Abstract
Mitochondrial function is altered in various pathologies, highlighting the crucial role mitochondria plays in maintaining cellular homeostasis. Mitochondrial structure undergoes constant fission and fusion in response to changing cellular environment. Due to this, analyzing mitochondrial structure could provide insight into the physiological state of the cell. In this protocol, we describe a method to analyze mitochondrial structure in body wall muscles in the nematode Caenorhabditis elegans, using both transgenic and dye-based approaches.
Keywords: C. elegans, Mitochondria, Calcium, Mitochondrial membrane potential, TMRE, Mitotracker
Background
Mitochondria are involved in ATP production, cellular respiration, calcium buffering and reactive oxidative species (ROS) metabolism ( Brookes et al., 2004 ). Mitochondrial structure and function are dynamic and closely linked, therefore analyzing mitochondrial structure can provide clues to the status of mitochondrial health (Sarasija and Norman, 2015). We developed two sets of protocols to assess mitochondrial structure in the body wall muscle of Caenorhabditis elegans. In the first protocol, we used transgenic ccIs4251 strain in which GFP is targeted to the matrix of the body wall muscle mitochondria to visualize the mitochondria ( Fire et al., 1998 ). In the second protocol, we used mitochondrially targeted dyes, MitoTrackerTM Red CMXRos and tetramethylrhodamine ethyl ester (TMRE) to determine the structural integrity of the body wall muscle mitochondria. Normally, animals used in in-vivo imaging are anesthetized, however anesthetizing the animals could lead to mitochondrial morphological changes ( Han et al., 2012 ), complicating data analysis. Our protocols allow for the in-vivo imaging of mitochondrial structure in live, un-anaesthetized nematodes.
Materials and Reagents
100 mm, 60 mm Petri dishes (Kord-Valmark Labware Products, catalog numbers: 2900, 2901)
Glass Pasteur pipettes (Krackeler Scientific, catalog number: 6-72050-900)
15-ml centrifuge tubes (Globe Scientific, catalog number: 6285)
22 x 22 mm coverslip (Globe Scientific, catalog number: 1404-10)
1.5 ml Micro Centrifuge tube (CELLTREAT Scientific, catalog number: 229443)
50 ml conical tubes (Corning, catalog number: 430829)
15 ml conical tubes (Corning, Centristar, catalog number: 430791)
C. elegans strains including strain SD1347, ccIs4251 [(pSAK2) myo-3p::GFP::LacZ::NLS + (pSAK4) myo-3p::mitochondrial GFP + dpy-20(+)] ( Liu et al., 2009 ) and OP50 (Caenorhabditis Genetics Center (CGC), University of Minnesota)
Deionized water (dH2O)
Polybead polystyrene 0.10 μm microspheres (Polysciences, catalog number: 00876-15)
Agarose (RPI, catalog number: A20090-500.0)
Clear nail polish (generic)
Carl ZeissTM ImmersolTM Immersion Oil (ZEISS, catalog number: 444960-0000-000)
Sodium chloride (NaCl) (Fisher Scientific, catalog number: BP358-10)
Agar (Fisher Scientific, catalog number: BP1423-2)
Bacto peptone (BD, BactoTM, catalog number: 211677)
Calcium chloride dihydrate (CaCl2·2H2O) (Fisher Scientific, catalog number: C79-500)
Magnesium sulfate heptahydrate (MgSO4·7H2O) (Fisher Scientific, catalog number: BP213-1)
Cholesterol (Fisher Scientific, catalog number: C314-500)
Potassium phosphate dibasic (K2HPO4) (Fisher Scientific, catalog number: BP363-1)
Potassium phosphate monobasic (KH2PO4) (Fisher Scientific, catalog number: P285-500)
Sodium phosphate dibasic anhydrous (Na2HPO4) (Fisher Scientific, catalog number: BP332-1)
Bleach (generic, plain)
Sodium hydroxide (NaOH) (Fisher Scientific, catalog number: BP359-500)
Bacto tryptone (BD, BactoTM, catalog number: 211705)
Bacto yeast extract (BD, BactoTM, catalog number: 212750)
MitoTracker Red CMXRos (Thermo Fisher Scientific, InvitrogenTM, catalog number: M7512)
Tetramethylrhodamine, Ethyl Ester, Perchlorate (TMRE) (Thermo Fisher Scientific, InvitrogenTM, catalog number: T669)
Standard worm (NGM) plates (see Recipes)
Sterile solutions (see Recipes)
-
Sterile stocks for NGM (see Recipes)
1 M CaCl2
1 M MgSO4
1 M K2HPO4
1 M KH2PO4
1 M KPO4 pH 6.0
M9 buffer (1 L) (see Recipes)
Bleach solution (see Recipes)
10 N NaOH (see Recipes)
MitotrackerTM Red CMXRos stock (see Recipes)
TMRE stock (see Recipes)
Equipment
Single channel pipettes (Rainin, models: PR-10, PR-20, PR-200, PR-1000)
Finnpipette II Multichannel pipettes (Fisher Scientific, model: FisherbrandTM FinnpipetteTM II, catalog number: 21377830)
20 °C Incubator (Percival Scientific, model: I-41NL)
Centrifuges (Eppendorf, models: 5415 D, 5415 R; Thermo Fischer Scientific, Thermo ScientificTM, model: IEC Centra CL2)
Zeiss SteREO Discovery.V8 microscope with SCHOTT Ace® I light source for maintaining (ZEISS, model: SteREO Discovery.V8)
Zeiss SteREO Discovery.V12 microscope with SCHOTT Ace® I light source and X-Cite® Series 120 Fluorescence Illuminator for transgenic selection (ZEISS, model: SteREO Discovery.V12)
Zeiss AxioObserver microscope with Andor Clara CCD camera and X-Cite® Series 120 Fluorescence Illuminator for imaging (ZEISS, model: Axio Observer)
PYREX® Griffin beakers (Corning, catalog number: 1000-PACK)
PYREX® Reusable Media Storage Bottles (Fisher Scientific)
Software
MetaMorph® Microscopy Automation & Image Analysis Software (Molecular Devices)
ImageJ (https://imagej.nih.gov/ij/)
Microsoft Office 2011 Excel (Microsoft Corporation, Redmond, USA)
-
GraphPad Prism software package (GraphPad Software Inc., San Diego, USA)
Part I: Determining mitochondrial structure using transgenic lines
Procedure
-
Growth and synchronization of nematode population
Transfer ccIs4251 L4 transgenic larvae, which express mitochondria-targeted GFP (mtGFP), onto a fresh E. coli (OP50) seeded NGM plate. Use at least three 60 mm or two 100 mm plates for each strain.
Incubate the animals at 20 °C for 3-4 days until plates contain a large number of eggs and gravid nematodes.
Wash the eggs and nematodes off the plates and transfer them using glass Pasteur pipettes into individual 15-ml centrifuge tubes for each strain using about 4 ml of M9 per plate of nematodes. Spin these down for 3 min at 6,180 × g and aspirate out the M9, retaining just the worms and eggs pellet. Add 3-4 ml of the bleach solution to each tube and vortex at full speed for 15 sec every 2 min for 6 min. Add M9 to fill each tube and spin at 6,180 × g for 1 min. Repeat the wash with M9 three times and move the worm eggs/carcass pellet to a fresh half-filled 15 ml tube of M9.
-
Synchronize the hatchlings by nutating at 20 °C for anywhere between 16-48 h (make sure that within an experiment all strains are synchronized for similar amounts of time). Due to lack of food, all the animals will arrest in their growth at L1 stage of larval growth. Spin these tubes down at 6,180 × g for 1.5 min and transfer them to individual OP50 seeded NGM plates and keep at 20 °C. Once the L1 animals are on OP50 seeded NGM plates, these animals will reach the L4 larval stage at ~42 h, and day 1 of adulthood at ~66 h.
Note: We have observed no difference in phenotypes between animals age synchronized by starving for 24 h vs. 48 h.
-
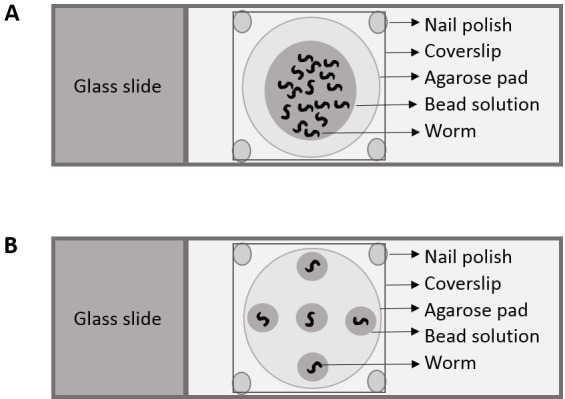
Mounting animals for imaging (Figure 1A)
Make 3% agarose solution in M9 and use this solution to make a ~18 mm diameter agarose pad on the top of a glass slide.
Add a 2 μl drop of polystyrene beads solution to this pad and pick approximately 20-30 animals onto this drop.
Gently place a coverslip on top of the animals and use a fingernail to gently nudge the coverslip about 0.5 mm to ‘roll’ the nematodes onto their back. Rolling the animals will allow them to be visualized dorsally or ventrally where a larger area of body wall mitochondria can be imaged versus when the animals are positioned laterally.
Now seal the edges of the coverslip with clear nail polish and the slide is ready for imaging on the Zeiss AxioObserver microscope equipped with an Andor Clara CCD camera using Metamorph software.
-
Imaging the mounted animals
Locate nematodes on the slide using the 10x objective and then switch over to the 63x oil objective to capture images.
-
Capture fluorescent images of body wall muscle mitochondria in the transgenic animals at 3 different points along their length.
Note: Ignore the vulval area in all animals, as the vulva is prone to deformation of body wall muscle mitochondria.
Imaging parameters such as the settings of the microscope and camera (magnification and exposure time (240 msec) should be kept the same during the imaging process).
Save these images and use them for data analysis under blinded conditions.
Figure 1. Cartoon depicting placement of nematodes for (A) Part I and (B) Part II.
Data analysis
Have someone other than the assayer/analyzer scramble the image file names and folders to obtain blinded conditions.
Open each image in Metamorph (ImageJ is a viable alternative) and characterize the mitochondria as linear, intermediate or fragmented, depending on the structural arrangement in > 50% of mitochondria (see Figure 2 for representative images).
-
Calculate the percentage mitochondria that exist as linear, intermediate and fragmented and analyze in chi-square tests using GraphPad Prism.
Notes:
Please refer to Sarasija and Norman, 2015 for representative data.
Analyze 15+ animals per strain for mitochondrial structure.
Mitochondrial morphological changes can happen due to various factors such as aging, drug treatments and genetic backgrounds.
Figure 2. Representative images of the three major mitochondrial phenotypic classifications in C. elegans body wall muscle mitochondria.
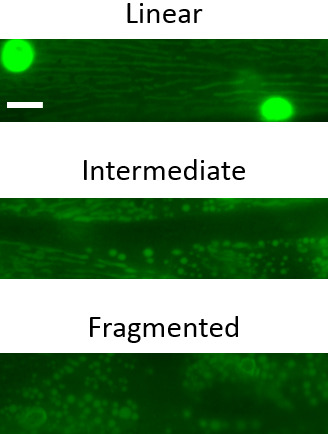
Scale bar represents 10 µm.
Part II: Determining mitochondrial structural and functional integrity using mitochondrial targeted fluorescent dyes
Procedure
-
Growth and synchronization of nematode population
Animals with mitoGFP, (ccIs4251) are grown and synchronized to L4 larval (for MitotrackerTM) or young adult (for TMRE) stage (~42 and ~54 h respectively, after putting down synchronized L1s on seeded NGM plates) for imaging as described in Part I Procedure A.
-
Staining the animals with mitochondrially targeted dyes
Wash animals off the plates along with OP50 into 15 ml centrifuge tubes.
Add MitotrackerTM or TMRE to a final concentration of 1 μg/ml or 200 nM in M9, respectively.
Nutate these animals in 1 μg/ml of MitotrackerTM or 200 nM of TMRE for 6 h at 20 °C in the dark.
Wash these animals with M9 four times and put them down on OP50 seeded NGM plates using the protocol described above.
-
Destain MitotrackerTM and TMRE by allowing the stained animals to forage on seeded plates overnight or for 1 h in the dark at 20 °C, respectively.
Note: Stain and destain animals on a staggering timescale so that each strain experiences similar staining and destaining times.
-
Mounting the animals for imaging (Figure 1B)
Make 3% agarose solution in M9 and use it to make ~18 mm diameter agarose pad.
Add five drops of polystyrene beads solution (~0.5 μl each drop); one at the center and 4 diametrically opposite to each other (Figure 1B).
Add one worm to each of the drops for 5 nematodes/slide. This will limit the impact of photobleaching.
Gently place a coverslip on top the animals and use a fingernail to gently nudge the coverslip about 0.5 mm to ‘roll’ the nematodes onto their back.
Now seal the edges of the coverslip with clear nail polish and the slide is ready for imaging on the Zeiss AxioObserver microscope equipped with an Andor Clara CCD camera using Metamorph software.
-
Imaging the mounted animals
Locate nematodes on the slide using the 10x objective and then switch over to the 63x oil objective to capture images.
Capture the green fluorescent image of body wall muscle mitochondria in the transgenic animals at a point between the pharynx and vulva. Now switch over to the red fluorescence filter set and capture the dye staining in the same area. Each animal should have two images associated with it; the mitochondria as labeled by the transgenic mitochondrially targeted GFP and the mitochondria as labeled by the mitochondrially targeted dyes.
Move clockwise and go to the next worm on the slide, repeating imaging protocol.
Imaging parameters such as the settings of the microscope and camera (magnification and exposure time (240 msec) should be kept the same during the imaging process).
Save these images and use them for data analysis under blinded conditions.
Data analysis
Assayer/analyst should be blinded as explained in Part I Procedure D.
Open the images of animals stained with MitotrackerTM CMXRos in MetaMorph and TMRE in ImageJ.
For the MitotrackerTM labeled animals, use a binary system and denote each image of the mitochondria as stained or unstained with the dye (see Figure 3 for representative images).
For the TMRE labeling, using the freehand tool in ImageJ, demarcate the boundaries of the mitochondria using the transgenic mitochondrial GFP image and measure the fluorescence intensity of the GFP signal. Now use the same region of interest in the TMRE labeled image and determine the fluorescence intensity of the TMRE dye labeling. Make sure to subtract the respective background fluorescence from both images. Now determine TMRE to GFP fluorescence intensity for each animal and normalize this to the average fluorescence intensity ratio of wild type animals (see Figure 4 for representative images).
Analyze the distribution of MitotrackerTM stained vs. unstained animals using a Chi-square test and the TMRE to GFP fluorescence intensity ratios of each strain using one-way ANOVA with Tukey or Bonferroni post-test. Please refer to the paper ( Sarasija and Norman, 2015 ) for any clarifications.
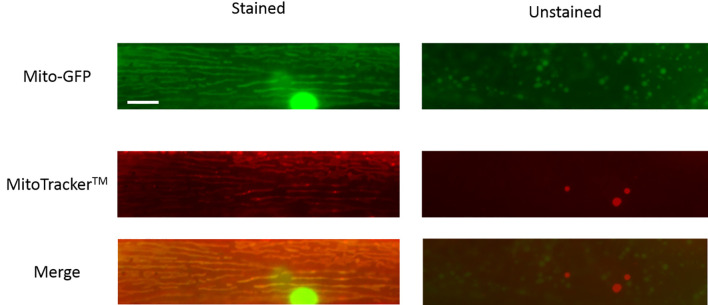
Figure 3. Representative images of C. elegans body wall muscle mitochondria that are stained or unstained by MitoTrackerTM.
Scale bar represents 10 µm.
Figure 4. Representative images of C. elegans body wall muscle mitochondria that are TMRE-labeled.
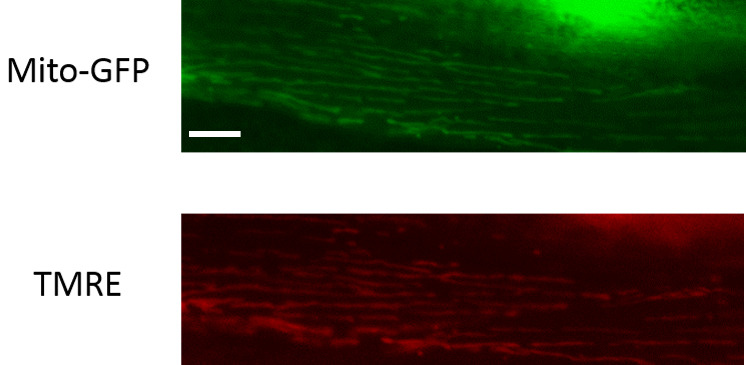
Scale bar represents 10 µm.
Notes
For further information on C. elegans growth and maintenance, please refer to the chapter on ‘Maintenance of C. elegans’ on WormBook available at http://www.wormbook.org/chapters/www_strainmaintain/strainmaintain.pdf.
Visualize these nematodes every day until they are imaged to make sure that they are not at the risk of starving. If the bacterial lawn on the plates look thin, nematodes need to be promptly moved to new OP50 seeded NGM plates. Starved animals should never be used for assaying.
The ccIs4251 strain expresses GFP that is targeted to the mitochondrial matrix and nucleus, which accounts for the presence of a large spherical structure in each muscle cell.
These protocols can be adapted to visualize the mitochondria in other tissues, like in the neurons. For example, jsIs609, a GFP transgene that is expressed in the mitochondrial matrix of mechanosensory neurons of C. elegans, can be used for this purpose ( Fatouros et al., 2012 ).
This protocol can be adapted to image animals at various stages. However, if mitochondrial dyes are to be used, then the animals will need to be incubated in the dyes accordingly, at the right time point. Also, it should be noted that the polystyrene beads will not immobilize animals at L1-L2 larval stages efficiently. If older animals need to be imaged, their eggs could pose an issue which could potentially be circumvented by focusing on imaging the region where eggs will not be present. Another option is to sterilize the animals using by moving them to 5-Fluoro-2’-deoxyuridine (FUDR) containing NGM plates. The latter solution might present problems since there is some concern in the field regarding the impact of FUDR on mitochondrial structure and function.
Please refer to Sarasija and Norman (2015) for representative data for dye staining analysis.
Recipes
Note: For additional information, please refer to He, 2011.
-
Standard Nematode Growth Media (NGM) plates
1 L 0.5 L 0.25 L NaCl 3 g 1.5 g 0.75 g Bacto-agar 17 g 8.5 g 4.25 g Bacto-peptone 2.5 g 1.25 g 0.625 g Store at 4 °C -
Sterile solutions
-
Autoclave on liquid cycle (45 min exposure), allow to cool to ~60 °C and then use sterile technique and add the following:
1 L 0.5 L 0.25 L 1 M CaCl2 1 ml 0.5 ml 0.25 ml 1 M MgSO4 1 ml 0.5 ml 0.25 ml 1 M KPO4 25 ml 12.5 ml 6.25 ml 5 mg/ml cholesterol 1 ml 0.5 ml 0.25 ml Swirl to mix thoroughly after each addition. After all additions are made, pour plates
To make liquid NGM, follow the same protocol but omit the addition of Bacto-agar (and obviously no need to pour into plates)
-
-
Sterile stocks for NGM
-
1 M CaCl2
In a beaker (sterile if possible), add:
110.9 g CaCl2
dH2O to 250 ml
Dissolve thoroughly and use vacuum filtration to sterilize, label, and store at room temperature
-
1 M MgSO4
In a beaker (sterile if possible), add:
120.3 g MgSO4
dH2O to 250 ml
Dissolve thoroughly and use vacuum filtration to sterilize, label, and store at room temperature
-
1 M K2HPO4
174.18 g bring up to 1 L with dH2O
-
1 M KH2PO4
136.01 g bring up to 1 L with dH2O and heat
-
1 M KPO4 pH 6.0
Measure out 132 ml of 1 M K2HPO4 and 868 ml of 1 M KH2PO4
Combine, and autoclave on liquid cycle (45 min exposure)
Store at room temperature
-
-
M9 buffer (1 L) (store at room temperature)
dH2O 1,000 ml NaCl 5 g KH2PO4 3 g Na2HPO4 6 g 1 M MgSO4 1 ml *add after autoclaving Split between 2 bottles: 500 ml each. Autoclave on liquid cycle (15 min exposure)
-
Bleach solution (store at room temperature)
25 ml final volume 50 ml final volume dH2O 18 ml 36 ml Bleach 7 ml 14 ml 10 N NaOH 400 μl 800 μl -
10 N NaOH
20 g NaOH
Bring up to 50 ml with dH2O
Store at room temperature
Notes:
Exercise CAUTION: This reaction is EXOTHERMIC!
50 ml conical should be fine, but make sure lid is tight, keep ice around to cool tube down to prevent conical breakage (expansion by heat).
-
MitotrackerTM Red CMXRos stock (make fresh/store at -20 °C for a week)
Add 1 ml of dimethylsulfoxide (DMSO) to a tube containing 50 µg of MitotrackerTM Red CMXRos to make a 50 µg/ml stock of MitotrackerTM Red CMXRos
Dilute this 50 fold in M9 for staining animals
-
TMRE stock (make fresh/store at -20 °C for a week)
Add 970.9 µl of DMSO to a tube containing 25 mg of TMRE to make a 50 mM stock solution
Dilute this stock 250 fold in M9 to get the desired concentration of 200 nM
Acknowledgments
The authors would like to acknowledge J. T. Laboy for her help in ordering and preparing reagents, and P. McKeown-Longo, Y. Tang, M. Barroso and their lab members for support. Some nematode strains were provided by the Caenorhabditis Genetics Center, which is funded by National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440). The Alzheimer’s Association (NIRG-09-132122) and NIH (GM088213) supported this work. The authors declare that there are no conflicts of interest or competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Brookes P. S., Yoon Y., Robotham J. L., Anders M. W. and Sheu S. S.(2004). Calcium, ATP, and ROS: a mitochondrial love-hate triangle. Am J Physiol Cell Physiol 287(4): C817-833. [DOI] [PubMed] [Google Scholar]
- 2. Fatouros C., Pir G. J., Biernat J., Koushika S. P., Mandelkow E., Mandelkow E. M., Schmidt E. and Baumeister R.(2012). Inhibition of tau aggregation in a novel Caenorhabditis elegans model of tauopathy mitigates proteotoxicity . Hum Mol Genet 21(16): 3587-3603. [DOI] [PubMed] [Google Scholar]
- 3. Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E. and Mello C. C.(1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature 391(6669): 806-811. [DOI] [PubMed] [Google Scholar]
- 4. Han S. M., Tsuda H., Yang Y., Vibbert J., Cottee P., Lee S. J., Winek J., Haueter C., Bellen H. J. and Miller M. A.(2012). Secreted VAPB/ALS8 major sperm protein domains modulate mitochondrial localization and morphology via growth cone guidance receptors. Dev Cell 22(2): 348-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. He F.(2011). Common worm media and buffers. Bio-protocol Bio101: e55. [Google Scholar]
- 6. Liu X., Long F., Peng H., Aerni S. J., Jiang M., Sanchez-Blanco A., Murray J. I., Preston E., Mericle B., Batzoglou S., Myers E. W. and Kim S. K.(2009). Analysis of cell fate from single-cell gene expression profiles in C. elegans . Cell 139(3): 623-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sarasija S. and Norman K. R.(2015). A γ-secretase independent role for presenilin in calcium homeostasis impacts mitochondrial function and morphology in Caenorhabditis elegans . Genetics 201(4): 1453-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stiernagle T.(2006). Maintenance of C. elegans. WormBook(Ed.). The C. elegans Research Community: doi/ 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]