ABSTRACT
UHRF1 (ubiquitin-like, containing PHD and RING finger domains 1) is an important epigenetic regulator that plays a part in DNA methylation, protein methylation and ubiquitination. It is also frequently overexpressed in many types of cancers, including cervical cancer, which is caused by human papillomavirus (HPV). In this study, we showed that UHRF1 was up-regulated in HPV oncogene E7 expressing cells and HPV-positive cervical cancer cells. We demonstrated that UHRF1 down-regulated the expression of UBE2L6 gene that encodes the ISG15-conjugating enzyme UbcH8. Overexpression of UHRF1 reduced UBE2L6 while knockdown UHRF1 elevated the expression of UBE2L6. We showed that UHRF1 regulated UBE2L6 gene by promoter hypermethylation in cervical cancer cells. Consistent with the functions of UHRF1, restored expression of UbcH8 induced apoptosis. These findings establish UBE2L6 as a novel target of UHRF1 that regulates the apoptosis function of UHRF1. Our studies suggest that UHRF1/ UbcH8 can be manipulated for therapy in cervical cancer.
KEYWORDS: HPV, E7, UHRF1, UbcH8, apoptosis
Introduction
Cervical cancer is one of the leading causes of cancer death in women worldwide. Human papillomavirus (HPV) has been detected in more than 99% of cervical cancers [1]. HPVs are 8000-base pair, double-stranded, circular DNA viruses that replicate in squamous epithelia. The high-risk HPVs encode two viral oncoproteins E6 and E7 that are required for cancer development. The HPV E7 oncoprotein inactivates pRb and releases E2F [2]. Free E2Fs activate the transcription of series of genes required for cell cycle progression and cell apoptosis [3].
UHRF1 (ubiquitin-like, containing PHD and RING finger domains 1, also known as Np95 and ICBP90), inverted CCAAT box-binding protein of 90 kDa, has been reported to be up-regulated by E2F-1 [4]. As an important epigenetic regulator, UHRF1 plays an essential role in DNA methylation [5], histone methylation [6,7,8] and protein ubiquitination [9]. DNA methylation often works together with histone modifications such as acetylation and methylation to regulate gene expression [10,11]. UHRF1 is the bridge protein which links DNA methylation and histone modifications and required for efficient maintenance of methylation [5,12]. It helps to tether DNMT1 (DNA-methyltransferase 1) to chromatin [5]. Epigenetic changes (DNA methylation and histone modifications) can induce silencing of critical genes allowing cancer cells to escape apoptosis and promote tumor progression [13,14]. UHRF1 knockdown inhibited cell proliferation and migration, and induced apoptosis [15]. Up-regulation of UHRF1 has been observed in a variety of cancers, such as lung cancer, bladder cancer, prostate cancer as well as cervical cancer [16,17,18,19]. The expression level of UHRF1 is closely related to clinical stage, metastatic and prognostic of bladder cancer and lung cancer [17,20].
UbcH8 (ubiquitin/ISG15-conjugating enzyme E2 L6, coded by gene UBE2L6), a member of the E2 ubiquitin conjugating enzymes, is also involved in ISGylation as E2 [21,22]. UBE2L6 is identified to be a gene significantly down-regulated by promoter hypermethylation in nasopharyngeal carcinoma (NPC) [23]. It catalyzes the covalent attachment of ubiquitin or ISG15 to abnormal or short-lived proteins for degradation. Similar to ubiquitylation, there are three kinds of enzymes in ISGylation pathway: E1-activating, E2-carrier and E3-ligase enzymes. The enzyme UBE1L (ubiquitin-activating enzyme E1-like) was shown to be the specific ISG15-activating enzyme [24]. Ablation of UbcH8 by RNAi suggested this to be the principal ISG15 E2 carrier [21]. Two E3 ubiquitin ligases, HERC5 (HECT domain and RLD 5) and TRIM25 (Tripartite Motif Protein 25), have been identified to conjugate ISG15 to protein substrates. ISG15 is considered to be covalently conjugated to cellular proteins through a sequential reaction similar to that of the ubiquitin conjugation system consisting of E1/E2/E3 enzymes. As an important molecule of ISGylation, ISG15 is one of the most strongly interferon (IFN)-induced genes and is also substantially induced by viral infection [5]. Significantly, ISGylation of HPV L1 capsid protein has a dominant-inhibitory effect on the infectivity of HPV-16 [25]. Proteomic studies have identified more than 300 cellular proteins that are targeted for ISGylation. Some of the ISGylation targets, such as CBX4 [26], PKM2 [27], STAT1 [28] and DYRK1A [29], are involved in apoptosis.
In this study, we showed that UHRF1 was up-regulated in HPV E7 expressing cells and cervical cancer cells. We demonstrated that UBE2L6 is a UHRF1 regulated gene. We further showed that UHRF1 regulated UBE2L6 gene by promoter hyper-methylation in cervical cancer cells. Consistent with the functions of UHRF1, restored expression of UbcH8 induced apoptosis. Our studies suggest that UBE2L6 is a novel target of UHRF1 that regulates its apoptosis function.
Results
The expressions of UHRF1 and UbcH8 are inversely correlated in HPV E7 expressing and cervical cancer cells
According to our previous RNA-seq results, up to 237 genes were differentially expressed between cells with and without HPV E7 [30]. Among these genes, UHRF1 was up-regulated while UbcH8 was down-regulated in HPV-16 E7 expressing cells (Figure 1A). As a dual E2 enzyme in ubiqutination and ISGylation, UbcH8 has been implicated in apoptosis as well as several other biological activities. Consistently, UHRF1 inhibits cell apoptosis in many cancers. In this study, we performed Real-time PCR in E7 expressing RPE1 cells (Figure 1A). The relative mRNA fold changes of UHRF1 and UbcH8 were consistent with RNA-seq results. UHRF1 was up-regulated 1.87-fold (compared to 2.39-fold by RNA-seq) and UbcH8 was down-regulated for 1.97-fold (compared to 2.37-fold by RNA-seq).
Figure 1.
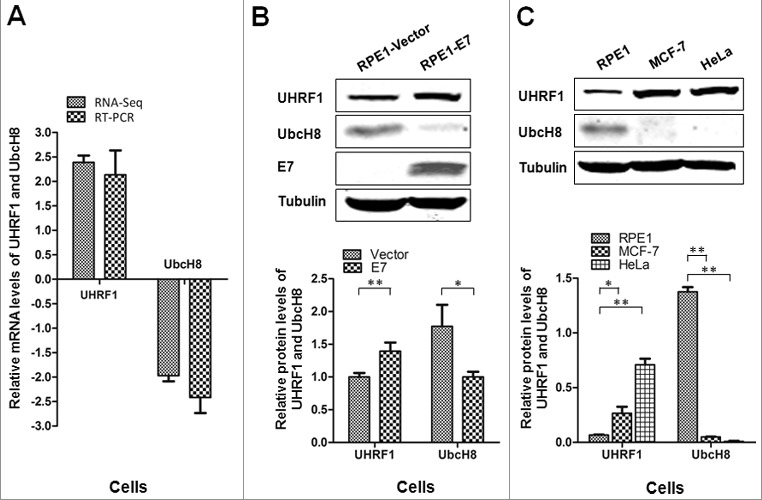
UHRF1 is up-regulated while UbcH8 is down-regulated in HPV-16 E7 expressing cells. (A) UHRF1 and UBE2L6 mRNA levels in NIKS cells determined by RNA-Seq and confirmed in RPE1-Vector and RPE1-E7 cells by Real time-PCR. (B) The steady-state levels of UHRF1 and UbcH8 proteins in RPE1 cells determined by Western blot. Lower panel showed quantification of UHRF1 and UbcH8 protein expressions. (C) The steady-state levels of UHRF1 and UbcH8 proteins in RPE1, MCF-7 and HeLa cells determined by Western blot. Lower panel showed quantification of UHRF1 and UbcH8 protein expressions. The data are mean ± SEM, for all panels: *, p < 0.05; **, p < 0.01. All data are representative of three independent experiments.
We then examined the steady-state levels of UHRF1 and UbcH8 in E7 expressing cells. As shown in Figure 1B, the protein level of UHRF1 was increased while the level of UbcH8 was decreased in E7 expressing cells compared with control cells. These results are consistent with our recent studies using PHKs and NIKS cells [30]. We further examined the steady-state levels of UHRF1 and UbcH8 in HPV-positive HeLa cells. As shown in Figure 1C, compared with RPE1 cells, the level of UHRF1 was higher while that of UbcH8 was lower in HeLa cells. Interestingly, the level of UHRF1 was also high and the level of UbcH8 was low in HPV-negative breast cancer MCF-7 cells (Figure 1C).
These results showed that the expressions of UHRF1 and UbcH8 were inversely correlated in HPV E7 expressing cells, cervical cancer cells, breast cancer cells.
UHRF1 regulates UbcH8 expression
As an important regulator of DNA CpG methylation that modulates gene expression, UHRF1 may down-regulate UbcH8. To test this possibility, we employed the RNAi approach by using two independent siRNAs. After transfection with si-UHRF1–1 and si-UHRF1–2 in RPE1 cells expressing HPV E7, the mRNA level of UHRF1 was down-regulated (Figure 2A, left panel). Consistent with the notion that UHRF1 down-regulates UbcH8, the mRNA level of UbcH8 was up-regulated (Figure 2A, left panel). Similarly, knockdown UHRF1 also led to down-regulation of UHRF1 mRNA and up-regulation of UbcH8 in HeLa cells (Figure 2A, right panel). Consistent with their mRNA expression, the steady state protein levels of UHRF1 and UbcH8 were also down- or up-regulated after UHRF1 siRNA transfection (Figure 2B). Next we transfected a plasmid encoding UHRF1 and examined the impact of UHRF1 ectopic expression on UbcH8 expression. As shown in Figure 2C, transient transfection of UHRF1, though only modestly increased UHRF1 protein level, resulted a significant reduction in the steady-state level of UbcH8. These results indicate that UHRF1 regulated UbcH8 expression.
Figure 2.
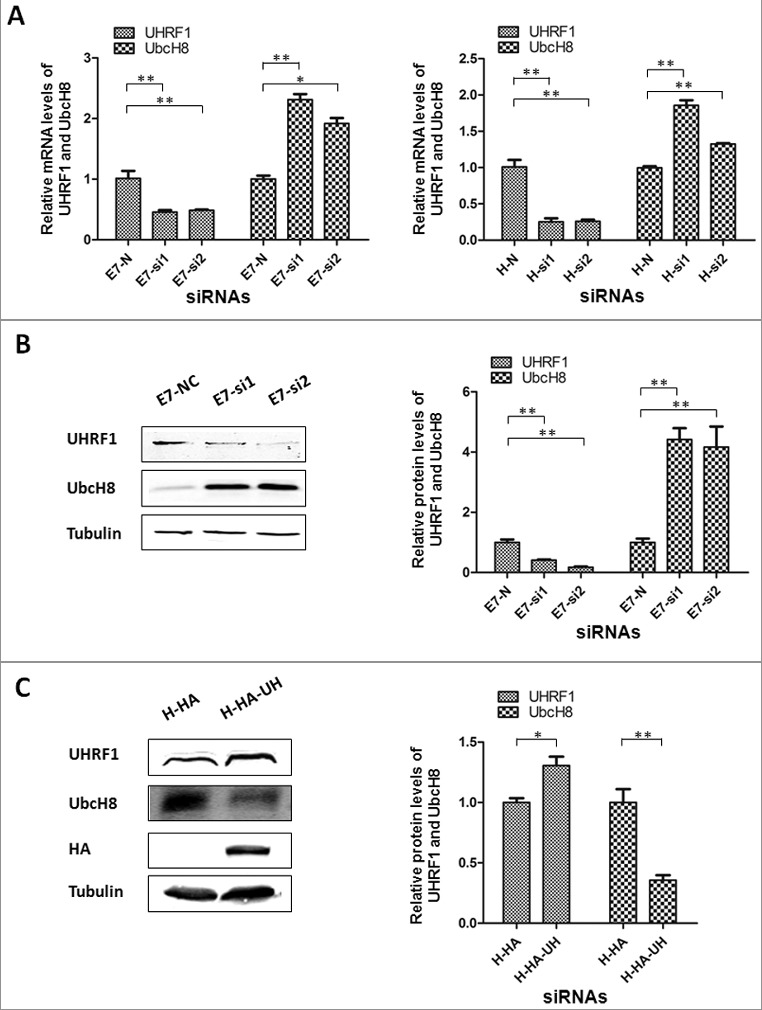
UHRF1 knockdown increases UbcH8 expression while its over-expression decreases UbcH8 expression. (A) The mRNA levels of UHRF1 and UBE2L6 were measured by Real time-PCR in RPE1 cells expressing E7 (E7) and HeLa cells after transfected with siRNAs targeting UHRF1. (B) The protein levels of UHRF1 and UbcH8 were measured by Western blot in E7 cells after transfected with siRNAs targeting UHRF1. Right panel, quantification of UHRF1 and UbcH8 protein expressions. (C) The protein levels of UHRF1 and UbcH8 were measured by Western blot in HeLa cells after transfection with plasmid encoding UHRF1. Right panel, quantification of UHRF1 and UbcH8 protein expressions. N, non-targeting siRNA; si1, siRNA-1 targeting UHRF1; si2, siRNA-2 targeting UHRF1. *, p < 0.05; **, p < 0.01.
UHRF1 binds to the UBE2L6 promoter and hypermethylates it in E7 expressing cells
UBE2L6 is reported as a gene significantly down-regulated by promoter hypermethylation in nasopharyngeal carcinoma (NPC) [23]. As reported, UHRF1 could bind to a CCAAT box on the promoter region of BRCA1 gene and involved in the regulation of its expression by methylation [31]. Meanwhile, we found several CCAAT boxes in the UBE2L6 promoter region. To examine whether UHRF1 binds to the UBE2L6 promoter, Chromatin Immunoprecipitation (ChIP) assay was performed to examine the loading of UHRF1 onto chromatin. The nearly 5000 bp promoter region of UBE2L6 gene from the start of transcription (+1), including two potential binding sites were illustrated in Figure 3A. As shown in Figure 3B, while little UHRF1 bound to the promoter region 1, it bound the promoter region 2 efficiently.
Figure 3.
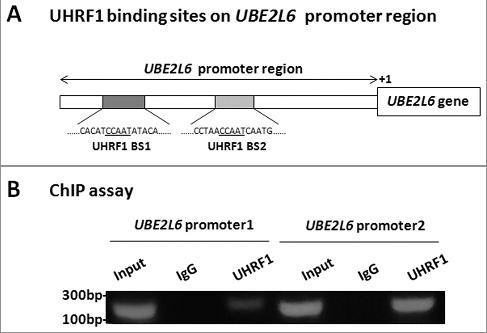
UHRF1 binds to the UBE2L6 promoter. (A) The 5000 bp promoter region of UBE2L6 gene. The two shadow areas indicate promoter regions, P1 (Promoter 1), −4616 to −4392, P2 (Promoter 2), −3456 to −3259. UHRF1 BS, UHRF1 binding site. (B) ChIP assay was performed using UHRF1 antibody in cultured RPE1 cells. The UHRF1-associated UBE2L6 promoter 1 and promoter 2 in the ChIP samples were detected by PCR with UBE2L6 promoter primers 1 and 2. Input, total DNA.
To explore whether UHRF1 methylates UBE2L6 promoter, we examined the status of UBE2L6 promoter methylation. Primers distinguishing unmethylated (U) and methylated (M) alleles were designed by using MethPrimer, a CpG island prediction program. Two CpG sites were identified in the UBE2L6 promoter that was highly prone to methylation (data not shown). The primers we designed for MSP (Methylation-specific PCR) were within these two CpG sites. MSP was performed in RPE1 cells expressing E7 or containing a vector to identity the methylation state of UBE2L6 promoter. As shown in Figure 4A, the UBE2L6 promoter was hypermethylated in E7 expressing cells. For the primer set used, methylation signal was stronger in E7 expressing cells. Significantly, the extent of UBE2L6 promoter methylation was reduced upon UHRF1 knockdown in RPE1 cells (Figure 4B). The potential mechanism by which UHRF1 regulated UBE2L6 gene expression by binding to CCAAT region and recruiting DNMT1 to CpG islands for promoter hypermethylation was showed in Figure 4C. These results indicated that UHRF1 regulated UBE2L6 gene expression by promoter hypermethylation in cervical cancer cells.
Figure 4.
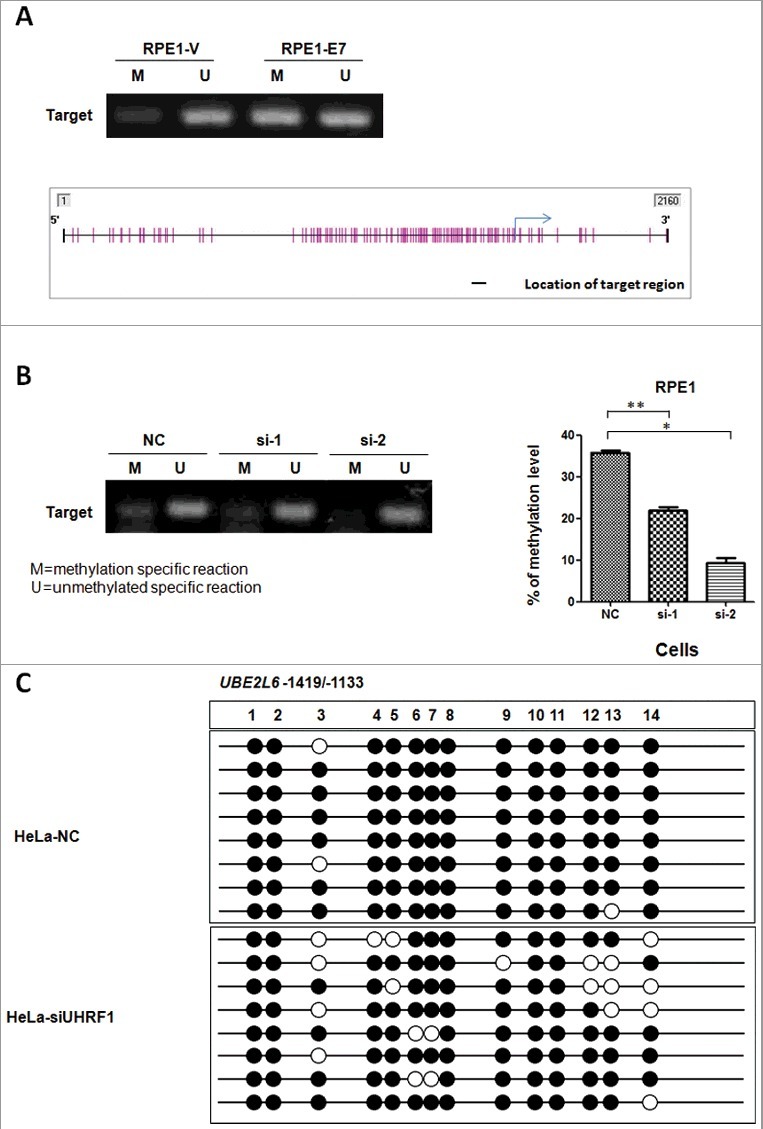
Elevated level of methylation on UBE2L6 promoter in E7 expressing cells. (A) MSP performed using extracts from RPE1-Vector and RPE1-E7 cells. M, Methylation specific primer. U, Unmethylated specific primer. The location of target region amplified by the primer set in predicted CpG islands were indicated with black short lines. The blue arrow indicated transcription start site (TSS) and the purple short vertical lines represented CpG sites. (B) MSP in E7 expressing cells after knockdown UHRF1 by methylation specific and unmethylated primer set. (C) BSP using primers in predicted CpG islands in HeLa cells before and after UHRF1 knock-down.
To directly detect the methylation status of UBE2L6 promoter, we examined the CpG dinucleotide methylation of 14 sites on UBE2L6 promoter region (Fig 4C) in HeLa cells by bisulfite sequencing PCR (BSP). As shown in Fig 4C, among all the 14 sites examined, although no changes were detected for methylation on sites 1–2, 8 and 10–11, methylation of sites 3–7, 9 and 12–14 were decreased after UHRF1 knockdown in HeLa cells. These results provided more evidence that UHRF1 epigenetically regulated UBE2L6 gene by promoter hypermethylation in cervical cancer cells.
UHRF1 inhibits apoptosis by regulating UBE2L6 expression
UHRF1 gene silencing promotes cell apoptosis in cervical squamous cell carcinoma CaSki cells [19] as well as several other types of cancer cells [15,32,33]. To determine the mechanism by which UHRF1 regulates apoptosis and the extent to which UbcH8 was involved, we knocked down UHRF1 by siRNA and examined DNA fragmentation, a hallmark of cellular apoptosis, by quantifying sub-G1 populations of cells using flow cytometry. As shown in Figure 5A (left panel), apoptotic cells increased after transfection with siRNAs targeting UHRF1 in HPV E7 expressing cells.
Figure 5.
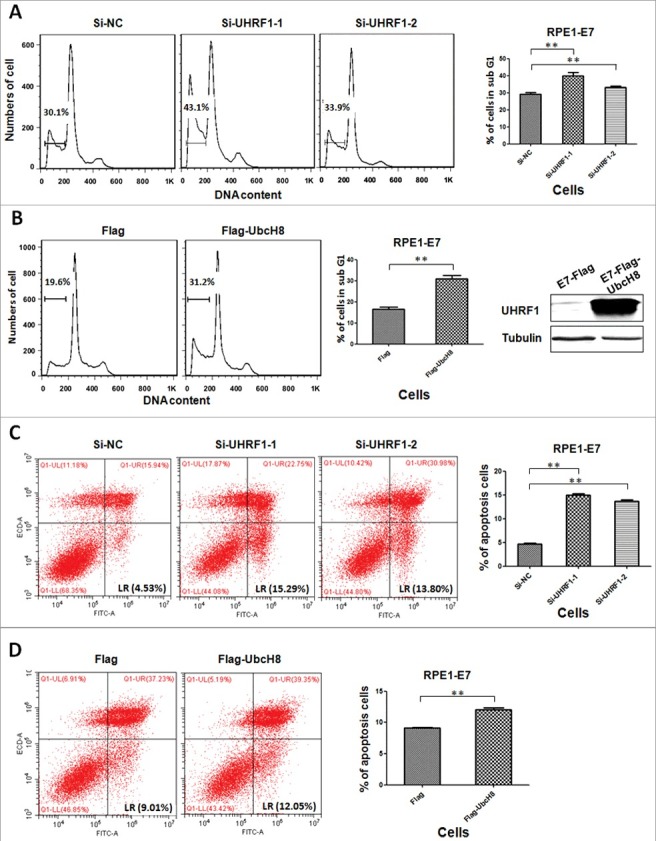
UHRF1 and UbcH8 regulate apoptosis. (A) RPE1-E7 cells were transfected with siRNAs targeting UHRF1 and treated with Etoposide. Cells were analyzed by flow cytometry after fixation and PI stain. Cells with sub-G1 DNA content were gated and quantified. (B) RPE1-E7 cells were transfected with plasmid encoding UbcH8, treated and analyzed as above. (C) RPE1-E7 cells were transfected with siRNAs targeting UHRF1 and treated with Etoposide. Cells were double stained with annexin V-FITC/PI and analyzed by flow cytometry. Cells that stained positive for annexin V-FITC and negative for PI were undergoing early stage apoptosis; cells that stained positive for both annexin V-FITC and PI were in the late stage of apoptosis or undergoing necrosis. Percentage of apoptotic cells (annexin V-FITC positive) was quantified. (D) RPE1-E7 cells were transfected with plasmid encoding UbcH8, treated and analyzed as above. Data of 3 independent experiments were summarized. *, p < 0.05 **, p < 0.01. NC, negative control.
If down-regulating UbcH8 is a mechanism by which UHRF1 inhibits apoptosis, UbcH8 overexpression should result in increased cellular apoptosis. To demonstrate the effect of overexpressing UbcH8 on apoptosis, we transfected RPE1 E7 expressing cells with plasmid encoding UbcH8 (Figure 5B, left panel), verified UbcH8 expression by Western blot and examined cellular apoptosis. As shown in Figure 5B, right panel, the population of apoptotic cells increased after transfection of UbcH8 plasmid. These results support our hypothesis that UbcH8 is a target of UHRF1 that mediates apoptosis.
To confirm these results, we used another method to detect cellular apoptosis, annexin V-FITC/PI apoptosis detection, which can distinguish early apoptotic cells from necrotic cells. Accordingly, RPE1 cells expressing HPV E7 were transfected with siRNAs targeting UHRF1, cells were stained with annixin V-FITC/PI and subjected to flow cytometry analysis. As shown in Figure 5C, consistent with DNA fragmentation analysis, apoptosis measured by annexin V-FITC/PI was increased after UHRF1 knockdown. Likewise and consistent with UbcH8 being a target for UHRF1, transfection of UbcH8 plasmid in E7 expressing cells increased cellular apoptosis (Figure 5D). Similar results were obtained in HeLa cells (Supplemental Figure).
Discussion
In this study, we found that UHRF1 was up-regulated while UbcH8 was down-regulated in HPV-16 E7 expressing cells and HPV-positive cervical cancer cells. We demonstrated that UHRF1 down-regulated the expression of UBE2L6 gene that encodes UbcH8. This study established UBE2L6 as a novel target of UHRF1. We also provided evidence that UHRF1 regulated UBE2L6 gene by promoter hypermethylation in cervical cancer cells. Significantly, UbcH8 contributes to UHRF1-mediated apoptosis. These results help understand the mechanism by which HPV induces cervical carcinogenesis and provide a potential candidate for therapy in cervical cancer.
Up-regulation of UHRF1 has been observed in cervical cancer as well as other cancers [16,17,18,19]. Consistently, UHRF1 gene silencing inhibits cell proliferation and promotes apoptosis in human cervical squamous cell carcinoma CaSki cells [19]. And UHRF1 promotes G1/S transition in HeLa cells [34]. It was suggested that the most accurate way to discriminate cervical high-grade lesions was the association of a suspect DNA profile and the presence of MIB-1- or UHRF1-positive cells in the upper two thirds of the epithelium [35]. On the other hand, expression of UHRF1 reduces the radiosensitivity of HeLa cells to gamma-irradiation [36]. Down-regulalion of UHRF1 and DNMT1 were involved in induction of apoptosis in human cervical cancer cells [37]. Mechanistically, in HeLa cells, the PHD domain of UHRF1 recognizes methylated histone H3 lysine 9 (H3K9) [38]. UHRF1 recruits G9a, which methylates H3K9, the latter in turn binds with higher affinity with the PHD domain of UHRF1 [39]. UHRF1 can also ubiquitinate histone H3 via its E3 ubiquitin ligase [38,40].
UHRF1 forms a complex with histone deacetylase 1 (HDAC1) and DNA methyltransferase 1 (DNMT1) via its SRA domain and represses the expression of several tumour suppressor genes including p16, BRCA1 and RB1 [16]. UHRF1 promotes cell growth and metastasis through repression of p16 in colorectal cancer [41]. UHRF1 recruited and cooperated with G9a to inhibit the p21 promoter activity, which correlated with elevated p21 protein level in UHRF1 siRNA-transfected HeLa cells [39]. UHRF1 is involved in the regulation of topoisomerase II alpha expression [42]. In our present study, UHRF1 regulated UBE2L6 gene by promoter hyper-methylation in cervical cancer cells. UbcH8 is a novel target identified for UHRF1 which promotes cellular apoptosis.
Apoptosis can be manipulated for cervical cancer therapy. The mechanism by which UbcH8 induces apoptosis is not known. Interestingly, UHRF1 depletion up-regulated the expression of PML and triggered extrinsic and intrinsic apoptotic pathways by promoting the expression of FasL/FADD, Bax, cytosolic cytochrome c, cleaved caspase-8, −9 and −3 and cleaved PRAP and by suppressing bcl-2 expression [43]. As the principal ISG15 E2 carrier, we speculate that UbcH8 facilitate ISG15 conjugation to cellular proteins that play important roles in apoptosis. Proteomic studies have identified more than 300 cellular proteins that are targeted for ISGylation, some of them, such as CBX4, PKM2, STAT1 and DYRK1A, are involved in apoptosis. It remains to be determined the specific proteins contribute to UbcH8-mediated apoptosis.
The innate immune system is the first line of defense against infectious agents that triggers the induction of antiviral type I interferon (IFN). To establish persistent infection and amplify genome, HPVs need to evade innate immune surveillance. ISG15 is one of the most strongly induced genes upon viral infection and type I IFN stimulation. ISG15 has drawn much attention as a potential regulator of the immune response and has been shown to mediate protection in a number of different viral infection models. UbcH8 serves as the predominant E2 enzyme in ISGylation. Significantly, ISGylation of HPV L1 capsid protein has a dominant-inhibitory effect on the infectivity of HPV-16. Future studies will examine the specific contribution of UbcH8 in HPV L1 ISGylation and cervical carcinogenesis.
In summary, our present study verified our recent finding of RNA-seq that indicates UHRF1 is up-regulated while UbcH8 is down-regulated in HPV16 E7 expressing cells. We found that UHRF1 regulated UBE2L6 gene by promoter hypermethylation in cervical cancer cells. Furthermore, their roles in cell apoptosis were also demonstrated in this work. These results establish UBE2L6 as a novel target of UHRF1 that mediates cell apoptosis. Therefore, our studies shed light on the mechanisms by which UHRF1 inhibit apoptosis in cervical cancer cells and provided a potential candidate for therapy in cervical cancer.
Materials and methods
Cell culture
The human telomerase reverse transcriptase-expressing human retinal pigment epithelium cells (RPE1) were maintained in a 1:1 dilution of DMEM-Ham's F-12 medium plus 10% FBS. RPE1 cells stably expressing HPV-16 E7 or containing the pBabe vector were established by retrovirus-mediated infection using the pBabe-puro-based retroviral construct. Cells were selected with (10.5 μg/mL) puromycin for 3 to 6 days, maintained in puromycin (6.5 μg/mL) and used within 15 passages. HeLa cells were maintained in DMEM medium plus 10% FBS.
Real-time PCR
Total RNA from RPE1, HeLa and the corresponding E7-expressing cells was isolated using an RNeasy kit (Qiagen) according to the manufacturer's instruction. cDNA was synthesized with a Superscript VILO cDNA synthesis kit (Invitrogen). iTaq Universal SYBR green Supermix (Bio-Rad) was used in a Bio-Rad CFX96 Touch Real-Time PCR detection system for quantitative real-time PCR (qRT-PCR). Data were analyzed using the threshold cycle (2−ΔΔCT) method. The primer sequences are listed in Supplemental Table S1.
Immunoblotting
Total cellular protein was prepared in lysis buffer (10 mM Tris [pH 7.4], 1% SDS, 1.0 mM sodium orthovanadate). The protein concentration was measured by the use of bicinchoninic acid (BCA) protein assay reagent (Pierce) and confirmed by Coomassie blue staining of membranes after blotting. Equal amounts of protein from each cell lysate were separated in an SDS polyacrylamide gel (PAGE) and transferred onto a nitrocellulose filter membrane (NC) membrane. Membranes were blotted with antibodies against UHRF1 (ab57083, abcam), UbcH8 (ab109086, abcam) and tubulin (Sigma; T-4026). Protein bands were detected using an Odyssey infrared imaging system (Li-COR, Lincoln, NE) and quantified using Image J (NIH).
Flow cytometry
RPE1 cells were treated with Etoposide (50 μM). At the end of the treatment period (24 h), the cells were detached, washed with PBS, resuspended in 1 mL of cold 70% (v/v) ethanol, and then stored at 4 °C for 24 h. After washed with PBS, the cells were stained with propidium iodide (PI) and 10 μg/mL ribonuclease (RNase) in PBS at 4 °C for 30 min away from light. The cells were washed and subjected to flow cytometric analysis of DNA content (FACScalibur, Becton Dickinson). Nuclei displaying hypodiploid (sub-G1) DNA contents were identified as apoptotic.
For annexin V-FITC apoptosis detection, RPE1 and HeLa cells were treated with 100 mM Etoposide for 24 h. At the end of the treatment period, apoptosis was assessed by using FITC annexin V Apoptosis Detection Kit (Biolegend) following the manufacturer's instructions. After staining with annexin V-FITC and propidium iodide (PI), the samples were analyzed using flow cytometer. When conjugated to a fluorochrome, annexin V targets and detects dying cells expressing PS on the reversed membrane surface. Combined with PI, annexin V may distinguish necrotic cells from apoptotic cells. Viable cells have low annexin V-FITC and low PI staining, apoptotic cells have high annexin V-FITC and low PI staining (lower-right quadrant), and necrotic cells have high annexin V-FITC staining and PI (upper-right quadrant). Bars on the graph (bottom) represent the percentage of apoptotic and necrotic cells.
siRNAs and transfection
Cells were transfected with a final concentration of 20 nM siRNA per target gene using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's instructions. For gene knockdown analysis, cells were harvested 48 h post-transfection and specific protein levels were analyzed by immunoblot. The siRNA duplexes were as follows: si-UHRF1–1 sense strand: 5’- AUGGUACAUUCCUUGGUGC-3’; si-UHRF1–2 sense strand: 5’- UUGUAGAUGCCAUCGUAGC-3’; Negative control siRNA sense strand: 5’-UUCUCCGAACGUGUCACGU-3’. Plasmid pFlagCMV2 -UbcH8 (Addgene) and the control were transfected into cells also using Lipofectamine 2000 (Invitrogen).
ChIP assay
The chromatin immunoprecipitation (ChIP) assay was performed using a ChIP assay kit from Millipore following the supplied protocol. Immunoprecipitations were performed using anti-UHRF1 or control IgG antibodies. PCR was performed with the primers designed from the sequences of the human UBE2L6 gene as follows: Predicted Promoter 1-sense, GAAGAGAGCCCTCATCAGAA; antisense, CCTTGTTGTGGGACTTTGGA. Predicted Promoter 2-sense, GCCTTTCTCCTA-CCACCTTTT; antisense, CCCTGTTTTACTTCTCCCTGAT.
Methylation specific PCR (MSP)
Genomic DNA was isolated using Quick-gDNA MicroPrep (Zymo, D3024) according to the manufacturer's instructions. For bisulfite treatment, 400–500 ng of DNA was used for each column using the EZ DNA Methylation-Gold Kit (Zymo, D5005). The methylation status of the UBE2L6 promoter region was determined by MSP. Primers distinguishing unmethylated (U) and methylated (M) alleles were designed by using MethPrimer (www.urogene.org/cgi-bin/methprimer/methprimer.cgi), a CpG island prediction program. The primer sequences designed for MSP were listed in Supplemental Table S2.
For the methylation specifc PCR reactions, in vitro-methylated genomic DNA treated with sodium bisulphite served as a positive methylation control; a water blank control was also included. For cases with borderline results, PCR analyses were repeated.
Bisulfite Sequencing PCR (BSP)
Genomic DNAs from HeLa cells were extracted and modified (with bisulfite) using the EZ DNA methylation-Gold kit (Zymo Research, D5005) according to manufacturers' protocols. Bisulfite-treated samples were then amplified by PCR using primers B-UBE2L6-sense TGTAATTTTAATATTTTGGGAGGT; B-UBE2L6-antisense TTTAAACACCAATTTATAATCTCAATAC. PCR products were cloned into pMD19 T-Vector (TaKaRa, 6013) and plasmids isolated from randomly picked colonies were sequenced.
Supplementary Material
Funding Statement
This work was supported by National Natural Science Foundation of China(NSFC) [grant number 81471944].
Competing financial interests
The authors declare no competing financial interests.
Acknowledgements
We thank Weifang Zhang, Lijun Qiao and Mei Qi for critical reading of the manuscript, members of our laboratories for technical assistance and helpful discussion. This work, including the efforts of Jason Chen, was funded by National Natural Science Foundation of China (Grant No. 81471944).
References
- [1].Wright J, Herzog T. Human papillomavirus: emerging trends in detection and management. Current Women's Health Reports. 2002;2:259–265. PMID:12150752 [PubMed] [Google Scholar]
- [2].Lee J, Russo A, Pavletich N. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature. 1998;391:859–865. doi: 10.1038/36038. PMID:9495340 [DOI] [PubMed] [Google Scholar]
- [3].Thurlings I, Martinez-Lopez L, Westendorp B, et al.. Synergistic functions of E2F7 and E2F8 are critical to suppress stress-induced skin cancer. Oncogene. 2017;36(6):829–839. PMID:27452520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Unoki M, Nishidate T, Nakamura Y, ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23:7601–7610. doi: 10.1038/sj.onc.1208053. PMID:15361834 [DOI] [PubMed] [Google Scholar]
- [5].Bostick M, Kim J, Esteve P, et al.. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. PMID:17673620 [DOI] [PubMed] [Google Scholar]
- [6].Cheng J, Yang Y, Fang J, et al.. Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein. J Biol Chem. 2013;288:1329–1339. doi: 10.1074/jbc.M112.415398. PMID:23161542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Rajakumara E, Wang Z, Ma H, et al.. PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol Cell. 2011;43:275–284. doi: 10.1016/j.molcel.2011.07.006. PMID:21777816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hashimoto H, Vertino P, Cheng X, Molecular coupling of DNA methylation and histone methylation. Epigenomics. 2010;2:657–669. doi: 10.2217/epi.10.44. PMID:21339843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Guan D, Factor D, Liu Y, et al.. The epigenetic regulator UHRF1 promotes ubiquitination-mediated degradation of the tumor-suppressor protein promyelocytic leukemia protein. Oncogene. 2013;32:3819–3828. doi: 10.1038/onc.2012.406. PMID:22945642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhang C, Li H, Wang Y, et al.. Epigenetic inactivation of the tumor suppressor gene RIZ1 in hepatocellular carcinoma involves both DNA methylation and histone modifications. J Hepatol. 2010;53:889–895. doi: 10.1016/j.jhep.2010.05.012. PMID:20675009 [DOI] [PubMed] [Google Scholar]
- [11].Wilson K, Cowart C, Rosen B, et al.. Characteristics Associated with HPV Diagnosis and Perceived Risk for Cervical Cancer Among Unmarried, Sexually Active College Women. J Cancer Educ Official J Am Association Cancer Educ. 2016. doi: 10.1007/s13187-016-1131-1. [DOI] [PubMed] [Google Scholar]
- [12].Sheng Y, Wang H, Liu D, et al.. Methylation of tumor suppressor gene CDH13 and SHP1 promoters and their epigenetic regulation by the UHRF1/PRMT5 complex in endometrial carcinoma. Gynecol Oncol. 2016;140:145–151. doi: 10.1016/j.ygyno.2015.11.017. PMID:26597461 [DOI] [PubMed] [Google Scholar]
- [13].Li L, Shu X, Wang Z, et al.. Epigenetic disruption of cell signaling in nasopharyngeal carcinoma. Chinese J Cancer. 2011;30:231–239. doi: 10.5732/cjc.011.10080. PMID:21439244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gronbaek K, Hother C, Jones P. Epigenetic changes in cancer. APMIS: Acta Pathol, Microbiol, et immunologica Scandinavica. 2007;115:1039–1059. doi: 10.1111/j.1600-0463.2007.apm_636.xml.x. PMID:18042143 [DOI] [PubMed] [Google Scholar]
- [15].Wan X, Yang S, Huang W, et al.. UHRF1 overexpression is involved in cell proliferation and biochemical recurrence in prostate cancer after radical prostatectomy. J Exp Clin Cancer Res CR. 2016;35:34. doi: 10.1186/s13046-016-0308-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Alhosin M, Sharif T, Mousli M, et al.. Down-regulation of UHRF1, associated with re-expression of tumor suppressor genes, is a common feature of natural compounds exhibiting anti-cancer properties. J Exp Clin Cancer Res CR. 2011;30:41. doi: 10.1186/1756-9966-30-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Unoki M, Kelly J, Neal D, et al.. UHRF1 is a novel molecular marker for diagnosis and the prognosis of bladder cancer. Br J Cancer. 2009;101:98–105. doi: 10.1038/sj.bjc.6605123. PMID:19491893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Babbio F, Pistore C, Curti L, et al.. The SRA protein UHRF1 promotes epigenetic crosstalks and is involved in prostate cancer progression. Oncogene. 2012;31:4878–4887. doi: 10.1038/onc.2011.641. PMID:22330138 [DOI] [PubMed] [Google Scholar]
- [19].Ge T, Yang M, Chen Z, et al.. UHRF1 gene silencing inhibits cell proliferation and promotes cell apoptosis in human cervical squamous cell carcinoma CaSki cells. J Ovarian Res. 2016;9:42. doi: 10.1186/s13048-016-0253-8. PMID:27431502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Unoki M, Daigo Y, Koinuma J, et al.. UHRF1 is a novel diagnostic marker of lung cancer. Br J Cancer. 2010;103:217–222. doi: 10.1038/sj.bjc.6605717. PMID:20517312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kim K, Giannakopoulos N, Virgin H, et al.. Interferon-inducible ubiquitin E2, Ubc8, is a conjugating enzyme for protein ISGylation. Mol Cell Biol. 2004;24:9592–9600. doi: 10.1128/MCB.24.21.9592-9600.2004. PMID:15485925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhao C, Beaudenon S, Kelley M, et al.. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-alpha/beta-induced ubiquitin-like protein. Proc Nat Acad Sci United States Am. 2004;101:7578–7582. doi: 10.1073/pnas.0402528101. PMID:15131269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhou X, Wei J, Chen F, et al.. Epigenetic downregulation of the ISG15-conjugating enzyme UbcH8 impairs lipolysis and correlates with poor prognosis in nasopharyngeal carcinoma. Oncotarget. 2015;6:41077–41091. doi: 10.18632/oncotarget.6218. PMID:26506425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yuan W, Krug R. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 2001;20:362–371. doi: 10.1093/emboj/20.3.362. PMID:11157743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Durfee L, Lyon N, Seo K, et al.. The ISG15 conjugation system broadly targets newly synthesized proteins: implications for the antiviral function of ISG15. Mol Cell. 2010;38:722–732. doi: 10.1016/j.molcel.2010.05.002. PMID:20542004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang X, Li L, Wu Y, et al.. CBX4 Suppresses Metastasis via Recruitment of HDAC3 to the Runx2 Promoter in Colorectal Carcinoma. Cancer Res. 2016;76:7277–7289. doi: 10.1158/0008-5472.CAN-16-2100. PMID:27864346 [DOI] [PubMed] [Google Scholar]
- [27].Zhu H, Wu J, Zhang W, et al.. PKM2 enhances chemosensitivity to cisplatin through interaction with the mTOR pathway in cervical cancer. Scientific Reports. 2016;6:30788. doi: 10.1038/srep30788. PMID:27492148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Koromilas A, Sexl V. The tumor suppressor function of STAT1 in breast cancer. Jak-Stat. 2013;2:e23353. doi: 10.4161/jkst.23353. PMID:24058806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Abbassi R, Johns T, Kassiou M, et al.. DYRK1A in neurodegeneration and cancer: Molecular basis and clinical implications. Pharmacol Therapeutics. 2015;151:87–98. doi: 10.1016/j.pharmthera.2015.03.004. [DOI] [PubMed] [Google Scholar]
- [30].Zhou Y, Zhang Q, Gao G, et al.. Role of WDHD1 in Human Papillomavirus-Mediated Oncogenesis Identified by Transcriptional Profiling of E7-Expressing Cells. J Virol. 2016;90:6071–6084. doi: 10.1128/JVI.00513-16. PMID:27099318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jin W, Chen L, Chen Y, et al.. UHRF1 is associated with epigenetic silencing of BRCA1 in sporadic breast cancer. Breast Cancer Res Treat. 2010;123:359–373. doi: 10.1007/s10549-009-0652-2. PMID:19943104 [DOI] [PubMed] [Google Scholar]
- [32].Ma J, Peng J, Mo R, et al.. Ubiquitin E3 ligase UHRF1 regulates p53 ubiquitination and p53-dependent cell apoptosis in clear cell Renal Cell Carcinoma. Biochem Biophys Res Commun. 2015;464:147–153. doi: 10.1016/j.bbrc.2015.06.104. PMID:26102039 [DOI] [PubMed] [Google Scholar]
- [33].Yan F, Wang X, Shao L, et al.. Analysis of UHRF1 expression in human ovarian cancer tissues and its regulation in cancer cell growth. Tumour Biol. 2015;36:8887–8893. doi: 10.1007/s13277-015-3638-1. PMID:26070868 [DOI] [PubMed] [Google Scholar]
- [34].Arima Y, Hirota T, Bronner C, et al.. Down-regulation of nuclear protein ICBP90 by p53/p21Cip1/WAF1-dependent DNA-damage checkpoint signals contributes to cell cycle arrest at G1/S transition. Genes Cells: Devoted to Mol Cellular Mech. 2004;9:131–142. doi: 10.1111/j.1356-9597.2004.00710.x. [DOI] [PubMed] [Google Scholar]
- [35].Lorenzato M, Caudroy S, Bronner C, et al.. Cell cycle and/or proliferation markers: what is the best method to discriminate cervical high-grade lesions? Hum Pathol. 2005;36:1101–1107. doi: 10.1016/j.humpath.2005.07.016. PMID:16226110 [DOI] [PubMed] [Google Scholar]
- [36].Li X, Meng Q, Fan S. Adenovirus-mediated expression of UHRF1 reduces the radiosensitivity of cervical cancer HeLa cells to gamma-irradiation. Acta Pharmacol Sin. 2009;30:458–466. doi: 10.1038/aps.2009.18. PMID:19270723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Krifa M, Alhosin M, Muller C, et al.. Limoniastrum guyonianum aqueous gall extract induces apoptosis in human cervical cancer cells involving p16 INK4A re-expression related to UHRF1 and DNMT1 down-regulation. J Exp Clin Cancer Res CR. 2013;32:30. doi: 10.1186/1756-9966-32-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Karagianni P, Amazit L, Qin J, et al.. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol Cell Biol. 2008;28:705–717. doi: 10.1128/MCB.01598-07. PMID:17967883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kim J, Esteve P, Jacobsen S, et al.. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res. 2009;37:493–505. doi: 10.1093/nar/gkn961. PMID:19056828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Citterio E, Papait R, Nicassio F, et al.. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol Cell Biol. 2004;24:2526–2535. doi: 10.1128/MCB.24.6.2526-2535.2004. PMID:14993289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wang F, Yang Y, Shi C, et al.. UHRF1 promotes cell growth and metastasis through repression of p16(ink(4)a) in colorectal cancer. Ann Surg Oncol. 2012;19:2753–2762. doi: 10.1245/s10434-011-2194-1. PMID:22219067 [DOI] [PubMed] [Google Scholar]
- [42].Hopfner R, Mousli M, Jeltsch J, et al.. ICBP90, a novel human CCAAT binding protein, involved in the regulation of topoisomerase IIalpha expression. Cancer Res. 2000;60:121–128. PMID:10646863 [PubMed] [Google Scholar]
- [43].Qin Y, Wang J, Gong W, et al.. UHRF1 depletion suppresses growth of gallbladder cancer cells through induction of apoptosis and cell cycle arrest. Oncol Rep. 2014;31:2635–2643. doi: 10.3892/or.2014.3145. PMID:24756644 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


