Abstract
Background and Purpose:
Candida albicans is the most common Candida species (sp.) isolated from fungal infections. Azole resistance in Candida species has been considerably increased in the last decades. Given the toxicity of the antimicrobial drugs, resistance to antifungal agents, and drug interactions, the identification of new antifungal agents seems essential. In this study, we assessed the antifungal effects of biogenic selenium nanoparticles on C. albicans and determined the expression of ERG11 and CDR1 genes.
Materials and Methods:
Selenium nanoparticles were synthesized with Bacillus sp. MSH-1. The ultrastructure of selenium nanoparticles was evaluated with a transmission electron microscope. The antifungal susceptibility test was performed according to the modified Clinical and Laboratory Standards Institute M27-A3 standard protocol. The expression levels of the CDR1 and ERG11 genes were analyzed using the quantitative real-time polymerase chain reaction (PCR) assay.
Results:
The azole-resistant C. albicans and wild type C. albicans strains were inhibited by 100 and 70 µg/mL of selenium nanoparticle concentrations, respectively. The expression of CDR1 and ERG11 genes was significantly down-regulated in these selenium nanoparticle concentrations.
Conclusion:
As the findings indicated, selenium nanoparticles had an appropriate antifungal activity against fluconazole-resistant and -susceptible C. albicans strains. Accordingly, these nanoparticles reduced the expression of CDR1 and ERG11 genes associated with azole resistance. Further studies are needed to investigate the synergistic effects of selenium nanoparticles using other antifungal drugs.
Key Words: Candida albicans, CDR1, ERG 11, Nanoparticles
Introduction
Over the past decade, Candida infections has dramatically increased among the high-risk patients [1]. The excessive use of antifungal agents due to the limited availability of antifungal medications leads to the emergence of drug resistance in pathogenic species [2]. Regardless of drug resistance, the long-term use of antifungal medications has irreparable side effects in patients, which can lead to death in some cases [3, 4].
The rate of azole resistance is on a growing trend in Candida species. Accordingly, there are extensive biochemical studies highlighting a significant diversity in the mechanisms conferring resistance to azoles. Some of these mechanisms include changes in the efflux system and mutation in ergosterol biosynthesis pathway genes [5]. The increased mRNA levels of Candida drug resistance gene family (CDR) and multi-drug resistance (MDR) have been associated with azole resistance [5, 6]. In addition, the overexpression of CDR1 and CDR2 in C. albicans is associated with cross-resistance to the azoles [7].
Ergosterol biosynthesis pathway is another pathway of azole resistance. The overexpression of ERG11 gene leads to an increase in the target drug. Therefore, a higher concentration of drug is required to react to all of enzyme molecules present in the cells, and thereby decrease the susceptibility to azole in Candida species [8]. The advent of this drug resistance necessitates the development of new antifungal agents to overcome the resistant strains.
Nanoparticles (NPs) are a new generation of antimicrobial agents that have been highly studied in the last two decades [9-11]. Among them, metal NPs and metal oxide have unique properties and are considered as antimicrobial agents [12]. Selenium is one of the essential elements for human health that improves the immune system and exerts anti-cancer effects. Nevertheless, there are problems with the pharmaceutical application of selenium for the treatment of infections. The nanotechnology recommends the use of this element for nutritional and pharmaceutical purposes [13].
On the other hand, selenium nanoparticles (Se NPs), as strong antioxidants, are much less toxic than selenium. Moreover, they have high power in scavenging free radicals; consequently, they can be used as a natural antioxidant [14]. Selenium sulfide has long been known as an antifungal agent used as anti-dandruff [15]. However, few studies have been published on the antifungal effects of Se NPs.
The expression of ERG11 and CDR genes have been identified as the mechanisms responsible for resistance to azole in Candida species. Regarding this and given the alterations in the structure of these two genes, the present study aimed to investigate the inhibitory effect of biogenic Se NPs on fluconazole-resistant C. albicans. In addition, the alternation in ERG11 and CDR1 gene expression was assessed in fluconazole-resistant C. albicans using the real-time polymerase chain (PCR) reaction technique.
Materials and Methods
Candida albicans strains
Fluconazole-resistant C. albicans (ATCC 76615) and fluconazole-susceptible C. albicans (ATCC 10231) were used to test antifungal susceptibility and gene expression by real-time PCR.
Preparation of stock nano-silver solution
The biosynthesis of Se NPs was accomplished using Bacillus sp. MSh-1 strain, which was previously isolated from the Caspian Sea (located in the northern part of Iran) and identified by 16S rDNA gene analysis technique (GenBank accession number: GU183144.1), based on a method described by Shakibaie et al. [16]. Briefly, selenium dioxide powder (Merk, Germany) was dissolved in sterilized distilled water, and then filtered by a 0.22 μm diameter pore. In the next step, 1 mL of this solution was added to 99 mL nutrient medium; subsequently, 1 mL of Bacillus sp. Msh-1 suspension was added.
After 14 h, the obtained solution was centrifuged for 10 min at 5000 rpm, and the supernatant was removed. Sediment was washed twice with 0.9% sodium chloride. The cells were then broken down by the liquid nitrogen and sonication (5 min/100 W). Subsequently, 4 ml of the above suspension was added to 2 ml ethanol and mixed vigorously. The tubes were centrifuged at 3000 rpm for 5 min and cooled for 24 h at 4°C to separate the two phases. After the gentle removal of the supernatant, the residual nanoparticles were washed with the chloroform, ethanol 70%, and distilled water, respectively.
Nanoparticle structure
The size and morphology of Se NPs were examined by using a transmission electron microscope (TEM, Zeiss Supra 55 VP TEM, operated at 100 KV). The chemical properties of nanoparticles were also characterized by the energy-dispersive X-ray spectroscopy technique [17].
Antifungal susceptibility test
Fungistatic activity and minimum inhibitory concentrations (MICs) of Se NPs against C. albicans strains were determined according to the recommend-dations stated in the Clinical and Laboratory Standards Institute M27-A3 and M27-S4 documents [18]. An RPMI-1640 medium buffered at pH 7.0 with 3-N-morpholino propanesulfonic acid was used as the culture medium. The inoculum size of C. albicans was 0.5-2.5×103 cells/mL.
The concentration of Se NPs in the dispersion ranged within 10-200 µg/mL. Subsequently, 100 μL of yeast suspension and 100 μL of each concentration of Se NPs were added to the respective well of microtiter plates. The microdilution plates inoculated at 35oC for 48 h. The lowest concentration of the Se NPs, inhibiting the visible growth of microorganisms, was considered as the MIC. C. parapsilosis ATCC 22019 standard strain was used as quality control.
RNA extraction and complementary DNA synthesis
We extracted RNA from the last dilution of the drug based on MIC, and also from the positive control sample. Afterwards, 6×108 cells of the fungal suspension were centrifuged for 2 min/12000 g, and the supernatant was removed. RNA was extracted using the RNA extraction kit (Gene, JET RNA purification kit, fermentase, Germany). After centrifugation, the sediment was stored at -20oC until use.
The complementary DNA (cDNA) was synthesized according to the manufacturer's recommendations using a cDNA synthesis kit (Fermentas, USA). The materials used for making cDNA included 0.9-1.5 μL RNA template, 1 μL of random hexamer primer, 9.5-10.1 μL DW, 4 μL 5x reaction buffer, 1 μL RiboLock RNase Inhibitor, 2 μL of 10 mM dNTP Mix, and 1 μL reverse transcriptase in a final volume of 20 μL.
Real-time polymerase chain reaction
Real-time PCR was performed using a Corbett Rotor-Gene 3000 (Corbett Robotics, Australia), and SYBR Premix Ex Taq II (BioFact, Daejeon, Korea). The sequence of primers and their characteristics are illustrated in Table 1. All PCR mixtures contained 10 μL SYBR Premix Ex Taq II (2×), 2 μL of first strand cDNA, 1 μL of each specific primers (4 pmol) and 6 μL of diethyl pyrocarbonate water in a final volume of 20 μL. The amplification was initiated at 95°C for 15 min, followed by 45 cycles of denaturation at 95°C for 30 sec, annealing at 60°C for 40 sec, and extension at 72°C for 30 sec.
Table 1.
Nucleotide sequences of CDR1 and ERG11 primers
| Name | nt | GC% | Tm | Sequence | GeneBank |
|---|---|---|---|---|---|
| CDR1-S | 20 | 50 | 53.65 | 5’GGTGCTAATATCCAATGTTGG’3 | X77589.1 |
| CDR1-AS | 20 | 50 | 50.22 | 5’GTAATGGTTCTCTTTCAGCTG’3 | |
| ERG11-S | 20 | 45 | 59.88 | 5’CAGAAAAGTGGCGTTGTTGA’3 | KM875729.1 |
| ERG11-AS | 20 | 45 | 59.69 | 5’GCAGCATCACGTTTCCAATA’3 |
The expression of all genes was normalized to the housekeeping gene β actin. Standard curves for each gene were established with five serially diluted cDNA, obtained from the cells grown to mid-logarithmic phase, using specific primers under the appropriate PCR conditions. Experiments under each condition were performed in duplicate, and negative control (water as template) was included in each run. The CDR1 and ERG11 gene expression was analyzed with REST software (2009; Version 2.0.7). The software uses the comparative Ct method (ΔΔCt) to analyze data.
Results
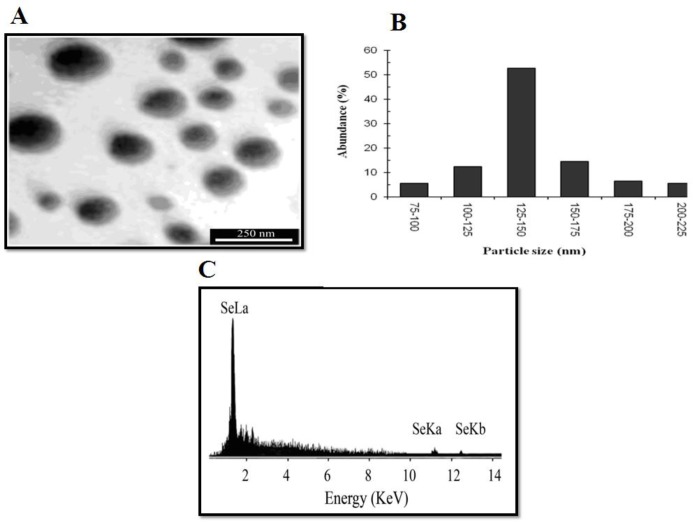
Spherical Se NPs were produced in a range of 80-200 nm by Bacillus sp. Msh-1 based on the transmission electron micrograph from pure Se NPs, and nanoparticles in the range of 125-150 nm were the most frequent ones. The energy-dispersive X-ray spectroscopy micro analysis of pure Se NPs showed three absorption peaks consisting of SeLα (1.37 keV), SeKα (11.22keV) and SeKβ (12.49 keV) (Figure 1). The MIC for fluconazole-resistant C. albicans and fluconazole-susceptible C. albicans in antifungal susceptibility test for Se NPs were 100 and 70 µg/mL, respectively.
Figure 1.
Synthesis and characterization of selenium colloidal nanoparticles synthesized by using Bacillus sp. MSh-1, A) Transmission electron micrograph of pure selenium nanoparticles, B) Particle size distribution histogram of the biogenic selenium nanoparticles, C) Energy-dispersive X-ray images of purified selenium nanoparticles
Fluconazole-resistant and -susceptible C. albicans strains were shown to reduce the expression of CDR1 and ERG11 genes after exposure to 70 and 100 µg/mL Se NPs, respectively. The results of CDR1 and ERG11 gene expression are displayed in Table 2. In fluconazole susceptible-C. albicans, CDR1 and ERG11 were down-regulated in the sample group by the mean factors of 0.067 and 0.038 expression rates, respectively. Similarly, in fluconazole-resistant C. albicans, CDR1 and ERG11 were down-regulated in the sample group by the mean factors of 0.022 and 0.027 expression rates, respectively.
Table 2.
Results of CDR1 and ERG11 genes expression in fluconazole-resistant and -susceptible C. albicans
| Candida species | Gene | Type | Reaction efficiency | Expression | Result |
|---|---|---|---|---|---|
| Fluconazole-resistant C. albicans | CDR1 | TRG | 0.69 | 0.022 | Down |
| ERG11 | TRG | 1.0 | 0.027 | Down | |
| Fluconazole-susceptible C. albicans | CDR1 | TRG | 0.64 | 0.067 | Down |
| ERG11 | TRG | 1.0 | 0.038 | Down |
TRG: target, REF: reference, Down: down-regulation
Discussion
The use of nanotechnology has increased in many medical fields, especially drug delivery [12]. The synthesis of nanoparticles by microorganisms and plant extracts is suggested as a suitable method, compared to the physical and chemical methods [13]. Unique properties, such as high surface-to-volume ratio and their nanoscale size, are the advantages of nanoparticles. This high surface-to-volume ratio of the nanoparticles provides more active sites for interacting with biological entities, such as cells [19].
To date, the antifungal effects of different nanoparticles, such as iron NPs, silver NPs, and gold NPs, have been studied [20-23]. The Se NPs increase the efficiency of glutathione peroxidase and thioredosin reductase [24] . Nano selenium is also reported as an antioxidant with reduced risk of direct toxicity on cells [25]. The antibacterial, antiviral, and antioxidant activities of Se NPs, in addition to their lower toxicity, have introduced them as interesting compounds in nano-biotechnology [26, 27].
In this study, the antifungal activity of Se NPs against C. albicans as a model for fungi was investigated. The results of the study showed that the Se NPs produced by Bacillus MSH-1 had antifungal activity against C. albicans. Limited studies have been carried out on the Se NPs. The anti-biofilm activity of biogenic Se NPs and selenium dioxide against the clinical isolates of Staphylococcus aureus, Pseudomonas aeruginosa, and Proteus mirabilis has been demonstrated [28].
Kazempour et al. investigated the antifungal activity of Se NPs, produced by Klebsiella pneumonia, against Aspergillus niger and C .albicans. They reported the Se NPs MIC values of 250 and 2000 µg/mL for A. niger and C. albicans, respectively [29]. In addition, Shakibaei et al. showed the antifungal effects of Se NPs synthesized by Bacillus species on Aspergillus fumigatus and C. albicans. In the mentioned study, the MICs of Se NPs against C. albicans and A. fumigatus were 70 and 100 μg/mL, respectively [30], which is consistent with our results.
Several genes associated with azole resistance (e.g., CDR1, CDR2, MDR1, ERG3, ERG6, ERG11, ERG9, RTA2, and NAG2) have been identified in Candida species. Each of these genes promotes the resistance of the organism to antifungal drugs with different molecular mechanisms [31-33]. All of these results indicate that Se NPs can be effective in the fungus with different mechanisms, compared to the conventional antifungal agents.
According to the results of this study, Se NPs could reduce the resistance of C. albicans to antifungal drug by decreasing the expression of the drug-related genes. In our study, the real-time PCR revealed the down-regulation of CDR1 and ERG11 genes for C. albicans exposed to Se NPs with concentrations of 70 and 100 μg/mL, respectively.
Conclusion
In conclusion, Se NPs reduced the expression of CDR1 and ERG11 genes. The down-regulation of these genes can decrease the resistance of Candida strains to azole. Common antifungal agents that are currently prescribed for the treatment of various fungal infections, have limitations, such as toxicity and drug resistance. According to the obtained results, the synthetized Se NPs can be an appropriate agent against C. albicans; moreover, they can facilitate overcoming the azole resistance. However, it is required to conduct more in vitro and in vivo investigations in this regard.
Acknowledgments
Hereby, we extend our gratitude to the Kerman University of Medical Sciences, Kerman, Iran, for their financial support.
Author’s contribution
S.A. A. M. obtained funding for the study. N. P., S.S., and S. H. prepared the manuscript, performed the experiments, and collected samples. S. R., S. KH. and M. K. performed the molecular experiments and prepared the manuscript.
Conflicts of interest
The authors declare no conflicts of interest. The authors are responsible for the content and writing of the paper.
Financial disclosure
This study was supported by the Research Deputy of Kerman University of Medical Sciences (Grant No: 94000091).
References
- 1.Khodavaisy S, Alialy M, Mahdavi Omran S, Habibi MR, Amri P, Monadi M, et al. The study on fungal colonization of respiratory tract in patients admitted to intensive care units of sari and Babol hospitals. Med J Mashhad Univ Med Sci. 2011;54(3):177–84. [Google Scholar]
- 2.Sardi JC, Scorzoni L, Bernardi T, Fusco-Almeida AM, Mendes Giannini MJ. Candida species: current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J Med Microbiol. 2013;62(1):10–24. doi: 10.1099/jmm.0.045054-0. [DOI] [PubMed] [Google Scholar]
- 3.Fisher BT, Zaoutis TE. Treatment of invasive candidiasis in immunocompromised pediatric patients. Pediatr Drugs. 2008;10(5):281–98. doi: 10.2165/00148581-200810050-00003. [DOI] [PubMed] [Google Scholar]
- 4.Vaezi A, Fakhim H, Khodavaisy S, Alizadeh A, Nazeri M, Soleimani A, et al. Epidemiological and mycological characteristics of candidemia in Iran: a systematic review and meta-analysis. J Mycol Med. 2017;27(2):146–52. doi: 10.1016/j.mycmed.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Alizadeh F, Khodavandi A, Zalakian S. Quantitation of ergosterol content and gene expression profile of ERG11 gene in fluconazole-resistant Candida albicans. Curr Med Mycol. 2017;3(1):13–9. doi: 10.29252/cmm.3.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lupetti A, Danesi R, Campa M, Del Tacca M, Kelly S. Molecular basis of resistance to azole antifungals. Trends Mol Med. 2002;8(2):76–81. doi: 10.1016/s1471-4914(02)02280-3. [DOI] [PubMed] [Google Scholar]
- 7.Khosravi Rad K, Falahati M, Roudbary M, Farahyar S, Nami S. Overexpression of MDR-1 and CDR-2 genes in fluconazole resistance of Candida albicans isolated from patients with vulvovaginal candidiasis. Curr Med Mycol. 2016;2(4):24–9. doi: 10.18869/acadpub.cmm.2.4.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li QQ, Skinner J, Bennett JE. Evaluation of reference genes for real-time quantitative PCR studies in Candida glabrata following azole treatment. BMC Mol Biol. 2012;13(1):22. doi: 10.1186/1471-2199-13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark LC, Dalkin B, Krongrad A, Combs GF Jr, Turnbull BW, Slate EH, et al. Decreased incidence of prostate cancer with selenium supplementation: results of a double-blind cancer prevention trial. Br J Urol. 1998;81(5):730–4. doi: 10.1046/j.1464-410x.1998.00630.x. [DOI] [PubMed] [Google Scholar]
- 10.Rayman MP. Selenium in cancer prevention: a review of the evidence and mechanism of action. Proc Nutr Soc. 2005;64(4):527–42. doi: 10.1079/pns2005467. [DOI] [PubMed] [Google Scholar]
- 11.Yang J, Huang K, Qin S, Wu X, Zhao Z, Chen F. Antibacterial action of selenium-enriched probiotics against pathogenic Escherichia coli. Dig Dis Sci. 2009;54(2):246–54. doi: 10.1007/s10620-008-0361-4. [DOI] [PubMed] [Google Scholar]
- 12.Panyam J, Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Adv Drug Deliv Rev. 2003;55(3):329–47. doi: 10.1016/s0169-409x(02)00228-4. [DOI] [PubMed] [Google Scholar]
- 13.Ratner MA, Ratner D. Nanotechnology: a gentle introduction to the next big idea. New Jersey: Prentice Hall Professional; 2003. [Google Scholar]
- 14.Singh KP, Gupta S. Nano-QSAR modeling for predicting biological activity of diverse nanomaterials. RSC Adv. 2014;4(26):13215–30. [Google Scholar]
- 15.Aggarwal K, Jain V, Sangwan S. Comparative study of ketoconazole versus selenium sulphide shampoo in pityriasis versicolor. Indian J Dermatol Venereol Leprol. 2003;69(2):86–7. [PubMed] [Google Scholar]
- 16.Shakibaie M, Khorramizadeh MR, Faramarzi MA, Sabzevari O, Shahverdi AR. Biosynthesis and recovery of selenium nanoparticles and the effects on matrix metalloproteinase‐2 expression. Biotechnol Appl Biochem. 2010;56(1):7–15. doi: 10.1042/BA20100042. [DOI] [PubMed] [Google Scholar]
- 17.Herzing AA, Watanabe M, Edwards JK, Conte M, Tang ZR, Hutchings GJ, et al. Energy dispersive X-ray spectroscopy of bimetallic nanoparticles in an aberration corrected scanning transmission electron microscope. Faraday Discuss. 2008;138:337–51. doi: 10.1039/b706293c. [DOI] [PubMed] [Google Scholar]
- 18.Wayne P. Clinical and laboratory standards institute: reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard; CLSI document M27-A3. CLSI. 2008;28:6–12. [Google Scholar]
- 19.Tran PA, Webster TJ. Selenium nanoparticles inhibit Staphylococcus aureus growth. Int J Nanomedicine. 2011;6:1553–8. doi: 10.2147/IJN.S21729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prucek R, Tuček J, Kilianová M, Panáček A, Kvítek L, Filip J, et al. The targeted antibacterial and antifungal properties of magnetic nanocomposite of iron oxide and silver nanoparticles. Biomaterials. 2011;32(21):4704–13. doi: 10.1016/j.biomaterials.2011.03.039. [DOI] [PubMed] [Google Scholar]
- 21.Jayaseelan C, Ramkumar R, Rahuman AA, Perumal P. Green synthesis of gold nanoparticles using seed aqueous extract of Abelmoschus esculentus and its antifungal activity. Ind Crops Products. 2013;45:423–9. [Google Scholar]
- 22.Kim KJ, Sung WS, Moon SK, Choi JS, Kim JG, Lee DG. Antifungal effect of silver nanoparticles on dermatophytes. J Microbiol Biotechnol. 2008;18(8):1482–4. [PubMed] [Google Scholar]
- 23.Seddighi NS, Salari S, Izadi AR. Evaluation of antifungal effect of iron-oxide nanoparticles against different Candida species. IET Nanobiotechnol. 2017;11(7):883–8. [Google Scholar]
- 24.Yost DA, Russell JC, Yang H. Non-metal colloidal particle immunoassay. Washington, DC: Patent and Trademark Office; 1990. [Google Scholar]
- 25.Wang H, Zhang J, Yu H. Elemental selenium at nano size possesses lower toxicity without compromising the fundamental effect on selenoenzymes: comparison with selenomethionine in mice. Free Radic Biol Med. 2007;42(10):1524–33. doi: 10.1016/j.freeradbiomed.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 26.Beheshti N, Soflaei S, Shakibaie M, Yazdi MH, Ghaffarifar F, Dalimi A, et al. Efficacy of biogenic selenium nanoparticles against Leishmania major: in vitro and in vivo studies. J Trace Elem Med Biol. 2013;27(3):203–7. doi: 10.1016/j.jtemb.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Shakibaie M, Shahverdi AR, Faramarzi MA, Hassanzadeh GR, Rahimi HR, Sabzevari O. Acute and subacute toxicity of novel biogenic selenium nanoparticles in mice. Pharm Biol. 2013;51(1):58–63. doi: 10.3109/13880209.2012.710241. [DOI] [PubMed] [Google Scholar]
- 28.Shakibaie M, Forootanfar H, Golkari Y, Mohammadi-Khorsand T, Shakibaie MR. Anti-biofilm activity of biogenic selenium nanoparticles and selenium dioxide against clinical isolates of Staphylococcus aureus, Pseudomonas aeruginosa, and Proteus mirabilis. J Trace Elem Med Biol. 2015;29:235–41. doi: 10.1016/j.jtemb.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 29.Kazempour ZB, Yazdi MH, Rafii F, Shahverdi AR. Sub-inhibitory concentration of biogenic selenium nanoparticles lacks post antifungal effect for Aspergillus niger and Candida albicans and stimulates the growth of Aspergillus niger. Iran J Microbiol. 2013;5(1):81–5. [PMC free article] [PubMed] [Google Scholar]
- 30.Shakibaie M, Salari Mohazab N, Ayatollahi Mousavi SA. Antifungal activity of selenium nanoparticles synthesized by bacillus species Msh-1 against Aspergillus fumigatus and Candida albicans. Jundishapur J Microbiol. 2015;8(9):e26381. doi: 10.5812/jjm.26381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cannon RD, Lamping E, Holmes AR, Niimi K, Tanabe K, Niimi M, et al. Candida albicans drug resistance another way to cope with stress. Microbiology. 2007;153(Pt 10):3211–7. doi: 10.1099/mic.0.2007/010405-0. [DOI] [PubMed] [Google Scholar]
- 32.Brumfield KM, Moroney JV, Moore TS, Simms TA, Donze D. Functional characterization of the Chlamydomonas reinhardtii ERG3 ortholog, a gene involved in the biosynthesis of ergosterol. PLoS One. 2010;5(1):e8659. doi: 10.1371/journal.pone.0008659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akins RA. An update on antifungal targets and mechanisms of resistance in Candida albicans. Med Mycol. 2005;43(4):285–318. doi: 10.1080/13693780500138971. [DOI] [PubMed] [Google Scholar]