ABSTRACT
Fetal stem cells are a unique type of adult stem cells that have been suggested to be broadly multipotent with some features of pluripotency. Their clinical potential has been documented but their upgrade to full pluripotency could open up a wide range of cell-based therapies particularly suited for pediatric tissue engineering, longitudinal studies or disease modeling. Here we describe episomal reprogramming of mesenchymal stem cells from the human amnion to pluripotency (AM-iPSC) in chemically defined conditions. The AM-iPSC expressed markers of embryonic stem cells, readily formed teratomas with tissues of all three germ layers present and had a normal karyotype after around 40 passages in culture. We employed novel computational methods to determine the degree of pluripotency from microarray and RNA sequencing data in these novel lines alongside an iPSC and ESC control and found that all lines were deemed pluripotent, however, with variable scores. Differential expression analysis then identified several groups of genes that potentially regulate this variability in lines within the boundaries of pluripotency, including metallothionein proteins. By further studying this variability, characteristics relevant to cell-based therapies, like differentiation propensity, could be uncovered and predicted in the pluripotent stage.
KEYWORDS: amnion, iPSC, episomal reprogramming, PluriTest, CellNet, RNA sequencing, microarray
Introduction
Human fetal stem cells are multipotent stem cells derived from solid extraembryonic/placental tissues and fluids – amnion, amniotic fluid, chorionic villi, umbilical cord, or umbilical cord blood. Populations of cells with epithelial, mesenchymal or hematopoietic phenotype can be isolated from these tissues and have shown potentials to be used in numerous clinical interventions [1–3], including tissue engineering [4–9], owing to their unique properties such as differentiation, tissue formation or immunomodulation. Amniotic membrane mesenchymal stem cells (AMSC) exhibit no tumorigenicity, on the contrary, they may possess anti-inflammatory and even anti-cancer properties by virtue of cytotoxic cytokine secretion [2,10]. Their baseline expression of markers typical for embryonic stem cells (ESC) was observed to be higher than in bone-marrow-derived mesenchymal stem cells (MSC) [11]. However, fetal stem cells still have a limited differentiation and proliferation capacity. Additionally, epigenetic aberrations have been detected in mesenchymal stromal cells at higher passages [12]. Therefore, cultured AMSC at a low passage may represent an ideal cell source for reprogramming into induced pluripotent stem cells (iPSC). Their mesenchymal properties may be favorable for reprogramming since acquisition of hyper-mesenchymal properties and delayed mesenchymal-to-epithelial transition (MET) increased efficiency of reprogramming [13]. Additionally, in mouse, CD73 was identified as a marker of an important intermediate in the reprogramming roadmap [14,15]. Furthermore, iPSC tend to retain a considerable portion of the epigenetic signature of their source cells which can skew their differentiation potential [16]. From the perspective of their commitment on the developmental scale, amniotic fetal stem cells occupy an intermediate stage between pluripotent and multipotent adult stem cells [10,17]. Using highly unspecialized cells might alleviate the epigenetic bias and therefore prove beneficial in addressing this phenomenon in iPSC.
Human pluripotent stem cells (PSC) could be a source of unlimited numbers of cells with the highest differentiation potential and therefore are very attractive from the perspective of the development of cell replacement therapies and tissue engineering applications. Efforts are underway to develop reproducible protocols for differentiating human pluripotent stem cells into a wide range of somatic cells. As an example, cardiac differentiation has been widely explored and numerous protocols published which lay ground for the concept of engineering cardiac tissue grafts (reviewed in Jackman et al., 2015 [18]; Sirabella et al., 2015 [19] and Budniatzky et al., 2014 [20]). A tissue engineering approach has recently been used to regenerate sciatic nerves in a rat model, with iPSC as a source of neural crest stem cells seeded into scaffolds [21]. PSC-derived neural crest stem cells were combined with intestinal organoids to create human intestinal tissue with a functional enteric nervous system [22]. These examples of PSC-based tissue engineering together with an emerging technology of organoid engineering demonstrate the potential of these technologies to be used in transplantations, research into physiology or drug and toxicological screening. Furthermore, clinical trials in cell replacement therapies for diseases like age-related macular degeneration, Parkinson's disease, diabetes, myocardial infarction and spinal cord injuries are currently ongoing or entering Phase I [23].
Generation of iPSC from amniotic membrane mesenchymal cells has been described previously, however, viral delivery of the reprogramming factors as well as undefined culture and medium components (such as fetal bovine serum (FBS), KnockOut™ Serum Replacement (KSR), mouse embryonic fibroblast feeder (MEF) layer) were used [24,25]. In our study, we attempted to derive and expand iPSC lines from AMSC of 4 different patients by means of non-integrating episomal reprogramming in xeno-free culture conditions. Previously, we described a similar approach of iPSC derivation from AFSC [26]. However, AFSC need to be obtained in the process of amniocentesis while AMSC can be derived from a small patch of the placental amnion that represents medical waste after examination by a pathologist. On the other hand, the advantage of AFSC as a source for reprogramming into iPSC is that the process can be initiated before birth (second trimester) and therefore any potential cell replacement therapies would be available earlier to the patient. Therefore, exploring both cell types as a cell source for reprogramming into iPSC is of value. AMSC can readily be used for producing iPSC lines for routine banking and for large population-level studies.
Results
Isolation and culture of amniotic membrane mesenchymal stem cells
Amnion mesenchymal stem cells were successfully extracted from around 6 cm2 patch of the placental amnion immediately after birth to maximize cell viability. The membranes were minced, mixed with digestion enzymes and the mixtures were blended in a controlled manner using the gentleMACS tissue dissociator. A single round of blending and a 30 min incubation at 37°C was optimal to maintain viability (data not shown) and resulted in a suspension of clumps and single cells. The clumps and single cells plated into regular tissue culture-treated vessels adhered in abundance in as early as 24h. They were allowed to proliferate for 2–3 passages before reprogramming into pluripotency. The morphology of the cells was a typical mesenchymal, elongated and phase-bright (Figure 1A). The medium used was EBM-2 basal medium supplemented with FBS, bFGF, EGF and IGF, a composition identical to the one previously used for amniotic fluid stem cells. The cells expressed markers of mesenchymal stem cells (MSC) CD44, CD73, CD166, CD105, CD90 and N-cadherin when measured by flow cytometry. The levels of N-cadherin were relatively low compared to other markers and in AMSC[3], it was mostly negative. The epithelial marker EpCAM was mostly negative or expressed only in low levels (Figure 1B, Table S1).
Figure 1.
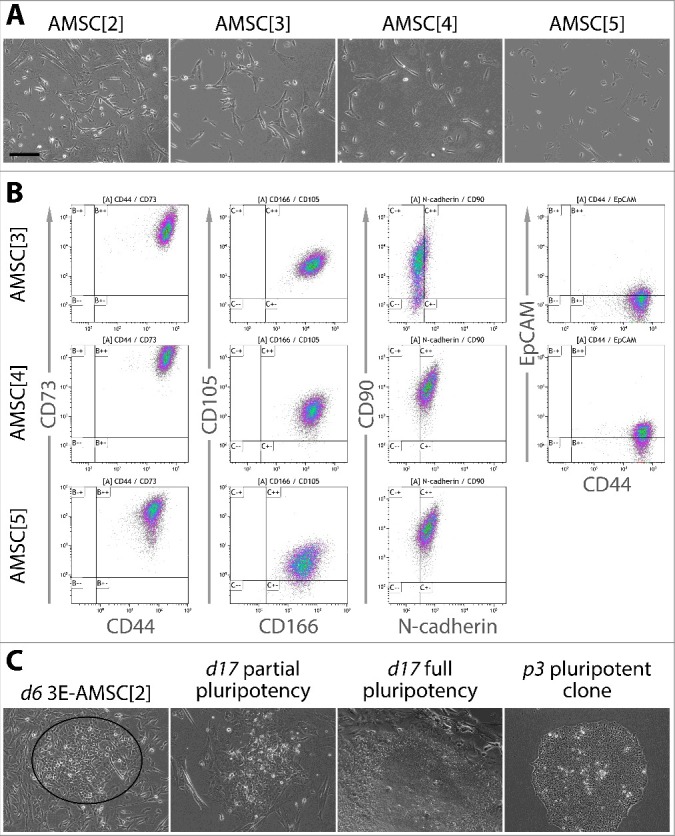
Morphology, characterization and reprogramming progression of the source AMSC. A. Cultured AMSC display a morphology of mesenchymal stem cells – elongated and spindle-shaped. B. Flow cytometric analysis of AMSC revealed expression of CD44, CD73, CD166, CD105, N-cadherin and CD90. Activation of the epithelial marker EpCAM was low. This expression profile is consistent with a mesenchymal stem cell phenotype. C. Morphological progression of reprogramming in AMSC transfected with 3 episomal reprogramming plasmids (3E). Days following transfection are denoted by “d” and the passage number following transfection by “p”. On day 6 (left), cells assuming cobblestone-like epithelial shape and undergoing MET were clearly discernible (ellipse). On day 17 (two middle pictures), partially and fully pluripotent cell colonies were both present, with partially pluripotent colonies being more abundant. Fully pluripotent colonies were expanded and clonal cultures established (right). Scale bar = 200 µm.
Reprogramming to pluripotency
Introduction of reprogramming plasmids into AMSC led to morphological changes representing the earliest stage of reprogramming as early as day 3 following transfection. The changes included loss of the spindle shape typical for MSC and acquisition of a “cobble stone” morphology, indicating MET. Later, a small fraction of the MET partially pluripotent colonies gave rise to colonies with characteristics of full pluripotency. These first appeared around day 14 and on day 17, they were ready for mechanical picking and clonal expansion (Figure 1C). Expanded clones were allowed to mature for around 15 passages, with a 3-day passaging interval, before carrying out detailed characterization.
Characterization of AM-iPSC by established methods
Profiling of the expression of ESC markers Oct3/4A, Nanog, Sox2, TRA-1-60, TRA-1-81, SSEA-1 and SSEA-4 by flow cytometry revealed a consistent expression pattern across AM-iPSC lines identical to the control line WA25. All markers except SSEA-1 were found to be expressed in nearly 100% of the cells (Figure 2, Table S2). The results of the flow cytometry analysis were corroborated by confocal imaging. Transcription factors Oct3/4A, Nanog and Sox2 localized to the nuclei of the cells and glycoproteins TRA-1-60, TRA-1-81 and SSEA-4 localized to the membrane (Figure 3).
Figure 2.
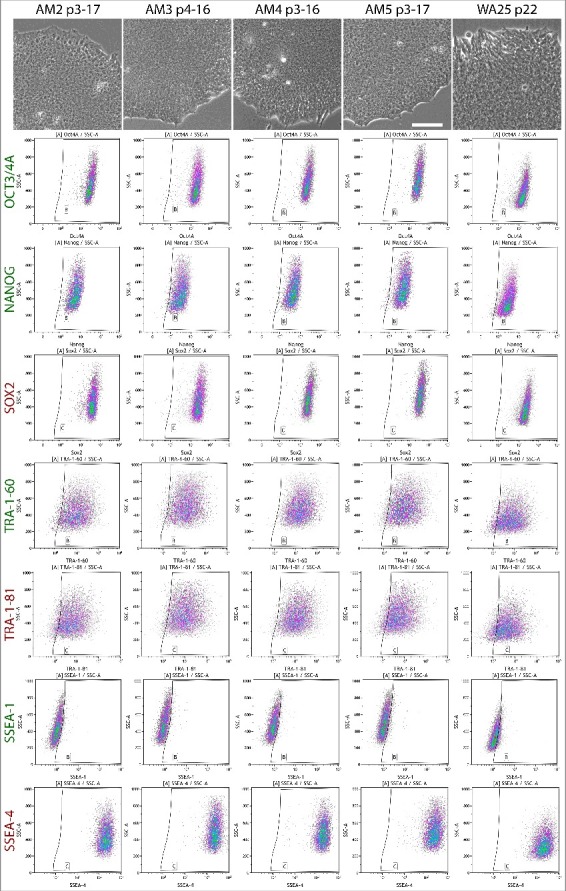
Flow cytometric analysis of the expression of ESC markers in AM-iPSC in their mature state grown in VTN-E8 conditions. Antibodies against markers Oct3/4A, Nanog, TRA-1-60 and SSEA-1 were conjugated to AlexaFluor 488 and Sox2, TRA-1-81 and SSEA-4 to AlexaFluor 647. All plots show the corresponding marker fluorescence on the horizontal axis on a log 10 scale and side scatter on a linear scale on the vertical axis. All 4 lines of AM-iPSC showed typical human primed pluripotent stem cell marker expression profile – Oct3/4A, Nanog, Sox2, TRA-1-60, TRA-1-81 and SSEA-4 being expressed in the majority of cells, while SSEA-1 is negative. The morphology of the corresponding lines is also shown, scale bar = 100µm.
Figure 3.
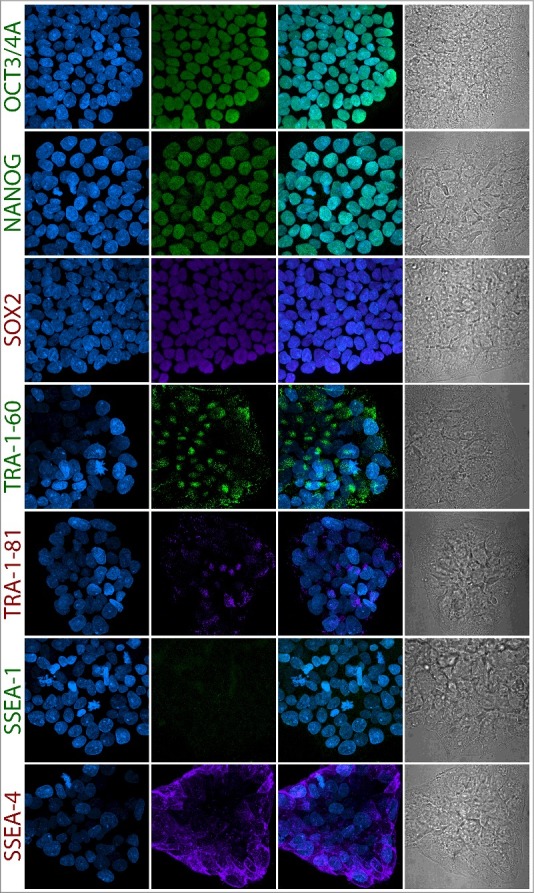
Confocal microscopy-based immunocytochemical analysis of the expression of ESC markers in AM-iPSC in their mature state grown in VTN-E8 conditions. Images of colonies of AM4 line are depicted on the figure. The images corroborate the flow cytometric analysis. The projections were reconstructed from the fluorescence signal spanning all of the individual scanned slices using the Maximum Intensity Projection algorithm in FIJI. The first column contains DAPI images, second column contains marker fluorescence, third column contains DAPI and marker overlay images and the fourth column contains a single-slice leftover transmitted light-based representation of the same colonies. Scale bar = 50µm.
Teratoma formation assay allows for a direct demonstration of the ability of cells to differentiate into tissues of all three germ layers. All AM-iPSC lines demonstrated their ability to form teratomas with endodermal (e.g. epithelia), mesodermal (e.g. cartilage or smooth muscle) and ectodermal (e.g. neural tissues) tissue derivatives present, confirmed by H&E-stained histological sections (Figure 4A). The proportions of endodermal, mesodermal and ectodermal derivatives were evaluated from the tissue sections by an expert pathologist. The germ layer with the lowest representation in the sections in all lines was mesoderm, the highest was ectoderm. The proportions were visibly skewed towards ectodermal tissues (Figure 4B), especially line AM3 that showed the highest variance of the percentages (Figure 4C). All lines displayed normal karyotypes (Figure 4D).
Figure 4.
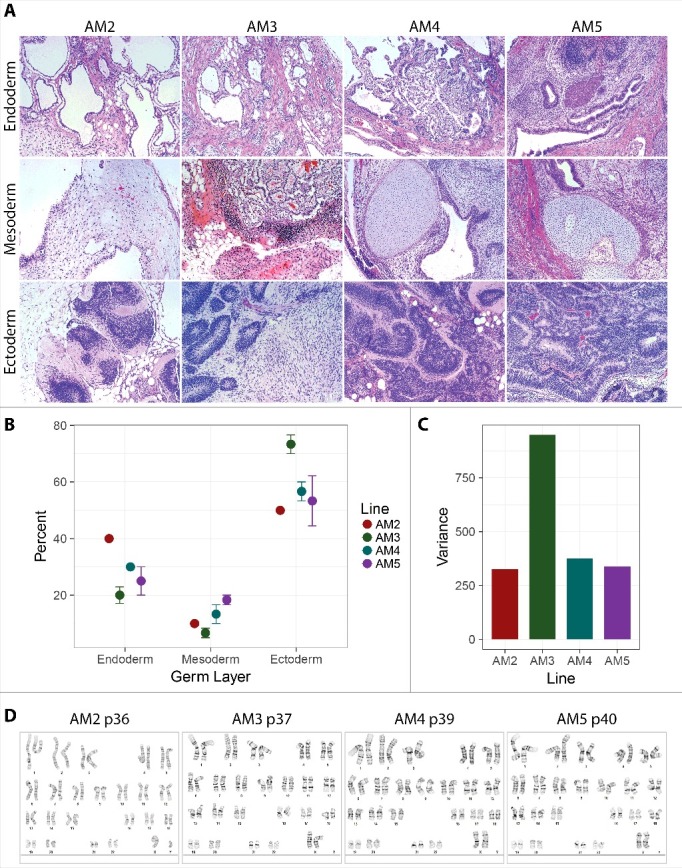
Teratoma formation capacity of AM-iPSC. Subcutaneous injection of AM-iPSC clumps into scid mice resulted in formation of teratomas. A. H&E-stained tissue sections revealed presence of tissues representative of all three germ layers. B. The percentages of the individual germ layer tissues were estimated by the pathologist. Mesoderm was the least abundant germ layer, while neuroectoderm was the most abundant. C. Variance of the percentages of the germ layer tissues was the highest for AM3 line. D. All lines showed normal karyotypes after a high number of passages in culture (p).
Evaluation of AM-iPSC pluripotency by computational methods
Profiling of the transcriptome by microarrays and RNA sequencing provides a rich source of information that can be used to confirm and evaluate pluripotency with a very high resolution. These methods represent an attractive alternative to the teratoma formation assay. We obtained global transcriptional profiles from the AM-iPSC lines, a control iPSC line MIRJT7i and a control ESC line WA25 using Illumina HT-12 v4 microarrays and RNA sequencing. We also included 3 samples of different lines, two of which are isogenic to AM3 and AM4, grown on VTN in KSR-based MEF-conditioned medium (KSR-CM), representing a traditional undefined medium. We then analyzed the microarray profiles using PluriTest which computes two classifiers from the data – pluripotency and novelty. High pluripotency scores and low novelty scores are required for queries to be deemed pluripotent. The PluriTest plot highlights the cluster area of the training set of samples, blue cloud represents differentiated cells, faint blue cloud partially pluripotent cells. Sample relations computed by PluriTest from the pre-processed data showed that AM5 line localized in a separate branch of the hierarchical clustering dendrogram, while KSR-CM lines were the most similar to the ESC control line (Figure 5A). All lines clustered close to the red cloud of the PluriTest plot and passed the test for pluripotency (Figure 5B). AM2 and AM3 lines had pluripotency scores below 20 and were therefore flagged as “further evaluate”. The pluripotency score of AM3 line was significantly lower than the average score of well-characterized WiCell control lines MIRJT7i (iPSC control) and WA25 (ESC control), while the pluripotency score of AM5 was significantly higher (Figure 5C). The opposite was true for the novelty scores, with AM3 having a significantly higher score relative to WiCell controls and AM5 a significantly lower score (Figure 5D). The pluripotency and novelty scores of the remaining lines, including KSR-CM lines, were not significantly different from the controls.
Figure 5.
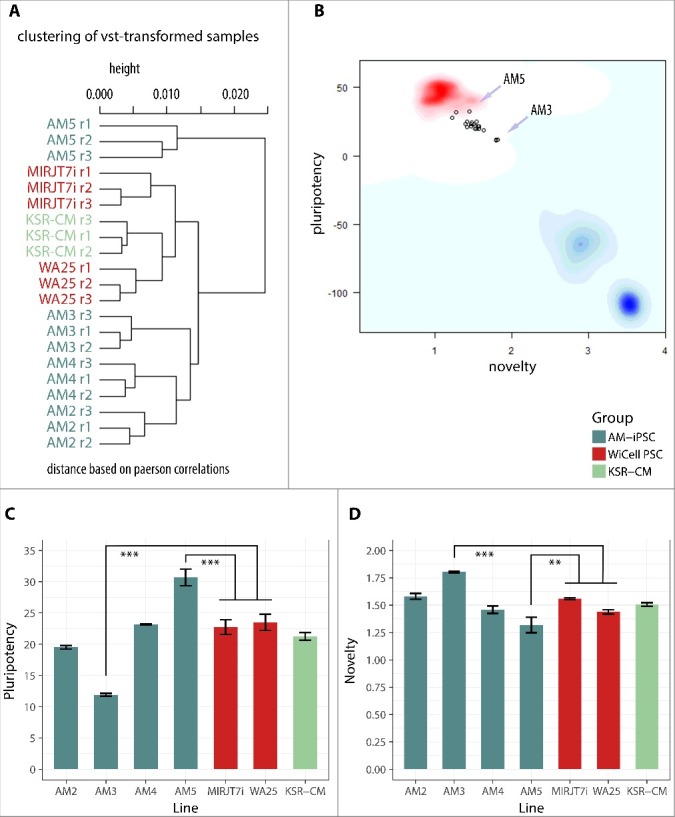
Analysis of the transcriptional profile of AM-iPSC by PluriTest. Control ESC line WA25 and iPSC line MIRJT7i (WiCell) were included. Also included were 3 iPSC lines grown in standard non-defined KSR-supplemented MEF-conditioned culture conditions (KSR-CM), two of which – AM3 and AM4 – were isogenic to AM3 and AM4 grown in defined VTN/E8 conditions. A. Hierarchical clustering of the lines showed AM5 having a distinct profile and KSR-CM lines being the most similar to the ESC line WA25. B. A plot of PluriTest classifiers “novelty” and “pluripotency”. The red cloud represents the cluster area of validated PSC lines (grown in standard KSR-CM culture condition). The blue cloud represents the cluster area of differentiated cells. The light blue cloud represents the cluster area of partially pluripotent stem cells. Each point represents a replicate of one line. All lines were identified as pluripotent but clustering outside of the red cloud. C. Pluripotency scores in individual lines, with AM5 scoring the highest and AM3 the lowest. D. Novelty scored in individual lines, with AM5 scoring the lowest and AM3 the highest. High pluripotency score and low novelty score render the transcriptional profile of AM5 the most similar to the typical pluripotent profile. ** p<0.01, *** p<0.001.
Another computational method of pluripotency evaluation from global transcriptional profiles is CellNet, recently adapted to accept RNA sequencing data as input. We performed RNA sequencing of the AM-iPSC lines (number of AM3 and AM4 replicates doubled) alongside the WiCell control lines and a larger set of KSR-CM lines. CellNet categorizes queries into cell/tissue types based on their transcriptional profile by identifying the relevant gene regulatory networks (GRN). All lines were categorized as “ESC”, with no apparent residual somatic cell/tissue signature, indicating undifferentiated status (Figure 6A). The GRN status scores was plotted for all queries relative to the score of the ESC training set (52 samples) and fibroblast training set (46 samples). All queries were similar to the ESC training scores with no statistically significant differences and distinct from fibroblast scores (Figure 6B). Furthermore, there was no statistically significant difference in the scores of VTN/E8- and VTN/KSR-CM-grown lines in terms of their similarity to the ESC training set (Figure 6C). These results confirm that the AM-iPSC lines are pluripotent.
Figure 6.
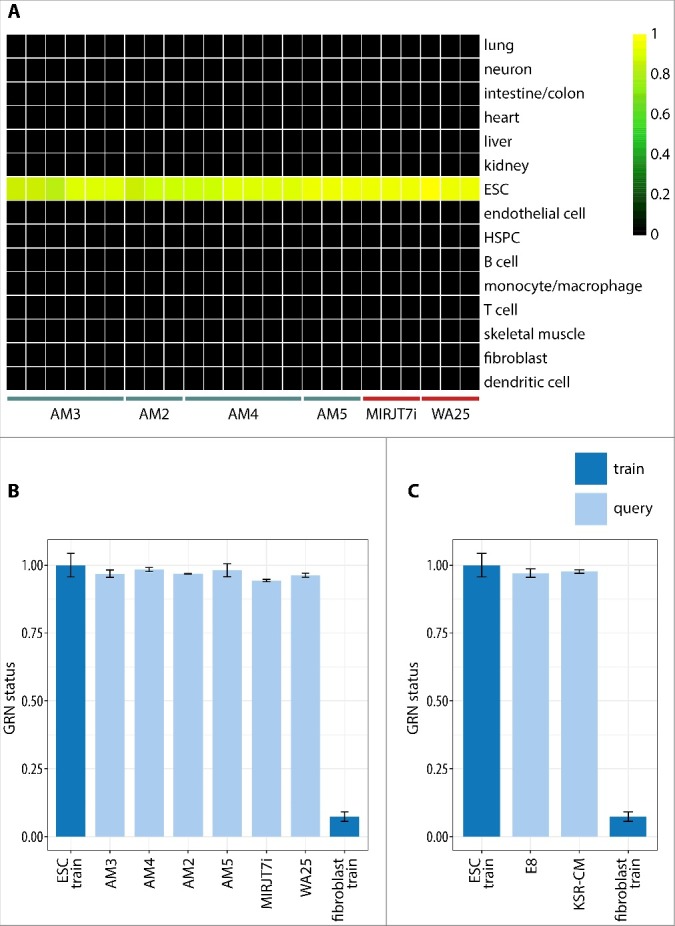
RNA sequencing-based gene regulatory network (GRN) analysis of the AM-iPSC transcriptome using CellNet. A. Heatmap showing classification of the samples based on the similarities of their expression profiles to reference cell/tissue type profiles. All AM-iPSC lines were classified as “ESC”, so did control iPSC line MIRJT7i and control ECS line WA25. Their undifferentiated status is demonstrated by the lack of transcriptional signature of other reference cell/tissue types. B. The heatmap was constructed from GRN classification scores that quantitate the extent to which the expression profile of a query sample is indistinguishable from the reference cell/tissue type's profile. All samples showed GRN classification scores similar to the ESC training scores, with no statistically significant differences, and distinct from fibroblast scores. C. No statistically significant differences in GRN classification scores were observed between VTN/E8 and VTN/KSR-CM-grown iPSC HSPC – hematopoietic stem and progenitor cells.
Transcriptional variability in pluripotent cell lines
The availability of the microarray and RNA sequencing data allows for further analysis of the transcriptional variability among pluripotent lines that are within the bounds of functional pluripotency. Differential gene expression analysis between PluriTest low-scoring AM3 and high-scoring AM5 line identified 123 genes with log fold change greater than 1.2 and false discovery rate (FDR)-adjusted P value below 0.02 (Figure 7A, Table S3). These genes were functionally annotated using the DAVID database (Table S5) and selected individual clusters of related genes were plotted. The identified genes belonged to metallothionein proteins, effectors of wnt signaling pathway, positive regulators of gene expression, actin genes or effectors of TGFβ signaling pathway (Figure 7B) and others. Out of the 123 genes, 28 are known pluripotency factors or were implicated as likely important for pluripotency as they belong to one of the three most pluripotency-associated W-dimensions of the PluriTest model (Figure 7C). The genes were as follows: GRPR, SFRP2, FGFR3, PLA2G3, ZIC3, PRDM14, GPC4, RPRM, PDPN, MT1F, CA2, ID1, FZD7, DNMT3B, COCH, FZD2, AFP, FAM46B, CYP26A1, PRODH, SHISA2, CACHD1, CKB, ZIC2, STC1, CRABP2, SFRP1 and IRX2.
Figure 7.
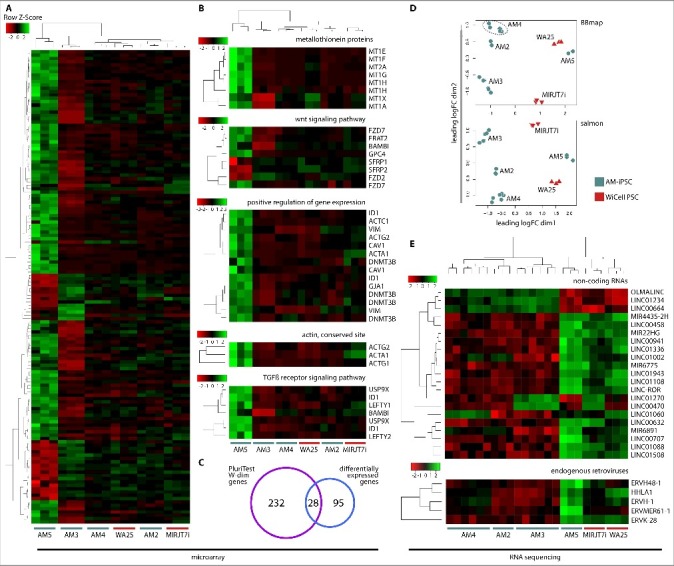
Differential expression analysis between high-scoring AM5 and low-scoring AM3 line.A. Heatmap of 123 most highly differentially expressed genes between AM5 and AM3 with a log-fold change of 1.2 and FDR-adjusted P value below 0.02 revealed a distinct transcriptional profile in both lines compared to AM2, AM4 and WiCell control lines. B. The list of differentially expressed genes was analyzed using DAVID and individual heatmaps were constructed to visualize differences in gene activity of the whole clusters of related genes. Genes with the best evidence of differential expression were metallothionein genes. Other genes were modulators of the wnt signaling pathways, positive regulators of gene expression, actin genes and effectors of the TGFß signaling pathway. C. From among the top 123 differentially expressed genes, 28 were found to belong to one of the three most pluripotency-associated W-dimensions of the PluriTest model. D. Transcriptional differences between AM5 and AM3 were further explored using RNA sequencing data and two analysis pipelines – BBmap + featureCounts method (alignment and counting) and salmon method (pseudoalignment-based quantitation). The dataset was filtered, normalized and multidimensional scaling plots were constructed for both analysis methods. Similar to the result of the microarray analysis, AM5 line showed distinct clustering from the other AM-iPSC lines. AM5 was the closest to the control ESC line WA25, especially as seen from the BBmap plot. E. Additional features identified as differentially expressed between PluriTest high-scoring AM5 and low-scoring AM3 line with a log fold change of ±1.6 and a false discovery rate-adjusted P value less than 1−06. Long non-coding RNA and microRNA were identified, as well as several endogenous retroviruses. Most of these features were found to be reinforced in AM5.
RNA sequencing-based differential gene expression analysis mostly corroborated the microarray analysis, though multidimensional scaling plots (MDS) showed a greater similarity between AM5 and WA25 ESC line. We chose 2 fundamentally different analysis pipelines for the RNA sequencing data – alignment-based (using BBmap, featureCounts and limma) and pseudoalignment-based (using salmon, tximport and limma) method (Figure 7D). The methods produced similar results. In addition to the metallothioneins identified by microarrays, MT1L, MT1M, MT1HL1, MT1P3 and MT2P1 genes were identified by RNA sequencing. 17 long non-coding (LINC) RNAs, such as LINC-ROR (regulator of reprogramming), 4 micro-RNAs and 5 endogenous retroviruses were also identified. Most of the non-coding RNAs and endogenous retroviral RNAs were reinforced in AM5 (Figure 7E, Table S4).
Discussion
Generating iPSC from fetal stem cells is an attractive way of making populations of cells with the broadest differentiation potential available perinatally for future cell-based therapies and banking for use later in life. Their clinical application, however, requires culture systems free of xeno-components. We recently showed that reprogramming of amniotic fluid stem cells (AFSC) into pluripotency and long-term iPSC culture is feasible in xeno-free conditions [26], demonstrating an example of using fetal stem cells for this purpose. However, novel prenatal genetic testing methods have reduced the availability of AFSC. Nevertheless, AFSC will continue to be an attractive cell source for reprogramming into pluripotency, mainly because conceptually this conversion can be carried out prenatally and allows for an autologous cell-based therapy to be available shortly after birth to e.g. correct a congenital defect, in case a fast intervention is necessary. Otherwise fetal cells with mesenchymal properties can be isolated from the placental amnion, circumventing the need for an amniocentesis. Since the placenta is part of the medical waste, autologous fetal cells can be derived from any newborn patient, not only for immediate cell-based therapies but also for banking purposes, longitudinal studies for basic research or disease modeling. A small patch of the amnion, after gentle and effective tissue dissociation, provides a sufficient starting population of cells for reprogramming.
AMSC can be readily cultured in conditions previously identified to be optimal for AFSC [5,6,26]. While AFSC require around 10 days of culture before they can be expanded for the first time, AMSC reach that point in around 5 days. Both cell types predominantly assume an MSC phenotype in culture as indicated by their morphology and the expression of MSC markers. AMSC remain an understudied cell source for reprogramming. While their unique properties show promise for a direct use in treatments [3,10], only a few studies explored the induction of pluripotency in these cells, albeit using integrative vectors or undefined culture conditions [24,25]. Here we show that AMSC can be reprogrammed into pluripotency using episomal plasmids and culture in chemically defined VTN/E8 conditions [27]. The process of full pluripotency acquisition took around 14 days. Flow cytometry and confocal imaging confirmed that the resulting iPSC (AM-iPSC) express pluripotency markers Oct3/4A, Nanog, Sox2 (transcription factors), TRA-1-60, TRA-1-81 and SSEA-4 (surface glycoproteins). We used Oct3/4 isoform A as it has been shown to be relevant to pluripotency [28]. Nanog was expressed in a majority of cells in all PSC lines, though its signal was visibly fainter (Figure 2, Table S2). This is a normal feature of human pluripotent stem cells that assume primed phenotype as opposed to naïve [29,30]. SSEA-1 is not expressed in human PSC [28,31] and served as a negative control marker.
Functional demonstration of pluripotency in-vivo is typically achieved by the teratoma formation assay in mice. Tissues of immunocompromised mice create an environment that allows human PSC to spontaneously differentiate into a variety of tissues whose germ layer origin can be inferred after histological inspection. All AM-iPSC lines produced teratomas with tissues representative of all 3 germ layers. The percentages of the individual germ layer representation in the teratomas were not balanced but rather ectodermal tissues were over-represented and mesodermal tissues under-represented. This could reflect differences in growth or presence of stimuli in mouse tissue microenvironment favoring ectodermal differentiation. Alternatively, a bias inherent to the primed phenotype of human PSC is possible [32], after all, their in-vivo counterpart cells represent the primitive ectoderm of early post-implantation embryos [33]. The teratoma germ layer profile of the AM3 line was the least balanced (highest variance). This could potentially hamper differentiation efforts of this line into tissues of the mesodermal lineage. Nevertheless, presence of derivatives of all 3 germ layer tissues in teratomas demonstrated functional pluripotency of all AM-iPSC lines in-vivo.
Teratoma formation assay has recently drawn criticism, primarily due to its notorious lack of standardization [34,35]. Furthermore, attention is given in the majority of studies to the presence of pertinent tissues in teratomas, while the quantitative representation of the tissues is rarely reported. Expression microarrays and RNA sequencing technology have become powerful tools for profiling transcriptional activity of the genes. Advances in statistical and bioinformatic analysis [36] of the resulting large datasets have led to development of methods of pluripotency confirmation that represent an alternative to the teratoma assay. While teratoma assay measures differentiation ability, alternative methods based on genomic and bioinformatic approaches have the ability to measure the levels of pluripotency directly and circumvent animal use. These approaches leverage machine learning and gene regulatory networks, two major examples of such are PluriTest [35] and CellNet [37,38]. PluriTest is based on microarray profiles (primarily Illumina HT-12 v3 and v4, soon to be expanded to RNA sequencing data) and returns two classifiers – pluripotency and novelty. The pluripotency score is a measure of the presence of a transcriptional signature associated with pluripotency, while the novelty score is a measure of a deviation from the established pluripotent model which could indicate abnormalities or unique signatures. All AM-iPSC lines passed the test and were deemed pluripotent, with AM2 (pluripotency score slightly lower than 20) and AM3 (markedly lower pluripotency and an elevated novelty score) marked as “further evaluate” instead of “pass”. On the other hand, AM5 showed a significantly higher pluripotency and lower novelty score compared to the controls. Based on the established methods, all AM-iPSC and control lines are pluripotent, therefore, the variability in scores observed by PluriTest intriguingly transpires within the bounds of pluripotency and will be explored further below. As shown by the two-dimensional PluriTest plot (Figure 5B), all lines cluster slightly off the red cloud that represents the cluster area of established ESC and iPSC lines. Previously, we have hypothesized that the chemically defined culture conditions can explain this [26] since the PluriTest training set was derived from cell lines grown on MEFs in KSR-based medium, a traditional growth condition of choice [35]. For a preliminary test of that hypothesis, we included samples from 3 lines reprogrammed and grown on VTN in KSR-based MEF-conditioned medium (KSR-CM), two of which were isogenic to AM3 and AM4. We created these lines (unpublished data) to measure the contribution of the culture medium rather than the presence of feeders. These lines, however, did not cluster in the red cloud either. In order to isolate RNA from pure populations of ESC/iPSC grown in traditional conditions, it is necessary to avoid their contamination with MEF feeder layer cells, however, it is not clear how MEFs were prevented from doing so in the original PluriTest report [35]. The method of discarding MEFs could potentially have contributed to the observed gap in PluriTest scores as some independent methods have involved culturing in suspension prior to RNA isolation [39]. Recently, a collection of well-characterized iPSC lines with a high genomic integrity from 222 ethnically diverse individuals was created [40]. The report on this collection, termed iPSCORE, included a PluriTest plot calculated from RNA sequencing data of 213 of those lines. At the time of the report publication as well as preparation of this manuscript, the detailed method of modification of the PluriTest algorithm to accept RNA sequencing data had not yet been published. However, the PluriTest plot in the report shows all iPSC lines clustering roughly in the same area as the lines presented here. The iPSCORE lines were derived from dermal fibroblast by Sendai virus reprogramming on MEFs. Fully reprogrammed colonies were then transferred to Matrigel in mTeSR-1 medium. It is possible that PSC cultured in feeder-free conditions normally yield scores that place them off the red cloud of the PluriTest plot.
An alternative bioinformatic method of assessing pluripotency in candidate lines is CellNet [37,38]. The tool leverages gene regulatory networks of cell or tissue types that were reconstructed from publicly available gene expression profiles. The majority of those were derived from in-vivo tissues as opposed to in-vitro cultured cells. CellNet is not limited to pluripotent cells but instead was trained to assess similarity to at least 14 other cell/tissue types. The tool was recently expanded to accept RNA sequencing data as input [38] so we sequenced RNA from AM-iPSC and control lines for use with CellNet. All lines tested showed “ESC” identity with no significant differences in classification scores. There was also no difference between VTN/E8 and VTN/KSR-CM-grown lines, suggesting that both conditions perform equally well in maintaining pluripotency. Our data also support the notion that the teratoma formation assay can be replaced by bioinformatic methods like PluriTest and CellNet for the purpose of confirming pluripotency. This would result in significant time and financial savings and it would eliminate the need for animal testing.
The availability of the microarray and RNA sequencing data allowed us to further explore the variability between the PSC lines tested. While CellNet deemed all lines equally pluripotent, PluriTest data identified AM5 to have a stronger pluripotency signature (closer to the red cloud) than the controls and AM3 to be weaker in this regard so we performed differential gene expression analysis between the two lines. Overall, the differential expression analysis calculated from the microarray data produced 28 genes that were shown to directly contribute to pluripotency scores in the PluriTest model [35]. Using microarrays and RNA sequencing can expand the network of contributors to maintenance of the pluripotent state, especially those that with a relatively lower level of transcription.
Both microarray and RNA sequencing analysis showed higher expression of metallothionein proteins in AM5. Metallothionein proteins are highly conserved, have low molecular weight and have important roles in metabolism of metals, especially zinc [41,42]. Transport of zinc into the nucleus was found to coincide with spontaneous differentiation of human ESC and metallothionein proteins were suspected to be the chaperones in that process [43]. However, very little is known about the role of metallothionein proteins in pluripotency. MT1G was previously included among genes identified in protein-protein interaction networks that are enhanced in PSC [44]. More recently, extracting genes that contribute to one of the three most pluripotency-associated W-dimensions in the PluriTest model yielded a list of 260 genes, including several well-established pluripotency regulators like Oct3/4, Lin28, Sox2, Dnmt3b [35]. On this list, MT1F was included, suggesting its potential role in pluripotency. To our knowledge, however, contribution to pluripotency maintenance by MT1G and MT1F has never been validated. Additionally, the levels of MT2A and MT1E were found to be somewhat lower in iPSC compared to isogenic ESC lines [45].
Further, the genomic analyses suggested that the wnt signaling pathway could be reinforced in the high-scoring AM5 line. The importance of FZD7 for pluripotency maintenance was previously demonstrated [46]. The wnt antagonists SFRP1 and 2 were downregulated. Our data also suggested a reinforced TGFβ signaling [47], important for pluripotency, in AM5.
RNA sequencing additionally identified several long non-coding (LINC) RNAs, microRNAs and human endogenous retrovirus (HERV) transcripts, most of which were more active in AM5. Among them was LINC-ROR (regulator of reprogramming) that was reported to be activated during reprogramming and among other LINCs, enriched in iPSC [48]. The role of human ERV transcripts in pluripotency has been documented. HERV-H was shown to be abundant in ESC, required for their identity [49-51] and a driver of LINC-ROR during reprogramming [52]. HERV-H LTR-associating 1 (HHLA1) was demonstrated to be a marker of a phenotype with a delayed and defective exit from pluripotency in subclones of some iPSC lines during neural differentiation [52]. AM5 line exhibited HHLA1 level comparable to the control WA25 ESC line. Remarkably, in subpopulations of human ESC and iPSC cultures, HERV-H confers properties of naïve pluripotency [51], a property that had not been observed in cultured human PSC without substantial modification of the culture conditions or forced expression of exogenous factors. Microarray analysis also suggested other known markers or regulators of pluripotency PODXL [53], PRDM14 [54] (included in pluripotency-associated W-dimension of PluriTest) and KLF4 as potential genes that could contribute to high scores of AM5.
Transcriptional differences between isogenic iPSC and ESC have been observed to be relatively small and the majority of differences between PSC lines are due to the genetic background [45]. Studying the transcriptional heterogeneity in iPSC lines could help with design of reliable methods of differentiation and positively influence the outcome of cell-based therapies, as differences can manifest themselves beyond the state of pluripotency. Computational methods of analyzing transcriptional signatures associated with pluripotency can prove to be powerful tools in this process. A major benefit of these tools as opposed to simply including a limited number of ESC controls in genomic experiments is the availability of a “built-in” control based on hundreds of well-established PSC lines. Here we list genes that represent potential candidates that differentiate between PluriTest high-scoring (higher similarity to a large set of well-characterized PSC, most of which were ESC) and low-scoring lines (lower similarity to well-characterized PSC). This could lead to development of strategies of correcting low-scoring lines either by modifying the reprogramming process or the iPSC themselves in their mature state, as more knowledge on pluripotency becomes available and as the differentiation protocols improve.
We demonstrate here a method of generating iPSC from human AMSC, an attractive cell source, in chemically defined conditions. AMSC can be easily isolated, can be obtained at birth from any patient, are highly undifferentiated and are especially suited for pediatric cell-based therapies either directly or reprogrammed to pluripotency. Together with analyzing the expression of ESC markers, we applied stringent methods of confirming pluripotency – the gold-standard teratoma formation assay and bioinformatic methods, capable of replacing the former. Given the high resolution of genomic and bioinformatic methods, optimal wiring of the pluripotent regulatory networks during reprogramming for a good balance between pluripotency maintenance and differentiation propensity can be identified in the future. Our future efforts will focus on linking these two properties with expression and epigenetic profiles in pursuit of maximized cell engineering outcomes. Single-cell RNA sequencing methods will help elucidate if reprogramming from fetal stem cells differs from reprogramming from commonly used cell types. Finally, studying the variability within the bounds of pluripotency could lead to discovery of novel members of the regulatory networks in addition to the known core factors, with potential candidates that include metallothionein proteins.
Materials and methods
Isolation and culture of amniotic membrane mesenchymal stem cells
Placentas were obtained within 3 hours following birth. A segment of the amnion of around 9 cm2 was cut and washed with phosphate buffered saline (PBS, 10010023, Thermo-Fisher Scientific, https://www.thermofisher.com/order/catalog/product/10010023) supplemented with antibiotic-antimycotic solution (15240062, Thermo-Fisher Scientific, https://www.thermofisher.com/order/catalog/product/15240062). The digestion of the membranes and extraction of single cells was achieved using gentleMACS tissue dissociation system – gentleMACS dissociator (130-093-235, Miltenyi Biotec, http://www.miltenyibiotec.com/en/research-areas/cancer-research/solid-tumor-cells/sample-preparation/gentlemacs-dissociator.aspx) and reagent kit (130-095-929, Miltenyi Biotec, http://www.miltenyibiotec.com/en/products-and-services/macs-sample-preparation/tissue-dissociators-and-tubes/tissue-dissociation-kits/tumor-dissociation-kit-human.aspx). The membranes were minced using a pair of scalpels to fine pieces and up to 1 g of the minced material was mixed with 4.7 ml of RPMI 1640 medium (32404014, Thermo-Fisher Scientific, https://www.thermofisher.com/order/catalog/product/32404014), dissociation enzymes from the reagent kit and transferred into C tubes (included in the kit). The tubes were mounted onto the dissociator and program “h_tumor_01” was run. Then the tubes were incubated at 37°C on a rocking platform for 30 min. A 70 µm strainer was used to obtain a single-cell suspension and the cells were centrifuged at 200 g for 5 min. Single cells were plated onto tissue culture-treated vessels at a density of around 105 cells/cm2. The culture medium comprised the following components: EBM-2 basal medium (CC-3156, Lonza, http://bio.lonza.com/uploads/tx_mwaxmarketingmaterial/Lonza_BenchGuides_Clonetics_Endothelial_Cell_Medium_Products.pdf), 15% fetal bovine serum (10439024, Thermo-Fisher Scientific, https://www.thermofisher.com/order/catalog/product/10439024), 20 ng/ml of basic fibroblast growth factor (bFGF, CYT-218, Prospec Bio, https://www.prospecbio.com/FGF-2_Human), 25 ng/ml of epidermal growth factor (EGF, CYT-332, Prospec Bio, https://www.prospecbio.com/EGF_Human_Pichia), 10 ng/ml of insulin-like growth factor (IGF, CYT-022, Prospec Bio, https://www.prospecbio.com/Long_R3_IGF1_Human). The cultures were passaged using TrypLE (12604013, Thermo-Fisher Scientific, https://www.thermofisher.com/order/catalog/product/12604013).
Reprogramming to pluripotency
Reprogramming of AMSC was initiated by means of transfection with non-integrating episomal plasmids published previously [55]. Plasmids used were as follows: pEP4 E02S EN2K (Oct4+Sox2, Nanog+Klf4; 20925 Addgene, http://www.addgene.org/20925), pEP4 E02S ET2K (Oct4+Sox2, SV40LT+Klf4; 20927 Addgene, http://www.addgene.org/20927), pCEP4 M2L (c-Myc+LIN28; 20926 Addgene, http://www.addgene.org/20926). Stab cultures were streaked on Luria agar plates containing 100 µg/ml of Ampicillin (A8351, Sigma-Aldrich). The plates were incubated for 8–12 hours at 37°C until individual bacterial colonies appeared. The colonies were then transferred into stirred, air exchange-allowing, bacterial culture bottles in Terrific Broth medium to expand the bacterial cells in suspension. Suspension cultures were maintained at 37°C for another 8–12 hours and while still in log-phase of growth, bacteria were harvested by centrifugation at 6000 g for 15 min.
Plasmid DNA was isolated from bacterial pellets using a commercial kit (12362, Qiagen, https://www.qiagen.com/us/shop/sample-technologies/dna/plasmid-dna/endofree-plasmid-kits/#orderinginformation) following the manufacturer's instruction. All three plasmids were transfected into the source cells using the Neon™ Transfection System (MPK5000, Thermo Fisher Scientific, https://www.thermofisher.com/order/catalog/product/MPK5000) using the Neon™ Transfection System 10 µL Kit (MPK1025, Thermo Fisher Scientific, https://www.thermofisher.com/order/catalog/product/MPK1025) according to the manufacturer's instruction. The plasmid amounts per 1 million transfected cells were 3.0µg of pEP4 EO2S ET2K, 3.0µg of pEP4 EO2S EN2K and 2.0µg of pCEP4 M2L. The transfection parameters were 950 V, 40 ms and 1 pulse. Following transfection, the cells were plated onto recombinant human vitronectin (VTN, 07180, StemCell Technologies, https://www.stemcell.com/products/vitronectin-xf.html)-coated 6-well plates and kept in the AMSC culture medium for the first 3–5 days. Then, the medium was replaced by the E8 medium [27] (05940, StemCell Technologies, https://www.stemcell.com/products/tesr-e8.html) supplemented with 100 µM sodium butyrate (04-0005, StemGent, https://www.stemgent.com/products/show/10). The medium was changed daily. Starting on day 14 following transfection, colonies of fully reprogrammed cells displaying morphology of PSC – compact colonies, well-defined borders, high nucleus-to-cytoplasm ratio, prominent nucleoli – were manually picked using a pipette while being observed under an inverted microscope, in a sterile culture cabinet. The picked colonies were dissociated into clumps by treatment with 0.5 mM ethylenediaminetetraacetic acid (UltraPure EDTA, 15575020, Thermo Fisher Scientific, https://www.thermofisher.com/order/catalog/product/15575020) in PBS for 3–4 minutes and an additional gentle trituration with a pipette tip. The clumps were plated into VTN-coated vessels with E8 medium without sodium butyrate. The clones were allowed to expand following the routine iPSC culture protocol outlined below. For the first several passages, if needed, differentiating colonies were manually scraped off using a pipette tip immediately before passaging, progressively producing pure cultures. A preliminary confirmation of pluripotency was achieved by flow cytometric analysis of TRA-1-60 and TRA-1-81 expression and back-up cultures of clones were frozen at early passages (procedures for flow cytometric analysis and freezing outlined below).
iPSC culture
Routine culture of PSC was performed on VTN coated vessels in E8 medium. Passaging was performed using the EDTA method [56]. Cultures were washed with 0.5 mM EDTA in PBS and incubated with EDTA/PBS for 5 min at room temperature (RT). EDTA solution was then aspirated, colonies washed off with E8 medium and further triturated to create a suspension of clumps of roughly 20–50 cells. Those were plated on freshly VTN coated plates with E8 medium.
For the purpose of cryopreservation, pellets of EDTA-harvested cell clumps were resuspended in CryoStor™ CS10 (07930 StemCell Technologies, http://www.stemcell.com/en/Products/All-Products/CryoStorCS10.aspx), transferred into cryovials and placed into Mr. Frosty™ containers (5100-0001, Thermo Fisher Scientific, https://www.thermofisher.com/order/catalog/product/5100-0001) providing 1°C/min cooling at −80°C. Following overnight incubation, the cryovials were transferred to liquid nitrogen storage.
Flow cytometry
Cell cultures were harvested using Accutase® (07920, StemCell Technologies, https://www.stemcell.com/products/accutase.html) – 7 min dissociation for AMSC and 10 min for iPSC. For the purpose of nuclear transcription factor staining, the cells were washed with PBS, fixed with 4% formaldehyde (28908, Thermo Fisher Scientific, https://www.thermofisher.com/order/catalog/product/28908) in PBS for 20 min at RT and permeabilized with BD Perm Buffer III (558050, BD Biosciences, http://www.bdbiosciences.com/us/applications/research/t-cell-immunology/th-1-cells/intracellular-markers/cell-signalling-and-transcription-factors/buffers/perm-buffer-iii/p/558050) for 30 min on ice. Staining of cells suspended in PBS + 2% FBS with directly labeled mouse anti-human monoclonal antibodies (listed below) was performed at 4°C for 45 min, followed by 3 washing steps with PBS + 2% FBS. For the purpose of surface antigen staining, the cells were washed with PBS + 2% FCS, incubated with the directly labeled mouse anti-human monoclonal antibodies listed below for 20 min at RT and subsequently washed. The cell suspension was analyzed using a FACSCanto II flow cytometer. AlexaFluor 488 was detected with 20mW 488 nm excitation-502LP-530/30BP. AlexaFluor 647 was detected with 17mW 633nm excitation-685LP-660/20BP. The resulting data analysis was performed using Kaluza 1.5a (Beckman Coulter, http://www.beckman.com/coulter-flow-cytometry/software/kaluza-analysis).
Confocal microscopy
For the purpose of immunocytochemical labeling of iPSC, the colonies were grown in E8 medium in VTN-coated 8-well chambered coverglass (155409, Thermo-Fisher Scientific, https://www.thermofisher.com/order/catalog/product/155409). For nuclear transcription factor staining, the wells were washed with PBS, fixed with 4% formaldehyde for 20 min at RT and permeabilized using BD Perm Buffer III. Incubation with directly labeled mouse anti-human monoclonal antibodies listed below was carried out at 4°C for 45 min. Three washing steps were performed with PBS + 2% FCS, each incubated for 10 min. For surface antigen staining, the wells were washed with PBS + 2% FCS. The cells were incubated with the directly labeled mouse anti-human monoclonal antibodies listed below for 20 min at RT and then washed with PBS + 2% FCS. The cells were then fixed with 4% formaldehyde in PBS for 20 min.
To amplify the fluorescent signal, staining the cells with secondary antibodies carrying fluorescent molecules matching the ones of the primary antibodies was performed in PBS + 2% FBS on ice for 20 min, followed by 3 washing steps. 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI, D1306, Thermo-Fisher Scientific, https://www.thermofisher.com/order/catalog/product/D1306?icid = fr-dapi-1) was added into the washing solution at a concentration of 800 nM and incubated for 5 min during one of the washing steps. Colonies were imaged using Ti-E with A1r-SI and N-STORM confocal microscope (Nikon, http://www.nikoninstruments.com/Products/Light-Microscope-Systems/Super-Resolution/N-STORM-Super-Resolution).
List of antibodies used in flow cytometry and immunohistochemistry experiments
Antibodies were purchased from the following sources: BD Biosciences (http://www.bdbiosciences.com/us/reagents/c/reagents), BioLegend http://www.biolegend.com/productstab), and Jackson Immunoresearch (https://www.jacksonimmuno.com/catalog/22).
Intracellular staining: monoclonal mouse anti-human AlexaFluor 488-conjugated Oct3/4A (561628, BD), AlexaFluor 488-conjugated Nanog (560791, BD), AlexaFluor 647-conjugated Sox2 (561593, BD). Surface staining: monoclonal mouse anti-human AlexaFluor 488-conjugated TRA-1-60 (330614, BioLegend), AlexaFluor 647-conjugated TRA-1-81 (330706, BioLegend), AlexaFluor 488-conjugated SSEA-1 (301910, BioLegend), AlexaFluor 647-conjugated SSEA-4 (330407, BioLegend). Isotype controls for these markers were matched in concentrations: AlexaFluor 488-conjugated mouse IgG1κ (isotype for Oct3/4A, Nanog; 557782, BD), AlexaFluor 647-conjugated mouse IgG1κ (for Sox2; 557783, BD), AlexaFluor 488-conjugated mouse IgMκ (for TRA-1-60; 401617, BioLegend), AlexaFluor 647-conjugated mouse IgMκ (for TRA-1-81; 401618, BioLegend), AlexaFluor 488-conjugated mouse IgG1κ (for SSEA-1; 400129, BioLegend), AlexaFluor 647-conjugated mouse IgG3κ (for SSEA-4; 401321, BioLegend).
Antibodies used in flow cytometric analysis of AMSC for the purpose of mesenchymal stem cell marker expression analysis were: mouse anti-human monoclonal IgG1κ isotype control (400102, BioLegend), CD44 (sc-59758, Santa Cruz), CD73 (344002, BioLegend), CD90 (328102, BioLegend), CD166 (343902, BioLegend), FITC-conjugated IgG1κ isotype control (21275513S, ImmunoTools), FITC-conjugated CD34 (21270341, ImmunoTools) and goat anti-mouse Cy2-conjugated secondary antibody (115-225-003, Jackson ImmunoResearch).
Secondary antibodies used in confocal imaging were as follows: F(ab′)2 fragment goat anti-mouse IgG+IgM, conjugated with Alexa Fluor 488 (115-546-068, Jackson ImmunoResearch) and Alexa Fluor 647 (115-606-068, Jackson ImmunoResearch).
Mesenchymal stem cell marker expression in AMSC was analyzed using flow cytometry using the following monoclonal mouse anti-human antibodies: Brilliant Violet 421-conjugated CD44 (103039, BioLegend), PE-Cy7-conjugated CD73 (344009, BioLegend), PE-conjugated CD166 (343903, BioLegend), Alexa Fluor 647-conjugated CD105 (323212, BioLegend), PE-conjugated N-cadherin (350806, BioLegend), Alexa Fluor 647-conjugated CD90 (328115, BioLegend), PE-Cy7-conjugated EpCAM (324221, BioLegend).
Teratoma formation assay
All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of South Alabama (protocol number 840976-2) and carried out at the vivarium of the Department of Comparative Medicine at the College of Medicine (Assurance Number A3288-01; accredited by the Association for Assessment and Accreditation of Laboratory Animal Care). iPSC colonies were harvested from the 6-well culture plates by EDTA as described above and resuspended in 1:1 mixture of E8 medium and Matrigel. Suspension of cell clumps containing between 500.000 to 1 million of cells were injected subcutaneously into both flanks of immunocompromised female scid-beige mice (Taconic Biosciences, http://www.taconic.com/mouse-model/scid-beige), 3 animals per line. 6 to 9 weeks post-injection, the teratomas were excised, fixed in formalin and processed for standard H&E histological analysis. The presence of tissues representative of all three germ layers was verified and scored by the pathologist. Representation percentages were estimated from 3 separate teratomas. Mean, standard deviation and variance was calculated and plotted in R statistical programming language using package “ggplot2” v2.2.1 [57].
RNA isolation, microarray profiling and RNA sequencing
RNA was extracted from iPSC cultures using RNeasy Plus Mini kit (74134, Qiagen, https://www.qiagen.com/us/shop/sample-technologies/rna-sample-technologies/total-rna/rneasy-plus-micro-and-mini-kits). Whole genome expression analysis was performed by the HudsonAlpha Institute for Biotechnology Genomic Services Laboratory (http://gsl.hudsonalpha.org/index) using the Human HT-12 v4 Expression BeadChip microarray system (Illumina, http://www.illumina.com/products/humanht_12_expression_beadchip_kits_v4.html). RNA sequencing was performed by the same laboratory using the sequencing platform HiSeq 2500 (Illumina, https://www.illumina.com/systems/sequencing-platforms/hiseq-2500.html). The sequencing library preparation was stranded and included reduction of ribosomal RNA. The libraries were sequenced as paired-end reads, length of 100. All samples passed quality control at the Genomic Services Laboratory – Quantification via Qubit® and Bioanalysis using the 2100 Bioanalyzer (Agilent) – and had an RNA Integrity Number of 10.
Bioinformatic confirmation of pluripotency by PluriTest and CellNet
IDAT files of raw microarray data (one file per replicate) were submitted for PluriTest [35] analysis through a web-based interface at www.pluritest.org.
Raw RNA sequencing data in form of FASTQ files were used for CellNet analysis, performed on a local workstation computer, as previously described [38], with minor modifications. The analysis pipeline is implemented in R statistical programming language on UNIX-like operating systems. We used Linux Mint KDE 18.1, R version 3.4.0, Bioconductor v3.4, RStudio v1.0.143, CellNet v0.0.0.9000. Salmon [58] v0.8.2 and its index corresponding to the same version was used. We compiled an example R script that contains all steps followed for the CellNet analysis presented in this manuscript (Supplemental Material).
Microarray data analysis
Initial pre-processing of the raw data was performed using GenomeStudio v2011.1 with the Gene Expression Module v1.9.0 to produce bead-level summary. The summary was imported into R/Bioconductor (versions above) using package “lumi” v2.28 [59] for further pre-processing by applying variance-stabilizing transformation and robust spline normalization. Differential expression analysis was performed by applying linear modeling using package “limma” v3.32.2 [60]. Heatmaps were drawn using package “gplots” v3.0.1 [61]. Functional annotation of the list of differentially expressed genes was performed using DAVID v6.8 at http://david.abcc.ncifcrf.gov/ [62,63]. Bar plots and line graphs were created using package “ggplot2”.
RNA sequencing data analysis
Quality control of the FASTQ files was performed using FastQC v0.11.5 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were aligned with BBmap [64] v36.92 to the human reference genome from ENSEMBL [65] (build GRCh38, primary assembly) guided by annotation GTF file GRCh38 release 87. Alignment SAM files were coordinate-sorted using Picard Tools v1.113-2 (http://broadinstitute.github.io/picard/) and converted to BAM format using SAMtools v0.1.19 [66]. Fragments were counted using featureCounts [67]. The counts were imported into R/Bioconductor. Multidimensional scaling and linear modeling-based differential expression analysis was performed with package “limma”. Heatmaps were drawn using package “gplots”. Venn diagram was created using package “VennDiagram” v1.6.17 [68]. Alternatively, Salmon 0.8.2 was used for transcript quantification in combination with package “tximport” [69] v1.4.0 for import into R and “limma”.
Statistical analysis
PluriTest's pluripotency score bar graph was plotted from means and standard errors of the means. CellNet score bar graphs were plotted from means and standard deviations. The measure of significance of difference in means was calculated using ANOVA and Tukey's Honest Significant Difference test. Teratoma tissue percentages and genomic analyses all entailed biological triplicates of samples.
Supplementary Material
Funding Statement
This work was supported by the Fonds Medizinische Forschung at the University of Zurich, Forschungskredit of the University of Zurich, The Swiss Society of Cardiology, The Swiss National Science Foundation under Grant [320030-122273] and [310030-143992], The 7th Framework Programme, Life Valve, European Commission under Grant [242008], the Olga Mayenfisch Foundation, the EMDO Foundation, the Start-up Grant 2012 of the University Hospital Zurich, Foundation of Scientific Research Zurich [STWF-17-013], and internal funding of the Mitchell Cancer Institute.
Abbreviations
- AFSC
amniotic fluid stem cells
- AM-iPSC / AM[N]
induced pluripotent stem cells derived from amniotic membrane stem cells, N is line/patient number
- AMSC
amniotic membrane stem cells
- bFGF
basic fibroblast growth factor
- DAPI
4′,6-diamidino-2-phenylindole
- E8
chemically defined medium for culture of human pluripotent stem cells [27]
- EDTA
ethylenediaminetetraacetic acid
- EGF
epidermal growth factor
- ESC
embryonic stem cells
- FASTQ
raw sequencing file format, contains sequence and quality
- FBS
fetal bovine serum
- IGF
insulin-like growth factor
- iPSC
induced pluripotent stem cells
- KSR
KnockOut™ Serum Replacement (Thermo-Fisher Scientific)
- MEF
mouse embryonic fibroblast feeder layer
- MET
mesenchymal-to-epithelial transition
- MSC
mesenchymal stem cells
- PBS
phosphate buffered saline
- PSC
pluripotent stem cells – embryonic and induced pluripotent stem cells
- VTN
recombinant human vitronectin
- GRN
gene regulatory networks
- FDR
false discovery rate
Disclosure of potential conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
This work was supported by the Fonds Medizinische Forschung at the University of Zurich, Forschungskredit of the University of Zurich, The Swiss Society of Cardiology, The Swiss National Science Foundation under Grant [320030-122273] and [310030-143992], The 7th Framework Programme, Life Valve, European Commission under Grant [242008], the Olga Mayenfisch Foundation, the EMDO Foundation, the Start-up Grant 2012 of the University Hospital Zurich, Foundation of Scientific Research Zurich [STWF-17-013], and internal funding of the Mitchell Cancer Institute.
References
- [1].Dziadosz M, Basch RS, Young BK. Human amniotic fluid: a source of stem cells for possible therapeutic use. Am J Obstet Gynecol. 2016;214:321–327. doi: 10.1016/j.ajog.2015.12.061. PMID:26767797 [DOI] [PubMed] [Google Scholar]
- [2].Niknejad H, Yazdanpanah G, Ahmadiani A. Induction of apoptosis, stimulation of cell-cycle arrest and inhibition of angiogenesis make human amnion-derived cells promising sources for cell therapy of cancer. Cell Tissue Res. 2016;363:599–608. doi: 10.1007/s00441-016-2364-3. PMID:26846225 [DOI] [PubMed] [Google Scholar]
- [3].Strom SC, Gramignoli R. Human amnion epithelial cells expressing HLA-G as novel cell-based treatment for liver disease. Hum Immunol. 2016;77:734–739. doi: 10.1016/j.humimm.2016.07.002. PMID:27476049 [DOI] [PubMed] [Google Scholar]
- [4].Schmidt D, Breymann C, Weber A, et al.. Umbilical cord blood derived endothelial progenitor cells for tissue engineering of vascular grafts. Ann Thorac Surg. 2004;78:2094–2098. doi: 10.1016/j.athoracsur.2004.06.052. PMID:15561042 [DOI] [PubMed] [Google Scholar]
- [5].Schmidt D, Achermann J, Odermatt B, et al.. Prenatally fabricated autologous human living heart valves based on amniotic fluid derived progenitor cells as single cell source. Circulation. 2007;116:I64–170. doi: 10.1161/CIRCULATIONAHA.106.681494. PMID:17846327 [DOI] [PubMed] [Google Scholar]
- [6].Schmidt D, Achermann J, Odermatt B, et al.. Cryopreserved amniotic fluid-derived cells: a lifelong autologous fetal stem cell source for heart valve tissue engineering. J Heart Valve Dis. 2008;17:446–455; discussion 455. PMID:18751475 [PubMed] [Google Scholar]
- [7].Weber B, Zeisberger SM, Hoerstrup SP. Prenatally harvested cells for cardiovascular tissue engineering: fabrication of autologous implants prior to birth. Placenta. 2011;32:S316–S319. doi: 10.1016/j.placenta.2011.04.001. PMID:21575988 [DOI] [PubMed] [Google Scholar]
- [8].Weber B, Emmert MY, Behr L, et al.. Prenatally engineered autologous amniotic fluid stem cell-based heart valves in the fetal circulation. Biomaterials. 2012;33:4031–4043. doi: 10.1016/j.biomaterials.2011.11.087. PMID:22421386 [DOI] [PubMed] [Google Scholar]
- [9].Kehl D, Weber B, Hoerstrup SP. Bioengineered living cardiac and venous valve replacements: current status and future prospects. Cardiovasc Pathol. 2016;25:300–305. doi: 10.1016/j.carpath.2016.03.001. PMID:27167776 [DOI] [PubMed] [Google Scholar]
- [10].Kang N-H, Hwang K-A, Kim SU, et al.. Potential antitumor therapeutic strategies of human amniotic membrane and amniotic fluid-derived stem cells. Cancer Gene Ther. 2012;19:517–522. doi: 10.1038/cgt.2012.30. PMID:22653384 [DOI] [PubMed] [Google Scholar]
- [11].Koike C, Zhou K, Takeda Y, et al.. Characterization of amniotic stem cells. Cell Reprogram. 2014;16:298–305. doi: 10.1089/cell.2013.0090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bork S, Pfister S, Witt H, et al.. DNA methylation pattern changes upon long-term culture and aging of human mesenchymal stromal cells. Aging Cell. 2010;9:54–63. doi: 10.1111/j.1474-9726.2009.00535.x. PMID:19895632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Liu X, Sun H, Qi J, et al.. Sequential introduction of reprogramming factors reveals a time-sensitive requirement for individual factors and a sequential EMT–MET mechanism for optimal reprogramming. Nat Cell Biol. 2013;15:829–838. doi: 10.1038/ncb2765. PMID:23708003 [DOI] [PubMed] [Google Scholar]
- [14].Lujan E, Zunder ER, Ng YH, et al.. Early reprogramming regulators identified by prospective isolation and mass cytometry. Nature. 2015;521:352–356. doi: 10.1038/nature14274. PMID:25830878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zunder E, Lujan E, Goltsev Y, et al.. A continuous molecular roadmap to iPSC reprogramming through progression analysis of single-cell mass cytometry. Cell Stem Cell. 2015;16:323–337. doi: 10.1016/j.stem.2015.01.015. PMID:25748935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kim K, Doi A, Wen B, et al.. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. PMID:20644535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Moschidou D, Mukherjee S, Blundell MP, et al.. Valproic acid confers functional pluripotency to human amniotic fluid stem cells in a transgene-free approach. Mol Ther. 2012;20:1953–1967. doi: 10.1038/mt.2012.117. PMID:22760542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Jackman CP, Shadrin IY, Carlson AL, et al.. Human cardiac tissue engineering: from pluripotent stem cells to heart repair. Curr Opin Chem Eng. 2015;7:57–64. doi: 10.1016/j.coche.2014.11.004. PMID:25599018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sirabella D, Cimetta E, Vunjak-Novakovic G “The state of the heart”: recent advances in engineering human cardiac tissue from pluripotent stem cells. Exp Biol Med Maywood NJ. 2015;240:1008–1018. doi: 10.1177/1535370215589910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Budniatzky I, Gepstein L. Concise review: reprogramming strategies for cardiovascular regenerative medicine: from induced pluripotent stem cells to direct reprogramming. Stem Cells Transl Med. 2014;3:448–457. doi: 10.5966/sctm.2013-0163. PMID:24591731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang A, Tang Z, Park I-H, et al.. Induced pluripotent stem cells for neural tissue engineering. Biomaterials. 2011;32:5023–5032. doi: 10.1016/j.biomaterials.2011.03.070. PMID:21514663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Workman MJ, Mahe MM, Trisno S, et al.. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat Med. 2016; doi: 10.1038/nm.4233. PMID:27869805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Trounson A, DeWitt ND. Pluripotent stem cells progressing to the clinic. Nat Rev Mol Cell Biol. 2016;17:194–200. doi: 10.1038/nrm.2016.10. PMID:26908143 [DOI] [PubMed] [Google Scholar]
- [24].Cai J, Li W, Su H, et al.. Generation of human induced pluripotent stem cells from umbilical cord matrix and amniotic membrane mesenchymal cells. J Biol Chem. 2010;285:11227–11234. doi: 10.1074/jbc.M109.086389. PMID:20139068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ge X, Wang I-NE, Toma I, et al.. Human amniotic mesenchymal stem cell-derived induced pluripotent stem cells may generate a universal source of cardiac cells. Stem Cells Dev. 2012;21:2798–2808. doi: 10.1089/scd.2011.0435. PMID:22530853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Slamecka J, Salimova L, McClellan S, et al.. Non-integrating episomal plasmid-based reprogramming of human amniotic fluid stem cells into induced pluripotent stem cells in chemically defined conditions. Cell Cycle. 2016;15:234–249. doi: 10.1080/15384101.2015.1121332. PMID:26654216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chen G, Gulbranson DR, Hou Z, et al.. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. PMID:21478862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Martí M, Mulero L, Pardo C, et al.. Characterization of pluripotent stem cells. Nat Protoc. 2013;8:223–253. doi: 10.1038/nprot.2012.154. PMID:23306458 [DOI] [PubMed] [Google Scholar]
- [29].Weinberger L, Ayyash M, Novershtern N, et al.. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat Rev Mol Cell Biol. 2016;17:155–169. doi: 10.1038/nrm.2015.28. PMID:26860365 [DOI] [PubMed] [Google Scholar]
- [30].Takashima Y, Guo G, Loos R, et al.. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. PMID:25215486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Pera MF, Reubinoff B, Trounson A. Human embryonic stem cells. J Cell Sci. 2000;113:5–10. PMID:10591620 [DOI] [PubMed] [Google Scholar]
- [32].Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. PMID:19497275 [DOI] [PubMed] [Google Scholar]
- [33].Dalton S. Linking the cell cycle to cell fate decisions. Trends Cell Biol. 2015;25:592–600. doi: 10.1016/j.tcb.2015.07.007. PMID:26410405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Müller F-J, Goldmann J, Löser P, et al.. A call to standardize teratoma assays used to define human pluripotent cell lines. Cell Stem Cell. 2010;6:412–414. doi: 10.1016/j.stem.2010.04.009. PMID:20452314 [DOI] [PubMed] [Google Scholar]
- [35].Müller F-J, Schuldt BM, Williams R, et al.. A bioinformatic assay for pluripotency in human cells. Nat Methods. 2011;8:315–317. doi: 10.1038/nmeth.1580. PMID:21378979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bian Q, Cahan P. Computational tools for stem cell biology. Trends Biotechnol. 2016;34:993–1009. doi: 10.1016/j.tibtech.2016.05.010. PMID:27318512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cahan P, Li H, Morris SA, et al.. CellNet: network biology applied to stem cell engineering. Cell. 2014;158:903–915. doi: 10.1016/j.cell.2014.07.020. PMID:25126793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Radley AH, Schwab RM, Tan Y, et al.. Assessment of engineered cells using CellNet and RNA-seq. Nat Protoc. 2017;12:1089–1102. doi: 10.1038/nprot.2017.022. PMID:28448485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Guo G, von Meyenn F, Santos F, et al.. Naive pluripotent stem cells derived directly from isolated cells of the human inner cell mass. Stem Cell Rep. 2016;6:437–446. doi: 10.1016/j.stemcr.2016.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Panopoulos AD, D'Antonio M, Benaglio P, et al.. iPSCORE: a resource of 222 iPSC lines enabling functional characterization of genetic variation across a variety of cell types. Stem Cell Rep. 2017;8:1086–1100. doi: 10.1016/j.stemcr.2017.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhang B, Georgiev O, Hagmann M, et al.. Activity of metal-responsive transcription factor 1 by toxic heavy metals and H2O2 in vitro is modulated by metallothionein. Mol Cell Biol. 2003;23:8471–8485. doi: 10.1128/MCB.23.23.8471-8485.2003. PMID:14612393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bell SG, Vallee BL. The metallothionein/thionein system: an oxidoreductive metabolic zinc link. Chem Bio Chem. 2009;10:55–62. doi: 10.1002/cbic.200800511. PMID:19089881 [DOI] [PubMed] [Google Scholar]
- [43].Wolford JL, Chishti Y, Jin Q, et al.. Loss of pluripotency in human embryonic stem cells directly correlates with an increase in nuclear zinc. PLoS ONE. 2010;5:e12308. doi: 10.1371/journal.pone.0012308. PMID:20808840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Müller F-J, Laurent LC, Kostka D, et al.. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. PMID:18724358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Choi J, Lee S, Mallard W, et al.. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat Biotechnol. 2015;33:1173–1181. doi: 10.1038/nbt.3388. PMID:26501951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Fernandez A, Huggins IJ, Perna L, et al.. The WNT receptor FZD7 is required for maintenance of the pluripotent state in human embryonic stem cells. Proc Natl Acad Sci. 2014;111:1409–1414. doi: 10.1073/pnas.1323697111. PMID:24474766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Beyer TA, Weiss A, Khomchuk Y, et al.. Switch Enhancers Interpret TGF-β and Hippo Signaling to Control Cell Fate in Human Embryonic Stem Cells. Cell Rep. 2013;5:1611–1624. doi: 10.1016/j.celrep.2013.11.021. PMID:24332857 [DOI] [PubMed] [Google Scholar]
- [48].Loewer S, Cabili MN, Guttman M, et al.. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. PMID:21057500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Santoni FA, Guerra J, Luban J. HERV-H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency. Retrovirology. 2012;9:111. doi: 10.1186/1742-4690-9-111. PMID:23253934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lu X, Sachs F, Ramsay L, et al.. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol. 2014;21:423–425. doi: 10.1038/nsmb.2799. PMID:24681886 [DOI] [PubMed] [Google Scholar]
- [51].Wang J, Xie G, Singh M, et al.. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature. 2014;516:405–409. doi: 10.1038/nature13804. PMID:25317556 [DOI] [PubMed] [Google Scholar]
- [52].Ohnuki M, Tanabe K, Sutou K, et al.. Dynamic regulation of human endogenous retroviruses mediates factor-induced reprogramming and differentiation potential. Proc Natl Acad Sci. 2014;111:12426–12431. doi: 10.1073/pnas.1413299111. PMID:25097266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Schopperle WM, DeWolf WC. The TRA-1-60 and TRA-1-81 Human Pluripotent Stem Cell Markers Are Expressed on Podocalyxin in Embryonal Carcinoma. Stem Cells. 2007;25:723–730. doi: 10.1634/stemcells.2005-0597. PMID:17124010 [DOI] [PubMed] [Google Scholar]
- [54].Chia N-Y, Chan Y-S, Feng B, et al.. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. PMID:20953172 [DOI] [PubMed] [Google Scholar]
- [55].Yu J, Hu K, Smuga-Otto K, et al.. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. PMID:19325077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Beers J, Gulbranson DR, George N, et al.. Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nat Protoc. 2012;7:2029–2040. doi: 10.1038/nprot.2012.130. PMID:23099485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Wickham H. ggplot2 [Internet]. New York, NY: Springer New York; 2009. [cited 2017July7]. Available from: http://link.springer.com/10.1007/978-0-387-98141-3 [Google Scholar]
- [58].Patro R, Duggal G, Love MI, et al.. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. PMID:28263959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinforma Oxf Engl. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224 [DOI] [PubMed] [Google Scholar]
- [60].Ritchie ME, Phipson B, Wu D, et al.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. PMID:25605792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Warnes G, Bolker B, Bonebakker L, et al.. gplots: various R programming tools for plotting data [Internet]. 2016. Available from: https://CRAN.R-project.org/package=gplots
- [62].Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. PMID:19131956 [DOI] [PubMed] [Google Scholar]
- [63].Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. PMID:19033363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Bushnell B. BBMap short read aligner. Univ Calif Berkeley Calif. [Internet] 2016; Available from: http://sourceforge.net/projects/bbmap [Google Scholar]
- [65].Yates A, Akanni W, Amode MR, et al.. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–D716. doi: 10.1093/nar/gkv1157. PMID:26687719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Li H, Handsaker B, Wysoker A, et al.. The Sequence Alignment/Map format and SAMtools. Bioinforma Oxf Engl. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinforma Oxf Engl. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- [68].Chen H. VennDiagram: generate high-resolution Venn and Euler plots, R Package [Internet]. 2016. Available from: https://CRAN.R-project.org/package=VennDiagram
- [69].Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research [Internet]. 2016. [cited 2017 Jul 7]; 4 Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4712774/ [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


