ABSTRACT
The epigenetic mechanisms underlying chemoresistance in cancer cells resulting from drug-induced reversible senescence are poorly understood. Chemoresistant ESC-like embryonal carcinoma PA1 cells treated with etoposide (ETO) were previously found to undergo prolonged G2 arrest with transient p53-dependent upregulation of opposing fate regulators, p21CIP1 (senescence) and OCT4A (self-renewal). Here we report on the analysis of the DNA methylation state of the distal enhancer (DE) and proximal enhancer (PE) of the Oct4A gene during this dual response. When compared to non–treated controls the methylation level increased from 1.3% to 12.5% and from 3% to 19.4%, in the DE and PE respectively. It included CpG and non-CpG methylation, which was not chaotic but presented two patterns in each enhancer. Discorrelating with methylation of enhancers, the transcription of Oct4A increased, however, a strong expression of the splicing form Oct4B was also induced, along with down-regulation of the Oct4A partners of in the pluripotency/self-renewal network Sox2 and Lin28. WB demonstrated disjoining of the OCT4A protein from the chromatin-bound fraction. In survival clones, methylation of the DE was considerably erased, while some remnant of methylation of the PE was still observed. The alternative splicing for Oct4B was reduced, Oct4A level insignificantly decreased, while the expression of Sox2 and Lin28 recovered, all three became proportionally above the control. These findings indicate the involvement of the transient patterned methylation of the Oct4A enhancers and alternative splicing in the adaptive regulation of cell fate choice during the p53-dependant dual state of reversible senescence in ESC-like cancer stem cells.
KEYWORDS: embryonal carcinoma, wt TP53, DNA damage, Oct4A, enhancer methylation, alternative splicing, cell senescence, transient pluripotency suppression
Introduction
The model of the drug-induced reversible senescence of germ cancer stem cells reveals an unusual behaviour of Pou5f1 (Oct4A) gene
Resistance of cancer stem cells (CSC) to genotoxic modalities is the main cause of disease relapse and mortality of cancer patients. In the last period this response is associated with induction of reversible stress-induced cell senescence of CSC, whose mechanisms and prediction of the outcome are far from understood [1]. As a model to study this phenomenon, we chose the ovarian embryonal carcinoma cell line PA1 (wt TP53), treated with etoposide (ETO), the inhibitor of Topoisomerase II. Embryonal carcinomas such as PA1 possess the features of, and are often used as, an analog for embryonal stem cells (ESC) [2]. Using this model we previously found that ETO treated PA1 cells entered massive G2-arrest, assumed flat morphology, and signalled the DNA damage response (DDR) for about 4–5 days in approximately 100% of cells (with cell nuclei marked by ϫH2AX and CHK2 foci), indicating cell senescence [3,4]. However the normal cell cycle did recover through clones escaping senescence. The most surprising fact was the finding of heterogeneous activation of opposite regulators of senescence (p21CIP1) and the core regulator of self-renewal/pluripotency (OCT4A) in these enlarged, flattened cells. This paradoxical duality for senescence and self-renewal was shown to be p53 dependent, as both p21CIP1 and OCT4A became down-regulated by siRNA p53 [4]. In turn OCT4A moderated the expression of p21CIP1. The re-growth of survival clones starting from day 7–10 was abolished by OCT4 mRNA silencing [3]. Interestingly, the stress-activated OCT4A protein accumulated in cell the nuclei but was mostly disjoined from binding to the chromatin fraction; in addition the alternative splicing of its gene was determined by RT-PCR [4].
Aim of the current study
The unusual expression and behavior of Oct4A (POU5F1) gene and protein in this model of reversible senescence prompted us to explore the methylation state of the Oct4A gene regulatory elements. In this study, the DNA methylation analysis of two regulatory enhancers, proximal (PE) and distal (DE) in control and ETO-treated cells, on day 3 and day 20 (post-recovery) was carried out. To verify previous findings of Oct4 expression, RT-qPCR analysis of Oct4A and Oct4B expression was undertaken as well as analysis of additional components of the self-renewal/pluripotency network Sox2 and Lin28 [5–7].
Results
The real-time PCR reveals up-regulation of Oct4A and Oct4B and down-regulation of Sox2 and Lin28 in Etoposide–treated cells, reversed in survival clones
Expression of Oct4A, Oct4B, Sox2 and Lin28 in control, 3 days and 20 days post ETO treatment was measured by RT-qPCR (Fig. 1). This analysis confirmed the previous results of semi-quantitative-PCR [4] of the expression profile change with induction of both enhanced OCT4A transcription but even more spectacular induction of its alternative splicing Oct4B transcript. Moreover, a several-fold decrease of expression of the direct and indirect partners of Oct4A in the pluripotency network, Sox2 and Lin28 after ETO were observed on day 3 (Fig. 1). The reduction of Sox2 and Lin28 when considered with the disjoining of Oct4A from the chromatin binding [4] indicates a loss of the pluripotency function of OCT4A. However, in the surviving clones on day 20 the transcription levels of Oct4A insignificantly decreased, while the amounts of its partners in the pluripotency network increased, all three becoming proportionally above the control levels (Fig. 1) indicating to restoration of the pluripotency and self-renewal enhancement.
Figure 1.
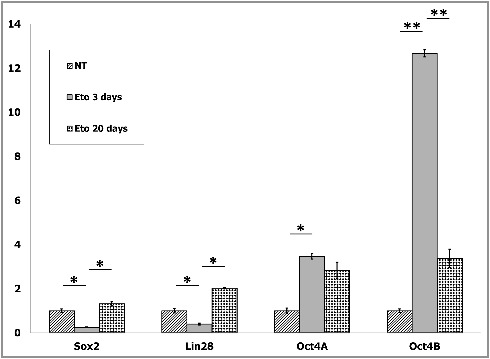
RT-qPCR shows that treatment of PA-1 cells with etoposide (ETO) leads on day 3 to bias of enhanced expression of the conventional and alternative splicing of POU5F1/Oct4 which is coupled to downregulation of the Oct4A partners in the pluripotency network Sox2 and Lin28. The discorrelation between the Oct4A, Sox2 and Lin28 is corrected, while the splicing variant Oct4B drops down, in the survival cells on day 20. Average numbers from two independent experiments are presented. Relative mRNA levels are shown after normalization against four housekeeping genes for every sample (for details see Material and Methods). mRNA levels in non-treated (NT) cells are arbitrary set as 1 and shown as folds after ETO treatment. Statistical significance is shown by a two-tailed T-test:*p<0.05; **p<0.01.
ETO induces moderate DNA methylation of distal enhancer (DE) and proximal enhancer (PE) of Oct4A gene, reversed in survival clones
DNA methylation of the Oct4A regulatory upstream elements was also examined in non-treated (NT) PA1 cells, 3 days post ETO treatment, during prolonged G2 arrest, and on day 20 post ETO treatment (Fig. 2). In the NT control, the enhancers were practically un-methylated. The moderate methylation of Oct4A enhancers, including CpG and non-CpG sites was observed on day 3 post ETO, increasing in DE from 1.3% to 12.5% and in PE from 3% to 19.4%. The methylation was not chaotically spread but targeted to specific locations. In particular, two distinctive methylation patterns were seen in each enhancer (marked on Fig. 2). Besides, we found some clones presenting CCWGG methylation in DE (Figs. 2 b and 3) without methylated CpGs. DE the CpG methylation in the proximity of the SOX2-OCT4 heterodimer binding site and methylation in the CCWGG epigenetic mark in the vicinity of the NANOG p1 binding sequence is observed (Supplemental Fig. 1). It is interesting to note that PE contains a cluster of 8 CCWGG (W = A/T) motifs with 6 of them corresponding to CCAGGs which represent targets for 5’-CAG-3’ methylation which predominantly occurs in ESC and oocytes [8,9]. In the PE, we observed a more pronounced methylation of CCWGG motifs as compared with DE. The pattern of the CpG and non-CpG methylation induced by ETO in the PE partially resembled much heavier methylated PE of differentiated peripheral blood leukocytes (Fig. 2). On day 20, in recovery clones we found methylation of DE and PE had been profoundly, but not fully, reduced and of the two distinctive methylation patterns were not observed anymore (Fig. 2). The observed residual CpG and CCWGG methylation in regulatory enhancers might reflect heterogeneity of recovering clonal population of PA1 cells (remnant senescent cells) as well as the drift of PE methylation in some clones.
Figure 2.
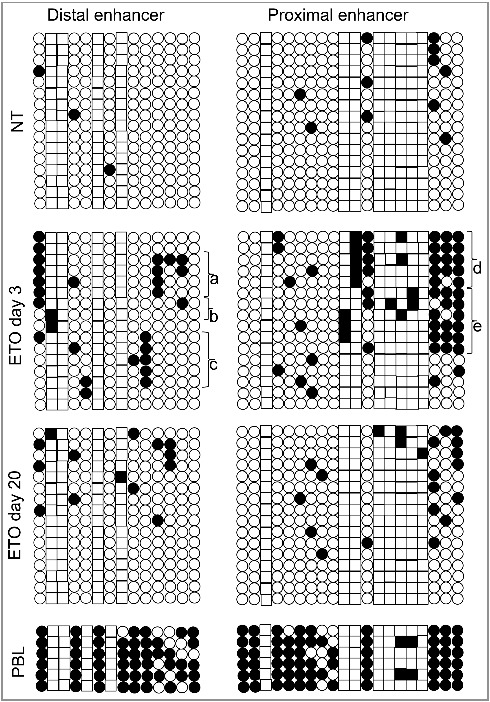
The results of bisulphite sequencing analysis of proximal and distal enhancers of Oct4A promoter regions. Each row of circles for a given amplicon represents the methylation status of CpGs or CCWGGs nucleotides (correspondingly, circles and squares) in one clone for that region. Number of columns corresponds to the number of analysed clones (16). Open circles and squares are un-methylated CpGs and CCWGGs, while the closed ones are methylated. (a) and (c) represent the two presumably distinctive methylation patterns of DE; (b) the two clones where only CCWGG methylation was detected; (d) and (e) correspond to two presumably distinctive methylation patterns of PE.
Figure 3.
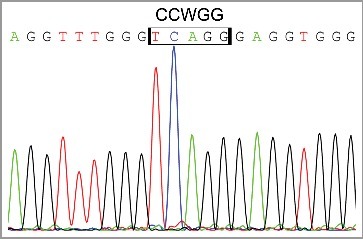
Fragment of sequencing chromatogram of bisulphite treated DNA demonstrating the presence of CCWGG methylation.
Discussion
The altered expression profile of the Oct4A gene induced by ETO is accompanied by an induction in alternative splicing forms Oct4B and Oct4B1, in addition Oct4-PG1 was also induced, all together more closely resembling the expression pattern seen in terminally differentiated peripheral blood leukocytes (PBL) [4]. Interestingly, the pattern of the enhanced methylation of the DE and particularly PE of Oct4A gene also tended towards the pattern of more heavily methylated PBL as seen on Fig. 2. This suggests the epigenetic state of PA1-ETO senescent cells is prepared for a shift in the fate commitment from pluripotency to differentiation. However, this state is unstable: when cells recovered clonogenic growth, the moderate methylation of enhancers became mostly erased, alternative splicing transcription of Oct4 gene went down, while the transcription of Oct4A partners in the self-renewal and pluripotency network Sox2 and Lin28 was recovered and even enhanced.
This interesting situation should be discussed in frames of the response of ESC-like germ cancer wt TP53 cells to genotoxic damage, which our PA1-ETO model represents. Firstly, ESC cells are known as not possessing the G1/S checkpoint and therefore are arrested after DNA damage directly in the G2/M damage checkpoint [10]. Secondly, in this checkpoint, the phosphorylation within the homeobox region of OCT4 by Aurora B kinase (AURKB) was shown negatively regulating its activity by interrupting sequence-specific DNA binding [11,12]. In accord with this data, we see in our model that inspite of the wt TP53 (which should arrest the DNA damaged somatic cells in G1), the PA1-ETO cells entirely entered the G2/M arrest, were positive there for AURKB (not shown), and disjoined OCT4A from the chromatin [4]. Thus the interruption of the positive feed-back loop of Oct4A itself and of its partners in the core self-renewal network has occurred.
How these obstacles may be related to transient moderate methylation of Oct4A enhancers in the drug-induced reversible cell senescence?
The methylation of Oct4 regulatory elements inducing alternative splicing of the Oct4B form has been previously reported [13]. As well, the recently revealed suppressive function of Oct4B by its non-coding RNA on translation of Oct4A may be mentioned [14], However, these data mostly refer to somatic tumors; in addition, DNA methylation of enhancers is regulated in cell-type specific manner [15]. Here, in the germ (oocyte) cancer cell model after ETO, along with some degree of enhancer methylation and shift to Oct4B alternative splicing, the surprising induction of enhanced transcription and accumulation of the OCT4A protein itself needs explanation, or at least a working hypothesis. Gene expression suppression by CpG methylation is well established [16,17] but as to non-CpG methylation found in our model, the situation is not unequivocal [18]. The finding of presumably two patterns of the methylation in two enhancers including both CpG and non-CpG sequences suggests a possibility of the operational switch of promoters for Oct4A to its splicing forms in the dual stage of senescence and self-renewal.We hypothesize that both Oct4A and Oct4B forms can be transcribed by shifts from one methylation pattern of Oct4 enhancers to another (may be, through a metastable CCWGG bridge) supporting the metastability of reversible senescence. We cannot exclude that two different Oct4A methylation patterns belong to different cells in the population, heterogenous for the OCT4/p21CIP1 expression ratio or even to two different epigenomes in the same G2-arrested cell.
Preference of this checkpoint, with the doubled DNA content, for arrest may be a prerequisite of cell fate divergence by bi-polarity of two epigenomes and sorting DNA damage/senescence from self-renewal in an asymmetric/symmetric two-step division noticed in this and similar models [19]. We also suggest that although transiently non-functional in the pluripotency network, the accumulated long-lived OCT4A protein, in addition to its p21CIP1 moderating function [3], retains an option to be operatively involved in the pluripotency network by an autocatalytic loop of its own and then attract the transcription of its network partners [5,20]. This could occur as soon as the DNA damage is repaired or sorted, the cell resumes division and enters G1, and the mechanism of keeping stressed OCT4A away from the chromatin binding (in G2 arrest) is withdrawn together with TP53 activation. It should be noted in addition, that activated p53 directly suppresses the promoter of the gateway to the pluripotent state, Nanog [21–23] whose transcription is however fluctuating [24,25]. Both p53-dependent mechanisms thus provide an option for a bi-stable switch between self-renewal/pluripotency and senescence/differentiation allowing the adaptive choice of individual cell fates.
In conclusion, we revealed in wtTP53 PA1-ETO cells the DDR response by the moderate two-patterned methylation of the distal and proximal Oct4A enhancers activates both conventional and alternative splicing forms of Oct4A transcripts, characterizing the reversible cell senescence of these ESC-like embryonal carcinoma cells.
Material and methods
Cell treatment, DNA extraction and bisulfite analysis
The treatment with ETO of PA1 cells was performed in the experiments reported earlier [1,2]. In brief, cells were treated with 8µM ETO for 20 h and afterwards cultivated and fed in normal medium without antibiotics. After discarding the medium with floating dead cells the PA1cells were detached from support and collected by centrifugation. Genomic DNA of ETO-treated and non-treated PA1 cells was obtained by overnight cell incubation in TES buffer containing 0.1% SDS and 100 µg/ml proteinase K at 55 °C with subsequent phenol/chloroform extraction and isopropanol precipitation. 2 µg of DNA in 50 µl of TE buffer was denatured during15 minutes in 0.3 M NaOH at 37°C. The denatured DNA was mixed with 550 µl of freshly prepared solution of 10 mM hydroquinone and 3 M sodium bisulfite at pH 5.0, and incubated under mineral oil at 50 °C for 12 h. Bisulfite treated DNA was desalted by isopropanol precipitation, desulfonated with 0.3 M NaOH for 5 min at room temperature, and precipitated with ethanol. Converted DNA was dissolved in 100 µl of water and stored at -20°C. 3 µl of precipitated DNA were used for each PCR. DNA of peripheral white blood cells (WBC) was used as a control for normal differentiated quiescent cells.
Bisulfite sequencing analysis
The bisulfite treated DNA was used to amplify the proximal and distal enhancers (PE, DE) of OCT4 promoter region with primers specific for bisulfite converted DNA: DE of OCT4, forward: 5'-AGGAGTTATTAGGAAAATGGGTAGTAG-3', reverse: 5'-TACCTTCTAAAAAAATAAATATCCC-3'. For PE of OCT4, forward: 5’-GGG GAG TTT AGG GTA GTT TTT TTG-3’, reverse: 5'-AAA CTA ACT AAA CCT CAA TTT CCC AA-3'. The PCR reaction was carried out using 2.5 units of homemade Taq polymerase in a final volume of 50 µl and the following cycling conditions: 5 minutes at 95 °C, followed by 35 cycles (30 seconds denaturation at 95 °C, annealing for 30 seconds at 62 °C and elongation at 72 °C for 1 minute. The PCR products were gel purified and cloned using TOPO TA cloning kit. To prevent clonal amplification of sequences, the competent cells transformed were plated immediately after heat shock; excluding shaking bacteria for 1 h. 16 clones for each PCR product were sequenced and analysed. The methylation analysis of enhancers was performed by alignment of sequenced clones against the genomic region corresponding to appropriate enhancer. The efficiency of cytosine to uracil conversion was estimated as the ratio of cytosine in a non-CpG context to total number of cytosine in the region. The clones with the efficiency of cytosine conversion less than 98% were omitted from the analysis.
DNA sequencing was performed using the ABI BigDyeTerminator Cycle Sequencing Kit v3.1 according to the manufacturer's instructions on a Gene Amp 9700 PCR machine and the sequences were detected on an ABI 3130XLGenetic Analyzer.
Real-time RT-PCR
Total RNA was extracted from PA-1 cells (2 × 106 cells) by using TRIZOL (Invitrogen). First-strand cDNA was synthesized using 2,5 µg of RNA, random hexamers and RevertAid™ M-MuLV Reverse Transcriptase (Fermentas, Lithuania) according to the manufacturer's protocols, and subsequently diluted with nuclease-free water (Fermentas) ten times. Real-time PCR was run on a ViiA7 (Applied Biosystems). Amplification mixtures (25 µl) contained 2µl template cDNA, 2xSYBR Green Master Mix buffer (12,5 µl) (Life Technologies, Carlsbad, CA, USA) and 2µM forward and reverse primer (Table 1). The cycling conditions comprised 10 min polymerase activation at 95oC and 40 cycles at 95oC for 15 sec and 60oC for 60 sec. A melt curve was also performed after the assay to check for specificity of the reaction. This consistent of 20 sec at 72o C followed by a ramp up of 1o step with 5-sec hold at each step. Every cDNA sample was normalized against four housekeeping genes: GAPDH, ACTB, B2M, and LRP10 using giNorm software [26]. The calculated gene expression stability coefficient M was applied to Q-PCR results.
Table 1. Forward and reverse primer sequences used in real-time RT-PCR
| Gene | Forward primer sequence | Reverse primer sequence | Reference |
|---|---|---|---|
| Oct4A | TCGCAAGCCCTCATTTCACC | GCCAGGTCCGAGGATCAAC | [27] |
| Oct4B | AGACTATTCCTTGGGGCCACAC | GGCTGAATACCTTCCCAAATAGA | [27] |
| Sox2 | GTACTGGCGAACCATCTCTGTG | CCAACGGTGTCAACCTGCATG | |
| Lin28 | CGGGCATCTGTAAGTGGTTC | CAGACCCTTGGCTGACTTCT | [28] |
| GAPDH | GGGTCTTACTCCTTGGAGGC | GTCATCCCTGAGCTAGACGG | [26] |
| ACTB | AATCTCATCTTGTTTTCTGCGC | AGTGTGACGTGGACATCCG | [26] |
| B2M | TCTCGCTCCGTGGCCTTAGC | GCCTACCTACTTTGGGTCTGTGT | [26] |
| LRP10 | ACCGCTGCAACTACCAGACT | ACGTCGACATTACTGGAAC | [26] |
Supplementary Material
Funding Statement
The study was supported by the Europe Social Fund Project, project No. 2013/0023/1DP/1.1.1.2.0/13/APIA/VIAA/037. The publishing costs are covered by the Riga Stradins University.
Acknowledgement
Dr. Bogdanova-Jatniece is acknowledged for sharing the sequences for Sox2 RT-qPCR. The authors thank Prof. MS Cragg for reading the manuscript. The study was supported by the Europe Social Fund Project, project No. 2013/0023/1DP/1.1.1.2.0/13/APIA/VIAA/037. The publishing costs are covered by the Riga Stradins University.
Disclosure of potential conflicts of interest
The authors disclose no conflict of interests.
References
- [1].Erenpreisa J, Salmina K, Cragg MS. Accelerated senescence of cancer stem cells: a failure to thrive or a route to survival? In: Dorszewska J, ed. Senescence – Physiology or Pathology. InTech; 2017. doi: 10.5772/intechopen.68582. [DOI] [Google Scholar]
- [2].Andrews PW, Matin MM, Bahrami AR, et al. Embryonic stem (ES) cells and embryonal carcinoma (EC) cells: opposite sides of the same coin. Biochem Soc Trans. 2005;33(6):1526. doi: 10.1042/BST20051526. PMID:16246161 [DOI] [PubMed] [Google Scholar]
- [3].Huna A, Salmina K, Erenpreisa J, et al. Role of stress-activated OCT4A in the cell fate decisions of embryonal carcinoma cells treated with etoposide. Cell Cycle. 2015;14(18):2969–2984. doi: 10.1080/15384101.2015.1056948. PMID:26102294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jackson TR, Salmina K, Huna A, et al. DNA damage causes TP53-dependent coupling of self-renewal and senescence pathways in embryonal carcinoma cells. Cell Cycle. 2013;12(3):430–441. doi: 10.4161/cc.23285. PMID:23287532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Boyer LA, Lee TI, Cole MF, et al. Core Transcriptional Regulatory Circuitry in Human Embryonic Stem Cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rizzino A. Sox2 and Oct-3/4: a versatile pair of master regulators that orchestrate the self-renewal and pluripotency of embryonic stem cells. Wiley Interdiscip Rev Syst Biol Med. 2009;1(2):228–236. doi: 10.1002/wsbm.12. PMID:20016762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Darr H, Benvenisty N. Genetic analysis of the role of the reprogramming gene LIN-28 in human embryonic stem cells. Stem Cells. 2009;27(2):352–362. doi: 10.1634/stemcells.2008-0720. PMID:19038789 [DOI] [PubMed] [Google Scholar]
- [8].Laurent L, Wong E, Li G, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20(3):320–331. doi: 10.1101/gr.101907.109. PMID:20133333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. PMID:19829295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Neganova I, Lako M. G1 to S phase cell cycle transition in somatic and embryonic stem cells. J Anat. 2008;213(1):30–44. doi: 10.1111/j.1469-7580.2008.00931.x. PMID:18638068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Brumbaugh J, Hou Z, Russell JD, et al. Phosphorylation regulates human OCT4. Proc Natl Acad Sci. 2012;109(19):7162–7168. doi: 10.1073/pnas.1203874109. PMID:22474382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Shin J, Kim TW, Kim H, et al. Aurkb/PP1-mediated resetting of Oct4 during the cell cycle determines the identity of embryonic stem cells. Elife. 2016;5:e10877. doi: 10.7554/eLife.10877. PMID:26880562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang Y, Meng L, Hu H, et al. Oct-4B isoform is differentially expressed in breast cancer cells: hypermethylation of regulatory elements of Oct-4A suggests an alternative promoter and transcriptional start site for Oct-4B transcription. Biosci Rep. 2011;31:109–115. doi: 10.1042/BSR20100033. PMID:20433421 [DOI] [PubMed] [Google Scholar]
- [14].Li D, Yang Z-K, Bu J-Y, et al. OCT4B modulates OCT4A expression as ceRNA in tumor cells. Oncol Rep. 2015;33(5):2622–2630. doi: 10.3892/or.2015.3862. PMID:25812694 [DOI] [PubMed] [Google Scholar]
- [15].Choi HW, Joo JY, Hong YJ, et al. Distinct Enhancer Activity of Oct4 in Naive and Primed Mouse Pluripotency. Stem Cell Reports. 2016;7(5):911–926. doi: 10.1016/j.stemcr.2016.09.012. PMID:28157483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321(6067):209–213. doi: 10.1038/321209a0. PMID:2423876 [DOI] [PubMed] [Google Scholar]
- [17].Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis. 2000;21(3):461–467. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10688866. Accessed December 5, 2017. doi: 10.1093/carcin/21.3.461. PMID:10688866 [DOI] [PubMed] [Google Scholar]
- [18].Jin J, Lian T, Gu C, et al. The effects of cytosine methylation on general transcription factors. Sci Rep. 2016;6(1):29119. doi: 10.1038/srep29119. PMID:27385050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Erenpreisa J, Salmina K, Belyayev A, et al. Survival at the brink: chromatin autophagy of tumour cells in response to genotoxic damage. In: Hayatt MA, ed. Autophagy: cancer, other pathologies, inflammation, immunity, infection, and aging. Elsevier; 2016. p. 275–294. [Google Scholar]
- [20].Lee J, Go Y, Kang I, et al. Oct-4 controls cell-cycle progression of embryonic stem cells. Biochem J. 2010;426(2):171–181. doi: 10.1042/BJ20091439. PMID:19968627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lin T, Chao C, Saito S, et al. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7(2):165–171. doi: 10.1038/ncb1211. PMID:15619621 [DOI] [PubMed] [Google Scholar]
- [22].Chambers I, Silva J, Colby D, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450(7173):1230–1234. doi: 10.1038/nature06403. PMID:18097409 [DOI] [PubMed] [Google Scholar]
- [23].Silva J, Nichols J, Theunissen TW, et al. Nanog Is the Gateway to the Pluripotent Ground State. Cell. 2009;138(4):722–737. doi: 10.1016/j.cell.2009.07.039. PMID:19703398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lahav G, Rosenfeld N, Sigal A, et al. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet. 2004;36(2):147–150. doi: 10.1038/ng1293. PMID:14730303 [DOI] [PubMed] [Google Scholar]
- [25].Kalmar T, Lim C, Hayward P, et al. Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. Goodell MA, ed. PLoS Biol. 2009;7(7):e1000149. doi: 10.1371/journal.pbio.1000149. PMID:19582141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. PMID:12184808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Asadi MH, Khalifeh K, Mowla SJ. OCT4 spliced variants are highly expressed in brain cancer tissues and inhibition of OCT4B1 causes G2/M arrest in brain cancer cells. J Neurooncol. 2016;130(3):455–463. doi: 10.1007/s11060-016-2255-1. PMID:27585657 [DOI] [PubMed] [Google Scholar]
- [28].Qiu C, Ma Y, Wang J, et al. Lin28-mediated post-transcriptional regulation of Oct4 expression in human embryonic stem cells. Nucleic Acids Res. 2010;38(4):1240–1248. doi: 10.1093/nar/gkp1071. PMID:19966271 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


