Abstract
The study of monozygotic twins discordant for Attention Deficit Hyperactivity Disorder can elucidate mechanisms that contribute to the disorder, which affects around 7% of children. First, using in vivo neuroanatomic imaging on 14 pairs of monozygotic twins (mean age 9.7, standard deviation 1.9 years), we find that discordance for the disorder is mirrored by differing dimensions of deep brain structures (the striatum and cerebellum), but not the cerebral cortex. Next, using whole blood DNA from the same twins, we find a significant enrichment of epigenetic differences in genes expressed in these ‘discordant’ brain structures. Specifically, there is differential methylation of probes lying in the shore and shelf and enhancer regions of striatal and cerebellar genes. Notably, gene sets pertaining to the cerebral cortex (which did not differ in volume between affected and unaffected twins) were not enriched by differentially methylated probes. Genotypic differences between the twin pairs – such as copy number and rare, single nucleotide variants- did not contribute to phenotypic discordance. Pathway analyses of the genes implicated by the most differentially methylated probes implicated GABA, dopamine and serotonin neurotransmitter systems. The study illustrates how neuroimaging can help guide the search for epigenomic mechanisms in neurodevelopmental disorders.
Introduction
Attention Deficit Hyperactivity Disorder (ADHD) affects around 7 – 10% of school age children, making it one of the most prevalent behavioral problems of childhood1. Although ADHD is highly heritable (twin studies h2 >0.7), no common single nucleotide variants have emerged with genome-wide significance and few candidate genes have been consistently replicated2, 3. Here, we aim to further progress into pathophysiological mechanisms through the study of monozygotic (MZ) twins discordant for ADHD.
How can monozygotic twins be discordant for a disorder that is as highly heritable as ADHD? Such MZ twins have identical demographic characteristics and highly similar genotypes and environments. Nonetheless, in fully discordant pairs, one twin shows the triad of impairing symptoms of inattention, hyperactivity and impulsivity that define ADHD, whereas the co-twin is free of symptoms. Here, we consider epigenetic and genotypic contributors to phenotypic discordance. Epigenetic changes, such as altered DNA methylation, correspond to changes in gene expression without changes in DNA sequence. Such differential methylation has been found in twins discordant for autism, depression, schizophrenia, and bipolar affective disorder4–8. The only previous epigenetic study of ADHD used a case-control design in unrelated singletons and found differential methylation of probes lying near VIPR2, a gene implicated in neurodevelopment9. Here, we extend this work to ask if epigenetic differences contribute to discordance for ADHD among MZ twins.
In addition to epigenetic contributions, genotypic changes may also affect phenotypic discordance. Although MZ twins have highly similar genotypes, they are not identical and can differ with regard to large gene duplications and deletions, i.e. copy number variants (CNVs). CNVs have sometimes, but not always, emerged as a source of discordance between MZ twins for several neuropsychiatric disorders, including autism and schizophrenia10–13. Thus, here we also address whether discordance for CNVs is associated with discordance for ADHD. Additional alterations such as functionally deleterious single nucleotide variants (SNVs) have been also recently implicated in neurodevelopmental disorders14, 15. Studies examining SNVs have mainly used a case-control rather than a MZ twin design, and none have considered ADHD. Here, we also consider DNA sequence variation as a potential driver of phenotypic discordance in twins.
The search for genetic and epigenetic mechanisms can be guided by brain-based phenotypes. Here, we focus on neuroanatomic differences tied to discordance for ADHD within twin pairs, defined in vivo through magnetic resonance imaging (MRI). It has been reported that there is a smaller caudate nucleus in the affected twin of MZ pairs discordant for ADHD16. We extend the search for disorder-related changes to other brain regions implicated in ADHD- the cerebellum, cerebral cortex and thalamus Anatomic anomalies in all of these regions has been reported among those with ADHD17–20. Additionally, interconnections between these regions form the basis for the large scale brain networks that support multiple cognitive functions that are disrupted in the disorder21.
We expect that some structures will mirror the diagnosis, differing between affected and unaffected twins (i.e., ‘discordant’ brain regions), whereas others will not differ (i.e., ‘concordant’ brain regions). Different brain regions show developmental differences in gene expression patterns. We hypothesize that epigenetic and/or genetic changes associated with discordance for ADHD will show enrichment in genes whose expression occurs in brain regions tied to discordance for the disorder. Conversely, genes expressed in structures that do not differ between affected and unaffected twins are not predicted to show such enrichment of epigenetic/genetic changes. This approach serves to augment confidence in the biological significance of epigenetic/genetic changes found in peripheral tissue samples by showing that they also mirror in vivo neuroanatomic changes in the same participants. Finally, we also conduct hypothesis-free analyses on our genome wide methylation, genotype, and exome sequencing data to define the biological pathways implicated by twin differences in genotype or epigenotype.
Materials and Methods
Participants
Discordance was defined by the presence of ADHD in one twin and absence of ADHD in the co-twin. Initially, we identified MZ twins believed by their parents to be discordant for ADHD through national support groups for ADHD and for families with twins. Exclusion criteria included cerebral palsy, psychiatric disorders other than Oppositional Defiant and Conduct Disorder, chronic medical or neurological disorders, pervasive developmental disorders, and Full Scale IQ <80. The parents of 364 MZ pairs were screened via telephone, and 334 pairs did not meet criteria. The reasons for exclusion were most commonly lack of sufficient discordance in ADHD symptoms, another primary diagnosis, lack of pervasive impairment due to ADHD or chronic medical conditions. Thirty pairs proceeded to assessment at the Clinical Center of NIH. Psychiatric diagnoses were based on the Diagnostic Interview for Children and Adolescents-Child, Adolescent, and Parent versions [DICA], revised)22. Fifteen of these MZ pairs were determined to be fully discordant for ADHD, the remainder were concordant or only partially discordant for ADHD. All 15 pairs provided DNA for genome wide methylation and genotype analysis. Whole exome sequencing required re-consenting, and this was obtained on eight of these pairs. All procedures were approved by the IRB of the NIH. Parents gave written consent, and children gave assent.
Brain imaging
T1-weighted neuroanatomic magnetic resonance imaging (MRI) was acquired using three-dimensional spoiled gradient-recalled echo in the steady state on a 1.5-T General Electric Signa scanner (Milwaukee, WI). Imaging parameters are given in the Supplementary Methods 1). Cerebral cortical, cerebellar and deep structure (caudate, putamen, thalamus) reconstruction and volumetric segmentation were performed using FreeSurfer version 5.3.0 (http://surfer.nmr.mgh.harvard.edu/). All segmentations were inspected by two raters; images on fourteen pairs of twins passed this quality control. As the brain volumes were correlated, we calculated the effective number of independent tests to which the actual tests performed were equivalent23 and set significance at P <0.007 (0.05/7.18, the number of effective tests).
Genome wide methylation mapping
Genomic DNA was extracted from whole blood and bisulfite conversion was performed using the EZ DNA Methylation-Gold kit (Zymo Research, Irvine, CA) following the manufacturer’s protocol (Supplementary Methods 2). The Illumina Infinium HumanMethylation450 BeadChip array was used to assess genome-wide methylation. The R package ChAMP was then used to convert raw intensity signals to β values, remove low quality probes, and perform BMIQ normalization for type I and II probes.24 Probes on sex chromosomes were removed, resulting in a final set of 472,685 probes. Hierarchical clustering was applied to these probes and the resulting dendrogram showed a high degree of clustering, confirming monozygosity, within 24 individuals in 12 MZ pairs as would be expected in MZ twins25, 26. Six samples, from three MZ twin pairs, were excluded from further analyses. Additional analyses suggest their dissimilarity resulted from quality control issues in DNA methylation assays (Supplementary Methods 2). No batch effects were detected (Supplementary Methods 3).
Analysis
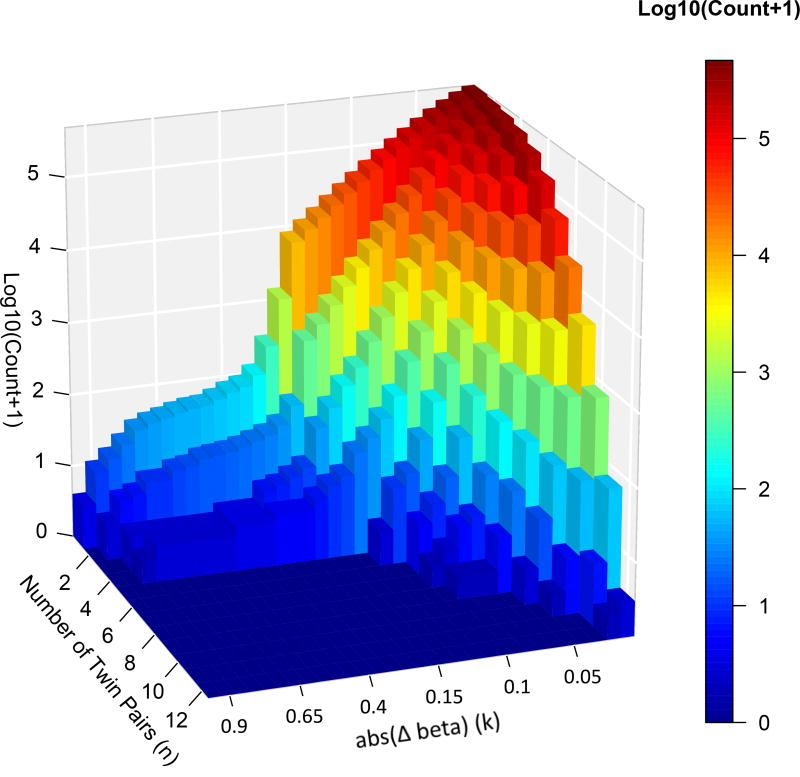
We hypothesize that methylation differences are likely to contribute to the pathophysiology of ADHD if either the differences are of a large magnitude or the methylation difference recurs across sets of twins. We implemented this hypothesis by incorporating two key variables (Δβ, n), defined as follows. We calculated the directed within-twin pair difference (affected β − unaffected β) and summarized in absolute magnitude, noting differences that were consistently positive (hypermethylated affected twin) or negative (hypomethylated in the affected twin) – abbreviated henceforth to Δβ. We binned these Δβ values using increments of 0.01 for β differences between 0.01 and 0.15, and increments of 0.05 for β differences between 0.15 and 0.95, the maximum observed difference. We then tabulated the number of differentially methylated probe sets detected for every combination of observed Δβ and number of twin sets (n) showing differences greater or equal to this Δβ (further explanation of the methods is given in Supplementary Methods 4). This provided a matrix with 173 observed probe sets (out of a possible 372), each occurring at a particular Δβ, n- see Figure 1. Thus, we conduct our analyses at multiple different thresholds, testing every possible combination of the magnitude of methylation differences and number of twin pairs showing this difference. This approach allows us to integrate these two key variables, without making assumptions about their relative importance and without imposing an arbitrary threshold for consideration.
Figure 1.
Histogram showing the log number of differentially methylated probes observed for every combination of Δβ and number of twin pairs.
Using the Illumina 450k methylation array profile we annotated the differentially methylated probes as falling in CpG islands, shores and shelves (4 kb genomic regions flanking CpG islands), or enhancers. We considered these different genomic regions given evidence that methylation of each region may have a different functional impact27. We then applied a one-tailed hypergeometric test to determine whether the 173 differentially methylated probe sets were overrepresented in any of these three genomic regions, compared to all probes (Supplementary Methods 5). To correct for testing 173 potentially correlated probe sets, we called significance at FWER<5%. To facilitate comparison with previous studies, we also analyzed data using the ‘rank-sum’ approach that has been widely used in psychiatric epigenetics4, 5, 28. This approach ranks probes according to the sum of the results of a paired t test (with low p values having a high rank) and the absolute magnitude of the mean methylation difference.
Identifying and testing of candidate of gene-sets
Our primary hypothesis was that there would be enrichment of differentially methylated probes associated with genes that are expressed in discordant, but not concordant brain regions. We thus formulated lists of genes highly expressed in these regions from the Human Brain Transcriptome29 (with a ceiling of 100 genes), focusing on genes expressed during early developmental stages, given the early onset of ADHD. We also devised lists of biological pathways implicated by genes associated with ADHD by previous studies2, 3, 30, 31 (Supplementary Methods 6). Finally, we created three “negative control” gene sets, which were not expected to show differential methylation profiles: gene sets pertaining to neurodegenerative (amyotrophic lateral sclerosis), pulmonary (chronic obstructive pulmonary disorder), and skeletal growth disorders. In total, we analyzed 39 candidate biological pathways or gene-sets.
To test for gene set enrichment, we mapped probes to their nearest gene, based on the genomic distance to the boundary of the protein coding locus (defined in the UCSC browser). We used the hypergeometric distribution to test whether the differentially methylated probes are significantly enriched in a candidate gene set for every given (Δβ, n) combination. A gene set was called significant when q <0.05. We repeated the above procedure for every combination of (Δβ, n) and counted the total number of significant calls for each candidate gene set across all combinations. The total number of significant calls required to pass the threshold for significant enrichment was determined through simulation (Supplementary Methods 7). To reach significance, the threshold for shore and shelf differentially methylated probes was ≥7 and the threshold for enhancer differentially methylated probes was ≥12 (both q = 0.043). A power analysis showed good power for the analyses pertaining to gene sets enriched by shore and shelf probes, and less power for those pertaining to enhancer probes (Supplementary Methods 8). Finally, we found no evidence that the gene set enrichment was confounded by cross-reactive probes, associated SNPs, or cell type heterogeneity in methylation analyses (Supplementary Methods 9 and 10).
Genes implicated by the probes with greatest differential methylation
We identified the genes implicated by small probe sets (<100 probes) which had either a large Δβ, n or both, thus implicating a manageably small number of associated genes for pathway analyses. These genes served as input for core Ingenuity Pathway Analyses. This software maps each gene to a corresponding gene object in the Ingenuity Knowledge Base. Enrichment of specific pathways was determined relative to the database, with Benjamini-Hochberg adjustment for multiple comparisons (q <0.05).
SNP arrays and CNV detection
Fifteen twin pairs were genotyped using the HumanOmniExpressExome BeadChip platform (964,193 SNPs), following the Illumina Infinium Assay protocol. The data were scanned by iScan and processed with the genotype module in GenomeStudio v2011.1 (Illumina, San Diego, CA). Samples with a call rate of <0.99 and SNPs with a GenTrain score of <0.7 were excluded from analysis. Copy number analysis was performed on the remaining 941,932 SNPs using CNVPartition v3.2 (Illumina, San Diego, CA) and Nexus Copy Number v7 (Biodiscovery, Inc, El Segundo, CA). Details of CNV definition are given in Supplementary Methods 11. We focused on CNVs that recurred across either affected or unaffected twins.
Exome sequencing to detect rare, functionally deleterious SNVs
We isolated exome DNA from whole blood genomic DNA and performed paired-end sequencing on the Illumina HiSeq 2500 (Illumina, San Diego, CA). We aligned sequence data using NovoAlign32 and removed PCR duplicates using SAMtools32. Data quality measures are given in Supplementary Methods 12. We compared the sequence of each pair of twins using programs designed to detect genetic differences between highly similar samples: Shimmer33, SomaticSniper34, and MuTect35. Genetic differences discovered by at least two programs were annotated for functional significance with Annovar36. Variants were classified as rare if they had a minor allele frequency of <1% in the 276 ClinSeq exomes with ≥10× coverage at that position (regardless of allele). We considered those rare, functionally deleterious SNVs which occurred only in affected or only in unaffected individuals.
Results
Clinical and neuroanatomic
The fifteen twin pairs who were fully discordant for ADHD had a mean age of 10.9 years (SD = 2.3 years) at first assessment. Twelve pairs (86%) were male, and all were of white, non-Hispanic ancestry. The affected twins had a mean of 7.8 (SD = 1.3) inattentive and 6.9 (SD = 1.6) hyperactive/impulsive symptoms. The unaffected twins had a mean of 1.2 (SD = 1.3) inattentive and 0.9 (SD = 1.4) hyperactive/impulsive symptoms (paired t-test for inattentive t(14) = 10.5, P <0.0001 and for hyperactive/impulsive symptoms t(14) = 10.6, P <0.00001). Comorbidity was confined to Oppositional Defiant Disorder, which was present in two affected twins. There was no twin difference in either general intelligence (affected twin mean IQ of 112, [SD = 15]; unaffected mean = 112 [SD = 12]; t(14) = 0.73, P = 0.47) nor in birth weight (unaffected, mean 2257 (SD 525) grams; affected 2196 (SD 455) grams: t(12)=1.11, p=0.29).
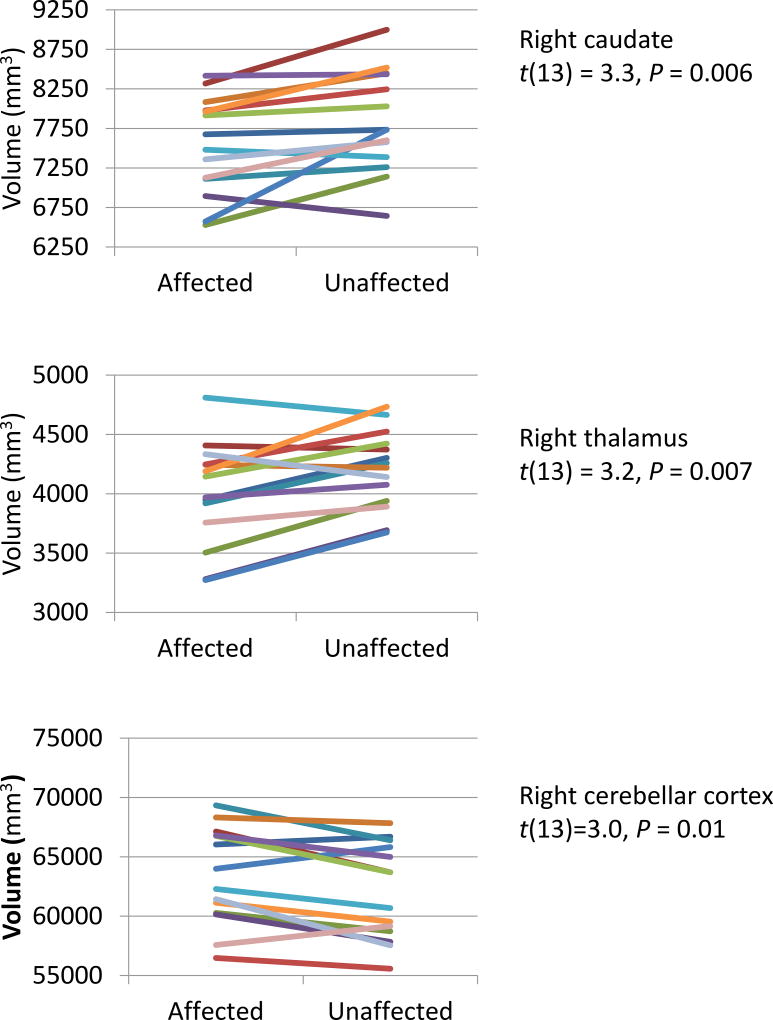
Within the 14 MZ twin pairs with neuroanatomic data, the affected twin had a significantly smaller (Bonferroni adjusted) right caudate (paired t = 3.31, P = 0.0055) and right thalamic nuclei (t = 3.2, P = 0.007) -Figure 2). By contrast, a larger right cerebellar cortex was associated with ADHD within twin pairs (t = 3.0, P = 0.01). The volumes of the right and left cerebral cortex did not differ between affected and unaffected twins (P >0.1).
Figure 2.
Brain structures showing neuroanatomic discordance in monozygotic twins. Each line connects the volumes for the brain structure indicated for a twin pair.
Epigenetic analyses
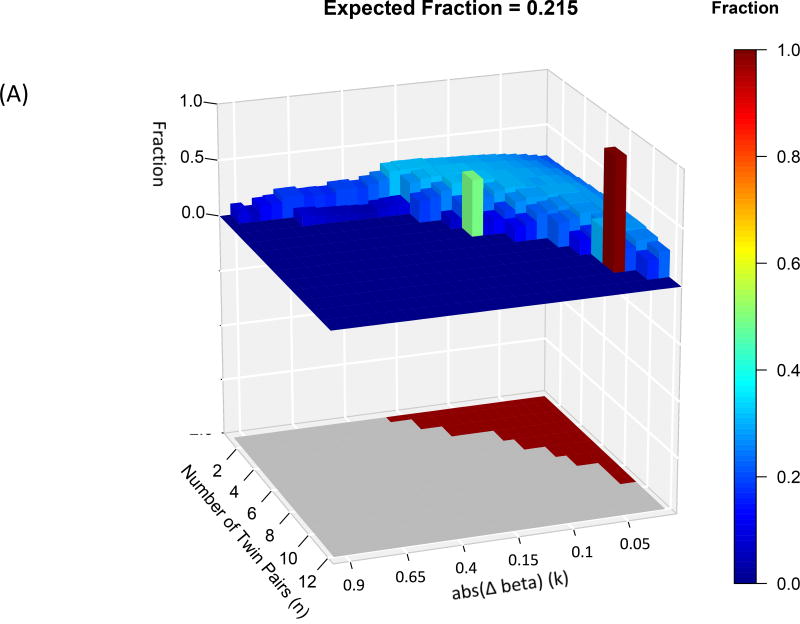
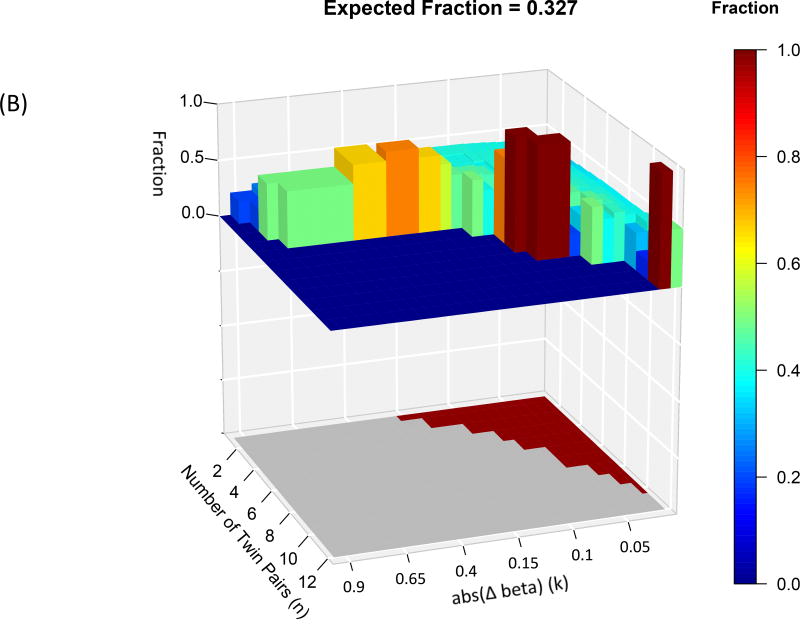
Differentially methylated probe sets were significantly enriched in shores and shelves and enhancers, but not in CpG islands (at FWER<0.05)- see Figure 3. Specifically, 68 of the 173 differentially methylated probe sets were enriched in shore and shelf regions and 67 differentially methylated probe sets were enriched in enhancer regions. We thus confined further analyses to shore/shelf and enhancer probes.
Figure 3.
Each graph has an upper 3D histogram showing the fraction of differentially methylated probes at each Δβ and N that lie in (A) enhancers; (B) shore and shelf; The lower 2D map indicates whether that differentially methylated probe set is significantly enriched (red) or not (grey). There was no significant enrichment of probes in CpG islands. Also note, differentially methylated probe sets with high fraction values may not attain significance due to the small numbers of probes in those sets.
We tested our primary hypothesis by determining whether these differentially methylated probe sets were enriched within genes expressed during early development in the discordant brain structures. Three of the brain gene sets pertaining to the discordant brain regions (striatum, thalamus and cerebellum) showed significant enrichment with differentially methylated shores and shelves, but not enhancer probes (see Methods; Table 1). Notably, we find that the gene sets pertaining to the concordant cerebral cortex were not enriched by differentially methylated probes. In further exploratory analyses, we calculated the Pearson correlations between probe methylation differences and volume differences in the discordant brain structures, and report significant probe-structure correlations (at p<0.001) in Table S8.
Table 1.
List of candidate gene sets tested, including the number of probes and number of differentially methylated probes associated with enhancer or shore and shelf regions.
| Gene-set name | N of genes |
N of probes associated with: |
N of significantly differentially methylated probes associated with: |
|||
|---|---|---|---|---|---|---|
| Enhancer regions |
SS* regions |
Enhancer regions |
SS* regions |
|||
| Brain gene-sets | Fetal cerebellum | 83 | 1136 | 1163 | 0 | 12 |
| Neonatal cerebellum | 100 | 1159 | 1175 | 0 | 9 | |
| Fetal striatum | 59 | 804 | 818 | 1 | 7 | |
| Neonatal striatum | 95 | 984 | 1378 | 0 | 4 | |
| Fetal mediodorsal nucleus thalamus | 100 | 949 | 968 | 0 | 10 | |
| Neonatal mediodorsal nucleus thalamus | 100 | 976 | 925 | 0 | 0 | |
| Fetal neocortex | 23 | 222 | 212 | 2 | 0 | |
| Neonatal neocortex | 13 | 109 | 95 | 1 | 0 | |
| ADHD gene-sets | Developmental growth involved in morphogenesis | 6 | 137 | 98 | 11 | 12 |
| System development | 34 | 742 | 551 | 3 | 9 | |
| Developmental growth | 8 | 233 | 119 | 13 | 9 | |
| Multicellular organismal development | 37 | 755 | 577 | 3 | 9 | |
| Generation of neurons | 16 | 404 | 244 | 14 | 8 | |
| Neuron differentiation | 15 | 396 | 237 | 14 | 7 | |
| Growth | 13 | 310 | 180 | 14 | 4 | |
| Neurological disease | 44 | 889 | 555 | 14 | 3 | |
| Neuron projection development | 13 | 381 | 215 | 15 | 2 | |
| Cation binding | 33 | 693 | 414 | 15 | 0 | |
| Cell projection organization | 14 | 414 | 230 | 12 | 0 | |
| Axonogenesis | 11 | 330 | 178 | 9 | 2 | |
| Glutamatergic synapse | 3 | 47 | 33 | 0 | 4 | |
| Keratinocyte proliferation | 3 | 51 | 28 | 0 | 2 | |
| Proteasome | 2 | 7 | 14 | 0 | 0 | |
| Ion binding | 36 | 765 | 648 | 7 | 0 | |
| Metal ion binding | 35 | 760 | 634 | 7 | 0 | |
| Calcium ion binding | 14 | 207 | 123 | 9 | 0 | |
| Hexokinase activity | 2 | 19 | 12 | 0 | 0 | |
| Microtubule cytoskeleton organization and biogenesis | 20 | 75 | 138 | 2 | 0 | |
| Golgi vesicle transport | 27 | 100 | 165 | 1 | 0 | |
| Regulation of cytoskeleton organization and biogenesis | 19 | 152 | 144 | 2 | 0 | |
| Spliceosome | 14 | 54 | 102 | 1 | 0 | |
| Nuclear hormone receptor binding | 11 | 110 | 166 | 0 | 0 | |
| Calcium channel activity | 22 | 292 | 317 | 3 | 3 | |
| Carboxylesterase activity | 13 | 57 | 44 | 1 | 0 | |
| Lipase activity | 19 | 124 | 91 | 2 | 0 | |
| ADHD CNV genes | 35 | 318 | 383 | 0 | 5 | |
| Control gene-sets | Skeletal development | 103 | 1303 | 1041 | 0 | 4 |
| Chronic obstructive pulmonary disease up | 157 | 630 | 1287 | 0 | 0 | |
| Amyotrophic lateral sclerosis ALS | 53 | 292 | 493 | 0 | 0 | |
Bold numbers indicate gene sets meeting the threshold of significance (at least 12 significant calls for enhancer gene sets and at least seven significant calls for shore and shelf gene sets).
SS= shore and shelf
We next examined 28 gene sets implicated in ADHD by previous research. Six of these ADHD gene sets showed enrichment of genes implicated by differentially methylated shore and shelf probes, and eight showed enrichment by differentially methylated enhancer probes. None of the ‘negative’ control gene-sets pertaining to non-ADHD related problems showed any enrichment (see Methods).
We repeated the analyses using the ‘rank-sum’ approach. (Supplementary Methods 13). In line with our main analyses, we found that the top ranked probes enriched both shore and shelf and enhancer regions (Supplementary Figure 5), but did not find any candidate gene set enrichment by these top ranked probes (top 100 ranked probes listed in Table S9).
Genetic variants
No CNVs occurred more than once across the affected twins, and there was no overall excess of CNVs in the affected twins. A comparison of the CNV burden between affected and unaffected twins is given in the Supplementary Table 7, Supplementary Figure 4.
No single nucleotide variants were detected that met our criteria of being (1) present in either only affected or only in unaffected twins; (2) recurrent; (3) designated as rare with adequate coverage by sequence data.
Pathway analyses
In exploratory, hypothesis free analyses, we mapped the 453 probes with the greatest differential methylation to the nearest gene – see Supplementary Table 10 Several of these genes are expressed in the discordant brain structures. These include neurodevelopmental genes, specifically homeobox genes (PAX6 and MEIS2), neural transcription regulators (BTB3D), and neurotrophins (NGFR).
Pathway analyses implicate neurotransmitter signaling
The genes implicated by the most differentially methylated probes were submitted to core pathway analyses. This approach identified significant enrichment (surviving Benjamini-Hochberg multiple testing correction; q <0.05) for signaling pathways within the brain (Table 2). For both shore and shelf and enhancer probes, GABA receptor signaling emerged as the most strongly implicated pathway. Other signaling pathways implicated by enhancer (though not shore and shelf) differentially methylated probes, included ERK (extracellular signal regulated kinase) and monoaminergic (serotonin, dopamine) neurotransmitter pathways.
Table 2.
Biological pathways that were overrepresented in the gene lists associated with the differentially methylated (A) shores and shelves regions, and (B) enhancer sites.
| (A) | ||
|---|---|---|
| Pathway | −log (BH p value) |
Gene names |
| GABA receptor signaling | 1.53 | ADCY9, MRAS, GABBR1, ADCY10, KCNH2 |
| Gs signaling | 1.53 | ADCY9, VIPR2, GLP1R, MRAS, ADD1, ADCY10 |
| ERK/MAPK signaling | 1.37 | YWHAG, ATF1, PPP2R5D, HIST1H3C, MRAS, RAC1, PIK3R5 |
| Breast cancer regulation by stathmin 1 | 1.37 | ADCY9, PPP2R5D, MRAS, RAC1, PIK3R5, ADCY10, TUBB |
| Superpathway of inositol phosphate compounds | 1.37 | SOCS3, ATP1A1, PPP2R5D, PIK3R5, MTMR7, INPP5A, SIRPA |
| IGF-1 signaling | 1.37 | SOCS3, YWHAG, NOV, MRAS, PIK3R5 |
| CDK5 signaling | 1.37 | ADCY9, PPP2R5D, NGFR, MRAS, ADCY10 |
| Gap junction signaling | 1.36 | ADCY9, NOV, MRAS, PIK3R5, ADCY10, TUBB |
| 3-phosphoinositide biosynthesis | 1.36 | SOCS3, ATP1A1, PPP2R5D, PIK3R5, MTMR7, SIRPA |
| Renin-angiotensin signaling | 1.34 | ADCY9, MRAS, RAC1, PIK3R5, ADCY10 |
| (B) | ||
|---|---|---|
| Pathway | −log (BH p value) |
Gene names |
| GABA receptor signaling | 2.49 | ADCY9, ADCY10, KCNH2 |
| Phospholipase C signaling | 2.40 | ADCY9, HDAC4, FCGR2A, ADCY10, PLD1 |
| Fc receptor-mediated phagocytosis in macrophages and monocytes | 2.09 | DOCK1, FCGR2A, PLD1 |
| Gs signaling | 1.79 | ADCY9, ADORA3, ADCY10 |
| Serotonin receptor signaling | 1.79 | ADCY9, ADCY10 |
| Cellular effects of sildenafil (Viagra) | 1.71 | ADCY9, ADCY10, KCNH2 |
| G-protein coupled receptor signaling | 1.57 | ADCY9, ADORA3, ADCY10, ADRA1B |
| CXCR4 signaling | 1.53 | DOCK1, ADCY9, ADCY10 |
| Gap junction signaling | 1.50 | ADCY9, NOV, ADCY10 |
| GPCR-mediated integration of enteroendocrine signaling exemplified by an L cell | 1.41 | ADCY9, ADCY10 |
| Endothelin-1 signaling | 1.39 | ADCY9, ADCY10, PLD1 |
| Leptin signaling in obesity | 1.37 | ADCY9, ADCY10 |
| Role of NFAT in cardiac hypertrophy | 1.35 | ADCY9, HDAC4, ADCY10 |
| Dopamine receptor signaling | 1.33 | ADCY9, ADCY10 |
| ERK/MAPK signaling | 1.30 | DOCK1, ELF2, YWHAG |
The minus log of the Benjamini-Hochberg (BH) corrected p value is given (values >1.3 indicate significance at adjusted P<0.05).
Discussion
This study leverages in vivo neuroanatomic imaging to inform the search for epigenetic and genetic changes that contribute to discordance for ADHD within MZ twin pairs. At the neuroanatomic level, the affected twin had a significantly smaller right striatum and thalamus, and a trend toward a larger cerebellum. Affected and unaffected twins did not differ in cerebral cortical volume. Among these twin pairs, differential methylation of shore and shelf and enhancer sites was associated with genes expressed during the early development of the striatum, thalamus, and cerebellum. Thus, as hypothesized, differentially methylated probes were enriched among genes expressed in discordant brain structures. Conversely, the cerebral cortex did not differ in volume between twins, and genes expressed in early cerebral cortical development did not show differential methylation. Hypothesis-free approaches using genome wide level methylation data implicated pathways pertaining to neurotransmitter signaling (mainly GABA) and genes expressed in discordant brain structures. Several candidate genes were also identified through our epigenetic analyses, including homeobox gene MEIS2 and the VIPR2 gene, implicated by the only other epigenetic study of ADHD9, 37. Finally, copy number variants and rare, deleterious SNVs did not emerge as a major driver of discordance in this small sample.
We extend prior reports of striatal volume differences between MZ twins discordant for ADHD by showing similar differences in the putamen, thalamus, and the cerebellum16. Each of these structures has been implicated in ADHD by previous studies38–42. Similar neuroanatomic divergence has also been reported in MZ twins discordant for other neuropsychiatric disorders, including autism, Alzheimer’s, and schizophrenia43–45. Of particular relevance are differences in cerebellar but not cerebral cortex volumes in MZ twins discordant for autism (Kates, Burnette et al. 2004). Autistic Spectrum Disorder and ADHD have a strong genetic overlap and share certain clinical features, such as early age of onset and male preponderance46, 47. Our finding adds neuroanatomic change in the cerebellum as another shared feature between Autistic Spectrum Disorder and ADHD.
Given that brain tissue is not available from twins with ADHD, nor from singletons, we linked peripheral differential methylation profiles with in vivo neuroanatomic changes. Others have more directly examined peripheral blood and brain tissue methylation patterns and found that the profiles are at least partly correlated48, 49. Additionally, epigenetic modifications have been linked with neurogenesis, brain development, and neurodevelopmental disorders50–53. In combination with data from previous studies, our findings bolster the case for assigning possible biological significance to our epigenetic findings.
Genes implicated by differential methylation
Notably, some candidate genes in ADHD – such as MEIS-2, BTBD3, NGRF, and VIPR2 – were also implicated by our differential methylation studies. Homeobox genes, pivotal in neurodevelopment, were strongly implicated by both highly differential methylation profiles and by expression in discordant brain structures (Table S10). Splice and nonsense mutations of the paired box homeotic gene-6 (PAX6), which encodes a transcriptional regulator involved in cerebellum and eye development,54 have been implicated in syndromes characterized by cerebellar deficits, intellectual disability and aniridia55. The PAX6 gene has rich interactions with another homeobox gene, Homeobox protein MEIS-2, which is both expressed in the striatum and differentially methylated in this study. Together, these genes act as transcriptional regulators of several genes (including EPH8A) in the developing midbrain56. The MEIS-2 gene has also been nominally associated with the severity of hyperactive-impulsive symptoms in a family-based study of ADHD37 and we found MEIS-2 associated probes to be hypermethylated in 10 of the affected twins (with Δβ>=0.02).
Others genes involved in brain development were also implicated by extreme differences in methylation profiles. The, BTB (POZ) domain containing 3 (BTBD3) gene acts as a key regulator of dendritic field orientation during development of sensory cortex and is expressed in the fetal cerebellum57. The nerve growth factor receptor (NGRF) gene is expressed in the cerebellum, and binds several neurotrophic factors. These neurotrophic factors are involved in neuronal survival, myelination, and synapse formation and are thus strong candidates for neuropsychiatric disorders58. Although NGFR was not associated with ADHD in a case-control study59 common variants in the gene have been associated with bipolar affective disorder, depression, and suicidality60–62.
We also find differential methylation of the VIPR2 gene, with hypermethylation occurring in three affected twins (with Δβ>=0.15), and this gene formed part of the gene-enriched Gs signaling pathways we report (q=0.03- see Table 2). A prior study, using salivary DNA, also found the VIPR2 gene to be differentially methylated9. We note however that we found hypermethylation, rather than hypomethylation as in the earlier study, and the probes in each study were at slightly different locations. Nonetheless, some genes implicated by differentially methylated probes are emerging as worthy of further evaluation.
Pathway analyses
The pathway analyses showed an enrichment of signaling pathways related to neurotransmission. GABA signaling pathways were implicated by both shore and shelf and enhancer differentially methylated probes. The GABA neurotransmitter pathway has rich interactions with dopaminergic systems that have long been thought to play a pivotal role in ADHD, and GABAergic genes appear enriched for CNVs in ADHD30, 63. Another implicated pathway, the CDK5 signaling pathway, also acts on dopamine signaling, amplifying signals through a positive feedback loop64. The ERK (extracellular signal regulated kinase) pathway is of particular interest, as genetic alterations in the pathways are emerging as a leading cause syndrome characterized by global intellectual impairment and constellations of motor and cognitive delays65.
Strengths and limitations
The MZ twins were ascertained through rigorous clinical assessment, which may have amplified the likelihood of causal events being present in the affected subjects. By the same token, such an “extreme” twin phenotype is very rare, so we may have failed to detect inherently infrequent genomic events such as rare, deleterious SNVs in such a small cohort.
Could differential methylation reflect discordance for factors other than ADHD, such as medication or comorbid disorders? Eight of the fourteen affected twins had taken psychostimulant medications as treatment for their ADHD; however, no associations between psychostimulants and methylation profiles have been found in prior work9. Non-disorder-specific neuropsychological differences are also unlikely to drive discordant methylation, as we did not find general intelligence differences between affected and unaffected twins. Contributions from other psychiatric disorders are unlikely, as the only comorbidity was oppositional defiant disorder, which was present in only two affected twins. Future studies would ideally include MZ twins who were concordant for health or concordant for ADHD, allowing firmer conclusions to be drawn about the specificity of the methylation changes we report.
Conclusion
The study illustrates how the integration of neuroimaging, genomics and epigenetics can reveal potential new pathophysiological mechanisms involved in the neurodevelopmental disorders.
Supplementary Material
Acknowledgments
The study was funded by the Intramural Research Programs of the National Human Genome Research Institute and the National Institute of Mental Health.
Footnotes
All authors declare no conflict of interest.
References
- 1.ADHD. Data and Statistics. http://www.cdc.gov/ncbddd/adhd/data.html. 2011, Accessed Date Accessed 2011 Accessed.
- 2.Hawi Z, Cummins T, Tong J, Johnson B, Lau R, Samarrai W, et al. The molecular genetic architecture of attention deficit hyperactivity disorder. Molecular Psychiatry. 2015 doi: 10.1038/mp.2014.183. [DOI] [PubMed] [Google Scholar]
- 3.Franke B, Faraone SV, Asherson P, Buitelaar J, Bau CHD, Ramos-Quiroga JA, et al. The genetics of attention deficit/hyperactivity disorder in adults, a review. Molecular Psychiatry. 2012;17:960–987. doi: 10.1038/mp.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong C, Meaburn EL, Ronald A, Price T, Jeffries A, Schalkwyk L, et al. Methylomic analysis of monozygotic twins discordant for autism spectrum disorder and related behavioural traits. Molecular Psychiatry. 2014;19(4):495–503. doi: 10.1038/mp.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cordova-Palomera A, Fatjo-Vilas M, Gasto C, Navarro V, Krebs M, Fananas L. Genome-wide methylation study on depression: differential methylation and variable methylation in monozygotic twins. Translational psychiatry. 2015;5(4):e557. doi: 10.1038/tp.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisher HL, Murphy TM, Arseneault L, Caspi A, Moffitt TE, Viana J, et al. Methylomic analysis of monozygotic twins discordant for childhood psychotic symptoms. Epigenetics. 2015;10(11):1014–1023. doi: 10.1080/15592294.2015.1099797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dempster EL, Pidsley R, Schalkwyk LC, Owens S, Georgiades A, Kane F, et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet. 2011:ddr416. doi: 10.1093/hmg/ddr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oh G, Wang S-C, Pal M, Chen ZF, Khare T, Tochigi M, et al. DNA modification study of major depressive disorder: beyond locus-by-locus comparisons. Biological psychiatry. 2015;77(3):246–255. doi: 10.1016/j.biopsych.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilmot B, Fry R, Smeester L, Musser ED, Mill J, Nigg JT. Methylomic analysis of salivary DNA in childhood ADHD identifies altered DNA methylation in VIPR2. Journal of Child Psychology and Psychiatry. 2015 doi: 10.1111/jcpp.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castellani CA, Awamleh Z, Melka MG, O'Reilly RL, Singh SM. Copy number variation distribution in six monozygotic twin pairs discordant for schizophrenia. Twin Research and Human Genetics. 2014;17(02):108–120. doi: 10.1017/thg.2014.6. [DOI] [PubMed] [Google Scholar]
- 11.Ono S, Imamura A, Tasaki S, Kurotaki N, Ozawa H, Yoshiura K-i, et al. Failure to confirm CNVs as of aetiological significance in twin pairs discordant for schizophrenia. Twin Research and Human Genetics. 2010;13(05):455–460. doi: 10.1375/twin.13.5.455. [DOI] [PubMed] [Google Scholar]
- 12.Bloom RJ, Kahler AK, Collins AL, Chen G, Cannon TD, Hultman C, et al. Comprehensive analysis of copy number variation in monozygotic twins discordant for bipolar disorder or schizophrenia. Schizophrenia Research. 2013;146(1–3):289–290. doi: 10.1016/j.schres.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruder CE, Piotrowski A, Gijsbers AA, Andersson R, Erickson S, de Ståhl TD, et al. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. The American Journal of Human Genetics. 2008;82(3):763–771. doi: 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506(7487):179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ronemus M, Iossifov I, Levy D, Wigler M. The role of de novo mutations in the genetics of autism spectrum disorders. Nature Reviews Genetics. 2014;15(2):133–141. doi: 10.1038/nrg3585. [DOI] [PubMed] [Google Scholar]
- 16.Castellanos FX, Sharp WS, Gottesman RF, Greenstein DK, Giedd JN, Rapoport JL. Anatomic brain abnormalities in monozygotic twins discordant for attention deficit hyperactivity disorder. American Journal of Psychiatry. 2003;160(9):1693–1696. doi: 10.1176/appi.ajp.160.9.1693. [DOI] [PubMed] [Google Scholar]
- 17.Nakao T, Radua J, Rubia K, Mataix-Cols D. Gray Matter Volume Abnormalities in ADHD: Voxel-Based Meta-Analysis Exploring the Effects of Age and Stimulant Medication. American Journal of Psychiatry. 2011;2011:24. doi: 10.1176/appi.ajp.2011.11020281. [DOI] [PubMed] [Google Scholar]
- 18.Valera EM, Faraone SV, Murray KE, Seidman LJ. Meta-analysis of structural imaging findings in attention-deficit/hyperactivity disorder. Biological Psychiatry. 2007;61(12):1361–1369. doi: 10.1016/j.biopsych.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 19.Frodl T, Skokauskas N. Meta-analysis of structural MRI studies in children and adults with attention deficit hyperactivity disorder indicates treatment effects. Acta Psychiatrica Scandinavica. 2012;125(2):114–126. doi: 10.1111/j.1600-0447.2011.01786.x. [DOI] [PubMed] [Google Scholar]
- 20.Xia S, Li X, Kimball AE, Kelly MS, Lesser I, Branch C. Thalamic shape and connectivity abnormalities in children with attention-deficit/hyperactivity disorder. Psychiatry Research: Neuroimaging. 2012;204(2):161–167. doi: 10.1016/j.pscychresns.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castellanos FX, Proal E. Large-scale brain systems in ADHD: Beyond the prefrontal-striatal model. Trends in Cognitive Sciences. 2012;16(1):17–26. doi: 10.1016/j.tics.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reich W. Diagnostic interview for children and adolescents (DICA) Journal of the American Academy of Child & Adolescent Psychiatry. 2000;39(1):59–66. doi: 10.1097/00004583-200001000-00017. [DOI] [PubMed] [Google Scholar]
- 23.Galwey NW. A new measure of the effective number of tests, a practical tool for comparing families of non - independent significance tests. Genetic epidemiology. 2009;33(7):559–568. doi: 10.1002/gepi.20408. [DOI] [PubMed] [Google Scholar]
- 24.Morris TJ, Butcher LM, Feber A, Teschendorff AE, Chakravarthy AR, Wojdacz TK, et al. ChAMP: 450k Chip Analysis Methylation Pipeline. Bioinformatics. 2014;30(3):428–430. doi: 10.1093/bioinformatics/btt684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dempster EL, Pidsley R, Schalkwyk LC, Owens S, Georgiades A, Kane F, et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet. 2011;20(24):4786–4796. doi: 10.1093/hmg/ddr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher HL, Murphy TM, Arseneault L, Caspi A, Moffitt TE, Viana J, et al. Methylomic analysis of monozygotic twins discordant for childhood psychotic symptoms. Epigenetics. 2015;10(11):1014–1023. doi: 10.1080/15592294.2015.1099797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature Reviews Genetics. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 28.Dempster EL, Wong CC, Lester KJ, Burrage J, Gregory AM, Mill J, et al. Genome-wide methylomic analysis of monozygotic twins discordant for adolescent depression. Biological psychiatry. 2014;76(12):977–983. doi: 10.1016/j.biopsych.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478(7370):483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elia J, Glessner JT, Wang K, Takahashi N, Shtir CJ, Hadley D, et al. Genome-wide copy number variation study associates metabotropic glutamate receptor gene networks with attention deficit hyperactivity disorder. Nat Genet. 2012;44(1):78–84. doi: 10.1038/ng.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams NM, Zaharieva I, Martin A, Langley K, Mantripragada K, Fossdal R, et al. Rare chromosomal deletions and duplications in attention-deficit hyperactivity disorder: a genome-wide analysis. The Lancet. 2010;376(9750):1401–1408. doi: 10.1016/S0140-6736(10)61109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hansen NF, Gartner JJ, Mei L, Samuels Y, Mullikin JC. Shimmer: detection of genetic alterations in tumors using next-generation sequence data. Bioinformatics. 2013;29(12):1498–1503. doi: 10.1093/bioinformatics/btt183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larson DE, Harris CC, Chen K, Koboldt DC, Abbott TE, Dooling DJ, et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28(3):311–317. doi: 10.1093/bioinformatics/btr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotech. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Research. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lasky-Su J, Neale BM, Franke B, Anney RJL, Zhou K, Maller JB, et al. Genome-wide association scan of quantitative traits for attention deficit hyperactivity disorder identifies novel associations and confirms candidate gene associations. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2008;147B(8):1345–1354. doi: 10.1002/ajmg.b.30867. [DOI] [PubMed] [Google Scholar]
- 38.Friedman LA, Rapoport JL. Brain development in ADHD. Current Opinion in Neurobiology. 2015;30:106–111. doi: 10.1016/j.conb.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 39.Shaw P, De Rossi P, Watson B, Wharton A, Greenstein D, Raznahan A, et al. Mapping the Development of the Basal Ganglia in Children With Attention-Deficit/Hyperactivity Disorder. Journal of the American Academy of Child & Adolescent Psychiatry. 2014;53(7):780–789. doi: 10.1016/j.jaac.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoodley CJ. Distinct regions of the cerebellum show gray matter decreases in autism, ADHD, and developmental dyslexia. Frontiers in systems neuroscience. 2014;8 doi: 10.3389/fnsys.2014.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivanov I. Morphological abnormalities of the thalamus in youths with attention deficit hyperactivity disorder. The American journal of psychiatry. 2010;167(4):397. doi: 10.1176/appi.ajp.2009.09030398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mackie S, Shaw P, Lenroot R, Pierson R, Greenstein DK, Nugent TF, 3rd, et al. Cerebellar development and clinical outcome in attention deficit hyperactivity disorder. American Journal of Psychiatry. 2007;164(4):647–655. doi: 10.1176/ajp.2007.164.4.647. [see comment] [DOI] [PubMed] [Google Scholar]
- 43.Rossi R, Pievani M, Järvenpää T, Testa C, Koskenvuo M, Räihä I, et al. Voxel-based morphometry study on monozygotic twins discordant for Alzheimer's disease. Acta Neurologica Scandinavica. 2015 doi: 10.1111/ane.12480. [DOI] [PubMed] [Google Scholar]
- 44.Suddath RL, Christison GW, Torrey EF, Casanova MF, Weinberger DR. Anatomical abnormalities in the brains of monozygotic twins discordant for schizophrenia. New England Journal of Medicine. 1990;322(12):789–794. doi: 10.1056/NEJM199003223221201. [DOI] [PubMed] [Google Scholar]
- 45.Pol HEH, Schnack HG, Mandl RC, Brans RG, van Haren NE, Baaré WF, et al. Gray and white matter density changes in monozygotic and same-sex dizygotic twins discordant for schizophrenia using voxel-based morphometry. Neuroimage. 2006;31(2):482–488. doi: 10.1016/j.neuroimage.2005.12.056. [DOI] [PubMed] [Google Scholar]
- 46.Ronald A, Simonoff E, Kuntsi J, Asherson P, Plomin R. Evidence for overlapping genetic influences on autistic and ADHD behaviours in a community twin sample. Journal of Child psychology and Psychiatry. 2008;49(5):535–542. doi: 10.1111/j.1469-7610.2007.01857.x. [DOI] [PubMed] [Google Scholar]
- 47.Taurines R, Schwenck C, Westerwald E, Sachse M, Siniatchkin M, Freitag C. ADHD and autism: differential diagnosis or overlapping traits? A selective review. ADHD Attention Deficit and Hyperactivity Disorders. 2012;4(3):115–139. doi: 10.1007/s12402-012-0086-2. [DOI] [PubMed] [Google Scholar]
- 48.Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13(6):R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaminsky Z, Tochigi M, Jia P, Pal M, Mill J, Kwan A, et al. A multi-tissue analysis identifies HLA complex group 9 gene methylation differences in bipolar disorder. Molecular psychiatry. 2012;17(7):728–740. doi: 10.1038/mp.2011.64. [DOI] [PubMed] [Google Scholar]
- 50.Lv J, Xin Y, Zhou W, Qiu Z. The epigenetic switches for neural development and psychiatric disorders. Journal of genetics and genomics. 2013;40(7):339–346. doi: 10.1016/j.jgg.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 51.Ma DK, Marchetto MC, Guo JU, Ming G-l, Gage FH, Song H. Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nature neuroscience. 2010;13(11):1338–1344. doi: 10.1038/nn.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pidsley R, Dempster E, Mill J. Brain weight in males is correlated with DNA methylation at IGF2. Molecular psychiatry. 2010;15(9):880–881. doi: 10.1038/mp.2009.138. [DOI] [PubMed] [Google Scholar]
- 53.Peña CJ, Bagot RC, Labonté B, Nestler EJ. Epigenetic signaling in psychiatric disorders. Journal of molecular biology. 2014;426(20):3389–3412. doi: 10.1016/j.jmb.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heins N, Malatesta P, Cecconi F, Nakafuku M, Tucker KL, Hack MA, et al. Glial cells generate neurons: the role of the transcription factor Pax6. Nature neuroscience. 2002;5(4):308–315. doi: 10.1038/nn828. [DOI] [PubMed] [Google Scholar]
- 55.Graziano C, D'Elia AV, Mazzanti L, Moscano F, Guidelli Guidi S, Scarano E, et al. A de novo nonsense mutation of PAX6 gene in a patient with aniridia, ataxia, and mental retardation. American Journal of Medical Genetics Part A. 2007;143A(15):1802–1805. doi: 10.1002/ajmg.a.31808. [DOI] [PubMed] [Google Scholar]
- 56.Nakamura T, Jenkins NA, Copeland NG. Identification of a new family of Pbx-related homeobox genes. Oncogene. 1996;13(10):2235–2242. [Google Scholar]
- 57.Matsui A, Tran M, Yoshida AC, Kikuchi SS, Mami U, Ogawa M, et al. BTBD3 controls dendrite orientation toward active axons in mammalian neocortex. Science. 2013;342(6162):1114–1118. doi: 10.1126/science.1244505. [DOI] [PubMed] [Google Scholar]
- 58.Sofroniew MV, Howe CL, Mobley WC. Nerve growth factor signaling, neuroprotection, and neural repair. Annual review of neuroscience. 2001;24(1):1217–1281. doi: 10.1146/annurev.neuro.24.1.1217. [DOI] [PubMed] [Google Scholar]
- 59.Ribases M, Hervas A, Ramos-Quiroga JA, Bosch R, Bielsa A, Gastaminza X, et al. Association study of 10 genes encoding neurotrophic factors and their receptors in adult and child attention-deficit/hyperactivity disorder. Biological Psychiatry. 2008;63(10):935–945. doi: 10.1016/j.biopsych.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 60.Gassó P, Ortiz AE, Mas S, Morer A, Calvo A, Bargalló N, et al. Association between genetic variants related to glutamatergic, dopaminergic and neurodevelopment pathways and white matter microstructure in child and adolescent patients with obsessive–compulsive disorder. Journal of affective disorders. 2015;186:284–292. doi: 10.1016/j.jad.2015.07.035. [DOI] [PubMed] [Google Scholar]
- 61.Scott LJ, Muglia P, Kong XQ, Guan W, Flickinger M, Upmanyu R, et al. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proceedings of the National Academy of Sciences. 2009;106(18):7501–7506. doi: 10.1073/pnas.0813386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kunugi H, Hashimoto R, Yoshida M, Tatsumi M, Kamijima K. A missense polymorphism (S205L) of the low - affinity neurotrophin receptor p75NTR gene is associated with depressive disorder and attempted suicide. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2004;129(1):44–46. doi: 10.1002/ajmg.b.30062. [DOI] [PubMed] [Google Scholar]
- 63.Levy F, de Leon J. Dopamine ADHD/OCD Theories: Is Glutamine Part of the Story? Neurotransmitter. 2015;2 [Google Scholar]
- 64.Nishi A, Bibb JA, Snyder GL, Higashi H, Nairn AC, Greengard P. Amplification of dopaminergic signaling by a positive feedback loop. Proceedings of the National Academy of Sciences. 2000;97(23):12840–12845. doi: 10.1073/pnas.220410397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krab LC, Goorden SM, Elgersma Y. Oncogenes on my mind: ERK and MTOR signaling in cognitive diseases. Trends in genetics. 2008;24(10):498–510. doi: 10.1016/j.tig.2008.07.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.