Abstract
Background
Oesophageal (OeC) and gastric (GC) cancer patients are treated with similar multimodal therapy and have poor survival. There remains an urgent clinical need to identify biomarkers to individualise patient management and improve outcomes. Therapy with immune checkpoint inhibitors has shown promising results in other cancers. Proposed biomarkers to predict potential response to immune checkpoint inhibitors include DNA mismatch repair (MMR) and/or Epstein–Barr virus (EBV) status. The aim of this study was to establish and compare EBV status and MMR status in large multi-centre series of OeC and GC.
Methods
EBV was assessed by EBV-encoded RNA (EBER) in situ hybridisation and MMR protein expression by immunohistochemistry (IHC) in 988 OeC and 1213 GC from multiple centres. In a subset of OeC, microsatellite instability (MSI) was tested in parallel with MMR IHC.
Results
Frequency of MMR deficiency (MMRdef) and MSI was low in OeC (0.8% and 0.6%, respectively) compared with GC (10.3%). None of the OeCs were EBER positive in contrast to 4.8% EBER positive GC. EBV positive GC patients were younger (p = 0.01), more often male (p = 0.001) and had a better overall survival (p = 0.012). MMRdef GC patients were older (p = 0.001) and showed more often intestinal-type histology (p = 0.022).
Conclusions
This is the largest study to date indicating that EBV and MMRdef do not play a role in OeC carcinogenesis in contrast to GC. The potential clinical usefulness of determining MMRdef/EBV status to screen patients for eligibility for immune-targeting therapy differs between OeC and GC patients.
Keywords: Oesophageal cancer, Gastric cancer, DNA mismatch repair, Microsatellite instability, Epstein–Barr virus
Highlights
-
•
In oesophageal cancer (OeC) the frequency of mismatch repair deficiency/microsatellite instability (MSI) was extremely low.
-
•
Epstein-Barr Virus infection does not play a role in OeC carcinogenesis.
-
•
Determination of EBV/MSI status for immunotherapy eligibility cannot be recommended for OeC patients.
-
•
Future trials involving immunotherapy require disease specific design and selection criteria.
1. Introduction
Oesophageal cancer (OeC) and gastric cancer (GC) are the eighth and fifth most common cancer worldwide, respectively, with an estimated total of 1,407,000 new cases and 1,123,000 deaths in 2012 [1]. The two main histological OeC subtypes are squamous cell carcinoma (SqC) and adenocarcinoma (AdC). The vast majority of GCs are adenocarcinomas.
In Europe, the standard of care for OeC and GC patients with locally advanced resectable disease is chemotherapy or chemoradiotherapy, followed by surgery [2], [3]. GC patients receive perioperative platinum/fluorouracil based chemotherapy. For OeC, patients with SqC are treated with preoperative chemoradiotherapy with carboplatin/paclitaxel. Patients with AdC receive perioperative platinum/fluorouracil or preoperative chemoradiotherapy. Nevertheless, survival remains poor, with 5-year overall survival between 36 and 47% [4], [5].
To date, few targeted therapy options are available to OeC/GC patients with metastatic disease: trastuzumab for HER2 positive disease [6] and ramucirumab, a VEGFR-2 antagonist without biomarker based patient selection [7], [8]. All other trials evaluating receptor tyrosine kinase or downstream signalling inhibitors in OeC/GC were unable to show a survival benefit [9]. There remains an urgent clinical need to identify biomarkers to individualise and improve OeC/GC patient management.
DNA mismatch repair (MMR) has been used as a predictive biomarker for PD1 inhibitor therapy response in multiple different cancer types, including colorectal cancer [10]. Evidence of Epstein-Barr virus (EBV) infection has been proposed as a potential marker for response to PD1/PDL1 inhibitors in GC [11]. Pembrolizumab, an antibody against PD1, was approved by the FDA for the treatment of unresectable or metastatic solid tumours, including OeC and GC, with mismatch repair deficiency (MMRdef) or microsatellite instability (MSI)-High [12].
The potential of immunotherapy in OeC was shown recently in phase 2 trials in non-selected oesophageal SqC and GC patients treated with nivolumab, a monocolonal antibody inhibiting PD1, in second line treatment [13], [14] and in a phase 3 trial in heavily pretreated non-selected Asian GC patients [15]. Furthermore, recent results from the phase 1b trials in patients with PD-L1 expressing OeC (KEYNOTE-028) and GC (KEYNOTE-012), showed promising activity of pembrolizumab in the metastatic setting [16], [17]. In metastatic colorectal cancer, a phase 2 study demonstrated the clinical benefit of pembrolizumab in patients with MMRdef [18].
In addition to the potential role of MMR proteins in selecting patients for immunotherapy, MMRdef has shown prognostic value [19] and seems to predict a poor response to fluorouracil based chemotherapy in colorectal cancer [20], [21]. It has been shown recently in MAGIC trial patients, that gastro-oesophageal cancer patients with MMRdef/MSI tumours treated with surgery alone survived longer compared with those treated with perioperative cytotoxic chemotherapy [22]. In OeC, MLH1 and MSH2 deficiency has been shown to be associated with poor prognosis in small series of SqC [23].
To date, the frequency of MMRdef/MSI in OeC cancer remains unclear because of the small sample size of studies. The reported frequency of MSI-High (MSI-H) ranges from 0 to 27%, but a number of previous studies did not distinguish between MSI-H and MSI-Low (MSI-L) (for an overview of all published studies on MMR and MSI in OeC, see Table 1). The recent study by The Cancer Genome Atlas (TCGA) did not find MSI in any of the 162 OeC [24]. With respect to the frequency of EBV infection in OeC, the majority of previous studies investigated SqC using different methodology, included relatively small number of patients and reported a frequency of EBV positivity from 0 to 36% (for an overview of all published studies on EBV in OeC, see Table 2). Thus, neither MSI/MMRdef nor EBV status has been investigated in large series of OeC using the same methodology and relating results to clinicopathological variables and patient survival.
Table 1.
Summary of published literature relating to the frequency of mismatch repair deficiency and microsatellite instability in oesophageal cancer.
| Authors | Year | Oesophageal cancer type | Total n | MMRdef n (%) | MSI-High n (%) | Method |
|---|---|---|---|---|---|---|
| TCGA [24] | 2017 | SqC | 90 | NI | 0 | PCR |
| AdC | 70 | 0 | ||||
| undiff | 2 | 0 | ||||
| Pandilla et al.[57] | 2013 | SqC | 60 | NI | 6 (10) | PCR |
| AdC | 30 | 2 (7) | ||||
| Farris et al.[38] | 2011 | SqC | 76 | 5 (7) | 5 (7) | IHC, PCR |
| Vasavi et al.[44] | 2010 | SqC | 45 | NI | 12 (27) | PCR |
| AdC | 5 | 1 (20) | ||||
| Matsumoto et al.[58] | 2007 | SqC | 62 | NI | 5 (8) | PCR |
| Falkenback et al.[48] | 2005 | AdC | 59 | 2 (3) | 2 (3) | IHC, PCR |
| Naidoo et al.[59] | 2005 | SqC | 100 | NI | 5 (5)a | PCR |
| Uehara et al.[23] | 2005 | SqC | 122 | 49 (40) | 6 (5)a | IHC |
| Evans et al.[47] | 2004 | AdC | 27 | 6 (22) | 0 | IHC, PCR |
| Araki et al.[45] | 2004 | SqC | 100 | NI | 0 | PCR |
| Hayashi et al.[60] | 2003 | SqC | 30 | NI | 1 (3) | PCR |
| Ikeguchi et al.[46] | 1999 | SqC | 20 | NI | 1 (5)a | PCR |
| Wu et al.[61] | 1998 | SqC | 92 | NI | 5 (5)a | PCR |
| Muzeau et al.[43] | 1997 | SqC | 20 | NI | 0 | PCR |
| AdC | 26 | 0 | ||||
| Gleeson et al.[62] | 1996 | AdC | 17 | NI | 1 (17) | PCR |
| Keller et al.[63] | 1995 | AdC | 15 | NI | 2 (13)a | PCR |
| Ogasawara et al.[64] | 1995 | SqC | 35 | NI | 21 (60)a | PCR |
| Meltzer et al.[65] | 1994 | SqC | 42 | NI | 1 (2)a | PCR |
| AdC | 36 | 2 (6)a |
Abbreviations: AdC, adenocarcinoma; SqC, squamous cell carcinoma; MMRdef, mismatch repair deficiency; MSI, microsatellite instability; PCR, polymerase chain reaction; IHC, immunohistochemistry; NI, not investigated; undiff, undifferentiated.
no distinction made between MSI-High and MSI-Low.
Table 2.
Summary of published literature relating to the frequency of Epstein-Barr virus in oesophageal cancer.
| Reference | Year | Oesophageal cancer type | Total n | EBV positive n (%) | Method |
|---|---|---|---|---|---|
| TCGA [24] | 2017 | SqC | 90 | 0 | Whole-exome sequencing |
| AdC | 70 | 0 | |||
| undiff | 2 | 0 | |||
| Genitsch et al.[34] | 2015 | AdC | 118 | 0 | EBER ISH |
| Farris et al.[38] | 2011 | AdC | 76 | 1 (1) | EBER ISH |
| Sunpaweravong et al.[36] | 2005 | SqC | 104 | 0 | EBER ISH |
| Wu et al.[39] | 2005 | SqC | 151 | 6 (4) | EBER ISH |
| undiff | 13 | 4 (31) | |||
| Awerkiew et al.[40] | 2003 | SqC | 23 | 8 (35) | PCR |
| AdC | 14 | 5 (36) | |||
| Yanai et al.[33] | 2003 | SqC | 34 | 0 | EBER ISH, PCR |
| Mizobuchi et al.[37] | 1997 | SqC | 41 | 0 | PCR |
| Wang et al.[35] | 1999 | SqC | 51 | 0 | EBER ISH, PCR |
| Wang et al.[41] | 1999 | SqC | 31 | 11 (36) | EBER ISH, PCR |
Abbreviations: AdC, adenocarcinoma; SqC, squamous cell carcinoma; EBER ISH, EBV-encoded RNA in situ hybridisation; PCR, polymerase chain reaction; undiff, undifferentiated.
The aim of this multi-centre study was to establish the EBV and MMR/MSI status in 988 OeC, including patients from the Medical Research Council (MRC) Oe02 trial [25], from Leeds (UK) and from Cologne (Germany) and relate the results to clinicopathological variables, survival and treatment interaction (preoperative chemo(radio)therapy). As patients with resectable OeC and GC are often treated using similar neoadjuvant therapy regimens and recruited into the same clinical trials across different countries or continents, we compared the frequency of EBV positivity and MMRdef in OeC with that of 1213 GC from Leeds (UK) and Yokohama (Japan).
2. Material and methods
2.1. General remarks
The definition whether a tumour is a gastric or oesophageal cancer is dependent on the macroscopic location of the bulk/epicentre of the tumour with respect to the gastro-oesophageal junction. Macroscopic images were not available to us for review as part of this study with the exception of the Japanese gastric cancer cases. In contrast to our Japanese colleagues who classify tumours as oesophageal, junctional or gastric, all other pathologists using the TNM classification categorise tumours as being either oesophageal or gastric. We therefore reviewed the macroscopic images from the Japanese junctional cancers to classify them as either oesophageal or gastric according to TNM rules. For all other cases, we have used the classification of the originally reporting pathologist.
2.2. Oesophageal cancer cohorts
2.2.1. UK MRC Oe02 trial
The Oe02 trial was a multi-centre phase 3 trial comparing preoperative chemotherapy (cisplatin + 5-fluorouracil) followed by surgery (CS group) to surgery alone (S group) in 802 OeC patients with locally advanced resectable disease, recruited from March 1992 to June 1998. Paraffin blocks of the resected primary tumour were collected retrospectively, and material from 443 patients was available for the present study (CS n = 212, S n = 231). Clinicopathological data which could not be established during the central pathology review were retrieved from pathology reports and the clinical trial database. The study was approved by the South East Research Ethics committee, London, UK, REC reference: 07/H1102/111.
2.2.2. Leeds Teaching Hospitals NHS Trust (LTHT), UK
The LTHT cohort included 223 OeC patients who underwent potentially curative surgery at the Department of Surgery, Leeds General Infirmary (Leeds, UK), between 1986 and 2006. A total of 83 patients had preoperative chemotherapy. Clinical and pathological data were retrieved from pathology reports, electronic patient hospital records and the Northern and Yorkshire Cancer Registry. The study was approved by the Leeds Research Ethics Committee (LREC No. CA01/122).
2.2.3. University Hospital Cologne (UHC), Germany
The UHC cohort included 322 OeC patients who underwent potentially curative surgery at the Department of Visceral Surgery, University of Cologne (Cologne, Germany), between 1999 and 2013. A total of 197 patients had preoperative chemotherapy. Clinical and pathological data were retrieved from pathology reports and electronic patient hospital records. The study was approved by the Ethics Committee at the University Hospital, Cologne (reference number: 09-232).
2.3. Gastric cancer cohorts
2.3.1. Leeds Teaching Hospitals NHS Trust, UK
The GC LTHT cohort included 799 patients who underwent potentially curative surgery at the Department of Surgery, Leeds General Infirmary (Leeds, UK) between 1970 and 2004. Eleven patients had preoperative chemotherapy. Demographical, clinical and pathological data were retrieved from pathological reports, electronic patient hospital records and the Northern and Yorkshire Cancer Registry. The study was approved by the Leeds Research Ethics Committee (LREC No. CA01/122).
2.3.2. Kanagawa Cancer Center Hospital (KCCH), Yokohama, Japan
The KCCH cohort included 414 patients with stage II-IV GC who underwent potentially curative surgery at the Kanagawa Cancer Center Hospital (Yokohama, Japan) between 2001 and 2010. None of the patients had preoperative chemotherapy, 202 patients were treated with chemotherapy after surgery. Demographical, clinical and pathological data were retrieved from pathological reports and patient hospital records. The study was approved by the Local Research Ethics Committee.
2.4. Methods
2.4.1. Cancer staging and histological subtyping
pT and pN stage was reported according to the Union for International Cancer control 6th and 7th edition of the TNM classification for OeC and GC, respectively.
The histological subtype of adenocarcinomas was established based on Lauren's classification [26]. According to Lauren's classification, signet-ring cell GCs were classified as diffuse-type cancer. As there is no category for mucinous cancers in the Lauren classification, such cancers were classified together with the mixed-type cancers which we used as a category for truly mixed-type cancers and cancers with indeterminate phenotype like the mucinous cancers. The histology type of the case, as stated in the pathology report, was used for statistical analyses.
2.4.2. Tissue microarray construction
Slides from all resection specimens were reviewed and a block with the highest tumour cell density was selected for tissue microarray (TMA) construction and/or marked for microdissection for DNA extraction (see below). The areas selected were representative of the overall histology of the case. The LTHT, KCCH, and Oe02 trial cases were reviewed by HG, LH and GH, together with local pathologists. The UHC cases were reviewed by AQ. A total of 962 OeCs (417, 223 and 322 patients from the Oe02, LTHT and UHC cohorts, respectively) and 1213 GCs (799 and 414 patients from LTHT and KCCH cohorts, respectively) were included in TMAs. TMA construction from the LTHT (OeC and GC) and Oe02 patient cohorts was performed using 0.6 mm tissue cores; 1.2 mm and 1 mm tissue cores were used for the UHC and KCCH cohorts, respectively.
2.4.3. Immunohistochemistry for mismatch repair proteins
MMR immunohistochemistry (IHC) data from previous studies were available for 230 KCCH [27] and 175 LTHT [28] GCs. Additional 184 KCCH and 624 LTHT GCs were stained as part of the present study.
TMA sections from the Oe02 trial cohort were stained for MLH1, MSH2, MSH6, PMS2, from the UHC cohort for MLH1, MSH2 and MSH6 and from the KCCH and LTHT cohort (OeC and GC) for MLH1 and MSH2. For details on antigen retrieval, primary antibodies, detection system, staining protocols see Table 1 in the supplementary material. For all cohorts, 3,3′-Diaminobenzidine (DAB) was used as a chromogen and haematoxylin as a counterstain.
A case was classified as MMR deficient (MMRdef) if tumour cell nuclei were negative for one or more MMR proteins in the presence of positively stained lymphocytes or fibroblasts as internal control. In the Oe02 trial cohort, 12 cases were negative for at least one MMR protein without positive internal controls on the TMA. For these cases, IHC was repeated on full sections. A case was classified as MMR proficient (MMRprof) if tumour cell nuclei, irrespective of the number or intensity, were positive for all MMR proteins tested.
2.4.4. EBV RNA in situ hybridisation
EBV data from a previous study were available for 437 LTHT and 216 KCCH GC [28]. Additional 362 LTHT and 198 KCCH GCs were stained as part of the present study. EBV status was determined on TMAs in the LTHT (OeC and GC), Oe02 and KCCH cohorts by EBV-encoded RNA (EBER) in situ hybridisation as previously described [29]. In the UHC cohort, a fluorescein-conjugated oligonucleotide probe in conjunction with a monoclonal anti-fluorescein antibody and DAB as chromogen (Leica Biosystems, Wetzlar, Germany) was used according to the instructions of the manufacturer. EBV positivity was defined as presence of staining in tumour cell nuclei, irrespective of the number of nuclei or intensity.
2.4.5. DNA extraction
DNA was extracted using a protocol based on the QIAmp DNA Micro Kit (Qiagen, Hilden, Germany) as previously described [30]. DNA concentration was measured by ND-100 Spectrophotometer (Labtech International) and adjusted to a final concentration of 1 ng/μl.
2.4.6. Assessment of microsatellite instability
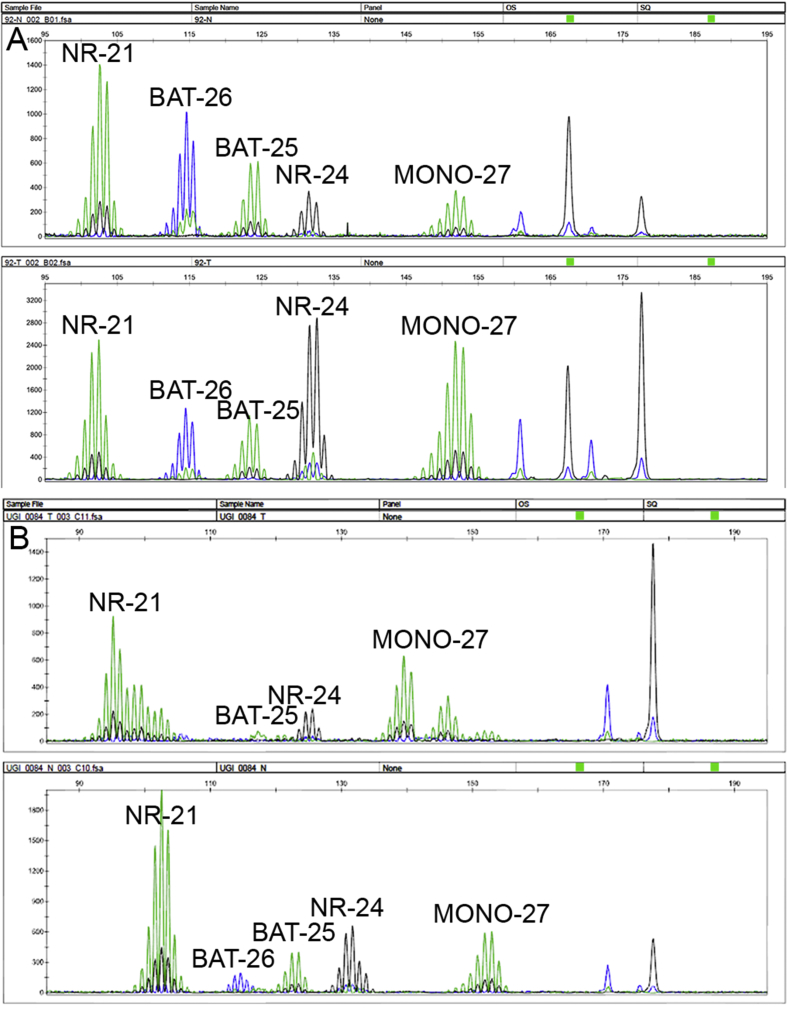
The MSI Analysis System, version 1.2 (Promega, Southampton, UK), was used for the detection of MSI in 419 Oe02 patients. This kit allows the simultaneous evaluation of 5 fluorescently labelled MSI markers: BAT-25, BAT-26, NR-21, NR-24 and MONO-27. PCR products were analysed using a 3100-Avant genetic analyser (Applied Biosystems, California, USA) as previously described [27]. Instability in two or more microsatellite loci was categorised as MSI-high (MSI-H) and in a single loci as MSI-low (MSI-L). Absence of MSI in all 5 markers and MSI-L were grouped as microsatellite stable (MSS) for further analyses following current guidelines [31].
2.4.7. Statistical analyses
All statistical analyses were performed using SPSS version 23 software (SPSS Inc., Chicago, III). The relationship between EBV or MMR status and clinicopathological variables (age, gender, depth of invasion (pT), lymph node status (pN), Lauren classification and neoadjuvant treatment) were assessed using chi-squared for categorical variables and Mann-Whitney U for continuous variables. LTHT and KCCH GC data were combined for the analysis of the relationship between EBV or MMR status and overall 5-year survival and differences were assessed using the log rank test. P values less than 0.05 were considered significant.
3. Results
3.1. EBV status
EBV data were available from 928 OeC patients (LTHT n = 223; Oe02 n = 383; UHC n = 322) and 1178 GC patients (LTHT n = 768; KCCH n = 410). All OeC were EBV negative. A total of 56 (4.8%) GC were EBV positive (LTHT: n = 30 [3.9%], KCCH: n = 26 [6.3%]). Supplementary figure 1 illustrates EBV staining in GC.
3.2. Microsatellite status and mismatch repair protein expression
MSI data were available from 362 OeC from the Oe02 cohort. A total of 57 (13.6%) cases had to be excluded due to repeated technical failures. A total of 356 (98.3%) OeC patients were classified as MSS, 4 (1.1%) OeC as MSI-L (3 AdC and 1 SqC) and 2 (0.6%) OeC as MSI-H (both AdC). Supplementary figure 2 shows a typical capillary electrophoresis output for a MSI-H OeC and a MSS OeC. For 306 patients, MMR IHC (MLH1, MSH2, MSH6 and PMS2) data and MSI testing results were available and showed 99.0% concordant results. We therefore decided to only use IHC for the remaining cohorts.
MMR expression data were available from a total of 916 OeC (LTHT n = 220; Oe02 n = 374; UHC n = 322). A sum of 43 (10.3%) and 3 (1.3%) OeC from the Oe02 and LTHT cohorts, respectively, were excluded due to technical failures. Seven (0.8%) OeC (5 AdC and 2 SqC) were classified as MMRdef (LTHT: 3 (1.4%) MLH1 deficient, Oe02: 1 (0.3%) MSH2 deficient, UHC: 3 (0.9%) MLH1 deficient). Patient clinicopathological variables and MMR status for OeC are summarised in Table 3. Owing to the very small number of MMRdef in OeC, it was not feasible to perform any statistical analysis with clinicopathological data or survival.
Table 3.
Mismatch repair status and clinicopathological variables in patients with oesophageal cancer.
| Clinicopathological variables | Mismatch repair proficient |
Mismatch repair deficient |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LTHT |
Oe02 |
UHC |
LTHT |
Oe02 |
UHC |
||||||||
| n | % | n | % | n | % | n | % | n | % | n | % | ||
| Sex | Male | 137 | 63.1 | 294 | 78.8 | 287 | 89.9 | 2 | 66.7 | 3 | 100 | ||
| Female | 80 | 36.9 | 79 | 21.2 | 32 | 10.1 | 1 | 33.3 | 1 | 100 | |||
| (y)pT(6) | T0 | 2 | 0.9 | 3 | 0.9 | ||||||||
| T1 | 32 | 14.7 | 27 | 7.2 | 63 | 19.7 | 1 | 33.3 | |||||
| T2 | 38 | 17.5 | 36 | 9.7 | 63 | 19.7 | 1 | 33.3 | |||||
| T3 | 136 | 62.7 | 301 | 80.7 | 185 | 58 | 2 | 66.7 | 1 | 100 | 2 | 66.7 | |
| T4 | 9 | 4.1 | 9 | 2.4 | 5 | 1.6 | |||||||
| (y)pN(6) | N0 | 83 | 38.2 | 123 | 33 | 122 | 38.2 | 1 | 100 | 3 | 100 | ||
| N1 | 133 | 61.3 | 250 | 67 | 197 | 61.8 | 3 | 100 | |||||
| unknown | 1 | 0.5 | |||||||||||
| Histological type | Adenocarcinoma | 165 | 76 | 275 | 73.7 | 319 | 100 | 2 | 66.7 | 3 | 100 | ||
| Squamous cell carcinoma | 49 | 22.6 | 87 | 23.3 | 1 | 33.3 | 1 | 100 | |||||
| Other | 3 | 1.4 | 11 | 2.9 | |||||||||
| Neoadjuvant treatment | Yes | 80 | 36.9 | 177 | 47.5 | 194 | 60.8 | 2 | 66.7 | 1 | 100 | 2 | 66.7 |
| No | 133 | 61.3 | 196 | 52.5 | 125 | 39.2 | 1 | 33.3 | 1 | 33.3 | |||
| unknown | 4 | 1.8 | |||||||||||
Abbreviations: LTHT, Leeds Teaching Hospital Trust; Oe02, oesophageal cancer trial 02 [25]; UHC, University Hospital Cologne.
MMR protein expression data were available from 1098 GC (LTHT n = 702; KCCH n = 396). A total of 113 (10.3%) cases were classified as MMRdef (LTHT: 70 (10.0%), KCCH: 43 (10.9%)). Supplementary figure 3 illustrates MMR protein expression in a MMRdef GC.
For 1063 GCs, both EBV and MMR data were available. A single GC from the LTHT cohort was MMRdef and EBV positive. This patient was male, 67 years old at the time of diagnosis and survived 17 years despite having an advanced intestinal-type GC (pT4, pN3) in the resected specimen.
3.3. Relationship of EBV status and MMR status with clinicopathological variables in patients with gastric cancer
Patients with EBV positive GC were younger (median [range] age EBV positive GC: 63 years (32–89 years) versus 68 years (14–96 years) in EBV negative GC, p = 0.01). A total of 48 (85.7%) patients with EBV positive GC were male compared with 8 (14.3%) of female patients (p = 0.001). EBV positive GC patients had a better overall 5-year survival compared with EBV negative GC patients (60.7% versus 41.7%; hazard ratio 1.72, 95% confidence interval 1.12–2.63 [p = 0.012]).
Patients with MMRdef GC were older (median [range] age MMRdef GC: 71 years [51–90 years] versus 68 years [24–96 years] in MMRprof GC, p = 0.001). A total of 77 (69.4%) MMRdef GC had intestinal-type histology compared with 20 (18.0%) with diffuse-type histology (p = 0.022). There was no difference in overall survival between MMRdef and MMRprof GCs (p = 0.383). There was no relationship with any other clinicopathological variables (Table 4).
Table 4.
Comparison of mismatch repair and EBV status with clinicopathological variables in patients with gastric cancer.
| Clinicopathological variables | Mismatch repair proficient |
Mismatch repair deficient |
p value | EBV negative |
EBV positive |
p value | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LTHT |
KCCH |
Total |
LTHT |
KCCH |
Total |
LTHT |
KCCH |
Total |
LTHT |
KCCH |
Total |
||||||||||||||||
| n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | ||||
| Gender | Male | 415 | 59 | 250 | 63 | 665 | 61 | 42 | 6 | 33 | 8 | 75 | 7 | 0.761 | 456 | 59 | 273 | 67 | 729 | 62 | 26 | 3 | 22 | 5 | 48 | 4 | 0.001 |
| Female | 214 | 30 | 102 | 26 | 316 | 29 | 28 | 4 | 10 | 3 | 38 | 3 | 281 | 37 | 110 | 27 | 391 | 33 | 4 | 1 | 4 | 1 | 8 | 1 | |||
| Unknown | 3 | 0 | 1 | 0 | 4 | 0 | 1 | 0 | 1 | 0 | 2 | 0 | |||||||||||||||
| (y)pT(7) | T1 | 83 | 12 | 34 | 9 | 117 | 11 | 5 | 1 | 3 | 1 | 8 | 1 | 0.074 | 105 | 14 | 37 | 9 | 142 | 12 | 4 | 1 | 2 | 0 | 6 | 1 | 0.794 |
| T2 | 69 | 10 | 52 | 13 | 121 | 11 | 2 | 0 | 5 | 1 | 7 | 1 | 75 | 10 | 58 | 14 | 133 | 11 | 5 | 1 | 4 | 1 | 9 | 1 | |||
| T3 | 179 | 25 | 52 | 13 | 231 | 21 | 26 | 4 | 3 | 1 | 29 | 3 | 210 | 27 | 52 | 13 | 262 | 22 | 9 | 1 | 3 | 1 | 12 | 1 | |||
| T4 | 301 | 43 | 214 | 54 | 515 | 47 | 37 | 5 | 32 | 8 | 69 | 6 | 348 | 45 | 236 | 58 | 584 | 50 | 12 | 2 | 17 | 4 | 29 | 2 | |||
| Unknown | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | |||||||||||||||||||
| (y)pN(7) | N0 | 206 | 29 | 70 | 18 | 276 | 25 | 22 | 3 | 13 | 3 | 35 | 3 | 0.722 | 242 | 32 | 82 | 20 | 324 | 28 | 13 | 2 | 4 | 1 | 17 | 1 | 0.931 |
| N1 | 123 | 18 | 80 | 20 | 203 | 18 | 19 | 3 | 6 | 2 | 25 | 2 | 155 | 20 | 83 | 20 | 238 | 20 | 6 | 1 | 5 | 1 | 11 | 1 | |||
| N2 | 146 | 21 | 91 | 23 | 237 | 22 | 14 | 2 | 8 | 2 | 22 | 2 | 152 | 20 | 96 | 23 | 248 | 21 | 7 | 1 | 7 | 2 | 14 | 1 | |||
| N3 | 156 | 22 | 111 | 28 | 267 | 24 | 15 | 2 | 16 | 4 | 31 | 3 | 189 | 25 | 122 | 30 | 311 | 26 | 4 | 1 | 10 | 2 | 14 | 1 | |||
| Unknown | 1 | 0 | 1 | 0 | 2 | 0 | 1 | 0 | 1 | 0 | |||||||||||||||||
| Lauren classification | Intestinal | 403 | 57 | 181 | 46 | 584 | 53 | 49 | 7 | 28 | 7 | 77 | 7 | 0.022 | 461 | 60 | 204 | 50 | 665 | 56 | 20 | 3 | 15 | 4 | 35 | 3 | 0.919 |
| Diffuse | 145 | 21 | 154 | 39 | 299 | 27 | 10 | 1 | 10 | 3 | 20 | 2 | 185 | 24 | 156 | 38 | 341 | 29 | 6 | 1 | 10 | 2 | 16 | 1 | |||
| Mucinous/mixed | 82 | 12 | 15 | 4 | 97 | 9 | 11 | 2 | 3 | 1 | 14 | 1 | 90 | 12 | 17 | 4 | 107 | 9 | 4 | 1 | 1 | 0 | 5 | 0 | |||
| Unknown | 2 | 0 | 3 | 1 | 5 | 0 | 2 | 1 | 2 | 0 | 7 | 2 | 9 | 1 | |||||||||||||
| Neoadjuvant treatment | Yes | 8 | 1 | 177 | 45 | 185 | 17 | 1 | 0 | 16 | 4 | 17 | 2 | 0.305 | 11 | 1 | 185 | 45 | 196 | 17 | 13 | 3 | 13 | 1 | 0.293 | ||
| No | 624 | 89 | 164 | 41 | 788 | 72 | 69 | 10 | 27 | 7 | 96 | 9 | 727 | 95 | 185 | 45 | 912 | 77 | 30 | 4 | 13 | 3 | 43 | 4 | |||
| Unknown | 12 | 3 | 12 | 1 | 14 | 3 | 14 | 1 | |||||||||||||||||||
Abbreviations: KCCH, Kanagawa Cancer Center Hospital; LTHT, Leeds Teaching Hospital Trust.
p-Values in bold are considered statistically significant.
A summary of the EBV, MMR and MSI status in each cohort is provided in Table 5.
Table 5.
Summary of EBV, mismatch repair and microsatellite instability status in oesophageal and gastric cancer.
| Molecular characteristics | OeC |
GC |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Oe02 |
LTHT |
UHC |
LTHT |
KCCH |
|||||||
| n = 443 | % | n = 223 | % | n = 322 | % | n = 768 | % | n = 410 | % | ||
| EBV | Negative | 383 | 100 | 223 | 100 | 322 | 100 | 738 | 96 | 384 | 94 |
| Positive | 0 | 0 | 0 | 0 | 0 | 0 | 30 | 4 | 26 | 6 | |
| MMR | Proficient | 373 | 100 | 217 | 99 | 319 | 99 | 632 | 90 | 353 | 89 |
| Deficient | 1 | 0 | 3 | 1 | 3 | 1 | 70 | 10 | 43 | 11 | |
| Microsatellite | Stable | 356 | 98 | NI | NI | NI | NI | ||||
| Instable-Low | 4 | 1 | NI | NI | NI | NI | |||||
| Instable-High | 2 | 1 | NI | NI | NI | NI | |||||
Abbreviations: EBV, Epstein-Barr Virus; GC, gastric cancer; KCCH, Kanagawa Cancer Center Hospital; LTHT, Leeds Teaching Hospitals NHS Trust; MMR, mismatch repair; MSI, microsatellite instability; MSS, microsatellite stable; OeC, oesophageal cancer; UHC, University Hospital Cologne; NI, not investigated.
4. Discussion
This is the largest gastro-oesophageal cancer study to date investigating MMR and EBV status in 988 OeC and 1213 GC. The extremely low frequency of MMR/MSI and lack of EBV infection in OeC relative to GC in our study confirms the recent TCGA results which investigated MSI and EBV in smaller series of 164 OeC [24] and 295 GC [11] using different methodologies.
All OeC were EBV negative which is consistent with the majority of previously published studies [32], [33], [34], [35], [36], [37]. Therefore, we can conclude now that EBV does not play a role in OeC carcinogenesis neither in SqC nor in AdC. A small number of previous studies reported an EBV positivity rate between 1 and 36% in OeC [38], [39], [40], [41]. This discrepancy is most likely related to different potentially less reliable methodology, such as PCR, which would also detect EBV in tumour-infiltrating lymphocytes [33] leading to false positive results. The present study used the generally accepted ‘gold standard’ EBER methodology. In our study, EBV positive GC patients had a significantly better overall survival compared with EBV negative patients which is consistent with results from other studies [42].
In the Oe02 cohort, we detected a very low frequency of MSI-H (0.6%) using the Bethesda microsatellite panel [31]. This result is consistent with the recent smaller TCGA study which found no MSI-H cases in 72 oesophageal AdC [24]. However, our result is in contrast to the literature reporting a frequency of MSI-H in OeC between 0 and 27% in SqC [43], [44], [45], [46] and 0–20% in AdC [22], [38], [43], [44], [47], [48]. Discrepancies in the frequency of MSI-H amongst studies could be related to different definitions of MSI-H [47], as well as differences in location [44] and number of microsatellite loci tested [46]. Recent studies in GC suggest that a mononucleotide and dinucleotide markers different to those included in the so-called Bethesda panel might improve accuracy and sensitivity of MSI testing in GC [49], [50].
There are few small studies reporting a MMRdef frequency of 3–40% in OeC mostly based on IHC of MLH1 and MSH2 [23], [38], [47], [48]. Some of the previous studies scores were based on staining intensity and cell proportions and classifying cases with weak staining and/or low percentages of positively stained tumour cells as MMRdef. Thus, when using our MMR scoring system where a case is classified as MMRprof, irrespective of the number of positive nuclei or staining intensity, the frequency of MMRdef in our study is comparable to previously published studies. Another potential reason for discrepant results in the literature could be the misclassification of AdC with a tumour bulk located in the stomach which extends into the GOJ as OeC. In contrast to the results from the MAGIC trial patients [22], there was no overall survival difference between MMRdef GC and MMRprof GC in our study. This is likely due to differences in disease stage, histological subtypes and age of GC patients in our study.
The frequency of MMRdef and EBV positivity in our GC cohort is consistent with the current literature [51], [52], [53]. As the same methodology was used to stain GC and OeC, our GC results also indirectly support the reliability of the low frequency of MMRdef and EBV in OeC in the present study. Furthermore, our results are comparable with results from a smaller study in the MAGIC trial patients comparing the frequency of MSI and MMRdef in GC and OeC [22].
Our study has some limitations. First, this is a retrospective study. Second, due to limited tissue availability, we were unable to perform IHC for all four MMR proteins in all cases, and we did not test all cases for MSI. However, evidence in the literature from GC found MMRdef was due to loss of MLH1 in 95.8% of cases, and deficiency in MSH6 and PMS2 was rare [51]. Similarly, a colorectal cancer study reported a positive predictive value and specificity of IHC for MMR proteins of 99.1% and 99.6%, respectively, compared with MSI [54]. Our own study showed that MSI status is in 99.0% of cases concordant with the MMR IHC status. Another potential limitation is our inability to determine the proportion of junctional (GOJ) AdC versus true oesophageal or true gastric AdC which might potentially be clinically relevant. This is related to the fact that detailed pre-chemotherapy endoscopic information regarding the location was not available for most cases. There are very few studies investigating EBV and MMRdef in GOJ cancer with inconsistent results most likely related to low sample sizes [22], [34], [55] or differences in defining the GOJ [56].
Our OeC findings suggest that OeC carcinogenesis is not associated with EBV infection and MMRdef/MSI does not appear to be an important underlying mechanism in OeC, neither SqC nor AdC. The use of EBV and/or MMR/MSI status to determine OeC patient eligibility for immunotherapy or adjuvant cytotoxic therapy cannot be recommended, and there remains the need to find alternative biomarkers for such therapy approaches in this patient population. The difference in the frequency of MMRdef and EBV infection between OeC and GC indicate not only pathophysiological differences in oesophageal and gastric carcinomas but might also have important implications for patient selection for future treatment and study planning. In contrast to the current practice of recruiting patients with GC or OeC into the same trials, trials involving immunotherapy require most likely disease specific different designs and selection criteria for patients with OeC.
Conflict of interest statement
DC received financial support from AstraZeneca, Amgen, Bayer, Celgene, Merrimack, Merck Serono, MedImmune and Sanofi. WA received financial support from Eli Lilly and Nestle. RL received financial support from Bayer. The remaining authors have no conflicts of interest to declare.
Acknowledgements
The authors thank N West, T Arai, Y Miyagi and Y Kameda for reviewing the slides to select cores for TMA construction.
This work was supported by Cancer Research UK [Oe02 trial cohort: C26441/A8944 to PI: HG]; The Pathological Society of Great Britain and Ireland to [HG]; Sasakawa Foundation UK [HG, TY]; Yorkshire Cancer Research [HG]; Kanagawa Standard Anti-cancer Therapy Support System (Japan) [TY] and The National Institute for Health Research Royal Marsden/Institute of Cancer Research Biomedical Research Centre (NIHR RM/ICR BRC) [DC, WA].
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ejca.2018.02.014.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
figs1.
figs2.
figs3.
References
- 1.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Canc. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Smyth E.C., Verheij M., Allum W., Cunningham D., Cervantes A., Arnold D. Gastric cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2016;27(Suppl. 5):v38–v49. doi: 10.1093/annonc/mdw350. [DOI] [PubMed] [Google Scholar]
- 3.Lordick F., Mariette C., Haustermans K., Obermannova R., Arnold D., Committee E.G. Oesophageal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2016;27(Suppl. 5):v50–v57. doi: 10.1093/annonc/mdw329. [DOI] [PubMed] [Google Scholar]
- 4.Alderson D., Cunningham D., Nankivell M., Blazeby J.M., Griffin S.M., Crellin A. Neoadjuvant cisplatin and fluorouracil versus epirubicin, cisplatin, and capecitabine followed by resection in patients with oesophageal adenocarcinoma (UK MRC OE05): an open-label, randomised phase 3 trial. Lancet Oncol. 2017;18(9):1249–1260. doi: 10.1016/S1470-2045(17)30447-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shapiro J., van Lanschot J.J.B., Hulshof M., van Hagen P., van Berge Henegouwen M.I., Wijnhoven B.P.L. Neoadjuvant chemoradiotherapy plus surgery versus surgery alone for oesophageal or junctional cancer (CROSS): long-term results of a randomised controlled trial. Lancet Oncol. 2015;16(9):1090–1098. doi: 10.1016/S1470-2045(15)00040-6. [DOI] [PubMed] [Google Scholar]
- 6.Bang Y.J., Van Cutsem E., Feyereislova A., Chung H.C., Shen L., Sawaki A. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376(9742):687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 7.Fuchs C.S., Tomasek J., Yong C.J., Dumitru F., Passalacqua R., Goswami C. Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): an international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet. 2014;383(9911):31–39. doi: 10.1016/S0140-6736(13)61719-5. [DOI] [PubMed] [Google Scholar]
- 8.Wilke H., Muro K., Van Cutsem E., Oh S.C., Bodoky G., Shimada Y. Ramucirumab plus paclitaxel versus placebo plus paclitaxel in patients with previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (RAINBOW): a double-blind, randomised phase 3 trial. Lancet Oncol. 2014;15(11):1224–1235. doi: 10.1016/S1470-2045(14)70420-6. [DOI] [PubMed] [Google Scholar]
- 9.Smyth E.C., Lagergren J., Fitzgerald R.C., Lordick F., Shah M.A., Lagergren P. Oesophageal cancer. Nat Rev Dis Prim. 2017;3:17048. doi: 10.1038/nrdp.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le D.T., Durham J.N., Smith K.N., Wang H., Bartlett B.R., Aulakh L.K. Mismatch-repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Food and Drug Administration . 2017. FDA approves first cancer treatment for any solid tumor with a specific genetic feature.https://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm560167.htm [Google Scholar]
- 13.Kudo T., Hamamoto Y., Kato K., Ura T., Kojima T., Tsushima T. Nivolumab treatment for oesophageal squamous-cell carcinoma: an open-label, multicentre, phase 2 trial. Lancet Oncol. 2017;18(5):631–639. doi: 10.1016/S1470-2045(17)30181-X. [DOI] [PubMed] [Google Scholar]
- 14.Janjigian Y.Y., Bendell J.C., Calvo E., Kim J.W., Ascierto P.A., Sharma P. CheckMate-032: phase I/II open-label study of safety and activity of nivolumab (nivo) alone or with ipilimumab (ipi) in advanced and metastatic (A/M) gastric cancer (GC) J Clin Oncol. 2016;34(15S) abst 4010. [Google Scholar]
- 15.Kang Y.K., Boku N., Satoh T., Ryu M.H., Chao Y., Kato K. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538–12, ATTRACTION-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390(10111):2461–2471. doi: 10.1016/S0140-6736(17)31827-5. [DOI] [PubMed] [Google Scholar]
- 16.Muro K., Chung H.C., Shankaran V., Geva R., Catenacci D., Gupta S. Pembrolizumab for patients with PD-L1-positive advanced gastric cancer (KEYNOTE-012): a multicentre, open-label, phase 1b trial. Lancet Oncol. 2016;17(6):717–726. doi: 10.1016/S1470-2045(16)00175-3. [DOI] [PubMed] [Google Scholar]
- 17.Doi T., Piha-Paul S.A., Jalal S.I., Mai-Dang H., Saraf S., Koshiji M. Updated results for the advanced esophageal carcinoma cohort of the phase 1b KEYNOTE-028 study of pembrolizumab. J Clin Oncol. 2016;34(15S) abst 4046. [Google Scholar]
- 18.Le D.T., Uram J.N., Wang H., Bartlett B.R., Kemberling H., Eyring A.D. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutchins G., Southward K., Handley K., Magill L., Beaumont C., Stahlschmidt J. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol. 2011;29(10):1261–1270. doi: 10.1200/JCO.2010.30.1366. [DOI] [PubMed] [Google Scholar]
- 20.Ribic C.M., Sargent D.J., Moore M.J., Thibodeau S.N., French A.J., Goldberg R.M. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349(3):247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sargent D.J., Marsoni S., Monges G., Thibodeau S.N., Labianca R., Hamilton S.R. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28(20):3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smyth E.C., Wotherspoon A., Peckitt C., Gonzalez D., Hulkki-Wilson S., Eltahir Z. Mismatch repair deficiency, microsatellite instability, and survival : an exploratory analysis of the Medical Research Council adjuvant gastric infusional chemotherapy (MAGIC) trial. JAMA Oncol. 2017;3(9):1197–1203. doi: 10.1001/jamaoncol.2016.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uehara H., Miyamoto M., Kato K., Cho Y., Kurokawa T., Murakami S. Deficiency of hMLH1 and hMSH2 expression is a poor prognostic factor in esophageal squamous cell carcinoma. J Surg Oncol. 2005;92(2):109–115. doi: 10.1002/jso.20332. [DOI] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas Research Network Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541(7636):169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allum W.H., Stenning S.P., Bancewicz J., Clark P.I., Langley R.E. Long-term results of a randomized trial of surgery with or without preoperative chemotherapy in esophageal cancer. J Clin Oncol. 2009;27(30):5062–5067. doi: 10.1200/JCO.2009.22.2083. [DOI] [PubMed] [Google Scholar]
- 26.Lauren P. The two histological main types of gastric carcinoma: diffuse and so called intestinal-type carcinoma. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 27.van Grieken N.C., Aoyama T., Chambers P.A., Bottomley D., Ward L.C., Inam I. KRAS and BRAF mutations are rare and related to DNA mismatch repair deficiency in gastric cancer from the east and the west: results from a large international multicentre study. Br J Canc. 2013;108(7):1495–1501. doi: 10.1038/bjc.2013.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin S.J., Gagnon-Bartsch J.A., Tan I.B., Earle S., Ruff L., Pettinger K. Signatures of tumour immunity distinguish Asian and non-Asian gastric adenocarcinomas. Gut. 2015;64(11):1721–1731. doi: 10.1136/gutjnl-2014-308252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zur Hausen A., van Rees B.P., van Beek J., Craanen M.E., Bloemena E., Offerhaus G.J. Epstein–Barr virus in gastric carcinomas and gastric stump carcinomas: a late event in gastric carcinogenesis. J Clin Pathol. 2004;57(5):487–491. doi: 10.1136/jcp.2003.014068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiss M.M., Hermsen M.A., Meijer G.A., van Grieken N.C., Baak J.P., Kuipers E.J. Comparative genomic hybridisation. Mol Pathol. 1999;52(5):243–251. doi: 10.1136/mp.52.5.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Umar A., Boland C.R., Terdiman J.P., Syngal S., de la Chapelle A., Ruschoff J. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96(4):261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Awerkiew S., zur Hausen A., Baldus S.E., Holscher A.H., Sidorenko S.I., Kutsev S.I. Presence of Epstein–Barr virus in esophageal cancer is restricted to tumor infiltrating lymphocytes. Med Microbiol Immunol. 2005;194(4):187–191. doi: 10.1007/s00430-004-0233-2. [DOI] [PubMed] [Google Scholar]
- 33.Yanai H., Hirano A., Matsusaki K., Kawano T., Miura O., Yoshida T. Epstein–Barr virus association is rare in esophageal squamous cell carcinoma. Int J Gastrointest Cancer. 2003;33(2–3):165–170. doi: 10.1385/IJGC:33:2-3:165. [DOI] [PubMed] [Google Scholar]
- 34.Genitsch V., Novotny A., Seiler C.A., Kroll D., Walch A., Langer R. Epstein–barr virus in gastro-esophageal adenocarcinomas – single center experiences in the context of current literature. Front Oncol. 2015;5:73. doi: 10.3389/fonc.2015.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J., Noffsinger A., Stemmermann G., Fenoglio-Preiser C. Esophageal squamous cell carcinomas arising in patients from a high-risk area of North China lack an association with Epstein–Barr virus. Canc Epidemiol Biomarkers Prev. 1999;8(12):1111–1114. [PubMed] [Google Scholar]
- 36.Sunpaweravong S., Mitarnun W., Puttawibul P. Absence of Epstein–Barr virus in esophageal squamous cell carcinoma. Dis Esophagus. 2005;18(6):398–399. doi: 10.1111/j.1442-2050.2005.00522.x. [DOI] [PubMed] [Google Scholar]
- 37.Mizobuchi S., Sakamoto H., Tachimori Y., Kato H., Watanabe H., Terada M. Absence of human papillomavirus-16 and -18 DNA and Epstein–Barr virus DNA in esophageal squamous cell carcinoma. Jpn J Clin Oncol. 1997;27(1):1–5. doi: 10.1093/jjco/27.1.1. [DOI] [PubMed] [Google Scholar]
- 38.Farris A.B., 3rd, Demicco E.G., Le L.P., Finberg K.E., Miller J., Mandal R. Clinicopathologic and molecular profiles of microsatellite unstable Barrett Esophagus-associated adenocarcinoma. Am J Surg Pathol. 2011;35(5):647–655. doi: 10.1097/PAS.0b013e31820f18a2. [DOI] [PubMed] [Google Scholar]
- 39.Wu M.Y., Wu X.Y., Zhuang C.X. Detection of HSV and EBV in esophageal carcinomas from a high-incidence area in Shantou China. Dis Esophagus. 2005;18(1):46–50. doi: 10.1111/j.1442-2050.2005.00423.x. [DOI] [PubMed] [Google Scholar]
- 40.Awerkiew S., Bollschweiler E., Metzger R., Schneider P.M., Holscher A.H., Pfister H. Esophageal cancer in Germany is associated with Epstein–Barr-virus but not with papillomaviruses. Med Microbiol Immunol. 2003;192(3):137–140. doi: 10.1007/s00430-002-0128-z. [DOI] [PubMed] [Google Scholar]
- 41.Wang L.S., Chow K.C., Wu Y.C., Li W.Y., Huang M.H. Detection of Epstein–Barr virus in esophageal squamous cell carcinoma in Taiwan. Am J Gastroenterol. 1999;94(10):2834–2839. doi: 10.1111/j.1572-0241.1999.01425.x. [DOI] [PubMed] [Google Scholar]
- 42.Camargo M.C., Kim W.H., Chiaravalli A.M., Kim K.M., Corvalan A.H., Matsuo K. Improved survival of gastric cancer with tumour Epstein–Barr virus positivity: an international pooled analysis. Gut. 2014;63(2):236–243. doi: 10.1136/gutjnl-2013-304531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muzeau F., Flejou J.F., Belghiti J., Thomas G., Hamelin R. Infrequent microsatellite instability in oesophageal cancers. Br J Canc. 1997;75(9):1336–1339. doi: 10.1038/bjc.1997.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vasavi M., Kiran V., Ravishankar B., Prabhakar B., Ahuja Y.R., Hasan Q. Microsatellite instability analysis and its correlation with hMLH1 repair gene hypermethylation status in esophageal pathologies including cancers. Canc Biomarkers. 2010;7(1):1–10. doi: 10.3233/CBM-2010-0135. [DOI] [PubMed] [Google Scholar]
- 45.Araki K., Wang B., Miyashita K., Cui Q., Ohno S., Baba H. Frequent loss of heterozygosity but rare microsatellite instability in oesophageal cancer in Japanese and Chinese patients. Oncology. 2004;67(2):151–158. doi: 10.1159/000081002. [DOI] [PubMed] [Google Scholar]
- 46.Ikeguchi M., Unate H., Maeta M., Kaibara N. Detection of loss of heterozygosity at microsatellite loci in esophageal squamous-cell carcinoma. Oncology. 1999;56(2):164–168. doi: 10.1159/000011959. [DOI] [PubMed] [Google Scholar]
- 47.Evans S.C., Gillis A., Geldenhuys L., Vaninetti N.M., Malatjalian D.A., Porter G.A. Microsatellite instability in esophageal adenocarcinoma. Canc Lett. 2004;212(2):241–251. doi: 10.1016/j.canlet.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 48.Falkenback D., Johansson J., Halvarsson B., Nilbert M. Defective mismatch-repair as a minor tumorigenic pathway in Barrett esophagus-associated adenocarcinoma. Canc Genet Cytogenet. 2005;157(1):82–86. doi: 10.1016/j.cancergencyto.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 49.Kim J.G., Shin S., Park J. Comparison between mononucleotide and dinucleotide marker panels in gastric cancer with loss of hMLH1 or hMSH2 expression. Int J Biol Markers. 2017;32(3):e352–e356. doi: 10.5301/ijbm.5000266. [DOI] [PubMed] [Google Scholar]
- 50.Park J., Shin S., Yoo H.M., Lee S.W., Kim J.G. Evaluation of the three customized MSI panels to improve the detection of microsatellite instability in gastric cancer. Clin Lab. 2017;63(4):705–716. doi: 10.7754/Clin.Lab.2016.161029. [DOI] [PubMed] [Google Scholar]
- 51.Setia N., Agoston A.T., Han H.S., Mullen J.T., Duda D.G., Clark J.W. A protein and mRNA expression-based classification of gastric cancer. Mod Pathol. 2016;29(7):772–784. doi: 10.1038/modpathol.2016.55. [DOI] [PubMed] [Google Scholar]
- 52.Kim H.S., Shin S.J., Beom S.H., Jung M., Choi Y.Y., Son T. Comprehensive expression profiles of gastric cancer molecular subtypes by immunohistochemistry: implications for individualized therapy. Oncotarget. 2016;7(28):44608–44620. doi: 10.18632/oncotarget.10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gonzalez R.S., Messing S., Tu X., McMahon L.A., Whitney-Miller C.L. Immunohistochemistry as a surrogate for molecular subtyping of gastric adenocarcinoma. Hum Pathol. 2016;56:16–21. doi: 10.1016/j.humpath.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 54.Engel C., Forberg J., Holinski-Feder E., Pagenstecher C., Plaschke J., Kloor M. Novel strategy for optimal sequential application of clinical criteria, immunohistochemistry and microsatellite analysis in the diagnosis of hereditary nonpolyposis colorectal cancer. Int J Canc. 2006;118(1):115–122. doi: 10.1002/ijc.21313. [DOI] [PubMed] [Google Scholar]
- 55.Chong I.Y., Cunningham D., Barber L.J., Campbell J., Chen L., Kozarewa I. The genomic landscape of oesophagogastric junctional adenocarcinoma. J Pathol. 2013;231(3):301–310. doi: 10.1002/path.4247. [DOI] [PubMed] [Google Scholar]
- 56.Hamilton S., Aaltonen L.E. IARC Press; Lyon: 2000. World Health organization classification of tumours. Pathology and genetics of tumours of the digestive system. [Google Scholar]
- 57.Pandilla R., Kotapalli V., Gowrishankar S., Chigurupati M., Patnaik S., Uppin S. Distinct genetic aberrations in oesophageal adeno and squamous carcinoma. Eur J Clin Invest. 2013;43(12):1233–1239. doi: 10.1111/eci.12163. [DOI] [PubMed] [Google Scholar]
- 58.Matsumoto Y., Nagasaka T., Kambara T., Hoshizima N., Murakami J., Sasamoto H. Microsatellite instability and clinicopathological features in esophageal squamous cell cancer. Oncol Rep. 2007;18(5):1123–1127. [PubMed] [Google Scholar]
- 59.Naidoo R., Ramburan A., Reddi A., Chetty R. Aberrations in the mismatch repair genes and the clinical impact on oesophageal squamous carcinomas from a high incidence area in South Africa. J Clin Pathol. 2005;58(3):281–284. doi: 10.1136/jcp.2003.014290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayashi M., Tamura G., Jin Z., Kato I., Sato M., Shibuya Y. Microsatellite instability in esophageal squamous cell carcinoma is not associated with hMLH1 promoter hypermethylation. Pathol Int. 2003;53(5):270–276. doi: 10.1046/j.1440-1827.2003.01478.x. [DOI] [PubMed] [Google Scholar]
- 61.Wu T.T., Watanabe T., Heitmiller R., Zahurak M., Forastiere A.A., Hamilton S.R. Genetic alterations in Barrett esophagus and adenocarcinomas of the esophagus and esophagogastric junction region. Am J Pathol. 1998;153(1):287–294. doi: 10.1016/S0002-9440(10)65570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gleeson C.M., Sloan J.M., McGuigan J.A., Ritchie A.J., Weber J.L., Russell S.E. Ubiquitous somatic alterations at microsatellite alleles occur infrequently in Barrett's-associated esophageal adenocarcinoma. Cancer Res. 1996;56(2):259–263. [PubMed] [Google Scholar]
- 63.Keller G., Rotter M., Vogelsang H., Bischoff P., Becker K.F., Mueller J. Microsatellite instability in adenocarcinomas of the upper gastrointestinal tract. Relation to clinicopathological data and family history. Am J Pathol. 1995;147(3):593–600. [PMC free article] [PubMed] [Google Scholar]
- 64.Ogasawara S., Maesawa C., Tamura G., Satodate R. Frequent microsatellite alterations on chromosome 3p in esophageal squamous cell carcinoma. Cancer Res. 1995;55(4):891–894. [PubMed] [Google Scholar]
- 65.Meltzer S.J., Yin J., Manin B., Rhyu M.G., Cottrell J., Hudson E. Microsatellite instability occurs frequently and in both diploid and aneuploid cell populations of Barrett's-associated esophageal adenocarcinomas. Cancer Res. 1994;54(13):3379–3382. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.