statistically significant differences from untreated cells.
Crystal structures of the original and re-engineered meganucleases
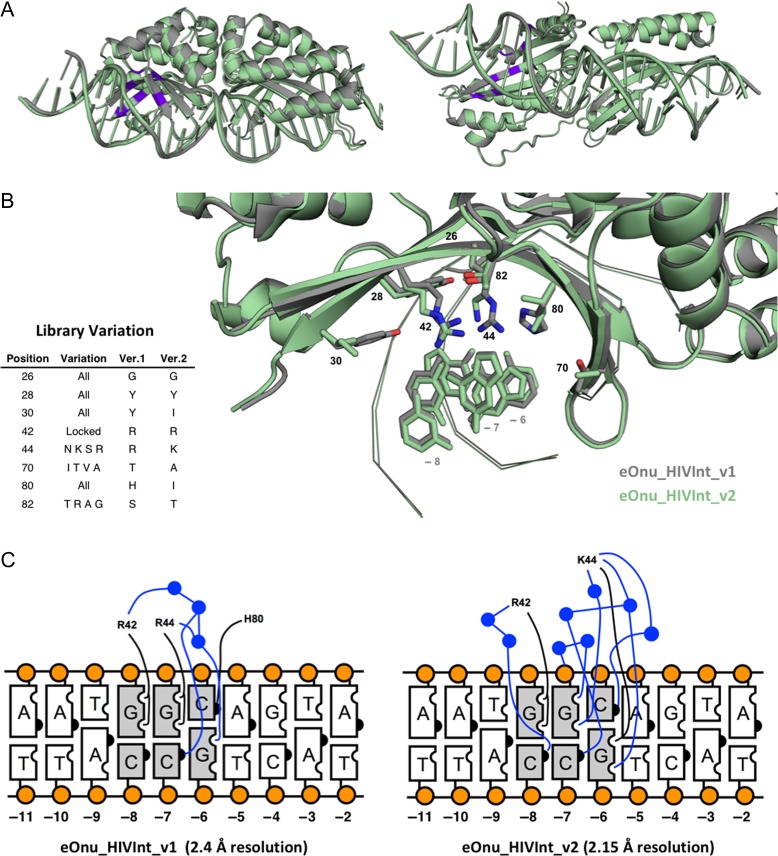
In order to better understand the behaviors of our original and redesigned enzymes, we solved the crystal structures of the stand-alone meganuclease for both the original (eOnu_HIVInt_v1) and improved-specificity (eOnu_HIVInt_v2) enzymes to 2.4 and 2.15 Å resolutions, respectively (Table I). As expected, a superposition of the two structures (Fig. 10A) indicates little to no structural changes outside of the amino acid positions varied (α-carbon RMSD of 0.217 Å, as calculated by alignment using Pymol) (Schrodinger LLC). A striking feature of the original HIVInt meganuclease (eOnu_HIVInt_v1) was the presence of two adjacent tyrosine residues (at residues 28 and 30) in the protein DNA interface with their sidechain rotamers pointing sideways rather than downward towards the DNA. A glycine residue at position 26 creates the necessary space for these large sidechains to fit. This presents a flat hydrophobic surface at the DNA interface, with the hydroxyl groups of the tyrosine residues available for interaction with waters, but not the individual DNA bases (Fig. 10B). All possible amino acids were allowed in these three positions in the redesign library, yet a tyrosine sidechain was again selected for position 28 and a glycine at position 26.
Table I.
X-ray data and refinement statistics
| eOnu HIVInt v1 | eOnu HIVInt v2 | |
|---|---|---|
| PDB ID | 5V0Q | 5T8D |
| Data collection | ||
| Space group | P 212 121 | P 212 121 |
| Cell dimensions | ||
| a, b, c (Å) | 39.69, 74.42, 164.86 | 39.70, 75.00, 165.16 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50.0–2.40 | 50.0–2.15 |
| Rmerge | 0.101 (0.800) | 0.093 (0.618) |
| I/σI | 26.8 (3.1) | 29.3 (3.2) |
| Completeness (%) | 99.6 (99.9) | 99.9 (99.2) |
| Redundancy | 12.2 (11.8) | 12.6 (11.0) |
| Refinement | ||
| No. reflections | 19 890 | 27 694 |
| Rwork (Rfree) | 18.56 (22.98) | 18.22 (22.95) |
| No. complex in ASU | 1 | 1 |
| No. atoms | ||
| Protein | 2374 | 2335 |
| DNA | 1066 | 1066 |
| Active site cations | 2 | 3 |
| Water | 63 | 261 |
| B-factor | 45.6 | 34.3 |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.003 | 0.007 |
| Bond angles (°) | 0.632 | 0.998 |
| Ramachandran | ||
| Preferred (%) | 96.86 | 95.83 |
| Allowed (%) | 3.14 | 4.17 |
| Outliers (%) | 0 | 0 |
Fig. 10.
Structures of the HIVInt engineered meganucleases. (A) Superposition of the crystal structures of eOnu_HIVInt_v1 (gray) and eOnu_HIVInt_v2 (light green), shown from the front and from the bottom. The eight amino acid positions included in our engineering library are highlighted purple. (B) A closer view of the amino acid positions in the library, colored the same as in (A). The provided table lists the residue numbers of the eight positions considered for variation in our library, what variation was incorporated at each position, and the final amino acids present at those positions in the original (version 1) and final (version 2) enzymes. The DNA bases at positions −8, −7 and −6 are shown for reference, but the rest of the DNA is shown in ribbon representation or hidden for simplicity. (C) Contact map of direct (black lines) and water-mediated (blue lines) contacts made to the bound DNA at base pairs −6, −7 and −8. Water molecules are illustrated by blue circles
The second tyrosine at position 30 was substituted with an isoleucine in the version 2 enzyme. The remaining space left in the absence of the second tyrosine sidechain is filled with an extensive network of ordered water molecules. The higher resolution of the eOnu_HIVInt_v2 structure (2.15 Å) allows for the analysis of many more water molecules in the DNA binding interface than are visible in the eOnu_HIVInt_v1 structure (2.4 Å). The arginine at position 42 was maintained for its base-specific contact to the guanine base at position −8 of the bound DNA (Fig. 10C). The arginine at position 44 of the version 1 enzyme was allowed to vary to a handful of sidechains (N, K, S, R), and a lysine was selected from the library for the version 2 enzyme. This new lysine sidechain makes a direct contact to the guanine of the −6 base pair and is a central player in a network of new water-mediated contacts to the base pairs at both positions −6 and −7. The direct contact made by His80 to the cytosine at position −6 in the eOnu_HIVInt_v1 structure is replaced with a water-mediated contact through the new Lys44 sidechain (Fig. 10C). Lastly, the amino acids at positions 70 and 82 retain their small sidechains, which do not make any base-specific contacts to the bound DNA. These residues either pack against the deoxyribose sugars of the bound DNA or participate in the hydrogen-bonding network within the protein–DNA interface.
Discussion
Antiviral gene disruption strategies and research
Chronic HIV infection is an attractive application for endonuclease therapy, in no small part because of the severity of the HIV disease and the tremendous worldwide health burden imposed by this virus. HIV is typically transmitted via exposure to blood or other infectious bodily fluids, after which it infects susceptible cells via the CD4+ receptor along with an obligate co-receptor, typically CCR5. These receptor requirements define a subset of immune cells including long-lived CD4+ T cells, and after entering the cell HIV integrates into the host cell genome, thus forming a life-long reservoir from which it can reactivate at any time. The unique biology of HIV presents two opportunities for endonuclease-mediated gene editing. In the first, endonucleases of various classes have been used to disrupt the cellular co-receptor, CCR5, thus rendering treated cells resistant to HIV infection (Cannon and June, 2011; Cornu et al., 2015). This approach has advanced into preclinical and clinical trials using zinc-finger nucleases to treat peripheral blood T cells or hematopoietic stem cells. The human trials have been particularly encouraging, with a remarkable reduction in HIV viremia and reconstitution of the immune system with modified CCR5 cells (Tebas et al., 2014). In the second approach, an endonuclease could be used to directly disrupt the integrated provirus within infected cells, thus eliminating the viral reservoir. Again, several classes of endonucleases have shown the ability to disrupt integrated HIV within infected cells, some of which have recently progressed into initial animal studies (Qu et al., 2013; Stone et al., 2013; Kaminski et al., 2016). As noted above, any endonuclease being contemplated for clinical use will need to be rigorously optimized.
For such clinical applications, high enzyme activity is important, but perhaps even more critical is extreme specificity for the desired target sequence. Off-target genomic activity could have various deleterious effects, most dramatic of which might be tumorigenesis, which could occur if off-target activity leads to disruption of tumor suppressors, activation of oncogenes, perturbations in insulators facilitating chromatin domain modifications, or other dysregulatory processes. Although not directly addressed here, nucleases for clinical use must also be free of toxic effects, which may not always relate directly to off-target DNA cleavage effects. Our data presented in this article, however, suggest that at least for the HIV-specific megaTALs studied here, recognition of off-target DNA sequences is the main driver of cellular toxicity. By minimizing these off-target effects, nuclease tolerability can be dramatically increased.
As mentioned in the introduction to this study, redesign and optimization of meganuclease specificity and function is especially challenging, due to the complex, non-modular interactions across its protein-DNA interface that dictate its sequence specificity and affinity. The extent of the engineering effort required depends on the specific application envisioned for a given endonuclease. For purely in vitro investigations, e.g. to evaluate the effect of gene knockout on a cellular phenotype, some degree of off-target activity may be tolerable, particularly if adequate controls and confirmatory approaches are available. In other cases, engineered endonucleases are being actively investigated as possible clinical therapeutics, and in this case highly specific enzymes are required. Engineered meganucleases and megaTALs are being evaluated as a possible means to disrupt genes with dominant-negative deleterious effects, such in SCID-X1(Aiuti and Roncarolo, 2009; Touzot et al., 2014), or XP-C in xeroderma pigmentosa (Dupuy et al., 2013). In the infectious disease field, there is substantial interest in using nucleases to disable viruses causing persistent infections, such as hepatitis B virus (Weber et al., 2014), herpes simplex virus (Aubert et al., 2016) and HIV (De Silva Feelixge et al., 2016).
Repacking a local region of the protein–DNA interface and resulting subtle alteration of DNA binding energy contribute to improvement of engineered meganuclease cleavage specificity
The original HIV-specific megaTAL we developed, eOnu_HIVInt_v1, displayed good cleavage activity towards its targeted HIV sequence (Sedlak et al., 2016), but was associated with significant cellular toxicity (this work). Further characterization using the tethered flow cytometric cleavage activity assay demonstrated that the meganuclease domain of the fusion megaTAL tolerates many possible single nucleotide substitution in its 22 base pair target sequence, particularly across base pair positions −6, −7 and −8 in the 5′ DNA half-site. The tethered flow assay also demonstrated that the meganuclease could also cleave select predicted human off-target sites, presumably leading to the toxicity observed, and making it an unsuitable choice for further development for therapeutic applications. We hypothesized that structure-guided redesign of the meganuclease domain of the fusion HIV-specific megaTAL could improve the overall balance of properties of DNA affinity and specificity, resulting in a superior, better-tolerated reagent.
Our enzyme redesign efforts benefitted from the availability of a high resolution crystal structure of the parent wild-type enzyme, I-OnuI (PDB ID 3QQY). Based on this, we were able to identify amino acid residues likely to interact with the specific DNA base pairs showing the least degree of selectivity (positions −6, −7 and −8) by eOnu_HIVInt_v1. By fully randomizing several of these amino acids, and allowing additional, more limited variation at others, we were able to create a manageably sized library of enzyme variants to select and ultimately characterize using our tethered flow cytometric assay. The best-performing redesigned enzyme, eOnu_HIVInt_v2, displays extensive reorganization of the immediate contacts between the revised amino acid residues and their underlying DNA base pairs, resulting in an increase (from 7 to 9) in the total number of observable contacts to the potential hydrogen-bond acceptors and donors located at base pair positions −6, −7 and −8 (Fig. 10C). The distribution of these contacts shifted significantly, from mostly direct interactions between protein side chains and DNA bases (5 of 7) in eOnu_HIVInt_v1 to mostly water-mediated interactions (6 of 9) in eOnu_HIVInt_v2. Providing that the overall complementarity and number of satisfied H-bond partners in the interface is maintained or improved, a reliance upon water-mediated contacts is fully capable of supporting highly sequence-specific readout of DNA base pairs, as has been documented both for meganucleases (Chevalier et al., 2003) and for other DNA binding proteins, as originally illustrated for the Trp repressor (Joachimiak et al., 1994). However, this apparent shift to a slightly greater reliance on ordered water molecules during DNA binding may be part of the reason why DNA binding affinity is slightly reduced, to the ultimate benefit of improve sequence specificity as summarized below.
The specificity of the re-engineered meganuclease is improved not only across the several base pairs in the 5′ half of the DNA target that were the target of structure-based mutation and re-selection, but also more broadly across many additonal base pair positions (illustrated by comparing their ‘one-off’ specificity profiles (Figs 4 and 7A, respectively) and the substantial reduction in activity towards several closely related off-targets in the human genome (Fig. 7B)). This change in behavior is accompanied by an approximate 3-fold loss in cognate target affinity as compared to the original enzyme, which both versions of the enzyme binding the intended target with sub-nanomolar KD values (160 and 420 pM, respectively). Both versions of the meganuclease display similar abilities to discriminate between the desired target sequence vs. the unrelated DNA sequence—corresponding to an ‘specificity index’ of binding (calculated as the ratio of KD-unrelated/KD-OnTarget) of ~5 for both enzymes (Fig. 8B, lower panels). Thus, to whatever extent the change in DNA binding behavior is coupled to the alteration of the enzyme's cleavage activity (if at all), it would appear to be correlated only with a relatively small, but significant reduction in overall DNA binding energy, rather than with altered DNA binding specificity.
We believe that the observed reduction in overall DNA binding affinity (and the corresponding reduction in overall DNA binding energy) is in fact related to the increased sensitivity of the enzyme towards base pair substitutions at many positions across the target site. Recognition specificity of meganucleases can be realized either during binding of the DNA target, or during catalysis, or both (Stoddard, 2014; Jacoby et al., 2017). In the case of one very well-studied enzyme (I-AniI), specificity towards the two halves of the target is actually divided between binding specificity and cleavage specificity (Thyme et al., 2009). The I-OnuI meganuclease enzyme (which served as the initial nuclease scaffold for both of the engineered nucleases in this study), appears to display cleavage specificity that is largely realized during catalysis (as evidenced by experiments that compare binding and cleavage specificity, measured across the central four base pairs of its target site (Lambert et al., 2016)).
The physical basis by which overall substrate binding energy can be closely coupled to catalytic specificity is relatively well understood and was particularly well articulated in a series of basic enzymatic studies in the 1960s and 70s (Jencks, 1975, 1981). Catalytic rate enhancement is known to be a product of the reduction in the energy barrier separating the initial substrate complex from the reaction transition state, and reductions in the magnitude of that barrier are known to be paid for by substrate binding energy. Changes in overall ground state binding energy can easily produce a differential effect on reaction rates measured across a panel of closely related substrates, by altering the overall magnitude of the catalytic energy barrier such that more (or fewer) substrate analogues are able to effectively clear that barrier during hydrolysis.
The most important characteristic for a redesigned enzyme is improved performance in the desired application. For the megaTAL containing our redesigned nuclease (eOnu_HIVInt_v2_7.5mT), improved performance was observed in all major respects. The meganuclease component showed markedly improved specificity for the desired vs. off-target DNA sites. This was also observed when the full megaTAL was used to treat HIV-containing HEK293T or SupT1 cells. In both cases, on-target activity was detected, but off-target cleavage fell to levels indistinguishable from untreated cells. This improved specificity was also manifest in the tolerability of the enzymes. In both cell types, the viability of treated cells approached that of untreated controls, a marked contrast to the severe toxicity observed with the first-generation megaTAL. These results also imply, at least in this case, that the bulk of the observed toxicity of the first-generation megaTAL was due to off-target protein/DNA interactions rather than other mechanisms. If generalizable, this finding is encouraging for future endonuclease design efforts, as it implies that rigorously optimized enzymes are likely to be well-tolerated in clinical applications.
Placing these results in the context of engineering other types of gene targeting nucleases
Strategies to engineer gene-targeting endonucleases, to improve the balance of properties between enzyme specificity and off-target cleavage activity, are an important area of research. Here, we have described structure-based engineering strategies to redesign a bipartite megaTAL nuclease system to improve the balance of DNA affinity and cleavage and thereby improve the enzyme's DNA cleavage specificity profile.
The reprogramming of DNA recognition is required for all gene-editing nuclease systems. Several efficient gene targeting nuclease platforms now exist, including one RNA-guided system (CRISPR/Cas9), three protein-guided systems (zinc finger nucleases (ZFNs), TAL effector nucleases (TALENs), and homing endonucleases, hereafter termed ‘meganucleases’) and fusion proteins such as TAL effector-meganucleases (megaTALs). These technologies and gene targeting proteins were recently reviewed in (Corrigan-Curay et al., 2015; Porteus, 2016). RNA-guided CRISPR/Cas9 endonucleases, being very simple to design, are broadly employed for basic biological research (reviewed in (Rose et al., 2017)) and are also being developed for targeted gene therapy and the creation of genetically engineered therapeutic cells (reviewed in Koo and Kim, 2017). ZFNs and TALENs (both of which are modular, protein-guided nucleases) require more effort to create, but also enjoy widespread use for a variety of applications (Huang et al., 2012; Schiffer et al., 2012; Hofer et al., 2013; Weber et al., 2014; Khalili et al., 2015; Benjamin et al., 2016; De Silva Feelixge et al., 2016; Spragg et al., 2016). Engineered meganucleases are the most difficult to create, but are also used for genome engineering applications (Gao et al., 2010; Antunes et al., 2012; Boissel et al., 2013; Chan et al., 2013; D'Halluin et al., 2013; Djukanovic et al., 2013; Menoret et al., 2013). Engineered meganucleases are often fused at their N-termini to TAL effector repeats; the resulting ‘megaTAL’ nucleases form highly specific gene-targeting nucleases (Boissel et al., 2013; Takeuchi et al., 2014; Wang et al., 2014; Sather et al., 2015).
Although they display very different molecular compositions, structures, and mechanisms, each of these nuclease systems employs a bipartite functional organization in which separate molecular domains or subunits contribute differentially to DNA recognition, binding and cleavage. For each platform, deliberate alteration of different regions of the gene-targeting molecule results in dramatic effects on their specificity and function. For example, while the RNA component of CRISPR/Cas9 largely dictates its specificity, the Cas9 protein forms many additional interactions with DNA that modulate overall DNA binding affinity and strand-separation activity, as well as enforcing the recognition of the 3′ Protospacer-Adjacent Motif (PAM) nucleotide sequence. Incorporation of point mutations in the REC domain of Cas9 can alter the overall specificity of the enzyme by making formation of a DNA/RNA bubble and subsequent DNA cleavage more sensitive to individual base pair mismatches between the DNA target and the guide RNA (Kleinstiver et al., 2016; Slaymaker et al., 2016).
ZFNs and TALENs also exhibit a molecular ‘division of labor’ during DNA recognition and cleavage. Both are obligate heterodimers that independently recognize separate halves of a desired DNA target site. Each protein chain contains multiple modular protein repeats that dictate DNA binding specificity, flanked by a non-specific nuclease domain. The site of cleavage is dictated primarily by the positioning of the protein repeats on the DNA target. As is the case for the CRISPR/Cas9 system, nuclease activity can be altered and optimized by mutations and alterations within the nuclease domain (many of which enforce the requirement for protein heterodimerization) (Miller et al., 2007), within the DNA-contacting repeat regions (Gersbach and Perez-Pinera, 2014; Richter et al., 2016), or within the protein linker regions that connect the two (Miller et al., 2011).
Concluding remarks: suggestions for technical modifications
As described above, the use of surface display technologies, coupled to flow cytometric selections for DNA cleavage activity (Fig. 1), is quite powerful in terms of (i) throughput and (ii) the ability to select and amplify rare clones that display desired cleavage specificity and activity. However, the reliance on a system in which the enzyme and substrate are physically tethered to one another (a requirement in order to avoid functional ‘cross-talk’ between separate clones in the yeast population) can confound the isolation of constructs with optimal properties, for two reasons, leading to compromised affinity and/or activity that then have to be rescued in a rather undesirable post-selection process of enzyme optimization.
First, the act of tethering enzyme and substrate (Fig. 1A) can allow the identification of constructs that are compromised in their substrate binding affinity. In the example described in this article, this issue has been minimized by performing iterative rounds of selections in the presence of elevated salt concentration (+100 mM KCl), and the resulting construct, which does in fact bind its cognate target site slightly less tightly than the initial construct, is found to still display a sub-nanomolar KD value against that target. The incorporation of a high-salt selection step has generally been found to discourage the weakest binders from interacting productively with the DNA substrate, and they are largely eliminated from the final sorted populations.
If we observe that our final engineered enzyme displays compromised binding affinity (either via lack of activity in the high-salt sorting step or lack of activity in a subsequent in vitro, non-tethered cleavage assay), we can generate a new library designed solely for the purpose of increasing DNA binding affinity. This usually corresponds to the introduction of positively charged sidechains (lysine or arginine) in positions where they will make contacts only to the negatively charged DNA backbone. This method can increase the overall binding affinity of the enzyme while not interfering the specific contacts made to the individual bases of the target sequence. We have consistently observed improved DNA binding for clones isolated from this type of affinity-focused library.
However, the introduction of too much additional positive charge and overall DNA-binding affinity runs the risk of creating an enzyme with increased non-specific DNA binding activity. To address this, we are also employing a protocol to perform cleavage selections in the presence of salmon sperm DNA (typically added to ~0.5 nM concentration) as a non-specific competitor. We now use this strategy both as an additional component in our selections or as a final test of the clones isolated from the DNA-affinity library selections.
Second, even when maintaining binding affinity and appropriate recognition fidelity as described above, the same tethered activity selection steps can potentially lead to reduced catalytic efficiency, by virtue of maintaining a high local enzyme concentration, over a long time-course of cell staining, near the labeled DNA substrate. This result was observed in the construct generated in this study, corresponding to a roughly 5× to 10× reduction in cleavage rate. This effect can be counteracted at the time of initial cleavage selections, both through the use of non-optimal (reduced) pH during the cleavage-dependent staining, and by reduction in the cell staining time.
Finally, the magnitude and effect all of these technical hurdles are reduced by minimizing the difference between the starting wild-type meganuclease target specificity and the sequence of the desired target site. With the identification of new meganuclease scaffolds for engineering (hundreds of identifiable meganuclease constructs with novel and highly diverged target sites are found in microbial sequence databases) the ease, efficiency and output of such efforts should continue to improve significantly.
Supplementary Material
Acknowledgments
We thank members of the Stoddard and Jerome laboratories at the Fred Hutchinson Cancer Research Center and the staff of the center's shared flow cytometry core, for expert assistance and advice.
Supplementary data
Supplementary data are available at Protein Engineering, Design & Selection online.
Funding
This work was supported by the National Institute of General Medical Sciences [Grant R01 GM105691 to B.L.S.], the National Institutes of Health [U19 AI096111-05 and UM1 AI126623-01 to K.R.J.] and [P30 AI 027757 to King Holmes (K.R.J. co-investigator)], by the Fred Hutchinson Cancer Research Center and by Bluebird Bio, Inc. The Berkeley Center for Structural Biology, where the data for our crystal structures was collected, is supported in part by the National Institutes of Health, National Institute of General Medical Sciences and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences under Contract no. DE-AC02-05CH11231.
References
- Adams P.D., Afonine P.V., Bunkoczi G. et al. (2010) Acta Crystallogr. D Biol. Crystallogr., 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiuti A. and Roncarolo M.G. (2009) Hematology Am. Soc. Hematol. Educ. Program, 682–689. [DOI] [PubMed] [Google Scholar]
- Andersen J.L., DeHart J.L., Zimmerman E.S., Ardon O., Kim B., Jacquot G., Benichou S. and Planelles V. (2006) PLoS Pathog., 2, e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes M.S., Smith J.J., Jantz D. and Medford J.I. (2012) BMC Biotechnol., 12, 86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K., Bordoli L., Kopp J. and Schwede T. (2006) Bioinformatics, 22, 195–201. [DOI] [PubMed] [Google Scholar]
- Arnould S., Chames P., Perez C. et al. (2006) J. Mol. Biol., 355, 443–458. [DOI] [PubMed] [Google Scholar]
- Aubert M., Madden E.A., Loprieno M. et al. (2016) JCI Insight, 1, e88468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter S.K., Lambert A.R., Scharenberg A.M. and Jarjour J. (2013) Methods Mol. Biol., 978, 45–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter S.K., Scharenberg A.M. and Lambert A.R. (2014) Methods Mol. Biol., 1123, 191–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin R., Berges B.K., Solis-Leal A., Igbinedion O., Strong C.L. and Schiller M.R. (2016) Hum. Genet., 135, 1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissel S., Jarjour J., Astrakhan A. et al. (2013) Nucleic Acids Res., 42, 2591–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissel S. and Scharenberg A.M. (2015) Methods Mol. Biol., 1239, 171–196. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M. and Usadel B. (2014) Bioinformatics, 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosque A. and Planelles V. (2011) Methods, 53, 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon P. and June C. (2011) Curr. Opin. HIV AIDS, 6, 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon P.M., Wilson W., Byles E., Kingsman S.M. and Kingsman A.J. (1994) J. Virol., 68, 4768–4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certo M.T., Gwiazda K.S., Kuhar R. et al. (2012) Nat. Methods, 9, 973–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y.S., Takeuchi R., Jarjour J., Huen D.S., Stoddard B.L. and Russell S. (2013) PLoS One, 8, e74254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Wen F., Sun N. and Zhao H. (2009) Protein Eng. Des. Sel., 22, 249–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier B., Turmel M., Lemieux C., Monnat R.J. and Stoddard B.L. (2003) J. Mol. Biol., 329, 253–269. [DOI] [PubMed] [Google Scholar]
- Cornu T.I., Mussolino C., Bloom K. and Cathomen T. (2015) Adv. Exp. Med. Biol., 848, 117–130. [DOI] [PubMed] [Google Scholar]
- Corrigan-Curay J., O'Reilly M., Kohn D.B. et al. (2015) Mol. Ther., 23, 796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Halluin K., Vanderstraeten C., Van Hulle J. et al. (2013) Plant Biotechnol. J., 11, 933–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva Feelixge H.S., Stone D., Pietz H.L., Roychoudhury P., Greninger A.L., Schiffer J.T., Aubert M. and Jerome K.R. (2016) Antiviral Res., 126, 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djukanovic V., Smith J., Lowe K. et al. (2013) Plant J., 76, 888–899. [DOI] [PubMed] [Google Scholar]
- Doyon J.B., Pattanayak V., Meyer C.B. and Liu D.R. (2006) J. Am. Chem. Soc., 128, 2477–2484. [DOI] [PubMed] [Google Scholar]
- Dupuy A., Valton J., Leduc S. et al. (2013) PLoS One, 8, e78678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine E.J., Cradick T.J., Zhao C.L., Lin Y. and Bao G. (2014) Nucleic Acids Res., 42, e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Smith J., Yang M. et al. (2010) Plant J., 61, 176–187. [DOI] [PubMed] [Google Scholar]
- Gentleman R.C., Carey V.J., Bates D.M. et al. (2004) Genome Biol., 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersbach C.A. and Perez-Pinera P. (2014) Expert Opin. Ther. Targets, 18, 835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D. and Schiestl R.H. (2007) Nat. Protoc., 2, 35–37. [DOI] [PubMed] [Google Scholar]
- Goecks J., Nekrutenko A. and Taylor J., Galaxy Team (2010) Genome Biol., 11, R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschin D.Y., Waite A.J., Katibah G.E., Miller J.C., Holmes M.C. and Rebar E.J. (2010) Mackay J.P. and Segal D.J. (eds),. Engineered Zinc Finger Proteins: Methods and Protocols. Humana Press, Totowa, NJ, pp. 247–256. [Google Scholar]
- Hofer U., Henley J.E., Exline C.M., Mulhern O., Lopez E. and Cannon P.M. (2013) J. Infect. Dis., 208, S160–S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia Y., Bale J.B., Gonen S. et al. (2016) Nature, 535, 136–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Zhu Z., Lin S. and Zhang B. (2012) J. Genet. Genomics, 39, 421–433. [DOI] [PubMed] [Google Scholar]
- Jacoby K., Lambert A.R. and Scharenberg A.M. (2017) Nucleic Acids Res., 45 doi:10.1093/nar/gkw1864, first published on February 17 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jencks W.P. (1975) Adv. Enzymol. Relat. Areas Mol. Biol., 43, 219–410. [DOI] [PubMed] [Google Scholar]
- Jencks W.P. (1981) Proc. Natl. Acad. Sci. USA, 78, 4046–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachimiak A., Haran T.E. and Sigler P.B. (1994) EMBO J., 13, 367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski R., Chen Y., Fischer T., Tedaldi E., Napoli A., Zhang Y., Karn J., Hu W. and Khalili K. (2016) Sci. Rep., 6, 22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalili K., Kaminski R., Gordon J., Cosentino L. and Hu W. (2015) J. Neurovirol., 21, 310–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z. and Joung J.K. (2016) Nature, 529, 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo T. and Kim J.S. (2017) Brief Funct. Genomics, 16, 38–45. [DOI] [PubMed] [Google Scholar]
- Lambert A.R., Hallinan J.P., Shen B.W. et al. (2016) Structure, 24, 862–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. and Salzberg S.L. (2012) Nat. Methods, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber M.R. (2010) Annu. Rev. Biochem., 79, 181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011) EMBnet J., 17 doi:10.14806.ej.17.1.200. [Google Scholar]
- Menoret S., Fontaniere S., Jantz D. et al. (2013) FASEB J., 27, 703–711. [DOI] [PubMed] [Google Scholar]
- Miller J.C., Holmes M.C., Wang J. et al. (2007) Nat. Biotechnol., 25, 778–785. [DOI] [PubMed] [Google Scholar]
- Miller J.C., Tan S., Qiao G. et al. (2011) Nat. Biotechnol., 29, 143–148. [DOI] [PubMed] [Google Scholar]
- Montefiori D.C. (2009) Methods Mol. Biol., 485, 395–405. [DOI] [PubMed] [Google Scholar]
- Morgan M., Anders S., Lawrence M., Aboyoun P., Pages H. and Gentleman R. (2009) Bioinformatics, 25, 2607–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. et al. (1997) Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Porteus M. (2016) Annu. Rev. Pharmacol. Toxicol., 56, 163–190. [DOI] [PubMed] [Google Scholar]
- Procko E., Berguig G.Y., Shen B.W. et al. (2014) Cell, 157, 1644–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X., Wang P., Ding D. et al. (2013) Nucleic Acids Res., 41, 7771–7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2013) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria. ISBN: 3-900051-07-0, http://www.R-project.org/. [Google Scholar]
- Richter A., Streubel J. and Boch J. (2016) Methods Mol. Biol., 1338, 9–25. [DOI] [PubMed] [Google Scholar]
- Roberts R.J., Belfort M., Bestor T. et al. (2003) Nucleic Acids Res., 31, 1805–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R.J., Vincze T., Posfai J. and Macelis D. (2015) Nucleic Acids Res., 43, D298–D299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano Ibarra G.S., Paul B., Sather B.D. et al. (2016) Mol. Ther. Nucleic Acids, 5, e352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose P.W., Prlic A., Altunkaya A. et al. (2017) Nucleic Acids Res., 45, D271–D281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sather B.D., Romano Ibarra G.S., Sommer K. et al. (2015) Sci. Transl. Med., 7, 307ra156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer J.T., Aubert M., Weber N.D., Mintzer E., Stone D. and Jerome K.R. (2012) J. Virol., 86, 8920–8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S. and Eliceiri K.W. (2012) Nat. Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger LLC The PyMOL Molecular Graphics System, Version 1.7. Schrodinger, LLC. [Google Scholar]
- Sedlak R.H., Liang S., Niyonzima N. et al. (2016) Sci. Rep., 6, 20064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X. and Zhang F. (2016) Science, 351, 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spragg C., De Silva Feelixge H. and Jerome K.R. (2016) Curr. Opin. HIV AIDS, 11, 442–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard B.L. (2014) Mobile DNA, 5, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone D., Kiem H.-P. and Jerome K.R. (2013) Curr. Opin. HIV AIDS, 8, 217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi R., Choi M. and Stoddard B.L. (2014) Proc. Natl. Acad. Sci., 111, 4061–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi R., Lambert A.R., Mak A.N.-S., Jacoby K., Dickson R.J., Gloor G.B., Scharenberg A.M., Edgell D.R. and Stoddard B.L. (2011) Proc. Natl. Acad. Sci., 108, 13077–13082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebas P., Stein D., Tang W.W. et al. (2014) N. Engl. J. Med., 370, 901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyme S.B., Akhmetova L., Montague T.G., Valen E. and Schier A.F. (2016) Nat. Commun., 7, 11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyme S.B., Jarjour J., Takeuchi R., Havranek J.J., Ashworth J., Scharenberg A.M., Stoddard B.L. and Baker D. (2009) Nature, 461, 1300–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touzot F., Hacein-Bey-Abina S., Fischer A. and Cavazzana M. (2014) Expert Opin. Biol. Ther., 14, 789–798. [DOI] [PubMed] [Google Scholar]
- Vouillot L., Thelie A. and Pollet N. (2015) G3 (Bethesda), 5, 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhou X.Y., Xiang P.Y., Wang L.L., Tang H., Xie F., Li L. and Wei H. (2014) PLoS One, 9, e108347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber N.D., Stone D., Sedlak R.H., De Silva Feelixge H.S., Roychoudhury P., Schiffer J.T., Aubert M. and Jerome K.R. (2014) PLoS One, 9, e97579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werther R.A., Hallinan J.P., Lambert A.R. et al. (2017) Nucleic Acids Res. in press. doi:10.1093/nar/gkx544. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.