ABSTRACT
Clostridium difficile is a leading cause of nosocomial infections, causing disease that ranges from mild diarrhea to potentially fatal colitis. A variety of surface proteins, including flagella, enable C. difficile colonization of the intestine. Once in the intestine, toxigenic C. difficile secretes two glucosylating toxins, TcdA and TcdB, which elicit inflammation and diarrheal disease symptoms. Regulation of colonization factors and TcdA and TcdB is an intense area of research in C. difficile biology. A recent publication from our group describes a novel regulatory mechanism that mediates the ON/OFF expression of co-regulated virulence factors of C. difficile, flagella and toxins. Herein, we review key findings from our work, present new data, and speculate the functional consequence of the ON/OFF expression of these virulence factors during host infection.
Keywords: Clostridium difficile, flagella, toxins, recombination, RecV
Discovery of flagellum and toxin phase variation in C. difficile
Clostridium difficile infections (CDI) are a cause of significant morbidity and mortality in industrialized countries. Diarrheal disease caused by CDI ranges from mild inflammatory diarrhea to pseudomembranous colitis, a severe condition characterized by pathologic lesions on the mucosal surface of colonic tissue. Antibiotic treatment is the leading risk factor for CDI, because the antibiotics disrupt the intestinal microbial community that is usually protective against CDI.1 In response to certain bile salts in the intestine, C. difficile spores germinate into actively growing vegetative cells.2,3 The vegetative bacteria secrete the protein toxins TcdA and TcdB, which are largely responsible for the inflammation, intestinal pathology, and diarrheal disease symptoms seen in CDI.4
C. difficile surface proteins mediate adherence to other microbial species, mucus, and intestinal cells within the colon for colonization.5 The peritrichous flagella produced by C. difficile aid in motility and modulate colonization in an animal model of infection.6 In addition, TcdA and TcdB production is linked to flagellum biosynthesis.7-10 SigD (σD), an alternative sigma factor encoded within the early stage flagellar gene operon (flgB operon), is essential for transcription of late stage flagellar genes, and also positively regulates transcription of the toxin genes (Fig. 1).11,12 Therefore, factors that regulate expression of the flgB operon not only control bacterial motility, they are also likely to impact toxin production and therefore virulence of C. difficile. The 5′ untranslated region (UTR) of the flgB operon mRNA contains a riboswitch (Cd1) specific to the nucleotide second messenger cyclic diguanylate (c-di-GMP).13,14 C-di-GMP binding to Cd1 causes premature transcription termination within the first 160 nucleotides of the 5′ UTR of the flgB operon mRNA, inhibiting flagellar gene expression, motility, and toxin production. Because the flgB 5′ UTR is 498 nucleotides, we postulated that an additional cis-acting regulatory element downstream of Cd1 could control flagellar and toxin gene expression.
Figure 1.
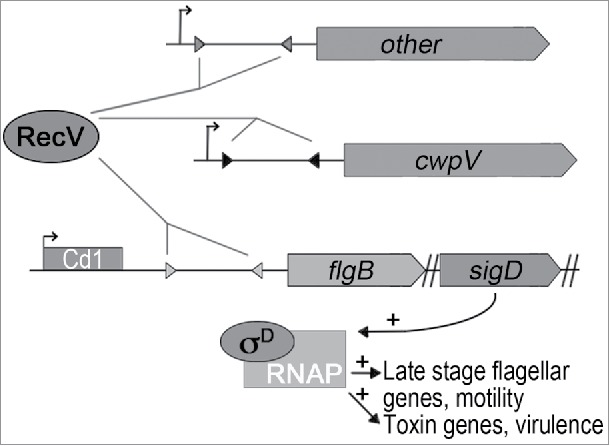
Diagram of the regulatory scheme for flagellar and toxin phase variation in C. difficile. A DNA invertible element, which we termed the “flagellar switch,” is located in the 5′ UTR of the flgB operon and controls transcription of structural and regulatory genes necessary for flagellum biosynthesis and motility. One orientation of the flagellar switch, but not the other, is permissive for downstream gene expression. In addition, the flagellar switch regulates toxin production by controlling the expression of sigD, which located in the flgB operon and encodes a sigma factor, σD, that promotes toxin gene transcription. RecV, a tyrosine recombinase, catalyzes DNA inversion in both directions at the flagellar and cwpV switches, and also impacts one or more unidentified genetic switches that affect colony morphology.
We determined that flagellum and toxin production is subject to phase variation via DNA inversion in C. difficile R20291,15 a representative 027 strain isolated in 2006 from an epidemic of CDI in the U.K.16 Between Cd1 and the first open reading frame of the flgB operon lies a 154 bp invertible DNA element flanked by 21 bp imperfect inverted repeats (Fig. 1). We termed the invertible element the “flagellar switch.” Based on genomes currently available on public databases, the flagellar switch sequence and flanking inverted repeats are conserved in all C. difficile genomes with flagellar biosynthesis genes. The flagellar switch is capable of inversion in at least two ribotypes, 027 and 017, under the conditions tested.
Our study showed that C. difficile with the flagellar switch oriented according to the published genome (FN545816.1) expresses and produces peritrichous flagella, engages in swimming motility, and secretes the glucosylating toxins, and is thus phase ON (“flg ON”). Conversely, bacteria with the sequence oriented in the inverse orientation are non-flagellated, non-motile, and attenuated for toxin secretion, and are thus phase OFF (“flg OFF”). The tyrosine recombinase RecV was determined to catalyze inversion of the flagellar switch, and mutation of recV in C. difficile showed that RecV is required for flagellar switch inversion. RecV was previously shown to control inversion of a genetic switch upstream of cwpV, which encodes a cell wall protein involved in autoaggregation and resistance to certain bacteriophage.17-19 Interestingly, the flagellar and cwpV switches have seemingly disparate sequences in the inverted repeats and surrounding DNA. The regulatory feature present in the flagellar switch that controls downstream flagellar gene expression has been elusive, but involves a mechanism operating post-transcription initiation. Below we discuss additional results and interpretations based on the original study.15
Mechanism of phase variable gene regulation
Phase variation occurs by multiple mechanisms in diverse mucosal bacterial pathogens, including site-specific recombination, general homologous recombination, slip strand mispairing, and DNA methylation.20 For site-specific recombination, a recombinase recognizes a specific DNA sequence and catalyzes strand exchange and DNA inversion. The orientation of the DNA sequence dictates whether or not a nearby gene is expressed. Classically, the invertible DNA element contains a promoter to transcriptionally control an adjacent gene or operon. The best-studied example of this mechanism is the Type I fimbrial biosynthetic operon in E. coli. Here, the invertible element fimS lies upstream of fimA, which encodes the fimbrial subunit. Within fimS is a promoter that drives transcription of fimA when properly oriented.21 Using a series of transcriptional reporters and growth conditions known to be permissive for flagellar gene expression, we found that the flagellar switch in C. difficile does not contain a promoter, and expression of the flgB operon relies on the upstream σA-dependent promoter.15
Phase variable expression of cwpV occurs through a post-transcriptional mechanism.17 In one orientation of the cwpV switch, the leader mRNA adopts a structure allowing transcriptional elongation into the cwpV coding sequence. In the opposite orientation, the leader mRNA forms an intrinsic terminator that causes premature termination of transcription, precluding transcription of the cwpV coding sequence and CwpV biosynthesis. To determine whether a similar cis-acting mechanism occurs via the flagellar switch, we evaluated the same transcriptional reporters in Bacillus subtilis, a spore-forming obligate aerobe, where we postulated that C. difficile-specific trans-acting regulatory factors would be absent. In contrast with results from C. difficile, we found that the flg ON and OFF reporters yielded equivalent activity in B. subtilis, indicating that no intrinsic transcription terminator is present in the flagellar switch. Northern blotting failed to detect evidence of premature transcript termination, suggesting that factor-dependent termination also does not occur.15 However, the low sensitivity of northern blotting using a Digoxigenin-labeled probe does not eliminate the possibility of factor-dependent termination.15
We speculate two possible mechanisms: small RNA-mediated silencing or Rho-dependent transcriptional termination. For the small RNA mechanism, based on the conditions tested, the non-coding RNA would need to be constitutively expressed and anneal to the flg OFF RNA, but not the flg ON RNA. Exclusion from the flg ON RNA could be due to RNA structure or the lack of a specific sequence when the switch RNA is in the ON orientation. The small RNA would likely promote the release of RNA polymerase from the transcription elongation complex from the flg OFF mRNA. It would be energetically unfavorable to produce a full length mRNA for the 23 kb flgB operon only to rapidly degrade it, so mechanisms involving destabilization of the mRNA or targeting of the mRNA for degradation are unlikely. For the second possible mechanism, Rho might recognize a sequence exclusive to the flg OFF RNA, or the flg ON RNA might form a structure that occludes Rho binding or translocation. Identifying a new post-transcription initiation regulatory mechanism for phase variation in C. difficile may reveal a widespread regulatory mechanism in other bacterial pathogens.
Strain dependent differences in flagellar phase variation
The C. difficile strain 630 (ribotype 012) was isolated in 1982 from a patient with severe CDI in Switzerland.22 Erythromycin-sensitive derivatives of 630, 630Δerm and 630E, are more amenable to currently available genetic tools and used most often to study C. difficile physiology and virulence.23,24 However, unlike the R20291 strain in which we could detect the flagellar switch in both ON and OFF orientations, only the ON orientation was detected in 630Δerm and in the 630 parent (data not shown).15 These results suggest that the flagellar switch is locked in these strains. The inability of the flagellar switch to invert to the flg OFF orientation in 630 and 630Δerm could be due to the shorter inverted repeats flanking the flagellar switches in these strains, with 20 bp instead of the 21 bp evident in all of the other available published genomes of C. difficile strains with a flagellar switch. Inverted repeat length has been demonstrated to affect recombination at the cwpV switch in several C. difficile strains.17 Reduced inverted repeat length could similarly prevent inversion of the flagellar switch in 630 and 630Δerm. For example, inverted repeat length might affect DNA supercoiling at the flagellar switch, inhibiting recombinase and/or accessory factor binding and subsequent catalysis of DNA inversion.25-27
Recent work suggests that the flagellar switch in 630 is capable of inversion from flg ON to OFF in some condition(s). Collery et al. found the flagellar switch in the OFF orientation in 630E (also referred to as JIR8094), another derivative of 630.28 To obtain 630E, C. difficile 630 was serially passaged an undisclosed number of times on non-selective agar medium.24 In the process, 630E acquired a non-motile and less toxigenic phenotype. We previously reported that ectopic expression of sigD in 630E was sufficient to rescue toxin production, consistent with mutations that affect expression of sigD in the flgB operon.11 Collery et al. inverted the flagellar switch to that in 630 and 630Δerm, which would presumably restore flagellar motility and toxin production.28,29 However, the 630E mutant strain remained non-motile and attenuated for production of both toxins.28 The lack of restored motility and toxinogenesis may be due to inversion of only 150 bp of the flagellar switch, whereas we experimentally determined that 154 bp comprise the flagellar switch.15 Alternatively, several other genetic differences between 630E and its 630 parent have been identified and may account for abrogated motility and toxin production in 630E strain. Repairing the additional identified genetic polymorphisms in 630E, such as those in genes encoding a topoisomerase, RNA helicase, or oligopeptide transporter,28 may restore flagellum and toxin biosynthesis. Ultimately, studying the differences between 630 and 630E may help reveal the mechanism by which the orientation of the flagellar switch modulates downstream gene expression.
RecV-dependent changes in C. difficile colony morphology
Differences in the expression of cell surface structures can affect how bacteria assemble into a colony, and changes to gross colony morphology can provide insight into virulence.30 The phase variable expression of cwpV influences colony morphology.18 C. difficile CwpV phase ON (“cwpV ON”) colonies exhibit a densely packed morphology with smooth edges, whereas cwpV phase OFF (“cwpV OFF”) bacteria yield dispersed, ruffled colonies. Other genes have also been implicated in colony morphology development.31,32 In pursuit of obtaining enriched flagellar phase variant populations, we observed smooth, circular (SC) colonies and rough, filamentous (RF) colonies, but colony morphology was not strongly associated with flagellar switch orientation.15 To identify the genes responsible for the SC and RF colony morphologies, we evaluated a panel of C. difficile mutants. We noted that a C. difficile R20291 sigD mutant, which is aflagellate and non-motile, can form both SC and RF colonies, indicating that colony morphology is independent of flagellum biosynthesis and motility (Fig. 2A). Mutating recV resulted in SC colonies in both flagellar phase locked ON and OFF strains, indicating that the RF colony morphology is independent of flagellar switch orientation but dependent on RecV (Fig. 2B). Furthermore, the RF colony morphology is independent of the cwpV switch orientation because recV mutants with the cwpV switch locked ON and OFF both yield SC colonies.
Figure 2.
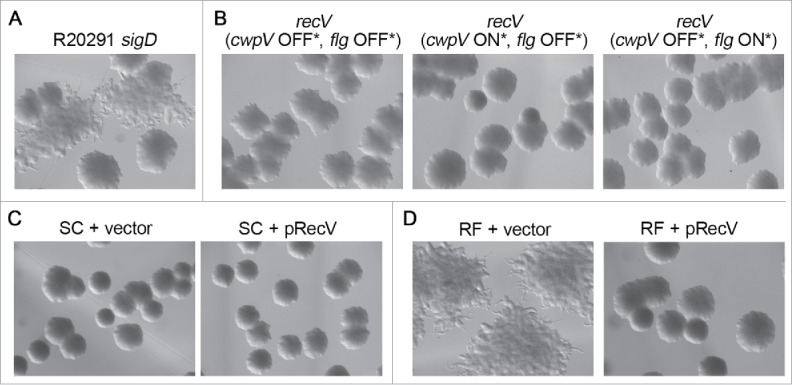
RecV controls a genetic switch responsible for the RF morphology in a σD-independent manner. Light microscopy images of C. difficile R20291 colonies grown on BHIS agar. (A) A R20291 sigD mutant develops smooth circular (SC) and rough filamentous (RF) colonies (B) All recV mutants yield SC colonies, regardless of flagellar and cwpV switch orientations. Switch genotypes are indicated in parentheses, with asterisks denoting phase-locked orientations. (C) Bacteria from SC colonies transformed with a plasmid for ectopic recV expression (pRecV, pRT1529) or the vector control (pRT1611) retain the SC colony morphology of the parent isolate. (D) Bacteria from RF colonies transformed with the vector control develop the parental RF colony morphology, while overexpression of recV (pRecV) results in conversion to the SC morphology.
It remains possible that RecV controls DNA inversion of another switch(es) controlling phase variable expression of genes mediating SC and RF colony morphology. If so, generating independent recV mutants could give rise to exclusively SC colonies (as observed) or RF colonies depending on the orientation of such a switch at the time of recV inactivation. To determine if RecV modulates colony morphology, we transformed C. difficile from SC and RF colonies with a plasmid allowing expression of recV or the vector control (Fig. 2C and 2D).15,33 Bacteria from SC and RF colonies bearing vector retained their respective colony morphologies, and bacteria from SC colonies expressing recV retained the SC colony morphology. In contrast, bacteria from RF colonies expressing recV yielded SC colonies. From these data, we surmise that RecV mediates recombination at a genetic switch upstream of genes responsible for the RF colony morphology.
Few studies have evaluated the ultrastructure of C. difficile colonies.34 Our data suggest that the production of a surface protein or polysaccharides is subject to RecV-dependent phase variation and impact colony morphology. This includes three different surface polysaccharides, named PSI-PSIII, as well as the export machinery and cell wall proteins that anchor these polysaccharides to the surface.35,36 Alternatively, RecV may indirectly affect colony morphology by mediating phase variation of a protein controlling the transcription, translation, or localization of a surface protein or polysaccharide. Two other invertible DNA sequences have been predicted based on the comparison of genomes of multiple C. difficile strains.37 These are located upstream of CDR20291_1514 and CDR20291_0685, which encode functional c-di-GMP phosphodiesterases.38,39 The nucleotide second messenger c-di-GMP has been shown to affect colony morphology in multiple bacteria,40,41 and may do so in C. difficile. The function and fitness contribution of the RecV-dependent phase variable surface proteins or polysaccharides are under investigation.
In vivo contribution of RecV-dependent phase variation
The role of phase variable C. difficile flagella and toxins during infection remains to be determined. Ideally, such studies would use phase-locked mutants, so that the inability to switch between flg ON and OFF states can be assessed. A recV mutant is phase-locked, but we refrained from evaluating this mutant in animal models of C. difficile disease because of the likely pleiotropic effects of the mutation. RecV controls inversion of both the flagellar and cwpV switches,15,17,18 and impacts at least one other locus. Thus, combinatorial genetic switch mutants through inactivation of recV would be required to assess the effect of each individual switch on C. difficile virulence. An alternative approach for generating phase-locked mutants is site-directed mutagenesis of the inverted repeats, a method successful for phase locking the fimbrial switch in E. coli,42 which would allow us to lock the flagellar switch without affecting inversion of other switches.
Gunther et al. found that the ability of uropathogenic Escherichia coli (UPEC) to phase vary type I fimbriae biosynthesis through inversion of fimS affects colonization.42 A fimS phase-locked OFF mutant was attenuated for colonization in a mouse model of urinary tract infection compared to wild type and fimS phase-locked ON strains, consistent with a known role for these fimbriae in UPEC virulence.42 Flagellar filaments contribute to C. difficile R20291 colonization during infection of mice,9 so flagellar phase locked C. difficile similarly may display colonization phenotypes consistent with the ON/OFF status of the flagellar switch and flagellum biosynthesis. However, the fitness benefit of adherence comes at a cost to flg ON bacteria: both flagellin and the glucosylating toxins are immunostimulatory. C. difficile flagellin stimulates host Toll-like receptor 5 (TLR5), which activates signaling pathways leading to the secretion of IL-8, a neutrophil chemokine, in epithelial cells.43,44 The glucosylating toxins stimulate apoptosis, necrosis, or pyroptosis depending on the host cell type and model of infection.45-48 Batah et al. found that flagellin and the toxins synergistically elicit a robust inflammatory response from the intestinal epithelium during infection of mice with R20291, whereas the individual antigens alone were not sufficient for eliciting such a response.49 Thus, while the flg ON state may be advantageous for establishing a C. difficile infection and disease, the flg OFF state may be selected at later stages of infection by avoidance of immune clearance.
The role for CwpV in C. difficile in the context of the anaerobic host intestine must also be considered. CwpV promotes bacterial autoaggregation in vitro, suggesting a role for CwpV in host colonization.18 CwpV also promotes resistance to bacteriophage predation by reducing phage adsorption and phage tail spike DNA injection.19 Thus, cwpV ON bacteria could resist phage attack in the intestine. Given the functional contributions of CwpV to C. difficile in vitro, the potential contribution of cwpV OFF bacteria to CDI is puzzling. Bacteria that are cwpV OFF may be less likely to generate an antibody response, and/or they could disperse from an unfavorable colonization site during infection.
Because RecV mediates inversion of multiple sequences, it is tempting to speculate that RecV coordinates their production, allowing synergism or antagonism between those factors. Considering flagella and CwpV together, these may have complementary roles in colonization, with a cwpV ON phenotype compensating for a lack of flagella in a flg OFF bacterium, and vice versa. Although antibodies against CwpV can be recovered after infection, CwpV might still be less immunostimulatory compared to flagellin and the toxins.50 In bacteria that are phase ON for both surface structures, the adherence of these bacteria could be greater than individually if they engage distinct receptors. However, in each case, the cost of immunogenicity will play a role in the overall survival of the respective bacteria.
Several other issues complicate the ability to predict the fitness outcome during infection. CwpV could alter flagellar function, although results in C. difficile 630 contradict this assertion: motility and flagellin production are unchanged in bacteria overexpressing CwpV.18 The SigD regulon (under the control of the flagellar switch) has several predicted and functionally characterized adhesins that may synergize with CwpV and flagella for colonization.12 Finally, additional RecV-dependent switch(es) may influence C. difficile virulence. Importantly, there is no evidence to date that RecV preferentially recognizes one switch orientation over the other for any of its targets, so presumably the abundance of phase ON and OFF of each target in a population relies on external selective pressures.
Concluding remarks
The discovery and characterization of phase variable production of flagella and toxins in C. difficile has provided new knowledge of a cis-acting regulatory feature for controlling these virulence factors, but has also left many unanswered questions.
Regulation at the flagellar switch occurs post-transcription initiation and requires an unidentified trans-acting regulatory factor. What is that factor, and how does it terminate flagellar gene expression in flg OFF, but not flg ON bacteria? RecV appears to control multiple switches. What is the phase variable surface protein or polysaccharide controlled by RecV and affecting colony morphology? What is the sequence that RecV recognizes among the switches? Is a single nucleotide deletion, as seen in 630E, sufficient to inhibit DNA inversion? Could an accessory factor(s) help RecV discriminate between its targets, leading to different rates of inversion? If so, what is the functional outcome to C. difficile physiology? Lastly, the contribution of RecV-mediated phase variation of flagella, toxins, autoaggregation, and phage resistance to fitness in a host has yet to be examined. Potential interactions between these phenotypes could substantially alter the course of CDI and transmission to a new host. Defining the mechanism controlling phase variable virulence factors in C. difficile could expose new targets for the development of therapeutic agents.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Robert W. McKee for helpful feedback on this manuscript.
Funding
This work was supported by NIH awards R01-AI107029 to R.T. and F31-AI120613 to B.R.A-F.
References
- [1].Theriot CM, Young VB. Interactions between the gastrointestinal microbiome and clostridium difficile. Annu Rev Microbiol. 2015;69:445-61. doi: 10.1146/annurev-micro-091014-104115. PMID:26488281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for clostridium difficile spores. J Bacteriol. 2008;190:2505-12. doi: 10.1128/JB.01765-07. PMID:18245298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Paredes-Sabja D, Shen A, Sorg JA. Clostridium difficile spore biology: Sporulation, germination, and spore structural proteins. Trends Microbiol. 2014;22:406-16. doi: 10.1016/j.tim.2014.04.003. PMID:24814671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Pruitt RN, Lacy DB. Toward a structural understanding of Clostridium difficile toxins A and B. Front Cell Inf Microbio. 2012;2:28. doi: 10.3389/fcimb.2012.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Janoir C. Virulence factors of Clostridium difficile and their role during infection. Anaerobe. 2016;37:13-24. doi: 10.1016/j.anaerobe.2015.10.009. PMID:26596863 [DOI] [PubMed] [Google Scholar]
- [6].Stevenson E, Minton NP, Kuehne SA. The role of flagella in Clostridium difficile pathogenicity. Trends Microbiol. 2015;23:275-82. doi: 10.1016/j.tim.2015.01.004. PMID:25659185 [DOI] [PubMed] [Google Scholar]
- [7].Dingle TC, Mulvey GL, Armstrong GD. Mutagenic analysis of the clostridium difficile flagellar proteins, FliC and FliD, and their contribution to virulence in hamsters. Infect Immun. 2011;79:4061-7. doi: 10.1128/IAI.05305-11. PMID:21788384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Aubry A, Hussack G, Chen W, KuoLee R, Twine SM, Fulton KM, Foote S, Carrillo CD, Tanha J, Logan SM. Modulation of toxin production by the flagellar regulon in clostridium difficile. Infect Immun. 2012;80:3521-32. doi: 10.1128/IAI.00224-12. PMID:22851750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Baban ST, Kuehne SA, Barketi-Klai A, Cartman ST, Kelly ML, Hardie KR, Kansau I, Collignon A, Minton NP. The role of flagella in clostridium difficile pathogenesis: ?Comparison between a non-epidemic and an epidemic strain. PLoS One. 2013;8:e73026. doi: 10.1371/journal.pone.0073026. PMID:24086268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Barketi-Klai A, Monot M, Hoys S, Lambert-Bordes S, Kuehne SA, Minton N, Collignon A, Dupuy B, Kansau I. The flagellin FliC of clostridium difficile is responsible for pleiotropic gene regulation during in vivo infection. PLoS One. 2014;9:e96876. doi: 10.1371/journal.pone.0096876. PMID:24841151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].McKee RW, Mangalea MR, Purcell EB, Borchardt EK, Tamayo R. The second messenger cyclic Di-GMP ?regulates clostridium difficile toxin production by controlling expression of sigD. J Bacteriol. 2013;195:5174-85. doi: 10.1128/JB.00501-13. PMID:24039264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Meouche El I, Peltier J, Monot M, Soutourina O, Pestel-Caron M, Dupuy B, Pons JL. Characterization of the SigD regulon of C. difficile and its positive control of toxin production through the regulation of tcdR. PLoS One. 2013;8:e83748. doi: 10.1371/journal.pone.0083748. PMID:24358307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bordeleau E, Purcell EB, Lafontaine DA, Fortier LC, Tamayo R, Burrus V Cyclic di-GMP riboswitch-regulated type IV pili contribute to aggregation of Clostridium difficile. J Bacteriol. 2015;197:819-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Purcell EB, Tamayo R. Cyclic diguanylate signaling in Gram-positive bacteria. FEMS Microbiol Rev. 2016;40:753-73. doi: 10.1093/femsre/fuw013. PMID:27354347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Anjuwon-Foster BR, Tamayo R. A genetic switch controls the production of flagella and toxins in Clostridium difficile. PLoS Genet. 2017;13:e1006701. doi: 10.1371/journal.pgen.1006701. PMID:28346491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, Lawley TD, Sebaihia M, Quail MA, Rose G, et al.. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 2009;10:R102. doi: 10.1186/gb-2009-10-9-r102. PMID:19781061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Emerson JE, Reynolds CB, Fagan RP, Shaw HA, Goulding D, Fairweather NF. A novel genetic switch controls phase variable expression of CwpV, a Clostridium difficile cell wall protein. Mol Microbiol. 2009;74:541-56. doi: 10.1111/j.1365-2958.2009.06812.x. PMID:19656296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Reynolds CB, Emerson JE, la Riva de L, Fagan RP, ?Fairweather NF. The clostridium difficile cell wall protein CwpV is antigenically variable between strains, but exhibits conserved aggregation-promoting function. PLoS Pathog. 2011;7:e1002024. doi: 10.1371/journal.ppat.1002024. PMID:21533071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sekulovic O, Ospina Bedoya M, Fivian-Hughes AS, ?Fairweather NF, Fortier LC. The Clostridium difficile cell wall protein CwpV confers phase-variable phage resistance. Mol Microbiol. 2015;98:329-42. doi: 10.1111/mmi.13121. PMID:26179020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].van der Woude MW, Bäumler AJ. Phase and antigenic variation in bacteria. Clin Microbiol Rev. 2004;17:581-611. doi: 10.1128/CMR.17.3.581-611.2004. PMID:15258095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Abraham JM, Freitag CS, Clements JR, Eisenstein BI. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc Natl Acad Sci U S A. 1985;82:5724-7. doi: 10.1073/pnas.82.17.5724. PMID:2863818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeño-Tárraga AM, Wang H, et al.. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet. 2006;38:779-86. doi: 10.1038/ng1830. PMID:16804543 [DOI] [PubMed] [Google Scholar]
- [23].Hussain HA, Roberts AP, Mullany P. Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Δerm) and demonstration that the conjugative transposon Tn916ΔE enters the genome of this strain at multiple sites. J Med Microbiol. 2005;54:137-41. doi: 10.1099/jmm.0.45790-0. PMID:15673506 [DOI] [PubMed] [Google Scholar]
- [24].O'Connor JR, Lyras D, Farrow KA, Adams V, Powell DR, Hinds J, Cheung JK, Rood JI. Construction and analysis of chromosomal Clostridium difficile mutants. Mol Microbiol. 2006;61:1335-51. doi: 10.1111/j.1365-2958.2006.05315.x. PMID:16925561 [DOI] [PubMed] [Google Scholar]
- [25].Dove SL, Dorman CJ. The site-specific recombination system regulating expression of the type 1 fimbrial subunit gene of Escherichia coli is sensitive to changes in DNA supercoiling. Mol Microbiol. 1994;14:975-88. doi: 10.1111/j.1365-2958.1994.tb01332.x. PMID:7715458 [DOI] [PubMed] [Google Scholar]
- [26].Kelly A, Conway C, O Cróinín T, Smith SGJ, Dorman CJ. DNA supercoiling and the Lrp protein determine the directionality of fim switch DNA inversion in Escherichia coli K-12. J Bacteriol. 2006;188:5356-63. doi: 10.1128/JB.00344-06. PMID:16855224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Corcoran CP, Dorman CJ. DNA relaxation-dependent phase biasing of the fim genetic switch in Escherichia coli depends on the interplay of H-NS, IHF and LRP. Mol Microbiol. 2009;74:1071-82. doi: 10.1111/j.1365-2958.2009.06919.x. PMID:19889099 [DOI] [PubMed] [Google Scholar]
- [28].Collery MM, Kuehne SA, McBride SM, Kelly ML, Monot M, Cockayne A, Dupuy B, Minton NP. What's a SNP between friends: The influence of single nucleotide polymorphisms on virulence and phenotypes of Clostridium difficile strain 630 and derivatives. Virulence. 2016:1-15. [Epub ahead of print] doi: 10.1080/21505594.2016.1237333. PMID:27652799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ng YK, Ehsaan M, Philip S, Collery MM, Janoir C, Collignon A, Cartman ST, Minton NP. Expanding the repertoire of gene tools for precise manipulation of the Clostridium difficile genome: Allelic exchange using pyrE alleles. PLoS One. 2013;8:e56051. doi: 10.1371/journal.pone.0056051. PMID:23405251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Weiser JN, Austrian R, Sreenivasan PK, Masure HR. Phase variation in pneumococcal opacity: Relationship between colonial morphology and nasopharyngeal colonization. Infect Immun. 1994;62:2582-9. PMID:8188381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].la Riva de L, Willing SE, Tate EW, Fairweather NF. Roles of cysteine proteases Cwp84 and Cwp13 in biogenesis of the cell wall of Clostridium difficile. J Bacteriol. 2011;193:3276-85. doi: 10.1128/JB.00248-11. PMID:21531808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kirby JM, Ahern H, Roberts AK, Kumar V, Freeman Z, Acharya KR, Shone CC. Cwp84, a surface-associated cysteine protease, plays a role in the maturation of the surface layer of Clostridium difficile. J Biol Chem. 2009;284:34666-73. doi: 10.1074/jbc.M109.051177. PMID:19808679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Fagan RP, Fairweather NF. Clostridium difficile has two parallel and essential sec secretion systems. J Biol Chem. 2011; 286:27483-93. doi: 10.1074/jbc.M111.263889. PMID:21659510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lipovsek S, Leitinger G, Rupnik M. Ultrastructure of clostridium difficile colonies. Anaerobe. 2013;24:66-70. doi: 10.1016/j.anaerobe.2013.09.014. PMID:24120350 [DOI] [PubMed] [Google Scholar]
- [35].Willing SE, Candela T, Shaw HA, Seager Z, Mesnage S, Fagan RP, Fairweather NF. Clostridium difficile surface proteins are anchored to the cell wall using CWB2 motifs that recognise the anionic polymer PSII. Mol Microbiol. 2015;96:596-608. doi: 10.1111/mmi.12958. PMID:25649385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chu M, Mallozzi MJG, Roxas BP, Bertolo L, Monteiro MA, Agellon A, Viswanathan VK, Vedantam G. A clostridium difficile cell wall glycopolymer locus influences bacterial shape, polysaccharide production and virulence. PLoS Pathog. 2016;12:e1005946. doi: 10.1371/journal.ppat.1005946. PMID:27741317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Stabler RA, Valiente E, Dawson LF, He M, Parkhill J, Wren BW. In-depth genetic analysis of Clostridium difficile PCR-ribotype 027 strains reveals high genome fluidity including point mutations and inversions. Gut Microbes. 2010;1:269-76. doi: 10.4161/gmic.1.4.11870. PMID:21327033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bordeleau E, Fortier L-C, Malouin F, Burrus V. c-di-GMP turn-over in clostridium difficile is controlled by a plethora of diguanylate cyclases and phosphodiesterases. PLoS Genet. 2011;7:e1002039. doi: 10.1371/journal.pgen.1002039. PMID:21483756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gao X, Dong X, Subramanian S, Matthews PM, Cooper CA, Kearns DB, Dann CE. Engineering of Bacillus subtilis strains to allow rapid characterization of heterologous diguanylate cyclases and phosphodiesterases. Appl Environ Microbiol. 2014;80:6167-74. doi: 10.1128/AEM.01638-14. PMID:25085482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1-52. doi: 10.1128/MMBR.00043-12. PMID:23471616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Beyhan S, Odell LS, Yildiz FH. Identification and characterization of cyclic diguanylate signaling systems controlling rugosity in Vibrio cholerae. J Bacteriol. 2008;190:7392-405. doi: 10.1128/JB.00564-08. PMID:18790873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gunther NW, Snyder JA, Lockatell V, Blomfield I, Johnson DE, Mobley HLT. Assessment of virulence of uropathogenic Escherichia coli type 1 fimbrial mutants in which the invertible element is phase-locked on or off. Infect Immun. 2002;70:3344-54. doi: 10.1128/IAI.70.7.3344-3354.2002. PMID:12065472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Batah J, Denève-Larrazet C, Jolivot PA, Kuehne S, ?Collignon A, Marvaud JC, Kansau I. Clostridium difficile flagella predominantly activate TLR5-linked NF-κB pathway in epithelial cells. Anaerobe. 2016;38:116-24. doi: 10.1016/j.anaerobe.2016.01.002. PMID:26790921 [DOI] [PubMed] [Google Scholar]
- [44].Yoshino Y, Kitazawa T, Ikeda M, Tatsuno K, Yanagimoto S, Okugawa S, Yotsuyanagi H, Ota Y. Clostridium difficile flagellin stimulates toll-like receptor 5, and toxin B promotes flagellin-induced chemokine production via TLR5. Life Sci. 2013;92:211-7. doi: 10.1016/j.lfs.2012.11.017. PMID:23261530 [DOI] [PubMed] [Google Scholar]
- [45].Ng J, Hirota SA, Gross O, Li Y, Ulke-Lemee A, Potentier MS, Schenck LP, Vilaysane A, Seamone ME, Feng H, et al.. Clostridium difficile toxin-induced inflammation and intestinal injury are mediated by the inflammasome. Gastroenterology. 2010;139:542-552.e1-3. doi: 10.1053/j.gastro.2010.04.005. PMID:20398664 [DOI] [PubMed] [Google Scholar]
- [46].Xu H, Yang J, Gao W, Li L, Li P, Zhang L, Gong YN, Peng X, Xi JJ, Chen S, et al.. Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature. 2014;513:237-41. doi: 10.1038/nature13449. PMID:24919149 [DOI] [PubMed] [Google Scholar]
- [47].Warny M, Keates AC, Keates S, Castagliuolo I, Zacks JK, Aboudola S, Qamar A, Pothoulakis C, LaMont JT, Kelly CP. p38 MAP kinase activation by Clostridium difficile toxin A mediates monocyte necrosis, IL-8 production, and enteritis. J Clin Invest. 2000;105:1147-56. doi: 10.1172/JCI7545. PMID:10772660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Farrow MA, Chumbler NM, Lapierre LA, Franklin JL, Rutherford SA, Goldenring JR, Lacy DB. Clostridium difficile toxin B-induced necrosis is mediated by the host epithelial cell NADPH oxidase complex. Proc Natl Acad Sci USA. 2013;110:18674-9. doi: 10.1073/pnas.1313658110. PMID:24167244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Batah J, Kobeissy H, Bui Pham PT, Denève-Larrazet C, Kuehne S, Collignon A, Janoir-Jouveshomme C, Marvaud JC, Kansau I. Clostridium difficile flagella induce a pro-inflammatory response in intestinal epithelium of mice in cooperation with toxins. Sci Rep. 2017;7:3256. doi: 10.1038/s41598-017-03621-z. PMID:28607468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wright A, Drudy D, Kyne L, Brown K, Fairweather NF. Immunoreactive cell wall proteins of Clostridium difficile identified by human sera. J Med Microbiol. 2008;57:750-6. doi: 10.1099/jmm.0.47532-0. PMID:18480333 [DOI] [PubMed] [Google Scholar]


