Abstract
Objective
The aim of this study was to investigate co-located nasal Staphylococcus aureus and coagulase-negative staphylococci (CoNS) (mainly Staphylococcus epidermidis), recovered from healthy medical students in their preclinical year, prior to exposure to the healthcare environment, for the carriage of genes and genetic elements common to both species and that may contribute to S. aureus and methicillin-resistant S. aureus (MRSA) evolution.
Design
Prospective observational cross-sectional study. Carriage of antimicrobial resistance and virulence-associated genes in the absence of significant antibiotic selective pressure was investigated among healthy medical students from geographically diverse origins who were nasally co-colonised with S. aureus and CoNS. Clonal lineages of S. aureus isolates were determined.
Setting/participants
Dublin-based international undergraduate medical students.
Results
Nasal S. aureus carriage was identified in 137/444 (30.8%) students of whom nine (6.6%) carried MRSA (ST59-MRSA-IV (6/9), CC1-MRSA-V-SCCfus (3/9)). The genes mecA, fusB, ileS2, qacA/qacC and the arginine catabolic mobile element-arc were detected among colonising nasal staphylococci and had a significantly greater association with CoNS than S. aureus. The rate of co-carriage of any of these genes in S. aureus/CoNS pairs recovered from the same individual was <1%.
Conclusions
The relatively high prevalence of these genes among CoNS of the healthy human flora in the absence of significant antibiotic selective pressure is of interest. Further research is required to determine what factors are involved and whether these are modifiable to help prevent the emergence and spread of antibiotic resistance among staphylococci.
Keywords: MRSA, antimicrobial resistance, nasal colonization, coagulase negative staphylococci, staphylococcus Aureus
Strengths and limitations of this study.
Global evaluation of antibiotic resistance gene carriage among staphylococci among healthy medical students in preclinical years through DNA microarray analyses.
Pairs of staphylococcal species were isolated from the same colonisation site (nares) of multiple participants to allow investigation of shared antibiotic resistance and virulence in the same human niche in a community setting.
A single-centre study design.
Coagulase-negative staphylococci was investigated only in students co-colonised with Staphylococcus aureus.
The study design did not facilitate follow-up of this cohort during clinical training.
Introduction
Staphylococcus aureus and S. epidermidis are significant colonisers of healthy human skin and nares and are among the leading causes of healthcare-associated infection. Morbidity, mortality and the financial burden associated with methicillin-resistant S. aureus (MRSA) infections are well documented. Furthermore, coagulase-negative staphylococci (CoNS) including S. epidermidis are reported reservoirs of antimicrobial-resistance genes and their associated mobile genetic elements, most notably the staphylococcal cassette chromosome (SCC) harbouring the mec gene (SCCmec).1
Twelve SCCmec types and numerous subtypes have been described among MRSA isolates to date. The more prevalent and diverse range of SCCs and SCCmec among CoNS further supports CoNS as a reservoir for antimicrobial resistance genes.1 The identification of SCC, SCCmec and SCC-associated elements with other antimicrobial and virulence genes and their epidemiological relationships among clinical staphylococci has advanced our understanding of the role of CoNS in the evolution of MRSA.2 For example, the fusidic acid resistance gene fusC is associated with SCCmec IV-SCC476 and other SCC-like elements have been identified in S. aureus, MRSA and CoNS and may contribute to MRSA emergence in countries with significant fusidic acid usage.3–5 Furthermore, the SCC-like arginine catabolic mobile element (ACME), which enhances acid tolerance, is abundant among clinical CoNS isolates, in particular S. epidermidis and S. haemolyticus.6 Among S. aureus, ACME has mainly been detected among isolates of the community-associated (CA) USA300 clone.7 CoNS are also a putative reservoir of the high-level mupirocin resistance encoding gene ileS2, which is also increasing among S. aureus/MRSA in healthcare and community environments related to horizontal gene transfer or expansion of specific clones.8 9
Increasingly, MRSA clones previously associated with the community such as clonal complex (CC) 1 are spreading to healthcare settings making the differentiation between healthcare-associated (HA) MRSA and CA-MRSA unclear.10 Therefore, detailed investigation of the genetic and phenotypic traits of colonising staphylococcal species in community settings is important to identify those with features that may contribute to their evolution into potentially successful and formidable HA clones. The aim of this study was to investigate co-located nasal S. aureus and CoNS (mainly S. epidermidis) recovered from healthy medical students in their preclinical year, prior to exposure to the healthcare environment, for the carriage of genes and genetic elements common to both species and that may contribute to S. aureus and MRSA evolution.
Methods
Study setting, participants and sample collection
This observational cross-sectional study was conducted at the Royal College of Surgeons in Ireland (RCSI) from December 2014 to January 2016. Nasal swabs (eSwab, Copan, Italy) were collected anonymously from undergraduate medical students. Eligible students were those attending the RCSI medical centre to submit a swab for mandatory MRSA screening in the week before they began their clinical attachments. In total, 444/450 eligible medical students (250 (56.3%) men, 194 (43.7%) women) participated in this study. All participants reported no previous hospital contact in the 6 weeks prior to recruitment. The student volunteers were from the second year of the undergraduate medical programme and, as such, all participants were domiciled in Ireland for a minimum of 2 years prior to recruitment. Data were collected anonymously from each participant, including age range, region of origin and previous healthcare contact. Ethical approval (approval number REC949) was obtained and informed consent was obtained from each participant.
Sample preparation
Swabs were processed to recover S. aureus (including MRSA) and pathogenic CoNS species using a modification of a published method.11 Swabs were enriched in brain heart infusion (BHI) supplemented with 6% (w/v) NaCl for 24 hours at 37°C followed by further enrichment in mannitol salt broth for 24 hours at 37°C. The enriched culture was diluted 1/1000 and 100 µL was spread onto SaSelect agar (Bio-Rad, Hercules, California, USA). Plates that yielded pink/orange colonies (presumptive S. aureus) were inspected for growth of colonies of relevant CoNS based on colony colour (eg, light pink colonies of various sizes, presumptive S. epidermidis; white/yellow colonies, S. haemolyticus, S. hominis, S. capitis, S. warneri, S. caprae, S. lugdunensis). Presumptive CoNS species and S. aureus were subcultured from these plates onto Columbia blood agar and identified by matrix-assisted laser desorption/ionisation (MALDI)–time-of-flight mass spectrometry using a MALDI Biotyper (Microflex LT, Bruker). Matched isolates (where S. aureus and a CoNS species were recovered from the same swab) were cryopreserved and stored at −20°C (Protect bacterial preserver beads (Technical Service Consultants, UK)).
Characterisation of S. aureus and CoNS isolates
Genomic DNA from S. aureus and CoNS isolates was extracted using enzymatic lysis using the buffers and solutions provided with the S. aureus Genotyping Kit 2.0 (Alere Technologies GmbH, Jena, Germany) and a DNeasy Blood and Tissue kit (Qiagen, Crawley, UK). Genetic characterisation of isolates was undertaken by DNA microarray profiling using the S. aureus Genotyping Kit 2.0 (Alere Technologies GmbH) as described previously.12 13 The kit detects 333 gene targets including staphylococcal antimicrobial-resistance, virulence, SCCmec and ACME-arc genes and assigns S. aureus isolates to multilocus sequence type (ST) or CCs. MRSA phenotype was confirmed in S. aureus and CoNS isolates positive for mecA by growth of pink (S. aureus) or colourless/white (CoNS) colonies on MRSASelect agar (Bio-Rad). When required, confirmation of carriage of fusidic acid resistance genes fusC, fusB and toxic shock syndrome toxin gene (tst1) were confirmed by PCR using the primers and conditions described by O’Neill et al 14 and Chen et al.15
Fusidic acid susceptibility testing
Fusidic acid MICs (Minimum Inibitory Concentrations) were determined by ETEST (bioMérieux, Marcy-l’Etoile, France) according to manufacturer’s instructions. Thirty S. aureus and CoNS isolates harbouring fusC or fusB were subcultured twice before testing. Results were interpreted according to EUCAST (http://www.eucast.org, assessed May 2015) susceptibility criteria.
Statistical analyses
Fisher’s exact test was used to analyse categorical variables (prevalence of genes) using GraphPad QuickCalcs online software. The significance of differences between groups was expressed as two-tailed P values. P values of ≤0.05 were considered statistically significant.
Patient and public involvement statement
The participants in this study were medical undergraduate students. They were invited to participate in this study when they underwent mandatory MRSA screening prior to commencement of clinical placements. As future clinicians, participants were broadly considered in the development of the research question. As nasal carriage of S. aureus and MRSA among healthcare staff contributes to transmission to patients in healthcare environments, medical students have an interest in contributing to this knowledge. Participants did not contribute to the study design. Participants were informed at recruitment that dissemination of the results would be through the scientific literature.
Results
Nasal carriage of staphylococcal species and regional distribution
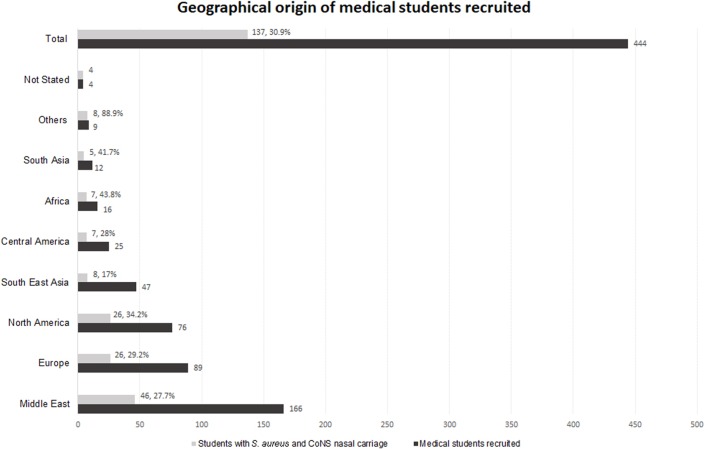
Thirty-one per cent (137/444) of students were positive for nasal carriage of S. aureus of whom 6.6% (9/137) were MRSA. Eighty-seven per cent (386/444) of students were positive for nasal carriage of CoNS (S. epidermidis (82% 364/444), S. haemolyticus (3% 14/444) or S. saprophyticus (2% 8/444) (table 1)). All students positive for S. aureus also carried S. epidermidis. Methicillin-resistant (MR)-CoNS were investigated in the S. aureus-positive cohort only of which 13.1% (18/137) were MR-CoNS. One student exhibited co-carriage of MRSA and MR-CoNS. The geographical region of origin of students harbouring S. aureus and CoNS is shown in figure 1. The Middle East, Europe and North America accounted for 68.6% of S. aureus carriers. For regions represented by ≥12 participants, the rate of nasal carriage of S. aureus varied geographically between 17% (South East Asia) and 44% (Africa).
Table 1.
Staphylococcus species recovered from 444 nasal swabs
| Staphylococcus species recovered | Total n, (%) (n=444) |
Methicillin-resistant phenotype n, (%) (n=137*) |
| Staphylococcus aureus | 137, (30.8) | |
| MRSA | 9, (6.6) | |
| S. epidermidis | 364, (81.9) | |
| MRSE | 17, (12.4) | |
| S. haemolyticus | 14, (3.1) | |
| MRSH | 0 | |
| S. saprophyticus | 8, (1.8) | |
| MRSS | 1, (0.73) | |
| Co-carriage species | ||
| S. aureus+S. epidermidis | 137, (30.8) | |
| MRSA+MR-CoNS | 1, (0.72) | |
| fusC-positive S. aureus+CoNS | 1, (0.72) |
*CoNS were investigated only in those positive for nasal S. aureus in the student cohort and not in all those recruited.
CoNS, coagulase-negative staphylococci; MRSA, methicillin-resistant S. aureus; MRSE, methicillin-resistant S. epidermidis; MRSH, methicillin-resistant S. haemolyticus; MRSS, methicillin-resistant S. saprophyticus.
Figure 1.
Geographical origin of medical students recruited. The geographical areas of origin of 444 medical students recruited to the study are shown (dark grey bars). Of those recruited, 137 were confirmed nasal Staphylococcus aureus and coagulase-negative staphylococci (CoNS) positive. The proportion of recruited students from each geographical origin with nasal S. aureus carriage are also shown (light grey bars).
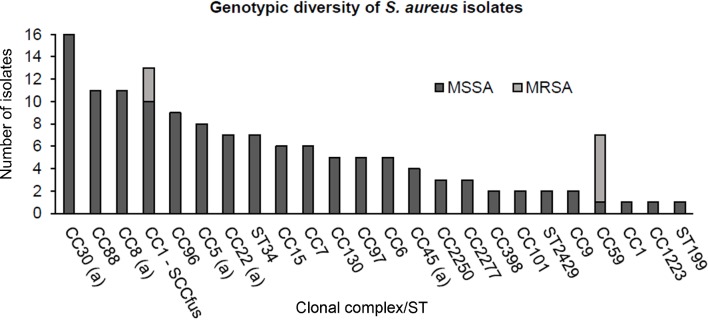
Clonal lineages among S. aureus isolates
The ST or CC distribution among 137 S. aureus isolates is shown in figure 2. Isolates belonged to a variety of CCs with 46/137 (33.5%) assigned to internationally disseminated CC5, CC8, CC22, CC30, CC45. A further 24/137 (17.5%) isolates belonged to CC1, CC59, CC88 or CC398.
Figure 2.
Genotypic diversity of 137 Staphylococcus aureus nasal isolates using DNA microarray analysis, including 128 methicillin-susceptible S. aureus (MSSA) (dark grey bars) and 9 methicillin-resistant S. aureus (MRSA) (light grey bars). Letter (a) indicates internationally disseminated clones into which staphylococcal cassette chromosome harbouring the mec gene (SCCmec) can integrate. CC, clonal complex; ST, sequence type.
SCCmec types and fusidic acid resistance among S. aureus and CoNS
Of the 333 staphylococcal genes detected by the microarray, the two most prevalent antibiotic resistance genes among nasal staphylococci were those encoding resistance to β-lactams and fusidic acid. The most common SCCmec type among nasal MRSA (n=9) and MR-CoNS (n=18) was SCCmec type IV (class B mec (mecA, DmecR1, ugpQ) and ccrA-2, ccrB-2). The nine MRSA isolates belonged to ST59-MRSA-IV (6/9) and CC1-MRSA-V-SCCfus (3/9) (class C mec (mecA, ugpQ) and ccrC and fusC (Q6GD50) and cassette chromosome recombinase (ccr) A-1, ccrB-1). Among 18 MR-CoNS identified (17 S. epidermidis and 1 S. saprophyticus), half harboured SCCmec type IV (8 S. epidermidis and the single S. saprophyticus). SCCmec types II, V and VII were identified in three, five and one of the remaining S. epidermidis isolates, respectively. Isolates from the one individual who exhibited nasal co-carriage of MRSA and MR-CoNS (S. epidermidis) both harboured SCCmec type IV (table 2).
Table 2.
Resistance/virulence genes detected among 137 co-located nasal Staphylococcus aureus/CoNS pairs
| Detected gene(s) | Phenotypic resistance/trait | No isolates positive n (%) |
P value | No S. aureus/CoNS pairs positive (n) | |
| S. aureus n=137 | CoNS n=137 | ||||
| Antibiotic resistance gene blaZ | β-lactam | 101 (73.7) | 92 (67.1) | 0.289 | 74 |
| fusB | Fusidic acid | 2 (1.5) | 27 (19.7) | 0.0002* | 0 |
| fusC† | Fusidic acid | 31 (22.6) | 25 (18.2) | 0.159 | 1 |
| mecA | Methicillin | 9 (6.5) | 18 (13.1) | 0.103 | 1 |
| ileS2 | Mupirocin | 1 (0.7) | 11 (8.0) | 0.005* | 1 |
| qac A and qac C | Quartenary ammonium salts | 3 (2.2) | 29 (21.2) | <0.0001* | 0 |
| tet(K) and tet(M) | Tetracycline | 13 (9.5) | 6 (4.4) | 0.152 | 0 |
| erm(C) | Macrolide/lincosamide | 6 (4.3) | 5 (3.6) | 1.000 | 0 |
| msr(A) | Macrolide | 2 (1.45) | 15 (10.9) | 0.002* | 1 |
| mph(C) | Macrolide | 0 | 15 (10.9) | <0.0001* | 0 |
| dfrS1 | Trimethoprim | 0 | 19 (13.8) | <0.0001* | 0 |
| vga | Streptogramin A | 1 (0.7) | 6 (4.3) | 0.120 | 0 |
| Virulence | |||||
| ACME-arc | pH tolerance | 1 (0.7) | 44 (32.1) | <0.0001* | 1 |
| tst1‡ | Toxic shock toxin | 33 (24.1) | 2 (1.5) | <0.0001* | 1 |
*Indicates a statistically significant result by Fisher’s exact test.
†Associated with SCC element (ccrA-1 and ccrB-1) in 13/137 S. aureus.
‡tst1 confirmed by PCR.
ACME, arginine catabolite mobile element; CoNS, coagulase-negative staphylococci.
In addition to the three CC1-MRSA-V isolates that carried SCCfus, the fusidic acid resistance genes fusC and fusB were identified in 28/128 (21.8%) and 2/128 (1.5%) of methicillin-susceptible S. aureus (MSSA) isolates, respectively. Ten of the 28 fusC-positive MSSA isolates belonged to CC1-MSSA-SCCfus, 11 were CC88-MSSA and 7 CC8-MSSA. All 10 CC1-MSSA-SCCfus isolates harboured a combination of SCCfus with the ccr genes, ccrA-1 and ccrB-1. The two fusB positive isolates belonged to CC5-MSSA and CC8-MSSA (table 2). Among MR-CoNS, 27.7% (5/18) S. epidermidis isolates carried fusC (two of them also carried ccr genes ccrA-1 ccrB-1) and 50% fusB (9/18, eight S. epidermidis and the one S. saprophyticus). Among methicillin-susceptible CoNS isolates, the fusC and fusB genes were identified in 20/119 (16.8%, 18 S. epidermidis and two S. saprophyticus) and 18/119 (15.1%, all S. epidermidis), respectively. One participant had nasal co-carriage of fusC-positive S. aureus (CC88-MSSA) and CoNS (S. epidermidis).
All SCCmec-positive staphylococci were confirmed to have an MRSA/MR-CoNS phenotype. However, there was poor correlation between fusC/fusB carriage and phenotypic fusidic acid resistance. Fusidic acid MICs for all fusC or fusB-positive S. aureus and CoNS isolates are shown in table 3. Phenotypic fusidic acid resistance was confirmed (based on EUCAST breakpoints, MICs ≥1 µg/mL) in 23/32 (71.8%) S. aureus and 20/38 (52.6%) CoNS nasal isolates harbouring either fusC or fusB (DNA microarray result confirmed by PCR). Eight nasal isolates (three S. aureus, five S. epidermidis) positive for fusB exhibited high-level fusidic acid resistance (MIC ≥32 µg/mL). Fusidic acid resistance was inducible in a further three S. aureus and seven S. epidermidis isolates following incubation with 0.01 µg/mL fusidic acid BHI agar.
Table 3.
Fusidic acid Minimum Inibitory Concentrations (MICs) for Staphylococcus aureus and CoNS
| MIC interpretation* | MIC ≤1 µg/mL S n (%) |
MIC ≥1 µg/mL R n(%) |
MIC ≥32 µg/mL HR n (%) |
| S. aureus (n=32) | 9 (28.1) | 20 (62.5) | 3 (9.3) |
| CoNS (n=38) | 18 (47.4) | 15 (39.5) | 5 (13.2) |
*Interpretation based on The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. V.7.1, 2017. http://www.eucast.org.
CoNS, coagulase-negative staphylococci; HR, high- level resistant; R, resistant; S, susceptible.
Other notable antimicrobial resistance genes among nasal S. aureus and CoNS
Apart from SCCmec element and fus genes, other antimicrobial genes detected among staphylococcal nasal flora were identified by DNA microarray. Tetracycline resistance genes, tet(K) or tet(M), were detected in 13/137 (9.5 %) of S. aureus isolates and 6/137 (4.3%) of the CoNS isolates. The quaternary ammonium compound resistance genes (qacA/qacC), encoding antiseptic resistance, were significantly more prevalent among CoNS isolates compared with S. aureus isolates (29/137 (21.2%) vs 2/137 (1.4%), P<0.0001). Significantly more CoNS than S. aureus isolates carried ileS2 encoding high-level mupirocin resistance (11/137 (8%) vs 1/137 (0.72%), P<0.01). However, none of these genes were common to S. aureus/CoNS pairs recovered from the same individual. The β-lactamase genes were abundant among S. aureus and CoNS; blaZ was present in 101/137 (73.72%) S. aureus isolates and 92/137 (67.1%) CoNS isolate and in 74/137 (54%) of individuals, these genes were common to S. aureus/CoNS pairs from the same nares. A summary of the antibiotic resistance genes found among S. aureus and CoNS is shown in table 2. The staphylococcal isolates were negative for all other antibiotic resistance genes spotted on the microarray.
Virulence genes among nasal S. aureus and CoNS
A single isolate, CC30-MSSA, was positive for the Panton-Valentine leucocidin genes (lukF/S-PV). Among nasal staphylococci, ACME-arc was significantly associated with CoNS compared with S. aureus (44/137 (32.1%) vs 1/137 (0.7%), P<0.0001). The toxic shock syndrome toxin gene tst1 was identified in 33/137 (24.1%) nasal S. aureus isolates. Unusually, DNA microarray identified tst1 in two S. epidermidis isolates and this was confirmed by PCR. ACME-arc was common to S. aureus/CoNS recovered from the nares in one individual only. One hundred and two (74.4%) S. aureus isolates encoded one or more enterotoxin genes. The enterotoxin gene cluster ((egc), containing seg, sei, SEM, sen, seo, seu) was the most prevalent (48/102, 47%) followed by seq/k (13/102, 12.7%) and sec/l (7/102, 6.8%). The staphylococcal isolates were negative for all other toxin genes spotted on the microarray.
Discussion
Studies of staphylococcal carriage and epidemiology among the healthy population in the absence of significant antibiotic pressure are important in identifying the potential for pathogenic evolution. To our knowledge, this is the first study to coinvestigate CoNS and S. aureus when recovered together from the nares of healthy preclinical medical students. The species distribution of nasal colonising CoNS was similar to other studies16 although the enrichment methods used here favoured S. aureus and S. epidermidis and may explain the low prevalence of other CoNS species. Our study revealed that, apart from the bla genes, which are abundant among staphylococci, the rates of co-carriage of antibiotic resistance genes in paired S. aureus/CoNS from the same individual were low in the community setting at <1%. Rates of simultaneous carriage of antimicrobial resistance among nasal staphylococci are likely to be higher under selective antibiotic pressure but few studies have investigated this among patients. One small study of hospitalised patients with nasal carriage of S. aureus and CoNS reported a rate of 12.5% patients carrying MRSA and MR-CoNS.17 However, the authors reported only two cases where simultaneous carriage of MR-CoNS and MRSA was detected and the strains involved carried different SCCmec types. Despite negligible detection of cospecies nasal carriage of these genes in medical students prior to healthcare exposure, based on antimicrobial resistance gene carriage by CoNS from this cohort, there is significant potential for mobilisation of genes to S. aureus that may enhance its pathogenic potential in the healthcare setting.
DNA microarray analyses revealed carriage of SCCmec, fusC, fusB, ileS2, qacA/qacC and ACME-arc among colonising nasal staphylococci in individuals with no previous healthcare exposure with greater prevalence among CoNS than S. aureus. This pattern among preclinical medical students supports CoNS as a reservoir with potential to subsequently accelerate antimicrobial resistance and pathogenicity among colonising S. aureus in clinical environments under antibiotic selective pressure.16 18 19
Despite considerable geographical distribution of the participants in this study, a S. aureus nasal carriage rate in the community of 30.8% was recorded. In this study, CC30, CC88 and CC8 were the most prevalent clones identified among nasal S. aureus. CC30 is among the internationally disseminated clones in which SCCmec has been acquired and is a successful colonising lineage, reported among HA and CA-MRSA. Among medical students, these MSSA isolates may therefore represent a significant pool for the uptake of SCCmec in a clinical setting. CC88 is frequently isolated in Australia but, in our study, the geographical background of isolates was mixed (including Middle East, Europe, South East Asia and Central America). CC8 is associated with MRSA infection and is globally disseminated.20 Although CC30, CC88 and CC8 were prevalent among community MSSA isolates in this study, among the relatively few MRSA recovered, none belonged to these CCs. Two CC/ST types detected among MRSA recovered from healthy medical students in this study were ST59-MRSA-IV and CC1-MRSA-V-SCCfusC. ST59 (Western Australian-MRSA-73) is a sporadic Australian strain and, apart from Panton Valentine Leukocidin (PVL) negativity, is indistinguishable from USA1000.21 In this study, the geographical background of these isolates was wide (Middle East, North America and South East Asia).
The identification of a significant reservoir of antibiotic resistance among medical students prior to healthcare exposure in subsequent clinical years highlights the need for effective infection prevention and control policies in relation to hand hygiene and surveillance. In the absence of antibiotic selective pressure, the colonising MRSA rate appears relatively stable and in this study was 2% (9/444), similar to rates reported elsewhere.22 However, a previous study among medical interns in China reported a nasal MRSA rate of 9.4% likely reflecting exposure to the healthcare environment.23 One study reported an increase in carriage rates of MR-CoNS from 14% among medical student preinternship to 29.28% among interns.24 Prevalence rates of MR-CoNS in recent community-based surveys are variable but rates of 16.5%19 and 17.2%25 are reported in similar cohorts to this study where, of those colonised with S. aureus, 13.1% carried MR-CoNS. SCCmec type IV, the smallest of the SCCmec elements, was the most prevalent type among MRSA and MR-CoNS here (66.6% and 50%). SCCmec IV has been detected in approximately 40% of methicillin-resistant S. epidermidis identified in humans.26 However, in this study, SCCmec type V was also represented among MRSA and MR-CoNS. While only one individual was colonised with MRSA and MR-CoNS in this study (both SCCmec type IV), the preponderance of SCCmec IV element in nasal MRSA and MR-CoNS suggests the potential for mecA gene transfer among these species even in the absence of selective pressure. The small size of this element, which has a low fitness cost, may enhance its dissemination potential.27
Fusidic acid resistance among S. aureus from healthy carriers in nine European countries in 2014 was reported to be <10%.28 However, we found a prevalence of 22.6% of fusC/fusB genes among healthy carriers. Fusidic acid resistance appears to correlate with increased use of this agent. For example, in New Zealand, where it is used as a first-line empiric agent for topical treatment of impetigo, prevalence rates of resistance in community S. aureus isolates increased from 17% in 1999 to 29% in 2013.29 In Europe, fusidic acid is combined with β-lactams for the treatment of staphylococcal bacteraemia, endocarditis and osteomyelitis30 and is used widely in the community for skin and soft tissue infections. A 2010 study of fusidic acid resistance among S. aureus clinical isolates showed Greece and Ireland to have the highest rates (52.5% and 19.9%).31 SCCfus has been identified in the CC1 background and more recently in other lineages such as ST239 and ST779.32–34 As highlighted here in the absence of significant antibiotic pressure in the community, it appears that this element is associated with MRSA and MSSA in the CC1 background. This genetic platform, particularly when associated with SCCmec in a composite element (SCCmec V+SCCfus) may enable the transfer of multidrug resistance in a single transfer event. The use of fusidic acid is unregulated in some countries and hence it may be used inappropriately in a community setting (eg, in short or discontinuous doses). Inappropriate use of fusidic acid may therefore favour coselection of methicillin resistance among S. aureus. In addition, in this study, 14/18 (77%) of MR-CoNS were positive for fusC or fusB. This association of resistances among the resident flora may provide further opportunity for dissemination of MRSA driven by fusidic acid selective pressure. Interestingly, a positive correlation between carriage of fusC/fusB and phenotypic resistance was observed in only 71.9% and 53.6% of S. aureus and CoNS, respectively. However, induction of gene expression with fusidic acid preincubation gave better correlation (82.2% and 76.3% correlation).
There were limitations to this study, which included a single-centred, relatively small study. Some nasally abundant CoNS species, for example, S. lugdunensis and S. hominis, were under-represented as the enrichment method favoured pathogenic staphylococci such as S. aureus and S. epidermidis. CoNS was investigated only in those co-colonised with S. aureus and therefore prevalence rates for genes among CoNS do not reflect the entire cohort. CCs and STs were determined only among S. aureus as the high rate of genetic recombination among CoNS makes strain typing unreliable. Although the microarray system used is reported effective for staphylococcal species other than S. aureus,35 some gene targets may be heterologous among staphylococci leading to false negatives. The study design did not facilitate follow-up of this cohort during clinical training which may have revealed further changes in gene carriage among colonising staphylococci. However, the multinational origin of the student body in our institution facilitated analysis of a relatively broad geographic cohort in a single study and emphasises the role that importation plays in S. aureus epidemiology. Unlike other studies of staphylococci in the healthy human nares, pairs of staphylococcal species originating from the same individual were investigated here for their resistance and virulence traits. These data support a low rate of transfer of antibiotic resistance between colonising staphylococcal species in the absence of healthcare contact. However, it is concerning that similar SCCmec and SCCfusC types in addition to ileS2, qacA/qacB and ACME are carried among CoNS and S. aureus in healthy individuals who will have subsequent roles in healthcare provision. Given the increasing emergence of HA-MRSA with features of community strains, further mobilisation of these elements under selective antibiotic pressure may enhance the transmission and success of S. aureus in the healthcare environment.
Supplementary Material
Acknowledgments
We acknowledge the support of the Mercer’s Medical Centre, RCSI and Ms Helen Barry, Chief Medical Scientist, St. James’ Hospital Dublin.
Footnotes
Contributors: PEB and DFH recruited students to the study. PEB conducted the laboratory work and drafted the manuscript. DFH and HH conceived the study and contributed to study design. ACS, PMK and DCC provided critical data interpretation and revised the drafted work. All authors contributed to the final approved draft.
Funding: PEB received funding for the study from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Brazil under the Science without Borders Program (Grant number 9172-13-0).
Competing interests: HH has received funding from Pfizer and Astellas outside the relevance of the submitted work. All other authors report no competing interests.
Patient consent: Obtained.
Ethics approval: Royal College of Surgeons in Ireland Ethics Commitee (REC)
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: All data for these analyses are included in the manuscript. No additional data are available.
References
- 1. Fluit AC, Carpaij N, Majoor EA, et al. Shared reservoir of ccrB gene sequences between coagulase-negative staphylococci and methicillin-resistant Staphylococcus aureus . J Antimicrob Chemother 2013;68:1707–13. 10.1093/jac/dkt121 [DOI] [PubMed] [Google Scholar]
- 2. Shore AC, Coleman DC. Staphylococcal cassette chromosome mec: recent advances and new insights. Int J Med Microbiol 2013;303:350–9. 10.1016/j.ijmm.2013.02.002 [DOI] [PubMed] [Google Scholar]
- 3. Hung WC, Chen HJ, Lin YT, et al. Skin commensal staphylococci may act as reservoir for fusidic acid resistance genes. PLoS One 2015;10:e0143106 10.1371/journal.pone.0143106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ellington MJ, Reuter S, Harris SR, et al. Emergent and evolving antimicrobial resistance cassettes in community-associated fusidic acid and meticillin-resistant Staphylococcus aureus . Int J Antimicrob Agents 2015;45:477–84. 10.1016/j.ijantimicag.2015.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baines SL, Howden BP, Heffernan H, et al. Rapid emergence and evolution of staphylococcus aureus clones harboring fusc-containing staphylococcal cassette chromosome elements. Antimicrob Agents Chemother 2016;60:2359–65. 10.1128/AAC.03020-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miragaia M, de Lencastre H, Perdreau-Remington F, et al. Genetic diversity of arginine catabolic mobile element in Staphylococcus epidermidis . PLoS One 2009;4:e7722 10.1371/journal.pone.0007722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Diep BA, Gill SR, Chang RF, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus . Lancet 2006;367:731–9. 10.1016/S0140-6736(06)68231-7 [DOI] [PubMed] [Google Scholar]
- 8. González-Domínguez M, Seral C, Potel C, et al. Genotypic and phenotypic characterization of methicillin-resistant Staphylococcus aureus (MRSA) clones with high-level mupirocin resistance. Diagn Microbiol Infect Dis 2016;85:213–7. 10.1016/j.diagmicrobio.2016.02.021 [DOI] [PubMed] [Google Scholar]
- 9. Bathoorn E, Hetem DJ, Alphenaar J, et al. Emergence of high-level mupirocin resistance in coagulase-negative staphylococci associated with increased short-term mupirocin use. J Clin Microbiol 2012;50:2947–50. 10.1128/JCM.00302-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Earls MR, Kinnevey PM, Brennan GI, et al. The recent emergence in hospitals of multidrug-resistant community-associated sequence type 1 and spa type t127 methicillin-resistant Staphylococcus aureus investigated by whole-genome sequencing: Implications for screening. PLoS One 2017;12:e0175542 10.1371/journal.pone.0175542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huber H, Giezendanner N, Stephan R, et al. Genotypes, antibiotic resistance profiles and microarray-based characterization of methicillin-resistant Staphylococcus aureus strains isolated from livestock and veterinarians in Switzerland. Zoonoses Public Health 2011;58:343–9. 10.1111/j.1863-2378.2010.01353.x [DOI] [PubMed] [Google Scholar]
- 12. Monecke S, Jatzwauk L, Weber S, et al. DNA microarray-based genotyping of methicillin-resistant Staphylococcus aureus strains from Eastern Saxony. Clin Microbiol Infect 2008;14:534–45. 10.1111/j.1469-0691.2008.01986.x [DOI] [PubMed] [Google Scholar]
- 13. Monecke S, Slickers P, Ehricht R. Assignment of Staphylococcus aureus isolates to clonal complexes based on microarray analysis and pattern recognition. FEMS Immunol Med Microbiol 2008;53:237–51. 10.1111/j.1574-695X.2008.00426.x [DOI] [PubMed] [Google Scholar]
- 14. O’Neill AJ, Larsen AR, Henriksen AS, et al. A fusidic acid-resistant epidemic strain of Staphylococcus aureus carries the fusB determinant, whereas fusA mutations are prevalent in other resistant isolates. Antimicrob Agents Chemother 2004;48:3594–7. 10.1128/AAC.48.9.3594-3597.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen HJ, Hung WC, Tseng SP, et al. Fusidic acid resistance determinants in Staphylococcus aureus clinical isolates. Antimicrob Agents Chemother 2010;54:4985–91. 10.1128/AAC.00523-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iravani Mohammad Abadi M, Moniri R, Khorshidi A, et al. Molecular characteristics of nasal carriage methicillin-resistant coagulase negative staphylococci in school students. Jundishapur J Microbiol 2015;8:e18591 10.5812/jjm.18591v2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Faria NA, Conceição T, Miragaia M, et al. Nasal carriage of methicillin resistant staphylococci. Microb Drug Resist 2014;20:108–17. 10.1089/mdr.2013.0197 [DOI] [PubMed] [Google Scholar]
- 18. Jamaluddin TZ, Kuwahara-Arai K, Hisata K, et al. Extreme genetic diversity of methicillin-resistant Staphylococcus epidermidis strains disseminated among healthy Japanese children. J Clin Microbiol 2008;46:3778–83. 10.1128/JCM.02262-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barbier F, Ruppé E, Hernandez D, et al. Methicillin-resistant coagulase-negative staphylococci in the community: high homology of SCCmec IVa between Staphylococcus epidermidis and major clones of methicillin-resistant Staphylococcus aureus . J Infect Dis 2010;202:270–81. 10.1086/653483 [DOI] [PubMed] [Google Scholar]
- 20. Jiménez JN, Ocampo AM, Vanegas JM, et al. CC8 MRSA strains harboring SCCmec type IVc are predominant in Colombian hospitals. PLoS One 2012;7:e38576 10.1371/journal.pone.0038576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Monecke S, Coombs G, Shore AC, et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus . PLoS One 2011;6:e17936 10.1371/journal.pone.0017936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abroo S, Hosseini Jazani N, Sharifi Y. Methicillin-resistant Staphylococcus aureus nasal carriage between healthy students of medical and nonmedical universities. Am J Infect Control 2017;45:709–12. 10.1016/j.ajic.2017.02.034 [DOI] [PubMed] [Google Scholar]
- 23. Ma XX, Sun DD, Wang S, et al. Nasal carriage of methicillin-resistant Staphylococcus aureus among preclinical medical students: epidemiologic and molecular characteristics of methicillin-resistant S. aureus clones. Diagn Microbiol Infect Dis 2011;70:22–30. 10.1016/j.diagmicrobio.2010.12.004 [DOI] [PubMed] [Google Scholar]
- 24. Baragundi MC, Solabannavar SS, Gokale SK, et al. Methicillin and multidrug resistant coagulase negative staphylococcal nasal carriage in medical students. J Commun Dis 2012;44:231–7. [PubMed] [Google Scholar]
- 25. Du X, Zhu Y, Song Y, et al. Molecular analysis of Staphylococcus epidermidis strains isolated from community and hospital environments in China. PLoS One 2013;8:e62742 10.1371/journal.pone.0062742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miragaia M, Thomas JC, Couto I, et al. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J Bacteriol 2007;189:2540–52. 10.1128/JB.01484-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Popovich KJ, Weinstein RA, Hota B. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clin Infect Dis 2008;46:787–94. 10.1086/528716 [DOI] [PubMed] [Google Scholar]
- 28. den Heijer CD, van Bijnen EM, Paget WJ, et al. Fusidic acid resistance in Staphylococcus aureus nasal carriage strains in nine European countries. Future Microbiol 2014;9:737–45. 10.2217/fmb.14.36 [DOI] [PubMed] [Google Scholar]
- 29. Williamson DA, Monecke S, Heffernan H, et al. High usage of topical fusidic acid and rapid clonal expansion of fusidic acid-resistant Staphylococcus aureus: a cautionary tale. Clin Infect Dis 2014;59:1451–4. 10.1093/cid/ciu658 [DOI] [PubMed] [Google Scholar]
- 30. Whitby M. Fusidic acid in septicaemia and endocarditis. Int J Antimicrob Agents 1999;12 Suppl 2:S17–S22. 10.1016/S0924-8579(98)00070-3 [DOI] [PubMed] [Google Scholar]
- 31. Castanheira M, Watters AA, Mendes RE, et al. Occurrence and molecular characterization of fusidic acid resistance mechanisms among Staphylococcus spp. from European countries (2008). J Antimicrob Chemother 2010;65:1353–8. 10.1093/jac/dkq094 [DOI] [PubMed] [Google Scholar]
- 32. Holden MT, Feil EJ, Lindsay JA, et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci U S A 2004;101:9786–91. 10.1073/pnas.0402521101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kinnevey PM, Shore AC, Brennan GI, et al. Emergence of sequence type 779 methicillin-resistant Staphylococcus aureus harboring a novel pseudo staphylococcal cassette chromosome mec (SCCmec)-SCC-SCCCRISPR composite element in Irish hospitals. Antimicrob Agents Chemother 2013;57:524–31. 10.1128/AAC.01689-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin YT, Tsai JC, Chen HJ, et al. A novel staphylococcal cassette chromosomal element, SCCfusC, carrying fusC and speG in fusidic acid-resistant methicillin-resistant Staphylococcus aureus . Antimicrob Agents Chemother 2014;58:1224–7. 10.1128/AAC.01772-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Argudín MA, Vanderhaeghen W, Butaye P. Diversity of antimicrobial resistance and virulence genes in methicillin-resistant non-Staphylococcus aureus staphylococci from veal calves. Res Vet Sci 2015;99:10–16. 10.1016/j.rvsc.2015.01.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.